Abstract
Objectives
Chikungunya (CHIK) has been classified as a communicable disease group IV in South Korea since late 2010. Based on this, we investigated the extent of imported cases of CHIK in dengue-suspected individuals returning from dengue-endemic regions.
Methods
A total of 486 dengue-suspected serum samples were screened for CHIK by enzyme-linked immunosorbent assay (ELISA) and reverse transcription-polymerase chain reaction (RT-PCR) analysis. Further RT-PCR-positive samples were used for the viral culture, and CHIK was subsequently confirmed by sequence analysis of the culture samples.
Results
Five out of 107 dengue-positive samples were found to be positive for CHIK and 15 out of 379 dengue-negative samples were found to be positive for CHIK by immunoglobulin M ELISA. Further, a CHIK virus was isolated from one of the two RT-PCR-positive sera by cell culture and confirmed by sequence analysis.
Conclusion
The present study documents the first evidence of travel-associated CHIK infection in South Korea. Considering the intense international traffic between countries, our finding emphasizes the urgent need for active patient and vector surveillance for timely response to reduce the introduction of CHIK in Korea.
Keywords: chikungunya, dengue, enzyme-linked immunosorbent assay, reverse transcription-polymerase chain reaction
1. Introduction
Chikungunya (CHIK) is a mosquito-borne febrile disease caused by the chikungunya virus (CHIKV), which belongs to the genus Alphavirus of the Togaviridae family [1]. The CHIKV has a single-stranded, positive-sense RNA genome, which consists of 5′-nsP1-nsP2-nsP3-nsP4-junction region-C-E3-E2-6K-E1-poly (A)-3′ [2]. The virus was first isolated from a febrile patient in Tanzania in 1953 and spread into Africa, Pacific, and South East Asian regions [3,4,5]. The virus has now been transmitted to more than 20 countries and is one of the most potential risks among the travel-associated diseases for travelers to tropical regions [6,7,8]. Since its re-emergence in the Indian Ocean Island in 2005, the virus was recognized as a global public health threat [9]. Three genotypes of CHIKV have been characterized based on their partial E1 sequences, which include Asian, East Central South African, and West African genotypes [10].
The CHIKV is transmitted to humans mainly by Aedes aegypti and Aedes albopictus mosquitoes. Clinical symptoms also resemble dengue, which include chills, fever, headache, myalgia, nausea, vomiting, and rashes [11]. In addition, overlap of disease-occurrence areas make it difficult to identify the exact pathogens involved, if the differential diagnostic methods are unavailable. In South Korea, dengue diagnosis began in 2001 at the Korea National Institute of Health (KNIH) and since then imported cases have been continuously reported [12,13]. Because no vaccines or therapeutic agents are available, early detection of the virus is important for timely vector control and prevention of viral dissemination into nonendemic regions. To cope with an abrupt re-emergence of CHIK in South East Asian regions, the Korean government designated CHIK as a communicable disease group IV in late 2010. However, before enacting this law, it was necessary to estimate the occurrence of CHIK cases introduced by patients with dengue-like diseases who had returned from dengue-endemic regions. In the present study, we first documented the travel-associated introduction of CHIKV into South Korea during 2009–2010, which stresses the importance of making modifications to the travel health policies.
2. Materials and Methods
2.1. Serum samples
A total of 873 serum samples were sent to the Division of Arboviruses at the KNIH for dengue diagnosis during 2009–2010. Among them 486 serum samples were tested for CHIK, in which 107 and 379 sera were positive and negative for dengue immunoglobulin M (IgM), respectively. This study was conducted as a routine laboratory diagnosis and did not require either informed consent or institutional review.
2.2. IgM enzyme-linked immunosorbent assay
All the collected serum samples were tested for the presence of CHIKV-specific IgM antibodies using the commercial enzyme-linked immunosorbent assay (ELISA) kit (NovaTec, Germany) according to the manufacturer's instruction. Based on absorbance at 450 nm, the NovaTec Units (NTUs) were calculated for each sample and samples having NTU > 11 were considered positive.
2.3. Multiplex reverse transcription-polymerase chain reaction
The viral RNA was extracted from serum samples by Viral Gene-spin DNA/RNA extraction kit (iNtRON Biotechnology, Gyeonggi-do, Korea) according to the manufacturer's protocol. The isolated RNA was reverse transcribed and amplified according to the one-step duplex reverse transcription-polymerase chain reaction (RT-PCR) assay developed for differential diagnosis of DENV and CHIKV [14]. In brief, two primer pairs (CHIK1 and 2, DEN1 and 2) targeting the CHIKV E1 gene and DENV C/prM gene were incorporated into the reaction and the exact pathogen was determined based on the fragment size (CHIKV: 205 bp; DENV: 511 bp). To exclude cross contamination by laboratory-reference CHIKV strain, we used DENV as a positive control in the RT-PCR assay (Figure 1). Further confirmation was achieved by PCR direct sequencing using the primers CHIK1 and CHIK2 (Macrogen, Seoul, Korea).
Figure 1.
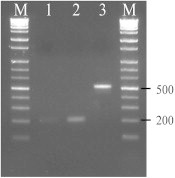
Detection of chikungunya virus (CHIKV) from a patient’s serum by duplex reverse transcription-polymerase chain reaction targeting both CHIKV and dengue virus (DENV). Lane 1, patient code 09-B09; lane 2, patient code 09-B36; lane 3, positive control (DENV), lane M, 1kb DNA ladder. The expected size of CHIKV and DENV fragment was 205 and 511 bp, respectively.
2.4. Virus isolation
Virus isolation from RT-PCR-positive serum was attempted using invertebrate C6/36 cells (ATCC, CRL 1660). Approximately 1 mL of 20-fold diluted serum was inoculated into C3/36 cells cultured in a 25-cm2 culture flask with minimal essential medium containing 5% fetal bovine serum and antibiotics. After absorption for 1 hour, the cells were washed once with phosphate-buffered saline and incubated at 28°C in a humidified CO2 incubator. The culture supernatant was collected 7 days after infection, clarified by brief centrifugation, and stored as small aliquots at −70°C for further analysis. The presence of virus in the culture supernatant was confirmed by RT-PCR and sequence analysis.
2.5. Nucleotide sequencing and phylogenetic analysis
The culture supernatant confirmed as CHIKV was further amplified using three primer pairs (9648F/10403R, 10145F/11158R, and 10959F/11770R) encompassing E2, E1, and 3′NC [15]. The RT-PCR analysis was performed using the SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA, USA) and DyNAzyme EXT DNA polymerase (Thermo). The resulting nucleotide sequences were assembled using the SeqMan program in the Lasergene software (version 8; DNASTAR, Madison, WI, USA) and the final sequence (approximately 1.9 kb in length) was identified by NCBI-BLAST search. Nucleotide sequences of 1044-nt long E1 partial genes from 21CHIKV isolates were selected for sequence comparison and phylogenetic analysis. Nucleotide and amino-acid sequences were aligned by CLUSTAL W algorithm using the BioEdit software (version 7.0). The neighbor-joining algorithm and the maximum composite likelihood model were used for tree construction by MEGA software (version 5.1) [16]. The reliability of the analysis was evaluated by a bootstrap test with 2000 replications. The o'nyong-nyong virus (GenBank ID: AF079456) was used as out-group in the tree.
3. Results
3.1. Laboratory testing
A total of 488 serum samples were tested for CHIKV infection. The results were summarized in Table 1 and Figure 1. Twenty serum samples were found to be ELISA positive and two serum samples were RT-PCR positive. It is notable that five cases were IgM positive for both CHIKV and DENV. Age distribution of CHIK-positive cases was four in teens, one in 20s, seven in 30s, one in 40s, two in 50s, one in 70s, and five cases are unclear. Travel destination of the cases was mainly South East Asia including Indonesia, Malaysia, the Philippines, Singapore, and Vietnam. The interval from onset of illness to serum sampling in CHKIV ELISA-positive cases was 4–32 days. As for two cases that were positive in RT-PCR analysis, possibility of cross contamination was excluded by sequence comparison with laboratory reference strain Ross (GenBank ID: AF490259).
Table 1.
Result of laboratory testing for Chikungunya
| Patient ID | Age | Days after |
Dengue |
Chikungunya |
Travel destination | ||
|---|---|---|---|---|---|---|---|
| Onset∗ | ELISA | RT-PCR | ELISA | RT-PCR | |||
| 09-A37 | IU† | IU | – | – | + | – | Philippines |
| 09-A63 | 33 | 7 | – | – | + | – | Indonesia |
| 09-A73 | 34 | IU | – | – | + | – | Indonesia |
| 09-B08 | 70 | 32 | – | – | + | – | Malaysia |
| 09-B09 | IU | IU | – | – | – | + | IU |
| 09-B36‡ | 57 | 26 | –– | –– | –+ | +– | Malaysia/Singapore |
| 09-C54 | 38 | IU | – | – | + | – | IU |
| 09-C77 | IU | 6 | – | – | + | – | Vietnam |
| 09-D14 | 22 | 9 | – | – | + | – | IU |
| 10-D052 | 11 | IU | – | – | + | – | IU |
| 10-D062 | 40 | IU | – | – | + | – | IU |
| 10-D094 | IU | IU | – | – | + | – | IU |
| 10-D100 | 36 | IU | – | – | + | – | IU |
| 10-D220 | 19 | 12 | + | – | + | – | Indonesia |
| 10-D226 | 56 | IU | – | – | + | – | IU |
| 10-D234 | 32 | 4 | + | – | + | – | Philippines |
| 10-D328 | IU | IU | + | – | + | – | East Timor |
| 10-D342 | 33 | 6 | – | – | + | – | IU |
| 10-D375 | 32 | IU | + | – | + | – | Philippines |
| 10-D445 | 12 | IU | – | – | + | – | IU |
| 10-D497 | IU | IU | + | – | + | – | Vietnam |
∗ Interval from the onset of illness to serum collection; †Information was unavailable. ‡ Virus was isolated from this patient’s acute-phase serum and the isolate was designated as KNIH/2009/77. ELISA = enzyme-linked immunosorbent assay; RT-PCR = reverse transcription-polymerase chain reaction.
3.2. Virus isolation and phylogenetic analysis
The two serum samples that were positive in RT-PCR analysis were subjected to virus isolation and one isolate was recovered from serum of a febrile patient (GenBank ID: 09-B36) who returned from Malaysia and Singapore. The isolate was further sequenced using primer pairs covering E2, E1, and 3′NS genes and named as KNIH/2009/77 (GenBank ID: KC8110970). The NCBI-BLAST search showed that the KNIH/2009/77 isolate has 98–99% sequence similarity with CHIKV isolates circulating in the South East Asia including Malaysia, Singapore, and Thailand. Similar to CHIKVs that have appeared since 2006 and had adaptive infectivity to Ae. albopictus mosquitoes, KNIH/2009/77 showed a typical amino-acid substitution (alanine to valine) at position 226 of the E1 protein (Figure 2). A phylogenetic tree was constructed based on partial sequences of the E1 gene of CHIKV isolates. The KNIH/2009/77 isolate was clustered within the Central/East/South Africans (CESA) genotype together with isolates from Malaysia and Singapore (Table 2, Figure 3).
Figure 2.
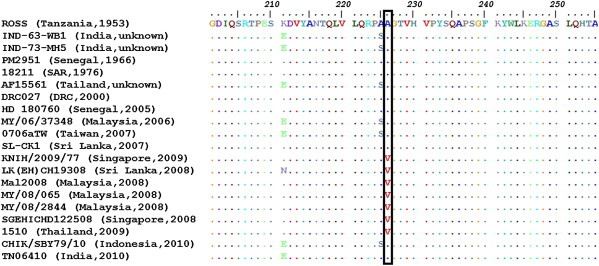
The alignment of amino-acid sequences of the E1 gene of chikungunya viruses. Amino-acid sequence substitution from alanine (A) to valine (V) was shown at the position 226. Country and isolation year are described in the parentheses of each virus name. SAR = South African Republic.
Table 2.
Details of the chikungunya viruses used for phylogenetic analysis
| Strain name | Geographic origin | Sampling year | GenBank ID |
|---|---|---|---|
| Ross | Tanzania | 1953 | AF490259 |
| lbH35 | Nigeria | 1964 | AF192893 |
| PM2951 | Senegal | 1966 | AF192891 |
| 18211 | SAR | 1976 | AF192903 |
| Ag41855 | Uganda | 1982 | AF192907 |
| DRC027 | DRC | 2000 | AY549577 |
| HD 180760 | Senegal | 2005 | HM045817 |
| MY/06/37348 | Malaysia | 2006 | FN295483 |
| 0706aTw | Taiwan | 2007 | EU192143 |
| SL-CK1 | Sri Lanka | 2007 | HM045801 |
| MY/08/065 | Malaysia | 2008 | FN295485 |
| SGEHICHD122508 | Singapore | 2008 | FJ445502 |
| LK(EH)CH19308 | Sri Lanka | 2008 | FJ513677 |
| MY/08/2844 | Malaysia | 2008 | FN295489 |
| Mal2008 | Malaysia | 2008 | GQ168719 |
| 1510 | Thailand | 2009 | JN661154 |
| CHIK/SBY79/10 | Indonesia | 2010 | AB678694 |
| TN06410 | India | 2010 | HM159389 |
| IND-63-WB1 | India | Unknown | EF027140 |
| IND-73-MH5 | India | Unknown | EF027141 |
| AF15561 | Thailand | Unknown | EF452493 |
DRC = Democratic Republic of Congo; SAR = South African Republic.
Figure 3.
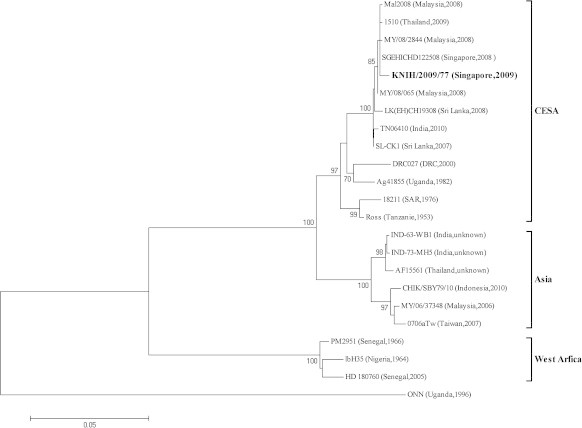
Phylogenetic analysis of the chikungunya virus based on the partial E1 gene (1044 nt) from 21 isolates. The tree was constructed by the neighbor-joining method (substitution model: maximum composite likelihood) using MEGA (version 5.1). The o'nyong-nyong virus (ONN, GenBank ID: AF079456) was used as out-group in the tree. The percentage of bootstrap values is shown at each node (2000 replications). CESA = Central/East/South African; DRC = Democratic Republic of Congo; SAR = South African Republic.
4. Discussion
In the present study, we investigated the extent of CHIKV infections in dengue-suspected patients who returned from travel. A total of 22 serum samples showed evidence of travel-related CHIK during 2009 and 2010. It was revealed that 4.5 % (17/379) of dengue-negative samples was CHIK-positive. Interestingly, results of ELISA showed that 4.7% (5/107) of dengue-positive cases were positive for CHIKV IgM in 2010. Generally, ELISA is just a screening test and a confirmatory testing such as plaque reduction neutralization test (PRNT) should be performed for differentiating CHIK and dengue. However, our laboratory had not set up the method at that time and it remains to be resolved later. We attempted to isolate the virus in the two RT-PCR-positive cases and one was recovered successfully from the serum collected within 2 days after onset of illness. A CHIKV infection has been categorized as a communicable disease group IV since late 2010 in South Korea. This functioned as a passive patient surveillance system based on compulsory case reporting by physicians. Since enacting the law, however, the number of requests for CHIK diagnosis from hospitals was only 25 cases by March 2013. It may be due to the low-level recognition of the disease in the medical community. From this viewpoint, the number of real CHIK might have been underestimated in South Korea. It is hard to differentiate dengue from CHIK with clinical symptoms alone. Therefore, we recommend serological and molecular testing of CHIKV as well as dengue to diagnose febrile patients who returned from tropical regions. It was revealed that mutant viruses with amino-acid substitution (alanine to valine at position 226) at the E1 gene have superior infectivity in Ae. albopictus than Ae. aegypti mosquitoes and are responsible for recent outbreaks in India, Malaysia, and Singapore [15,17,18]. An isolate, KNIH/2009/77, recovered in this study was also a mutant-type CHIKV that may have the potential to infect Ae. albopictus mosquitoes distributed in Korea. The possibility of importation and spread of CHIKV by acute febrile traveler was reported in 2007 in Italy [19]. Considering that Ae. albopictus mosquitoes are already distributed in Korea, we could not exclude an abrupt introduction of CHIKV as well as DENV.
In conclusion, we presented first evidence of travel-associated CHIKV infections in South Korea. This finding highlights the need to conduct laboratory and vector surveillance program for CHIKV as well as an urgent need to modify evidence-based travel health policies and recommendations.
Acknowledgments
The authors thank Mr Ryou Jung-Sang for his administrative work in importing CHIKV reference strain from NCPV in the United Kingdom. This study was supported by the National Institute of Health, Korea Centers for Disease Control and Prevention (Grant No. 4837-300-210-13).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Peters CDJ. Fields virology. 2nd ed. In: Fields BN, Knipe DM, editors. New York: Raven Press; 1990. Alphaviruses; p. 713–61.
- 2.Strauss EG, Strauss JH. The togaviruses and flaviviruses. In: Schlesinger SSM, editor. New York: Plenum Press; 1986. Structure and replication of the alphavirus genome; p. 35–90.
- 3.Robinson M.C. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. I. Clinical features. Trans R Soc Trop Med Hyg. 1955 Jan;49(1):28–32. doi: 10.1016/0035-9203(55)90080-8. [DOI] [PubMed] [Google Scholar]
- 4.Sudeep A.B., Parashar D. Chikungunya: an overview. J Biosci. 2008 Nov;33(4):443–449. doi: 10.1007/s12038-008-0063-2. [DOI] [PubMed] [Google Scholar]
- 5.Powers A.M., Logue C.H. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol. 2007 Sep;88(Pt 9):2363–2377. doi: 10.1099/vir.0.82858-0. [DOI] [PubMed] [Google Scholar]
- 6.Lanciotti R.S., Kosoy O.L., Laven J.J. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis. 2007 May;13(5):764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paramasivan R., Philip Samuel P., Thenmozhi V. Chikungunya virus isolated in Lakshadweep islands in the Indian Ocean: evidence of the Central/East African genotype. Jpn J Infect Dis. 2009 Jan;62(1):67–69. [PubMed] [Google Scholar]
- 8.Tanay A., Schwartz E., Bin H., Zeller H., Niedrig M., Dan M. Chikungunya fever in Israeli travelers returning from northwestern India. J Travel Med. 2008 Sep–Oct;15(5):382–384. doi: 10.1111/j.1708-8305.2008.00244.x. [DOI] [PubMed] [Google Scholar]
- 9.Ng L.C., Hapuarachchi H.C. Tracing the path of Chikungunya virus—evolution and adaptation. Infect Genet Evol. 2010 Oct;10(7):876–885. doi: 10.1016/j.meegid.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Powers A.M., Brault A.C., Tesh R.B., Weaver S.C. Re-emergence of chikungunya and o'nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol. 2000 Feb;81(Pt 2):471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 11.Jupp P.G., McIntosh B.M. The arboviruses: epidemiology and ecology. In: Monath T.P., editor. vol. II. CRC Press; Boca Raton: 1988. pp. 137–158. (Chikungunya Virus Disease). [Google Scholar]
- 12.Jeong Y.E., Kim Y.H., Cho J.E., Han M.G., Ju Y.R. Identification of dengue type 1 virus (DENV-1) in Koreans traveling abroad. Public Health Res Perspect. 2011 Jun;2(1):34–40. doi: 10.1016/j.phrp.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J.-H., Lee D.-W. Dengue fever in South Korea, 2006–2010. Emerg Infect Dis. 2012 Sep;18(9):1525–1527. doi: 10.3201/eid1809.111811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dash P.K., Parida M., Santhosh S.R. Development and evaluation of a 1-step duplex reverse transcription polymerase chain reaction for differential diagnosis of chikungunya and dengue infection. Diagn Microbiol Infect Dis. 2008 Sep;62(1):52–57. doi: 10.1016/j.diagmicrobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Schuffenecker I., Iteman I., Michault A. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006 Jul;3(7):e263. doi: 10.1371/journal.pmed.0030263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011 Oct;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsetsarkin K.A., Vanlandingham D.L., McGee C.E., Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007 Dec;3(12):e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar N.P., Joseph R., Kamaraj T., Jambulingam P. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J Gen Virol. 2008 Aug;89(Pt 8):1945–1948. doi: 10.1099/vir.0.83628-0. [DOI] [PubMed] [Google Scholar]
- 19.Rezza G., Nicoletti L., Angelini R. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007 Dec;370(9602):1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]


