Abstract
DNA unwinding and polymerization are complex processes involving many intermediate species in the reactions. Our understanding of these processes is limited because the rates of the reactions or the existence of intermediate species is not apparent without specially designed experimental techniques and data analysis procedures. In this chapter we describe how pre-steady state single-turnover measurements analyzed by model-based methods can be used for estimating the elementary rate constants. Using the hexameric helicase and the DNA polymerase from bacteriophage T7 as model systems, we provide stepwise procedures for measuring the kinetics of the reactions they catalyze based on radioactivity and fluorescence. We also describe analysis of the experimental measurements using publicly available models and software gfit (http://gfit.sf.net).
Keywords: Hexameric helicase, Replication, DNA unwinding, T7 bacteriophage, DNA polymerase, DNA synthesis, Strand displacement, Primer extension, gfit, global regression analysis
1. Introduction
Helicases are motor proteins that use the chemical energy of NTP hydrolysis to separate the strands of double stranded nucleic acids (1–4). Often helicases work in association with other proteins, such as the DNA polymerase or single strand binding proteins to perform their function with a greater efficiency. Characterization of the nucleic acid unwinding activity involves measurement of the rate and the processivity of the helicase-catalyzed unwinding reaction. These unwinding parameters can be used as basic handles to understand the mechanism of unwinding and the role of the helicase in a particular biological process. Measurement of the unwinding rate of helicases as a function of DNA duplex stability provided insights into the active or passive nature of the helicase-catalyzed reaction (5). Measurement of the unwinding rate of phage T7 hexameric helicase in the presence of the DNA polymerase provided insights into the synergistic action of the two motor proteins in DNA replication (6).
In this chapter, we describe assays to measure DNA unwinding catalyzed by the helicase, and DNA polymerization catalyzed by the helicase and polymerase proteins using the hexameric ring-shaped T7 gp4 helicase as our model system. We outline procedures to fit the kinetic data to specific models that provide kinetic parameters such as the rate of DNA unwinding and the rate of nucleotide incorporation during DNA polymerization.
The product of bacteriophage T7 gene gp4 is a ring shaped protein (7–9) that has both DNA unwinding and primase activities (10). T7 gp4 can move along single stranded (ss) (11) or double stranded (ds) (12) DNA and separate DNA strands using the energy of dTTP hydrolysis. The protein can also synthesize short (4–5 -mer) RNA primers using ATP and CTP at priming sites on the lagging DNA strand. T7 gp4 unwinds dsDNA using the strand exclusion model wherein one strand of the dsDNA is threaded into the central channel of the helicase and the other strand is excluded as the helicase unwinds the dsDNA (13,14). This strand exclusion model is accepted generally for ring shaped helicases (15–17). The unwinding rate of T7 gp4 depends on dsDNA stability and that its speed of DNA unwinding is slower than its speed of translocation along ssDNA (5). The other key component of T7 replication complex is gp5 DNA polymerase (18–20). Interestingly, the rate of DNA unwinding by T7 gp4 is accelerated by the polymerase and approached the ssDNA translocation rate of the helicase (6). Accurate measurements of kinetic rates of translocation, unwinding, and polymerization provide both quantitative and qualitative insights about the DNA replication mechanism.
1.1. Methods for measuring reaction kinetics
Unwinding and polymerization pathways comprise many interacting reaction steps such as substrate binding, catalysis, and conformational changes. The large number of these steps makes their identification and characterization increasingly difficult. Therefore, one prerequisite of a successful experimental design is its ability to decouple the pathway: to study a part of the pathway in isolation or while controlling the influence of the other parts. Most clear results are produced by experiments that measure kinetics of one fully decoupled reaction step.
One strategy to maximize decoupling enabled by recent technological advances involves monitoring reactions occurring with single molecules. These techniques have been successfully applied to both DNA unwinding and polymerization (21–28). Unfortunately, only parts of DNA processing pathways can be currently observed at single molecule level, which brings us back to conventional measurement techniques that integrate signals originating from billions of enzyme molecules. During the course of reaction, as the system reaches a steady state, molecules of enzyme become distributed along the reaction pathway populating all of the reaction species and participating in all reactions simultaneously. Combined signal from such heterogeneous mixture is hard to interpret in terms of individual reaction rates.
These problems are overcome by using pre-steady state and single round kinetic techniques. Pre-steady state kinetic measurements involve synchronization of the system – assembling the reaction mixture in a way that populates only one species of the pathway. This can be done, for example, by withholding a component required for the next reaction step. After the reaction in the synchronized mixture is started, but before it reaches a steady state, the measured signal can be attributed to a few steps that follow the synchronization point.
The pre-steady state period of reaction is quite short due to a natural tendency of molecular systems to lose synchronization. To extend it and to be able to characterize more reaction steps in one experiment, single round conditions can be used. Single round conditions effectively prevent the system from reaching a steady state by allowing each molecule of enzyme transform no more than one molecule of substrate. In case of an unwinding reaction, this can be achieved by adding a helicase trap at the time of initiation. An excess of ssDNA can capture free helicase molecules from solution preventing them from re-binding to new substrate molecules. Pre-steady state single round approaches enhance our ability to decouple unwinding and polymerization pathways to measure the rates of their reaction steps.
1.1.1. Unwinding kinetics
DNA unwinding rates have been measured using both bulk and single molecule techniques (5,6,29). Single round conditions simplify interpretation of a bulk kinetic measurement result. Helicases can processively unwind stretches of dsDNA longer than their binding site. Therefore an unwinding reaction can be described as an n-step process and for experiments conducted under single round conditions, the results can be fit with a stepping equation (30,31). Such analysis of unwinding kinetics data for dsDNA substrates of different lengths can be used for estimating helicase stepping rate and size and for assessing processivity of the helicase; i.e., how far the helicase moves along the DNA before it falls off.
1.1.1.1. Assembly of helicase on the DNA substrate
Ring-shaped helicases assemble around ssDNA, and assembly is usually a slow step (relative to the rate of DNA unwinding). Therefore, to measure the unwinding rate (rather than the assembly rate) and to synchronize the reactions, it is important to preassemble the helicase on the DNA substrate prior to reaction start. T7 gp4, like other ring-shaped helicases, binds to the DNA only in the presence of its nucleotide substrate (dTTP in the case of T7 gp4). This makes the preassembly of the helicase on the DNA substrate, without reaction occurring during the assembly period, challenging. Based on our finding that T7 gp4 forms hexamers and binds DNA in the absence of Mg(II), we arrived at the following assembly procedure (this might be applicable to other helicases that require the presence of NTP to bind DNA). We preassemble the helicase on the DNA by adding dTTP, but by leaving out Mg(II). In the absence of Mg(II) and in the presence of added EDTA (to chelate contaminating divalent metal ions), T7 gp4 does not hydrolyze dTTP or unwind DNA (32).
1.1.1.2. DNA substrates and DNA unwinding kinetics
Two types of the unwinding assays are described: discontinuous gel-based radiometric assay and continuous stopped-flow fluorescence assay. They are both all-or-none unwinding assays where unwinding rates are obtained from the kinetics of end product formation, i.e., the kinetics of the appearance of the fully unwound DNA. Since the unwinding rate of T7 gp4 is fast, the kinetics are measured using a rapid quenched-flow or a stopped-flow apparatus (Fig. 2.1.) that allow mixing in the millisecond time scales. A typical DNA unwinding substrate for the hexameric helicase consists of a fork DNA. Two short DNA strands (top and bottom) are annealed to generate a duplex region (40 bp, here) and ssDNA overhangs at one end. A 5′ ssDNA overhang (dT35) in the top strand is needed for helicase binding (the DnaB family helicases that are 5′-3′ helicases), and the 3′ ssDNA overhang (dT15) in the bottom strand is required for strand exclusion during unwinding (Fig. 2.2.). In the gel-based assay, one of the DNA strands is radiolabeled, so the radioactive fork substrate and the ssDNA product can be quantified after they are resolved by native PAGE. The kinetics of DNA unwinding is fit to obtain the unwinding rate. In the fluorescence-based assay, a fluorescent dye (fluorescein) is incorporated at the 5′ end of the bottom strand (Fig. 2.3.). A run of three guanosines at the 3′ end of the top strand quenches fluorescein fluorescence when the substrate is duplexed. When helicase unwinds the dsDNA and the top strand is displaced away from the dye, the fluorescence increases. The time dependent increase in fluorescence is measured continuously in a stopped-flow apparatus.
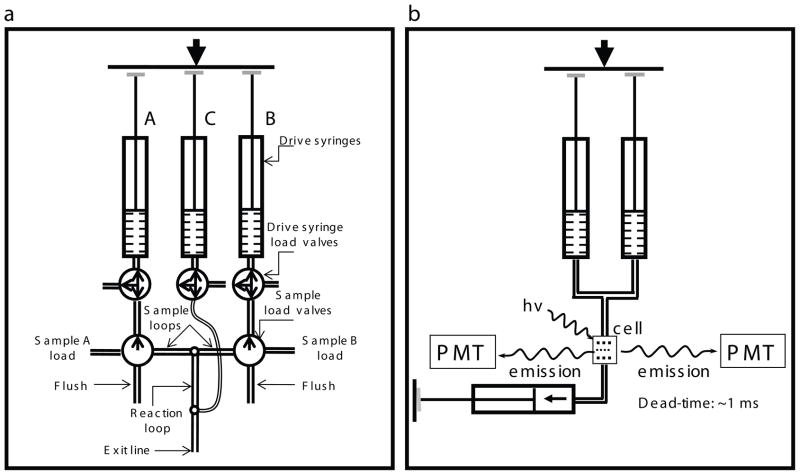
Fig. 2.1.
Instrumental designs for the rapid kinetic studies. a) Chemical quenched-flow RQF-3 (www.kintek-corp.com, figure kindly provided by Prof. K.A. Jhonson). Sample A and Sample B are loaded in sample loops from the load ports via a three way valve. Upon firing the instrument, solution A and B are forced through the delay line by water from the drive syringes A and B, and then reactants are mixed in the valve to start the reaction. The reaction mix flows through the selected delay line and mixed with the quench solution from syringe C after predetermined time intervals. The quenched sample is collected into a tube from the exit line. Different length delay lines are selected through an eight – way valve for reaction times in the range of 2–100 msec by selecting different reaction loops. b) Stopped-flow instrument. The KinTek stopped flow has a stable light source and sensitive detection system that can measure absorbance and fluorescence simultaneously. There are two channels for fluorescence detection, and three drive syringes, but in normal mode of operation two syringes are connected (those shown) and used to drive mixing of two reactants A and B into the observation cell. Reaction time is under computer control allowing times from few msec to several minutes. The instrument dead-time is determined as outlined in www.kintek-corp.com.
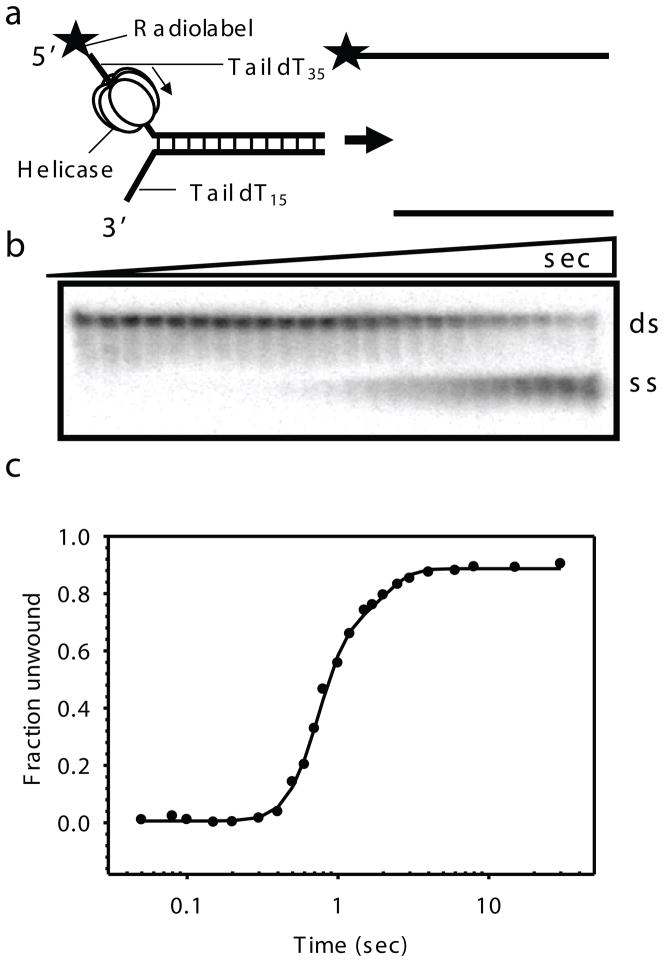
Fig. 2.2.
Gel-based radiometric assay for DNA unwinding. a) The DNA unwinding fork substrate design with radiolabeled top strand. T7 gp4 assembles on the top strand and moves in the 5′ to 3′ direction to unwind the dsDNA substrate. b) Representative native gel showing the ds and ss DNA resolved as a function of reaction time. Time points here are 0, 0.05, 0.08, 0.1, 0.15, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 1, 1.2, 1.5, 1.7, 2, 2.5, 3, 4, 6, 8, 15, 30 s (for 40ds duplex). c) Kinetics of ds40 unwinding at 18°C in reactions containing T7 gp4, dTTP, MgCl2 and SSB as the trap. The kinetics is fit to the gfit unwinding model (unwinding.m), which provides kf and s, from which the average rate of unwinding (kf × s) was determined. A good fit for the 40ds DNA unwinding data here gave parameters; A = 0.829, kf = 9.89, minD = 0, s = 6.34 and F0 = 0.002 and hence an average rate of unwinding ~ 62.7 bp/s.
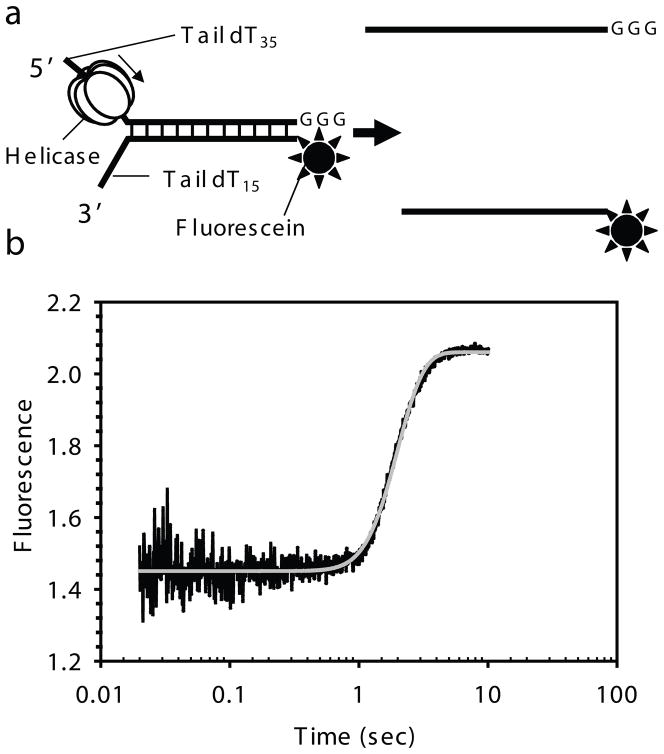
Fig. 2.3.
Fluorescence-base stopped-flow assay for DNA unwinding. a) DNA unwinding fork substrate design with fluorescein in the lower strand and GGG at the 3′ end in the top strand. T7 gp4 moves in the 5′ to 3′ direction to unwind the dsDNA substrate. b) Representative kinetic trace showing the unwinding of the fork DNA by the time-dependent increase in fluorescence. The kinetics is fit to the gfit unwinding model (unwinding.m), which provides kf and s, from which the average rate of unwinding (kf × s) was calculated. A good fit for the 40ds DNA unwinding data in the presence of top ssDNA trap was obtained at N = 2 and hence gave parameters for the two phases; A1 = 0.862, A2 = 0.137, kf1 = 4.488, kf2 = 1.413, minD = 0, s = 5.395 and F0 = 0.995 and hence an average rate of unwinding for the fast population is ~ 24.2 bp/s and for the slow population is ~7.6 bp/s.
1.1.2. Polymerization kinetics
DNA polymerization rates can be estimated from the individual nucleotide incorporation rates measured using transient state kinetic methods. DNA polymerases incorporate hundreds to thousands of nucleotides during polymerization in a template-dependent manner, adding one dNMP to the primer at a time and moving with a step-size of one nucleotide. Each nucleotide is added at a different rate that depends on several factors, not all well understood, one of which is the sequence context around the base to be added. The nucleotide addition rate can be determined accurately using a combination of rapid kinetics, product analysis on a high resolution sequencing gel, and data analysis. The rapid kinetic methods providing milli seconds time resolution capture the formation and decay of intermediate products, the sequencing gels can resolve the DNA products with a single base resolution, and data analysis extracts the single nucleotide incorporation rate and the polymerase off-rates from the observed kinetics of primer elongation.
1.1.2.1. DNA synthesis kinetics
DNA polymerase extends a primer annealed to a template DNA by utilizing dNTPs as substrates. When the template is single stranded, the polymerase can copy the template without the helicase, but when the template is double stranded, the polymerase requires the helicase to unwind the dsDNA. T7 DNA polymerase requires T7 gp4 to catalyze strand displacement DNA synthesis. The rate of DNA unwinding by the helicase with concomitant DNA synthesis can be measured by the unwinding assays described above using a replication fork substrate. Alternatively, the kinetics of the reaction can be measured by following the primer extension reaction. We describe methods to obtain the rate of each nucleotide addition in the primer extension reaction. Replication fork substrates are made by annealing the top and bottom ssDNAs to generate a duplex region (40 bp, here) and two ssDNA overhangs. A third strand, a primer (24 mer, here) is annealed to the bottom strand (of a defined sequence) to create a primer/template junction (Fig 2.4.). The primer is either radiolabeled or fluorescein-labeled (at the 5′-end) to follow the primer extension kinetics. Reaction products are separated by PAGE with single-base resolution to measure the amount of each intermediate.
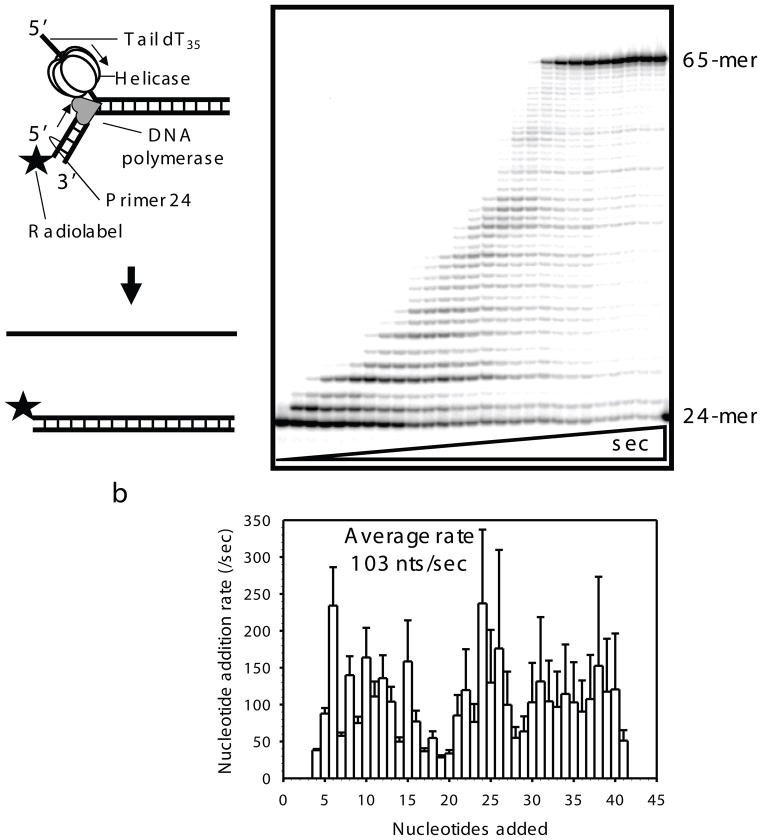
Fig. 2.4.
Strand displacement DNA synthesis by helicase-polymerase replisome. a) T7 DNA polymerase and T7 gp4 are assembled on the replication fork substrate with a radiolabeled DNA primer. T7 gp4 on top strand moves to unwind the dsDNA and DNA polymerase extends the primer in the 5′ to 3′ direction by duplicating the bottom strand. Sequencing gel shows progressive strand displacement DNA synthesis activity of the replisome. Time points here are 0, 0.004, 0.006, 0.008, 0.01, 0.015, 0.02, 0.03, 0.04, 0.05, 0.06, 0.08, 0.1, 0.12, 0.15, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.75, 1, 1.5, 2, 3, 4 s (for 41 nt base extensions). The 24 mer primer is elongated to 65 nt runoff product. b) Rate constants of individual nucleotide addition plotted against nucleotide added (rates of first three nucleotide addition are not plotted here as they are always high probably due to premelting of duplex with helicase binding). Rate constants are estimated by fitting the kinetics of individual DNA product synthesis to the polymerization model of gfit (polymerase_ni.m). Errors are calculated from the global fits to the polymerization model.
1.2. Analysis of reaction kinetics measurements
Even the most advanced experimental techniques cannot fully decouple all steps involved in unwinding and polymerization process. Most experimental observations arise not from one but from multiple simultaneously occurring reactions. Such results do not directly provide a value for any reaction rate or even a confirmation that the reaction step actually takes place. This information can be extracted from the results by applying model-based (regression) analysis that has the following goals: a) to test if the proposed model is in agreement with the observations, b) to estimate parameters of the model and c) to estimate their confidence intervals.
In this chapter we describe analysis of unwinding and polymerization data using gfit, an open source program (http://gfit.sourceforge.net) (33). Although current version of gfit runs within MATLAB environment and uses computational models written in MATLAB language. Programming, however, is only involved in creating new models; all analysis tasks in gfit using existing models are performed through graphical user interface and do not require any knowledge of programming or MATLAB environment. In the following sections we discuss details of the analysis procedure, computational problems that it presents, and the ways to address these problems.
1.2.1. Steps involved in model-based analysis
Analysis starts with creating a computational model based on existing knowledge about the system. The model should be able to compute (simulate) a predicted result of each experiment. To perform computations, the model uses two kinds of inputs: experimental conditions, known values from the experimental protocol (e.g., incubation time, nucleotide concentration) and model parameters, usually unknown, intrinsic properties of the system (e.g., translocation step size, rate of nucleotide incorporation). Although, for any given experiment, conditions and parameters are easily distinguishable, the same variable (e.g., enzyme concentration) may appear as a condition in one experiment and as a parameter in another.
Correctness of a model can never be demonstrated conclusively. However, given certain parameter values, a model may produce simulations closely matching (fitting) experimental measurements. Testing if the model is consistent with all the experimental observations, a key step in data analysis, is performed by global curve fitting (optimization). Failure to find a good set of parameter values usually means that the model is not accurate and requires structural changes. Finding a good fit not only demonstrates consistency, but also provides estimates of parameters. Such result does not conclude the analysis because the estimated parameter values carry little meaning without an indication of their uniqueness and an estimate of their confidence intervals. Restarting optimization many times from randomly selected positions in the parameter space may lead to discovery of alternative parameter sets fitting the data, an indication of low confidence. Underdetermined parameters may stem from an overly complicated model or from insufficient experimental data. The problem can be solved by simplifying the model, by adding explicit constraints to parameters (e.g., equating two related rate constants, sharing a parameter between multiple experiments), or by providing more experimental results. Global analysis of experiments that highlight different aspects of system’s behavior provides most rigorous validation of the model and allows estimation of its parameters with highest confidence.
1.2.2. Software for model-based analysis
Although procedures for model-based data analysis are well-established, their practical applications to biological systems present several computational challenges and places rigid requirements on the analysis software. The main function of the software is to provide methods for statistical analysis that operate on experimental data and computational models supplied by the researcher. Biological research projects are highly dynamic. Therefore the software should make it easy to modify the model, to include new experiments into global analysis, to change statistical weights, to use same parameter value for multiple experiments, etc. The desired flexibility can be achieved by developing a project-specific analysis program (34), although our experience shows that this approach requires frequent modifications of the program code, which slow down the analysis and lead to programming errors.
Computational models are the central parts of the analysis. The models have to simulate a wide range of biological processes and experimental procedures. Ordinary differential equations (ODEs) is currently the most common method to model biology. However, defining a model strictly as a set of ODEs is unacceptable because some systems have to be simulated with a floating number of ODEs (e.g., unwinding and polymerization, as discussed below), others – by solving ODEs several times while changing initial concentrations to replicate pre-incubation, dilution, and mixing performed as part of the experimental procedure, yet others may require completely different computational techniques. Since simulation is usually the slowest step in the analysis process, it should be performed as efficiently as possible. An analysis procedure may involve 103 to 108 simulations with different experimental conditions and parameters. To carry out the analysis without human intervention, each output of the model has to be directly comparable with the result of the corresponding experiment. In short, simulations require accuracy, flexibility, and efficiency. These requirements can be best met by writing models in a general purpose programming language (e.g. C++, Python, MATLAB) making it possible to accurately capture all details of the mechanism and experimental design and using most efficient algorithms.
Considering the above requirements, model-based analysis of experiments in this chapter was performed using gfit, software that solves the general case of this problem (33). Since gfit uses MATLAB scripts as models, it can be used for analysis of any type of experimental data – kinetic, thermodynamic, or any other. Details about writing gfit models are beyond the scope of this chapter. Instead, we described how the existing models can be used for data analysis. Models and sample datasets used in this chapter, more detailed documentation and more modeling examples can be found at gfit website: http://gfit.sourceforge.net.
1.2.3. Data analysis using gfit
Experimental data in gfit is represented by a collection of experiments. Each experiment may include multiple variables, which store numerical information. A variable may contain a scalar (single number), a vector (a column of numbers), or a matrix (arrays with more than two dimensions are also supported, but do not appear in this chapter). gfit uses variables for communicating with models. Input variables are used for sending a model the data it needs to perform a simulation; simulation results are received from the model as output variables. Since every model has its own special requirements for input data and produces different types of results, the model has to describe its input and output variables, their names, dimensional relationships, and other properties. By reading model description, gfit learns how the model should be run and what kind of simulations to expect from it.
Same information is used by gfit for importing experimental data. During import, the data is checked against the requirements of the currently selected model, to determine whether it is suitable for simulation and for fitting. If a critical piece of information is missing, or if data violates some of the model’s requirements, it is rejected. For example, it is an error not to include variable time in an unwinding experiment, because unwinding.m model lists this variable as a requirement. It is also an error to include a vector of 10 numbers as time, and a vector of 11 numbers - as F, because the model stipulates that the two variables should be vectors of equal length.
Experimental data is imported to gfit from a spreadsheet by copying a block of data into clipboard. Many experiments arranged side-by-side to each other can be imported in one operation. Each experiment must have a name starting with a letter and containing any characters thereafter. All names of experiments should appear in the top row of the imported block. Variables for each experiment should occupy spreadsheet cells directly below or below to the right of experiment’s name. Names and other properties of variables should be according to model’s definitions. To obtain names of variables defined by the current model, select menu Model → Copy data headers. This command places into the clipboard a sample header for one experiment. Paste contents of clipboard into an empty spreadsheet. Note that the header contains all variables defined by the model; not all of them have to appear in actual experimental data. Variable’s data occupy rectangular blocks of spreadsheet cells. The top left corner of each block should appear immediately below the variable’s name. The number for a scalar variable may also occupy the cell immediately to the right from its name. In that case, a colon, “:”, should be added to the name.
Experimental data is imported from clipboard by selecting menu Data → Paste-add Data. If import is successful, gfit generates parameters required for simulation and fitting of imported experiments. For each parameter, the name, optimization flag, lower bound constraint, current value, and upper bound constraint are shown. To simulate experiments, gfit combines data from parameters and experimental conditions and sends it to the model. The way each parameter is used for simulation of each experiment can be defined by researcher by selecting Combine Parameters and Separate Elements from Parameters menu or after right-clicking parameter name. The same parameter can be used in one or in many experiments, in one or in many input variables, and, if the input variable is an array, in multiple positions of the array.
Checked optimization flag indicates that the parameter should be optimized during fitting. During fitting, parameter value may vary between lower and upper bound constraints, which can be adjusted by researcher.
1.2.4. Computational model for unwinding
Unwinding can be modeled as a process involving multiple steps of equal size, s, and rate, kf. The only observable product, ssDNA, is produced at the last step. Its appearance is simulated by the model unwinding.m as incomplete gamma function (30,31,35). This method is both computationally efficient and allows using continuous (not only integers) number of steps. To account for heterogeneity in helicase population, the model calculates the sum of N unwinding processes. Other parameters of the model that can be estimated by fitting include unwinding amplitude, A, background signal, F0, and minimal stable duplex length, minD. It should be noted that the estimated step size s is strongly affected by enzyme’s heterogeneity and thus, maybe inaccurate.
1.2.5. Computational model for polymerization
Polymerization is another example of a multi-step process with the number of steps, N-1, equal to the length of the DNA template. At each step, the polymerase can either add a nucleotide to the growing chain with rate kf, or dissociate from the substrate, rate kd. The process is modeled as N ODEs. Since intermediate polymerization species can be resolved on a gel, a vector concentration is measured for each time point. The result of the experiment is therefore a matrix of size M by N, where M is the number of time points. To avoid writing dedicated models for each template length in gfit it is possible to write a universal model, polymerase_ni.m, that always simulates the correct number of steps according to the experimental data.
Forward and dissociation rates at each step do not have to be the same. Therefore, for each simulation the model requires N-1 -long vectors for kf and kd. Accordingly, for each experiment gfit generates N-1 parameters for each of the variables. By default, these parameters are linked (forced to keep the same value) into single parameters kf and kd. It is a researcher’s option to either separate the parameters completely, or to group them in any desired fashion. For example, one could try fitting the data while linking the kf-s according to the incorporated base type – kfG, kfT, kfA, kfC.
2. Materials
2.1. Proteins
-
2.1.1
T7 gp4 (gp4A′) was purified as described (36). The molar concentration of the protein in 8M Guanidine hydrochloride is obtained by UV absorption (extinction coefficient at 280 nm 0.0836 μM−1cm−1) (see Note 1).
-
2.1.2
T7 gp5 (D5A, D7A) was purified as described (37). The molar concentration of gp5 in 8M Guanidine hydrochloride was obtained by UV absorption (extinction coefficient at 280 nm of 0.13442 μM−1cm−1) (see Note 1).
-
2.1.3
E. coli thioredoxin enzyme was purchased from Sigma-Aldrich (Catalog number T0910).
-
2.1.4
If needed purify E. coli SSB as described by Lohman et al. (38) (see Note 1).
2.2. Instruments
-
2.2.1
Criterion cell (BioRad) for native PAGE.
-
2.2.2
Preparatory vertical gel slab electrophoresis (Hoefer Scientific Instruments SE-600 series) for DNA purification.
-
2.2.3
Elutrap electroeluter for DNA purification (Hoefer Scientific Instruments).
-
2.2.4
Sequencing gel unit (Sequi Gen GT, BioRad)
-
2.2.5
RQF3 rapid quenched-flow instrument from KinTek corporation (Fig. 2.1a.).
-
2.2.6
SF-2004 Stopped-flow instrument KinTek corporation (Fig. 2.1b.).
-
2.2.7
Typhoon scanner and Phosphorimager screens and from Molecular Dyanamics.
2.3. Software requirements for quantification of the data
-
2.3.1
ImageQuant 5.0 or later required for quantitation of scanned data
-
2.3.2
MATLAB v6.5 SP 1or later required for running simulations
-
2.3.4
MATLAB Optimization Toolbox required for regression analysis
2.4. Reagents
-
2.4.1
Deionized water of 18.3 Ohm conductivity (Milli – Q water) for all the solution preparation and reactions.
-
2.4.2
Oligonucleotides from IDT (Coralville, IA) or any other source.
-
2.4.3
BT1, BT2 Elutrap membranes for Elutrap electroeluter (Whatman).
-
2.4.5
Snake venom phoshodiesterase I (Catalog number P4506), DNase I, 8 M Guanidine hydrochloride, BSA can be purchased from Sigma-Aldrich.
-
2.4.5
5′ and 6′ carboxy -Fluorescein succinimidyl ester (Molecular Probes).
-
2.4.6
Radiolabeled nucleotides (Perkin Elmer).
-
2.4.7
Sephadex G-25 spin columns for oligonucleotides (Roche). The gel filtration matrix can also be purchased and then packed in 1.5 ml tube spin columns available from BioRad.
-
2.4.8
Make Deoxy and ribo nucleotides (Sigma-Aldrich) solutions in Tris buffer and adjust pH to 7.5. Estimate concentration of the solution using respective molar extinction coefficients and store at −20°C.
-
2.4.9
Polynucleotide kinase (New England Biolabs).
-
2.4.10
0.5 mM EDTA (pH 8.0) stock solution, stored at 22°C.
-
2.4.11
1 M Tris buffer, pH 7.6, stored at 22°C.
-
2.4.12
1 M Magnesium chloride stock solution, stored at 22°C.
-
2.4.13
20% SDS stock solution, stored at 22°C.
-
2.4.14
5 M Sodium chloride stock solution, stored at 22°C.
-
2.4.15
5 M Sodium hydroxide stock solution, stored at 22°C.
-
2.4.16
Replication buffer: 50 mM Tris-Cl, pH 7.6, 40 mM NaCl, 10% glycerol. A 10X stock can be made and stored at room temperature in tightly capped tube.
-
2.4.17
Acrylamide/bis acrylamide mixture 40% (w/v) 19:1 cross linking (Amresco, Ultrapure grade), stored at 4°C.
-
2.4.18
Loading dye 12x stock for gel based unwinding assays: 0.25% (w/v) Bromophenol blue, 40% sucrose, 12% (w/v) SDS. It is stored at 4°C and brought to room temperature before use to allow SDS to dissolve back in the solution.
-
2.4.19
Loading dye for sequencing gels: 95% (v/v) Formamide, 0.025% (w/v) Bromophenol blue, 10 mM EDTA. It is stored at −20°C. Avoid contact with Formamide.
2.5. Sample data and models
A zip archive containing all models and data used in this chapter can be downloaded from http://gfit.sourceforge.net. The archive also includes readme.txt file with most current information. The data files are in tab/newline-delimited format. The files can be opened in a text editor, but it is more convenient to view them in a spreadsheet application.
3. Methods
T7 gp4-catalyzed DNA unwinding activity requires a fork substrate (Fig. 2.2 and 2.3). When the activity of helicase and polymerase is measured, the unwinding substrate is further modified to anneal a primer to the bottom strand to create a primer/template junction for DNA polymerase binding (Fig. 2.4). All the proteins are preassembled on the DNA substrate in the presence of dTTP, but in the absence of Mg(II). Reaction are started as ‘standing start’ by adding Mg(II) (see Note 7). The rapid quenched-flow methods for gel-based DNA unwinding assays (31) and strand displacement DNA synthesis for polymerase-helicase in replisome (6) are described here. A high throughput stopped flow fluorescence-based DNA unwinding assay for helicase is also explained (42).
3.1. DNA substrate assembly for the radiometric assays
Purify the oligodeoxynucleotides by preparatory 7M Urea-PAGE/TBE gel.
Identify the major DNA band by UV shadowing and excise it out with a clean blade (see Note 2).
Cut the gel band to ~1mm by 1mm pieces and elute the oligonucleotide from the gel pieces by electroelution on Elutrap electroeluter using BT1 and BT2 membranes and 1x TBE buffer (see Note 2). Carry out electroelution at 120 volts. BT2 membrane is permeable to oligonucleotides but BT1 is not so the eluted oligodeoxynucleotides is retained in the chamber between the two membranes at the positive electrode side.
Collect eluted fraction in every 2–3 hrs, 3–4 times. Precipitate the DNA by ethanol/salt followed by 70% ethanol wash to desalt the pellet.
Resuspend the air dried pellet in TE pH 8.0 buffer for the storage.
Determine the oligodeoxynucleotide concentrations after digestion with snake venom phosphodiesterase I using absorption at 260 nm and individual base extinction coefficient values at 260 nm (39,40) (see Note 3).
Radiolabel the 5′ end of the top strand (for gel based helicase unwinding assays) or primer (for DNA synthesis assays) with γ 32P-ATP and polynucleotide kinase at 37°C for 30 min. followed by incubation at 65°C for 20 min. to denature the PNK enzyme (NEB catalogue).
Clean the labeled DNA by Sephadex G-25 spin column (Roche Scientific). Check the purity of the labeled DNA by separating labeled oligonucleotide from the free label on PEI cellulose F TLC (Merck)/0.4M potassium phosphate buffer pH 3.4. Expose the TLC to phosphorimager screen and scan the phosphorimage after an appropriate time. If amount of free radiolabel is high, then do another round of clean up with Sephadex G-25 (see Note 4).
Assemble the DNA substrates by mixing equal molar amounts of the DNAs in replication buffer, incubate the mix at 95°C for 10 min. followed by slow cooling (2–3 hrs) to room temperature (see Note 5 and Fig. 2.2).
3.2. DNA Substrate assembly for the fluorescent assays
Label the 5′ end of the bottom strand (for unwinding experiment) or of primer (for DNA synthesis experiment) which has a 6 carbon linker terminated with an amino group, with 5′ and 6′ carboxy -Fluorescein succinimidyl ester (see Note 6). Carry out the labeling in carbonate buffer using procedure from Molecular Probes. Labeled oligodeoxynucleotides can also be purchased from IDT.
Purify the labeled oligodeoxynucleotides by a long preparatory 7M Urea-PAGE/TBE gel followed by electroelution as described in Section 2.4.
Anneal the DNA substrate by mixing the dye-labeled bottom strand with the complementary portion of the top strand with or with the primer as described in Section 2.4. (Fig. 2.3).
Purify the annealed substrate complex by long preparatory non-denaturating PAGE followed by the electroelution.
Determine concentrations of the substrates after digestion with phosphodiesterase in combination with DNase I. Standardize the extinction coefficients of the unlabeled substrates with 0.1 M NaOH and use it to get the concentration of the labeled substrate (absorbance at 260 nm in 0.1 N NaOH) (41).
3.3. Gel-based DNA unwinding assays for hexameric helicases
Set up the temperature to 18°C (or desired temperature) in the circulatory cooling water bath attached to the rapid quenched-flow instrument. Consult the information brochure of the quenched-flow instrument (Fig. 2.1a.) available on Kintek corporation web site (www.kintek-corp.com) for operation details of the instrument.
Clean the two drive syringes and the quench syringe thoroughly with water. Fill up the two drive syringes with water avoiding any air bubble and the middle quench syringe with the 100 mM EDTA quench. Keep the top three valves in closed position for this procedure.
Wash the two sample loops by connecting 10ml syringes filled with water in load ports A and B in ‘load’ mode while having the exit line connected to the vacuum. Rinse the sample loops with methanol same way in ‘load’ mode followed by connecting the exit line with vacuum line till dry.
Rinse all the reaction loops (1 to 8) one by one with water in ‘flush’ mode through flush tubes, by changing the position of the reaction loop valve 1 through 8 and having the exit line connected to vacuum. Rinse all the loops with methanol while having them connected to the vacuum till dry.
Turn on the motor and then the system control panel. Type ‘RUN’ when ‘_’ appears on the screen and then press enter. Type ‘0’ (no) for ‘USE CONSTANT QUENCH VOLUME’ and then again type ‘0’ (none) for ‘ENTER 2ND QUENCH DELAY (SEC)’. This mode of operation is preferred as chances of experimental errors are less in this operation. Quench solution volumes vary when loops 1 to 6 are used and so the quench solution has to be pre added in the sample collection tubes to ensure same quench volume in all the samples. Instrument should be calibrated for the quench volumes of each loop and difference in the volumes should be known to work in this mode.
Select ‘ADJUST POSITION’ from the main menu followed by selecting ‘+’ for the downward movement and to bring down the ramp by pressing start button on the motor. Lower the ramp to just the top of the drive and quench syringe pistons. Similarly ‘-’ can be selected to bring the ramp up if needed to readjust it or to refill the drive and quench syringes during the experiment.
Mark 1.5 ml reaction tubes according to the sample time points add required amount of 100 mM EDTA quench in the tube depending on loop to be used for that sample. Put 23 μ1 of 12x loading dye for final 1(w/v) % SDS, 20% glycerol and 0.25% (w/v) bromophenol blue in the reaction. Puncture lids of the tubes to allow the exit line of the RQF during reaction for the sample collection.
Select ‘QUENCH FLOW RUN’ from the main menu and type a time in seconds and press ‘enter’. Set the valves in ‘fire’ mode, hold the 1.5 ml tube below with collection line in the tube through the lid hole. Press ‘G’ button to fire. Two to three of blank fires without reaction help forcing out air bubbles from the tubing and loops. Wash and dry the loop in ‘flush’ mode for the reactions.
Make a solution with 5 nM radiolabeled fork DNA substrate, 200 nM hexameric amount of gp4, 2 mM dTTP, and 5 mM EDTA in replication buffer (see Note 7 and 8).. Put solution in a 1 ml syringe avoiding any air bubble and connect to sample loading port A. Amount of solution made depends upon number of time points. Load this solution in the sample loop in ‘load’ position.
Prepare same amount of the other solution with 2 mM dTTP, 8 mM free MgCl2 (accounting for EDTA and dNTPs) and appropriate traps if required (3 μM top strand ssDNA or 6 μM dT90 the protein traps or 3 μM SSB the DNA trap). Put solution in a 1 ml syringe and hook up to second sample loading port same ways as above and load it in the sample loop in the ‘load’ position.
The two solutions are in the respective sample loops in equal volumes. Setup the knobs in ‘fire’ mode and fire from control panel as described above to rapidly mix the two solutions, 20 μl from each syringe in the reaction loop, to initiate the reactions. The reaction gets incubated according to the time set. Reactions are stopped by rapid mixing with 1.5 fold volume of a quenching solution of EDTA from the quench syringe. After sample collection, vortex and spin down the contents in the reaction tube. Wash the reaction loop with water followed by methanol to dry between each time point. Load 10 to 15 μl of the reaction on 10% native PAGE/1x TBE buffer pH 8.3 with 0.2 % SDS and resolve the products (see Note 9).
Peel off the gel from the plates and wrap the in clear cling wrap. Exposed the gels to the phosphorimager screen for appropriate times depending on counts and then scan the screen in a phosphorimage scanner.
-
Quantify the bands in the scanned image using the ImageQuant software. Calculate the fraction unwound for each time point by using the equation -
where SS is unwound single stranded DNA count, DS is the double stranded DNA substrate count, SS0 and DS0 are the respective counts for single stranded and double stranded DNA at 0 time.
Analyze the results as described in Section 3.7.2., to estimate unwinding rate and other parameters (Fig. 2.2).
3.4. Real time fluorescence DNA unwinding assay for hexameric helicase
Set up the temperature at 18°C (or desired temperature) in circulatory cooling water bath of SF-2004, stopped flow instrument (Fig. 2.1b.). Consult the information brochure of the stopped flow instrument available on www.kintek-corp.com for details about the instrument (see Note 8).
Fill the syringe chamber and reaction cell with 2% (v/v) RBS 35 detergent (Merck) and incubate for 30 minutes for cleaning in ‘load’ position of valves.
Rinse thoroughly with water to remove any traces of the detergent. Do this exercise by loading in ‘load’ position and removing in ‘fire’ position. Bring the valve to ‘load’ position at the end.
Turn on the mercury lamp and allow it to stabilize to 75V. Keep the cooling bath running for keeping the lamp at room temperature.
Set up the absorption wavelength to 480 nm. Insert a 515 nm cut-off long pass filter in the wavelength slot of the photomultiplier tube (PMT) path to be used. Adjust the slit width of light path to 2 mm (0.5mm/turn, 4 turns of the slit adjustment knob) for both the slits.
Document the reaction conditions for each syringe. Under ‘Instrument’, enter set time/channels and click OK. Set up the required parameters in the control panel; time 15 s (depending on the ds length to be unwound), PMT voltage in the range of 800–900 V, wave length 480 nm.
Use the replication buffer to do a mock fire (fire as described below) to clean up the water traces from reaction cell and syringes.
Prepare a solution of 20 nM DNA substrate (you can use as low as 5 nM) with fluorescein label at the 5′ end of the bottom strand, 200 nM gp4 (or desired concentration), 4 mM dTTP (or desired concentration), 6 mM EDTA, 2 μM BSA in replication buffer and load in one of the loading syringe (see Note 7 and 8). Incubate the mix for 15 min. at 18°C.
Make a solution with free concentration of 8 mM MgCl2, 2 μM BSA and 3 μM ssDNA or 3 μM E. coli SSB trap and load in the second loading syringe. Incubate the mix for 15 minutes at 18°C.
Click on ‘Adjust syringe drive’ in the ‘Control’ menu to bring the ramp down above the drive syringes.
Ensure now that the valve is in the ‘fire’ position and click on ‘Collect data’ button to fire the instrument for reaction to start. Upon firing, the instrument mixes the two solutions rapidly in the reaction chamber in equal volumes. The reaction mix is excited with 480 nm light and fluorescence is detected by a photomultiplier tube filtered through a 515 nm cut-off long pass filter.
Save all traces and average the overlaying ones for further analysis. Go to ‘Data analysis’, ‘Select traces’ and delete abnormal traces. Go to File menu, and save the data as ‘Stop flow data’ by clicking OK. Select ‘Data analysis’ and select ‘Average traces’ to get an average trace and save it.
Save the data as ASCII spreadsheet selecting for 1000 data points/trace. Copy the fluorescence data for each time point and paste in spreadsheet by clicking Data followed by choosing Text to columns, fixed width and save it in a spread sheet format.
Analyze the results as described in Section 3.7.2. to estimate unwinding rate and other parameters (Fig. 2.3).
3.5. DNA synthesis reactions on ds template by the helicase-polymerase
The kinetics of strand displacement DNA synthesis by T7 helicase-polymerase replisome is measured in the rapid quenched-flow instrument at 18°C (or desired temperature). Set up the instrument as described in Section 3.3.. Prepare the sample collection tubes with required amounts of EDTA according to the loops to be used.
Mix 50 μM E. coli Thioredoxin with freshly made 5 mM DTT in replication buffer for 5 min. at room temperature (22°C). Assemble T7 DNA polymerase by adding 10 μM of gp5 (in 1: 5 molar proportion to Thioredoxin) in the mix and incubating it at room temperature for 5 minutes (37). Store the T7 polymerase on ice till used (2–3 hrs) (see Note 10).
Assemble 1200 nM T7 gp4 hexamer on 600 nM fork DNA substrate (with a radiolabeled or fluorescein labeled primer annealed to the bottom strand) in the presence of 2 mM dTTP and 2 mM DTT, 2 mM EDTA in replication buffer on ice for 30 min. (You may use less polymerase and fork substrate as long as their concentrations is above the Kd value)
Add the polymerase-DNA mix to T7 gp4 mix and further incubate the solution at room temperature for 30 min. All the concentration mentioned here for replisome assembly are the final concentrations for the syringe.
Load the replisome-DNA complex solution in one of the sample loading syringe of the quenched-flow as described in Section 3.3..
Make solution with 0.2 mM each of dATP, dCTP and dGTP, MgCl2 (free concentration of 8 mM) and 6 μM dT90 trap (optional) in the replication buffer. Load this solution to the second sample loading syringe.
Start reactions by rapidly mixing equal volumes of the two solutions upon firing the instrument, and quench after various intervals with 300 mM EDTA from the quench syringe.
Dilute reactions with the sequencing dye (1:2 proportion of sample to sequencing dye for 20% GC content 40ds duplex) boil the samples for 10 min. at 95 C and load immediately on 24% acrylamide/7M urea sequencing gel with 1.5 x TBE buffer and run for 14 hrs at constant 90W. As a marker you can load sequencing dye with xylene cyanol in an end lane and run the gel till the xylene cyanol dye migrates to the end of plate. The 24-mer primer runs above this dye in 20–23% gel (see Note 11).
Let the gel cool down to the room temperature for 40 minutes. Carefully peel the gel from the plate on a cling wrap with the help of a wet flat spatula.
Cover the radioactive gel in the cling wrap from both the sides and then expose it to the phosphorimager screen for the required time and then scan the screen in a phosphorimager mode in the scanner. The fluorescent gel is directly scanned (best without a cling wrap) in the scanner in fluorescence mode at 526 nm Short Pass filter, Green 532 nm, Voltage 600 to 1000 V (depending on the intensity of the label) at normal sensitivity to get the image. Usually, 100 nM of fluorescein labeled substrate in the reactions give good counts when scanned at 800V for 100 micron size image
Quantify all visible DNA bands in the image by ImageQuant software (Fig. 2.4).
Paste counts data for different length bands in spreadsheet according to increasing time in columns and increasing band size in rows. Add up the counts for of all the bands in each lane in the gel (row for each time point) to get the total intensity. Get the fraction of the counts in every band by dividing its count by the total count of that lane on the gel.
Analyze the results as described in Section 3.7.3. to estimate nucleotide addition rates and polymerase dissociation rates for each base addition (Fig. 2.4).
3.7. Data analysis
3.7.1. Installation of gfit
gfit software can be downloaded from http://gfit.sourceforge.net. This website also contains most up-to-date installation instructions.
Unzip the downloaded archive to your hard disk. For this chapter we will assume location E:/. Folder E:/Mgfit will be created.
Start MATLAB.
Change MATLAB’s current directory to E:/Mgfit.
To start installation, type mgfit in MATLAB’s command line after ≫ and press Enter.
Respond ‘Yes’ to the query about adding E:/Mgfit to MATLAB’s path.
Restart MATLAB if requested.
After installation, the same command, mgfit, will bring up gfit user interface window.
3.7.2. Analysis of unwinding data
Start gfit by typing mgfit in MATLAB command line. The gfit window will appear.
Click on Model → Pick Model and select unwinding.m file (included in models and data archive) and click Open. The name of the model will appear in the model field and the status line will show empty data set.
Arrange experimental data in a spreadsheet as described in Section 1.2.3.. If using bookchapter’s sample data, open file unwinding_dataset.txt (included in models and data archive) in a spreadsheet application. If using your own data, use that file as an example.
-
To transfer the data to gfit, select the data and copy it into the clipboard. In gfit window, choose menu Data → Paste-add Data. If data import is successful, gfit shows a table of parameters that will be used for simulation or fitting of imported experiments. View data by clicking button Plot.
gfit window shows following parameters: amplitude (A), stepping rate (kf), step length (s), minimum duplex length (minD), background signal (F0), and number of enzyme populations (N).
Adjust starting values of parameters and optimization flags. If fluorescence data is used, set the flag for F0. If analyzing data for one DNA length, clear the flag for minD.
Click button Fit. During optimization, the iterations are shown in the main MATLAB window. If fitting is successful, the status line shows message fitting converged followed by the sum of squared deviations. Two extra columns appear in parameter table showing the optimized parameter values and their asymptotic confidence intervals (see Note 12 and 13). Goodness of fit may be improved by increasing the number of enzyme populations.
Export analysis results to spreadsheet. Select menu Analysis → Copy Report and paste table of parameters into spreadsheet. To export simulation results, select menu Data → Export to Matlab. This command creates structure DS in MATLAB’s workspace. Substructure DS.sim contains an array of experiments from which simulated variables can be accessed one-by-one and copied.
To improve the quality of fit and to test whether discovered optimal parameter values are unique, perform global search by selecting menu Analysis → Random Restart. To terminate the search click button Cancel (see Note 14).
3.7.3. Analysis of polymerization data
Start gfit by typing mgfit in MATLAB command line. gfit window will appear.
Select polymerase_ni.m model by choosing menu Model → Pick Model. The name of the model will appear in the model field.
Arrange experimental data in a spreadsheet as described in Section 1.2.3.. If using bookchapter’s sample data, open file polymerization_dataset.txt (included in models and data archive) in a spreadsheet application. If using your own data, use that file as an example.
-
To transfer the data to gfit, select the data and copy it into the clipboard. In gfit window, choose menu Data → Paste-add Data. If data import is successful, gfit shows a table of parameters that will be used for simulation or fitting of imported experiments. View data by clicking button Plot.
gfit window shows following parameters: base addition rate (kf), dissociation rate (koff), and starting concentration (C0).
Click button Fit. If a good fit cannot be found, polymerization may be occurring with non-uniform kf and koff rates. Allow each step to have a unique kf rate by right-clicking the parameter and selecting Parameters→ Separate elements option.
Search parameter space using Random Restart and export analysis results as described in previous section.
Acknowledgments
Authors would like to thanks the Patel lab members for proofreading the chapter and testing the model and the National Institute of Health grant GM55310.
Footnotes
Check all the protein preparations for nuclease activity to avoid anomalous results due to DNA substrate degradation.
BT1 membranes used for Elutrap are brittle and need extra care while handling. BT2 membranes need to be stored at 4°C and be moist all the time. Reverse the polarity of electrodes for about 30 s before taking a elute fraction of the oligonucleotide to dislodge any DNA sticking to the membranes. Use fresh gloves and blades for handling each oligonucleotide while cutting out the band form the preparatory gel and setting up the elution to prevent any intermixing and contamination due to handling. Do not overexpose the DNA band to UV light.
It is very important to get accurate oligonucleotide concentrations for preparing correctly annealed substrate for all the assays described here.
Sephadex G-25 spin column purification works well for all the oligonucleotides above 8 nt; however, the recovery and purity may vary depending on the secondary structure of the oligo. Other matrices like P-5, P-30 from BioRad or Sephadex G-50 could also be tried if needed. Use medium size particle grade matrix powder for making the spin column.
It is good to have GC proportion of the duplex region evenly distributed through the entire duplex region to avoid getting biased rates due to localized high or low GC patches. During oligonucleotide design avoid primase recognition site 3′CTG (on the top oligo) to prevent primase activity when not needed. Avoid freezing and thawing the annealed substrates to retain their proper form. It is best to make them fresh or if required store at 4 °C.
Do not have Tris or any other competing amine group in the labeling reactions with fluorescein for efficient labeling of the amine group. Fluorescein labeled DNA substrate should always be protected from light by using dark tubes for reactions and storage. Keep the stopped flow reaction chamber covered with aluminum foil to avoid outside light exposure of the dye.
In all the assays described here, it is important to keep a basal level of EDTA in protein assembly solution to chelate any contaminating magnesium ions present in buffers to prevent uncontrolled start of the reaction.
All the buffer solutions used should be filtered through the 0.22 micron filters when ever possible to avoid blockage or bacterial growth in the RQF or stopped flow instruments. Avoid air bubbles in tubes, loops and syringes in both the instruments when collecting data. A vacuum line is required for the RQF instrument for efficient flushing and drying of the loops. Do not connect the RQF instrument exit line to vacuum when valves are in ‘load’ position (reactants in the sample loading syringe) or in ‘fire’ mode during the experiment. Clean RQF and stopped flow instrument tubes before and after to prevent any blockage/buildup in the system. Never fire either instruments while having valves in ‘load’ position.
The quenched DNA unwinding reactions assays should be loaded immediately on the gel; if possible, as the experiment is being carried out, to avoid re-annealing of the complementary strands especially in reactions without trap.
Use fresh DTT for T7 DNA polymerase assembly. Store the assembled T7 DNA polymerase on ice until use (2 – 3 hrs) and do not use the frozen assembly.
Sequencing gels should be made with high purity acrylamide solutions and reagents. The gels should be run at high power to have temperatures around 45–50°C to get high resolution of the bands. Gel prepared with wedge spacer (0.25–0.4 mm here) gives good resolution for the bands of 20–80 mer size range. Sequencing gel loading dye to sample ratio should be increased if length or GC content of the duplex region of the DNA substrate is higher than mentioned here. Xylene cyanol dye interferes with the fluorescein intensity measurements and should not be used with fluorescent samples.
The parameters of unwinding can be estimated more reliably by globally fitting unwinding kinetics for substrates of different length and similar GC composition. Processivity is best determined from the gel-based assay by analyzing unwinding amplitude as a function of dsDNA length (31).
Fitting operation can be aborted at any time by clicking Cancel. If, for any reason, fitting did not complete successfully, the latest set of parameters will appear in the right column. To continue optimization, copy new parameter values to the starting ones by clicking column header ≪optimum or ≪value and click button Fit. Sometimes a successful fit can be improved simply by re-starting it.
Random restart method searches parameter space for the best fit by continuously reinitializing local optimization from randomly selected parameter values. The results of local optimizations are accumulated in file Mgfit/Temp/Optim_nnn.txt. The file can be opened in a spreadsheet and will appear as a table with each row representing parameter values at a local optimum. The first column of the table contains goodness of fit values. Parameters appear in the table in the order they were discovered. Sort the table by the first column to locate the best fits.
References
- 1.Lohman TM. Helicase-catalyzed DNA unwinding. J Biol Chem. 1993;268:2269–2272. [PubMed] [Google Scholar]
- 2.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 3.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 4.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 5.Donmez I, Rajagopal V, Jeong YJ, Patel SS. Nucleic acid unwinding by hepatitis C virus and bacteriophage t7 helicases is sensitive to base pair stability. J Biol Chem. 2007;282:21116–21123. doi: 10.1074/jbc.M702136200. [DOI] [PubMed] [Google Scholar]
- 6.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egelman EH, Yu X, Wild R, Hingorani MM, Patel SS. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 9.Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage t7. Mol Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 10.Tabor S, Richardson CC. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc Natl Acad Sci USA. 1981;78:205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DE, Narayan M, Patel SS. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J Mol Biol. 2002;321:807–819. doi: 10.1016/s0022-2836(02)00733-7. [DOI] [PubMed] [Google Scholar]
- 12.Rasnik I, Jeong YJ, McKinney SA, Rajagopal V, Patel SS, Ha T. Branch migration enzyme as a Brownian ratchet. EMBO J. 2008;27:1727–1735. doi: 10.1038/emboj.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahnert P, Patel SS. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J Biol Chem. 1997;272:32267–32273. doi: 10.1074/jbc.272.51.32267. [DOI] [PubMed] [Google Scholar]
- 14.Hacker KJ, Johnson KA. A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry. 1997;36:14080–14087. doi: 10.1021/bi971644v. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan DL, Davey MJ, O’Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J Biol Chem. 2003;278:49171–49182. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol. 2000;301:285–299. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- 17.Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W. Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB Helicase? Fluorescence energy transfer studies. J Biol Chem. 1998;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- 18.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 19.Modrich P, Richardson CC. Bacteriophage T7 Deoxyribonucleic acid replication in vitro. A protein of Escherichia coli required for bacteriophage T7 DNA polymerase activity. J Biol Chem. 1975;250:5508–5514. [PubMed] [Google Scholar]
- 20.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 21.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 22.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci USA. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 25.van Oijen AM. Single-molecule studies of complex systems: the replisome. Mol Biosyst. 2007;3:117–125. doi: 10.1039/b612545j. [DOI] [PubMed] [Google Scholar]
- 26.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Oijen AM. Cutting the forest to see a single tree? Nat Chem Biol. 2008;4:440–443. doi: 10.1038/nchembio0808-440. [DOI] [PubMed] [Google Scholar]
- 28.Tanner NA, Hamdan SM, Jergic S, Schaeffer PM, Dixon NE, van Oijen AM. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc Natl Acad Sci USA. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 31.Jeong YJ, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc Natl Acad Sci USA. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Picha KM, Patel SS. Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J Biol Chem. 1998;273:27315–27319. doi: 10.1074/jbc.273.42.27315. [DOI] [PubMed] [Google Scholar]
- 33.Levin MK, Hingorani MH, Holmes RM, Patel SS, Carson JH. Methods Mol Biol. Humana Press, Inc; 2009. Model-based global analysis of heterogeneous experimental data using gfit. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel SS, Bandwar RP, Levin MK. Transient-state kinetics and computational analysis of transcription initiation. In: Johnson KA, editor. The practical approach series/Kinetic analysis of macromolecules. Oxford University Press; 2002. [Google Scholar]
- 35.Lucius AL, Maluf NK, Fischer CJ, Lohman TM. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys J. 2003;85:2224–2239. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel SS, Rosenberg AH, Studier FW, Johnson KA. Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J Biol Chem. 1992;267:15013–15021. [PubMed] [Google Scholar]
- 37.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 38.Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 39.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kallansrud G, Ward B. A comparison of measured and calculated single- and double-stranded oligodeoxynucleotide extinction coefficients. Anal Biochem. 1996;236:134–138. doi: 10.1006/abio.1996.0141. [DOI] [PubMed] [Google Scholar]
- 41.Sjoback R, Nygren J, Kubista M. Absorption and fluorescence properties of fluorescein. Spectrochim Acta [A] 1995;51:7–21. [Google Scholar]
- 42.Donmez I, Patel SS. Coupling of DNA unwinding to nucleotide hydrolysis in a ring-shaped helicase. EMBO J. 2008;27:1718–1726. doi: 10.1038/emboj.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]