Abstract
Background: Salmonella and Shigella cause significant morbidity and mortality among children worldwide. Increased antimicrobial resistance results in greater burden of disease.
Materials and Methods: From 2005 to 2011, Salmonella and Shigella isolates collected from ill children at a major hospital in Yucatan, Mexico, were subjected to serotyping and antimicrobial susceptibility testing by disk diffusion and agar dilution. The identification of blaCTX, blaCMY, blaSHV, blaTEM, and blaOXA and qnr resistance genes was conducted by PCR and sequencing.
Results: Among 2344 children with acute gastroenteritis, salmonellosis decreased from 17.7% in 2005 to 11.2% in 2011 (p < 0.001). In contrast, shigellosis increased from 8.3% in 2010 to 12.1% in 2011. Compared to children with Salmonella, those with Shigella had significantly more bloody stools (59 vs 36%, p < 0.001), dehydration (27 vs 15%, p = 0.031), and seizures (11 vs 3%, p = 0.03). In Salmonella (n = 365), there was a significant decrease in resistance to ampicillin (43 to 16%, p < 0.001), trimethoprim–sulfamethoxazole (44 to 26%, p = 0.014), and extended-spectrum cephalosporins (27 to 10%, p = 0.009). Reduced susceptibility to ciprofloxacin in Salmonella rose from 30 to 41% (p < 0.001). All ceftriaxone-resistant isolates harbored the blaCMY-2 gene. qnr genes were found in 42 (36%) of the 117 Salmonella isolates with a ciprofloxacin MIC ≥ 0.125 μg/ml. Four were qnrA1 and 38 were qnrB19. Resistance to ampicillin (40%) and trimethoprim–sulfamethoxazole (58%) was common in Shigella (n = 218), but isolates remained fully susceptible to ceftriaxone and ciprofloxacin.
Conclusion: Illness from Salmonella has decreased while severe Shigella infections have increased among children with gastroenteritis in the Yucatan Peninsula. While Shigella resistance to clinically important antibiotics remained unchanged, resistance to most of these, except ciprofloxacin, declined in Salmonella. blaCMY-2 and qnr genes are common in Salmonella isolates.
Keywords: Salmonella, Shigella, incidence, antimicrobial resistance, beta-lactamase genes, blaCMY-2, qnr genes, Mexico
INTRODUCTION
Salmonella and Shigella are associated with a high burden of illness among children in the developing world (Niyogi, 2005; Woc-Colburn and Bobak, 2009; Kotloff et al., 2013). In Mexico, the disease patterns of these pathogens have undergone significant changes in the last decades. During the early 1970s, Shigella dysenteriae type 1 and Salmonella Typhi caused severe outbreaks among the local population. The epidemic strains of S. Typhi and S. dysenteriae were both multidrug resistant (MDR) to chloramphenicol, tetracycline, streptomycin, and sulfonamides, harbored on different plasmids (Datta and Olarte, 1974). Subsequently, S. dysenteriae infections decreased, while Shigella flexneri became the predominant species isolated from endemic infections. S. dysenteriae, nonetheless, was still prevalent among visitors to the Yucatan Peninsula during the late 1980s (Centers for Disease Control [CDC], 1988). Typhoid fever also sharply declined, while non-typhoidal Salmonella, extremely prevalent along the food chain, was identified as one of the main causes of gastroenteritis in hospitalized children. Salmonella Typhimurium, commonly carrying resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline, was the most common serotype encountered in Yucatan (Zaidi et al., 2006). Extended-spectrum cephalosporin (ESC)-resistant S. Typhimurium isolates first emerged in 2002, and rapidly spread throughout the state causing severe diarrhea and fatal systemic infections in infants (Zaidi et al., 2007).
In the past 25 years, the combination of the widespread use of oral rehydration solutions, rising antimicrobial resistance, and regional socioeconomic changes has modified the epidemiology and the impact of these enteric pathogens (von Seidlein et al., 2006; Parry and Threlfall, 2008). There is a clear need to update relevant epidemiologic data (Sansonetti, 2006) as there are few studies from Latin America. Monitoring for ESC and fluoroquinolone resistance is particularly important because these compounds are among the few therapeutic options available for severe Salmonella and Shigella infections. In this study, the main objectives were (1) to analyze trends in incidence and antimicrobial resistance of Salmonella and Shigella infections in children with gastroenteritis and/or systemic infections who sought medical attention at a major hospital in the Yucatan Peninsula, (2) to compare the clinical presentations of the ill children, and (3) determine the prevalence of extended-spectrum beta-lactamases, CMY-2 and Qnr in Salmonella and Shigella isolates.
MATERIALS AND METHODS
SETTING
The Hospital General O’Horan is a tertiary-care public hospital that receives patients from throughout the state of Yucatan and neighboring states in southeast Mexico. The Pediatrics Emergency Department has an active surveillance program for diarrheal pathogens, current since 2000, which includes children with mild to severe gastroenteritis as well as those with systemic infections. Stool cultures are systematically collected from children with diarrhea. Blood cultures are also routinely collected from children who appear septic or develop hematological, neurological, or other extraintestinal complications.
CLINICAL AND EPIDEMIOLOGICAL DATA
A trained nurse administered standardized questionnaires to collect demographic and clinical information on the childrens’diarrheal episodes.
MICROBIOLOGY
Stool samples were collected in sterile containers containing Cary–Blair media. Samples were inoculated onto XLD, Hektoen Enteric, and brilliant green agars as well as tetrathionate and Rappaport broths and incubated at 35°C for 18–24 h. Broths were subcultured to XLD and brilliant green agar plates and incubated for another 18–24 h. Identification of Salmonella and Shigella isolates were performed with conventional biochemical tests. Salmonella isolates were serotyped by the Kauffman–White scheme using commercial antisera (Becton Dickinson, Franklin Lakes, NJ, USA). All Shigella isolates were serogrouped; serotyping was performed on S. dysenteriae isolates only. Antimicrobial susceptibility testing was routinely performed for ampicillin, chloramphenicol, gentamicin, nalidixic acid, trimethoprim–sulfamethoxazole, ciprofloxacin, ceftriaxone, ceftazidime, and tetracycline by disk diffusion (CLSI, 2012a). Minimum inhibitory concentrations (MICs) for ciprofloxacin, azithromycin, ceftriaxone, nalidixic acid, and furazolidone were determined by agar dilution (CLSI, 2012b). Susceptibility and resistance breakpoints used for azithromycin were 16 and 32 μg/ml, respectively (Sjolund-Karlsson et al., 2011) and 2 μg/ml and 8 μg/ml for furazolidone (Rampling et al., 1990). Ceftriaxone and/or ceftazidime non-susceptible isolates were tested on a secondary panel of beta-lactam antimicrobials that included cefepime, cefotaxime, cefoxitin, imipenem, meropenem, amoxicillin–clavulanic acid, and piperacillin-tazobactam. According to resistance phenotypes, these isolates were also screened for AmpC beta-lactamase production with a combination disk method using cefoxitin (30 μg) and cefoxitin with boronic acid (30 μg + 300 μg; Jacoby et al., 2006).
GENETIC CHARACTERIZATION OF bla and qnr GENES
All ceftriaxone-resistant isolates (MIC ≥ 4) were tested for the presence of beta-lactamase genes. A multiplex PCR was used for the detection of genes encoding the CTX, SHV, TEM, and OXA enzymes according to previously published methods (Briñas et al., 2002; Edelstein et al., 2003). Detection of the blaCMY gene was performed using a previously described assay (Zhao et al., 2001). Salmonella isolates with ciprofloxacin MICs ≥ 0.125 μg/ml were tested for qnrA, qnrB, and qnrS genes following published methods (Robicsek et al., 2006).
Specific group primers used for the study are listed in Table 1. Control strains were kindly provided by Patrick McDermott (U.S. Food and Drug Administration) and Jesus Silva-Sanchez (Instituto Nacional de Salud Publica, Mexico).
Table 1.
Specific group primers used for PCR assays for beta-lactamase and qnr genes.
| PCR name | Primer name | Sequence (5′–3′) | Length (base) | Amplicon size (pb) |
|---|---|---|---|---|
| Multiplex TEM, SHV, CTX, OXA | tem-F | TTCTTGAAGACGAAAGGGC | 19 | 1150 |
| tem-R | ACGCTCAGTGGAACGAAAC | 19 | ||
| shv-F | CACTCAAGGATGTATTGTG | 19 | 702 | |
| shv-R | TTAGCGTTGCCAGTGCTCG | 19 | ||
| ctx-F | TTTGCGATGTGCAGTACCAGTAA | 23 | 544 | |
| ctx-R | CGATATCGTTGGTGGTGCCATA | 22 | ||
| oxa-F | TTCAAGCCAAAGGCACGATAG | 21 | 885 | |
| oxa-R | TCCGAGTTGACTGCCGGGTTG | 21 | ||
| CMY | cmy-F | GACAGCCTCTTTCTCCACA | 19 | 1000 |
| cmy-R | TGGAACGAAGGCTACGTA | 18 | ||
| Multiplex qnrA, qnrB and qnrS | qnrA-F | ATTTCTCACGCCAGGATTTG | 20 | 516 |
| qnrA-R | GATCGGCAAAGGTTAGGTCA | 20 | ||
| qnrB-F | GATCGTGAAAGCCAGAAAGG | 20 | 469 | |
| qnrB-R | ACGATGCCTGGTAGTTGTCC | 20 | ||
| qnrS-F | ACGACATTCGTCAACTGCAA | 20 | 417 | |
| qnrS-R | TAAATTGGCACCCTGTAGGC | 20 |
Amplified products were separated by gel electrophoresis on 2% agarose and stained with ethidium bromide. Sequencing was performed using both forward and reverse PCR primers on a ABI Prism 3130 XL Genetic Analyzer (Applied Biosystems). Sequences were analyzed using the National Center for Biotechnology Information’s BLAST network service (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences were compared to the original gene sequences reported in the GenBank database of accession numbers J01749, X91840, AY070235, EU432277, FJ460235 (Sutcliffe, 1979; Bauernfeind et al., 1996; Jacoby et al., 2008; Wiesner et al., 2009). Sequences for qnrA1 and qnrB19 were submitted under accession numbers KF517418 and KF517417, respectively.
DATA ANALYSIS
The chi-square test was used to detect resistance trends over time and differences in clinical presentations for each pathogen; a p value < 0.05 was considered significant. The odds ratio and its 95% confidence interval (CI) were calculated for the different clinical variables. Antimicrobial susceptibility results were analyzed with Whonet software version 5.6 (www.whonet.org).
ETHICS
The data was obtained from two different studies that were approved by the Hospital General O’Horan Research and Ethics Committee; written informed consent was obtained from the children’s guardians to collect stool samples and use the data for scientific purposes. For this analysis, clinical and microbiological data were entered on new databases; a unique numerical code was assigned to each patient and personal identifiers were removed to ensure anonymity.
RESULTS
EPIDEMIOLOGY
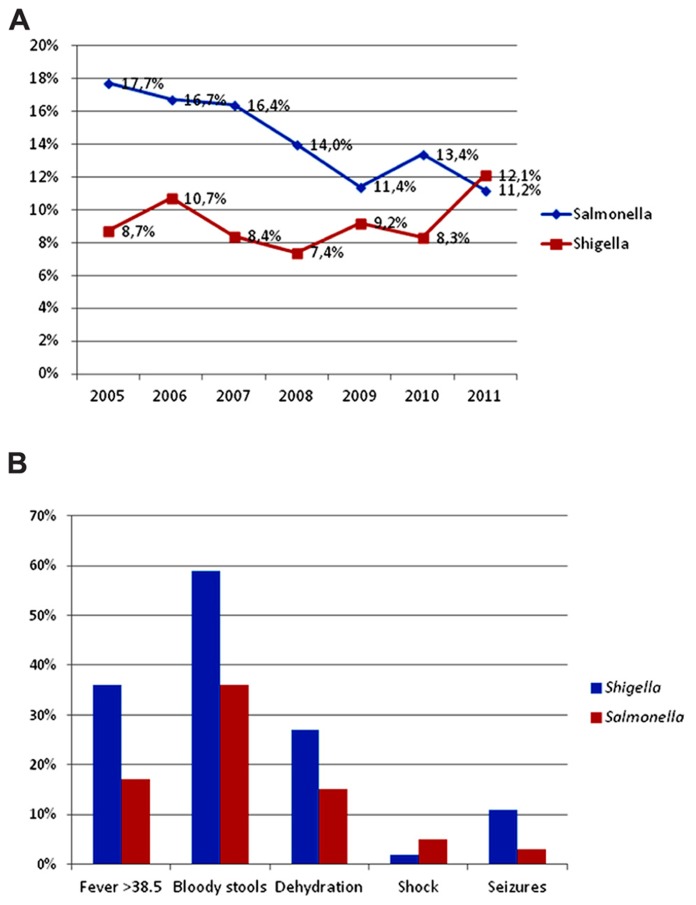
From January 2005 to December 2011, 2344 children under 10 years of age who were admitted to the hospital for acute gastroenteritis were included in the study. One child with fever and malaise without gastroenteritis, whose blood culture was positive for S. Typhi, was also included. Children came from 94 localities within the state of Yucatan and from 35 other cities or towns in the neighboring states. Eight hundred ninety-one (38%) were under 1 year of age, 1176 (54%) were between 1 and 4 years of age, and 186 (8%) were between 5 and 9 years of age. Salmonella gastroenteritis decreased from 17.7% of all cases in 2005 to 11.2% in 2011 (p < 0.001). Shigella infections increased during this period, with a precipitous rise from 8.3% in 2010 to 12.1% in 2011, but the difference was not statistically significant (Figure 1A). A total of 365 Salmonella and 218 Shigella isolates were collected. The five most commonly isolated Salmonella serovars from ill children remained constant; S. Typhimurium was consistently the top serovar (19–21%) followed by S. Agona (9%), S. Muenchen (6–7%), S. Muenster (6–11%), and S. Enteritidis (5–7%). The S. Typhi isolate was susceptible to all tested antibiotics. Similarly, there was no change in the distribution of Shigella species; S. flexneri was predominant (68–76%), followed by S. sonnei (16–17%), S. boydii (5–12%), and S. dysenteriae (2%). S. dysenteriae strains belonged to type 3 (two isolates) and type 2 (two isolates).
FIGURE 1.
Frequency and clinical presentation of Salmonella and Shigella infections in ill children at a state referral hospital inYucatan, Mexico, 2005–2011. (A) There was a statistically significant trend toward decreasing Salmonella gastroenteritis, from 17.7% in 2005 to 11.2% in 2011 (p < 0.001). In contrast, there was a precipitous increase in shigellosis from 8.3% in 2010 to 12.1% in 2011. (B) Children with Shigella had twice as much fever ≥38.5°C, bloody stools, dehydration, and almost four times as many seizures, compared to those with Salmonella. Children with salmonellosis, however, were more likely to be admitted with hypovolemic shock.
CLINICAL PRESENTATION
Prospectively obtained clinical data was available for 112 patients with Shigella and 118 patients with Salmonella. Compared to children with Salmonella, those with Shigella had more fever ≥ 38.5°C (36 vs 17%, p = 0.001, OR = 2.7, 95% CI = 1.4–5.3), bloody stools (59 vs 36%, p < 0.001, OR = 2.6, 95% CI = 1.5–4.6), dehydration (27 vs 15%, p = 0.031, OR = 2.0, 95% CI = 1.0–4.1), and almost four times as many seizures (11 vs 3%, p = 0.03, OR = 3.4, 95% CI = 0.98–13.0; Figure 1B).
ANTIMICROBIAL RESISTANCE
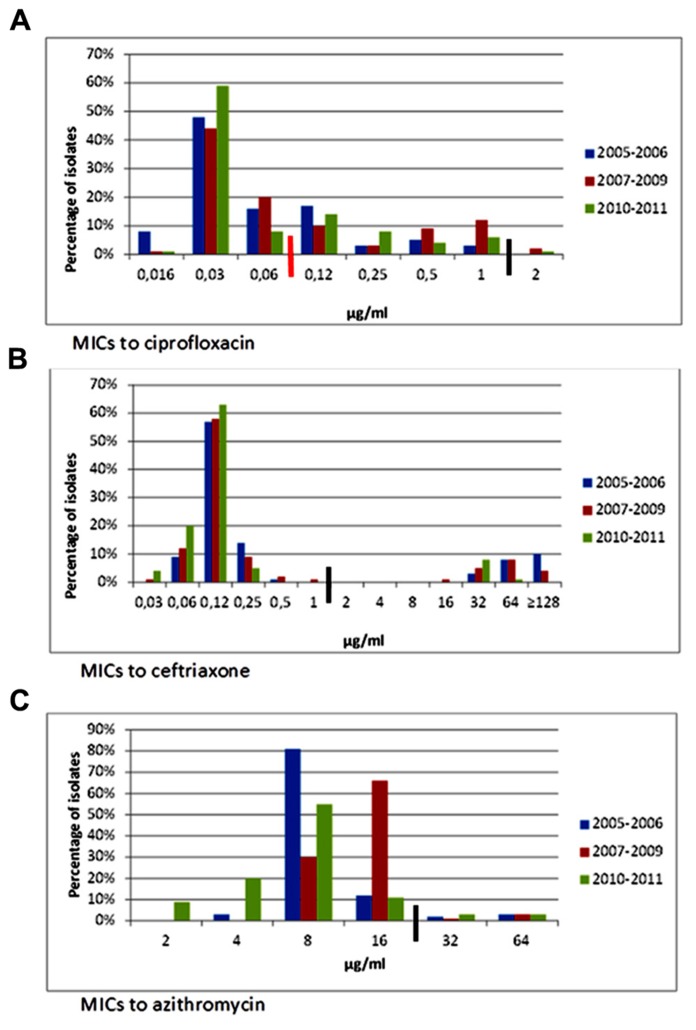
Ampicillin resistance in Salmonella decreased from 43% in 2005 to 16% in 2011 (p < 0.001), while trimethoprim–sulfamethoxazole resistance decreased from 44% to 26% (p = 0.014). Likewise, resistance to ceftriaxone and other ESC, most commonly observed in S. Typhimurium, decreased from 27% to 10% (p = 0.009). Trends for the different time periods throughout the study are shown on Table 2 and Figures 2A–C. MIC50 and MIC90 values for the whole study period were, respectively, 0.03 and 0.5 μg/ml for ciprofloxacin, 0.125 and 64 μg/ml for ceftriaxone, and 8 and 16 μg/ml for azithromycin.
Table 2.
Antimicrobial susceptibility in Salmonella isolates from ill children inYucatan, Mexico, 2005–2011. Percentage of resistance*.
| Year of isolation | AMP† | CRO‡ | CAZ‡ | AZT | CIP | NAL | CHL | FZD | GEN | STX§ | TET |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005–2006 (n = 147) | 33.3 | 19.7 | 19.7 | 4.8 | 0 | 29.2 | 33.4 | 30.8 | 12.2 | 38.1 | 52.4 |
| 2007–2009 (n = 138) | 25.4 | 18.1 | 18.1 | 3.6 | 2.2 | 37.7 | 31.9 | 28.4 | 9.4 | 34.1 | 45.7 |
| 2010–2011 (n = 80) | 20.0 | 8.8 | 8.8 | 5.0 | 1.2 | 31.3 | 33.8 | 28.7 | 7.5 | 28.8 | 42.5 |
AMP, ampicillin; AZT, azithromycin; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; CRO, ceftriaxone; FZD, furazolidone; GEN, gentamicin; NAL, nalidixic acid; STX, trimethoprim–sulfamethoxazole; TET, tetracycline.
Includes resistant and intermediate strains.
Difference from 2005 to 2011: p < 0.001.
Difference from 2005 to 2011: p = 0.009.
Difference from 2005 to 2011: p = 0.014.
FIGURE 2.
Minimum inhibitory concentrations to ciprofloxacin, ceftriaxone, and azithromycin in Salmonella isolates from ill children at a state referral hospital in Yucatan, Mexico, 2005–2011. Thick black lines indicate breakpoints for resistance. Number of isolates for each period was as follows: 2005–2006, n = 147; 2007–2009, n = 138, and 2010–2011, n = 80. (A) Ciprofloxacin MICs. Reduced susceptibility to ciprofloxacin in Salmonella rose from 28% in 2005–2006 to 33% in 2010–2011. Non-susceptibility (MIC = 2) first emerged in 2007 in a S. Anatum isolate and later appeared in S. Typhimurium multidrug-resistant, cephalosporin-resistant isolates. Black line is resistance breakpoint for isolates from stools; red line is breakpoint for systemic infections. CLSI breakpoints for susceptibility and resistance in isolates from stools are 1 and 4 μg/ml, respectively. Breakpoints for susceptibility and resistance in isolates from systemic infections are 0.06 and 1 μg/ml, respectively. (B) Ceftriaxone MICs. Resistance to ceftriaxone and other extended-spectrum cephalosporins significantly decreased from 19.7% during 2005–2006 to 8.8% in 2010–2011. Salmonella isolates were distributed into two distinct populations, one which was susceptible and the other resistant to ceftriaxone. CLSI breakpoints for susceptibility and resistance are 1 and 4 μg/ml, respectively. (C) Azithromycin MICs. No significant changes in azithromycin MICs were noted. Suggested breakpoints for susceptibility and resistance are 16 and 64 μg/ml, respectively (Sjolund-Karlsson et al., 2011).
Reduced susceptibility to ciprofloxacin (MIC ≥ 0.12 and ≤ 1 μg/ml) was found in 113 Salmonella isolates evenly distributed throughout the study period; 77 of these (68%) were resistant to nalidixic acid, and was most commonly seen in S. Typhimurium (34%), S. Enteritidis (16%), S. Albany (9%), S. Muenster (6%), S. Muenchen (5%), and S. Reading (5%).
The percentage of isolates with reduced susceptibility to ciprofloxacin rose from 30% in 2005 to 41% in 2011 (p < 0.001). The number of isolates with MIC ≥ 1 μg/ml, however, peaked during 2007–2009 (14%) and then decreased in 2010–2011 (7%), although the difference was not significant (Figure 2A). Isolates with ciprofloxacin MIC = 2 μg/ml were first detected in 2007 in S. Anatum, and subsequently, in two MDR, ESC-resistant S. Typhimurium strains (2008 and 2011), and one S. Enteritidis strain (2009).
Sixty-one ceftriaxone and ceftazidime resistant isolates were uniformly resistant to cefotaxime, cefoxitin, aztreonam, and amoxicillin/clavulanic acid; 52% were resistant to piperacillin/tazobactam. All isolates were susceptible to cefepime, imipenem, and meropenem, and were inhibited by cefoxitin in the presence of boronic acid. Before 2005, ESC-resistance was limited to S. Typhimurium; it was subsequently detected in S. Havana (2005), S. Anatum (2007), S. Agona (2008), S. Enteritidis (2009), and S. Weltevreden (2011).
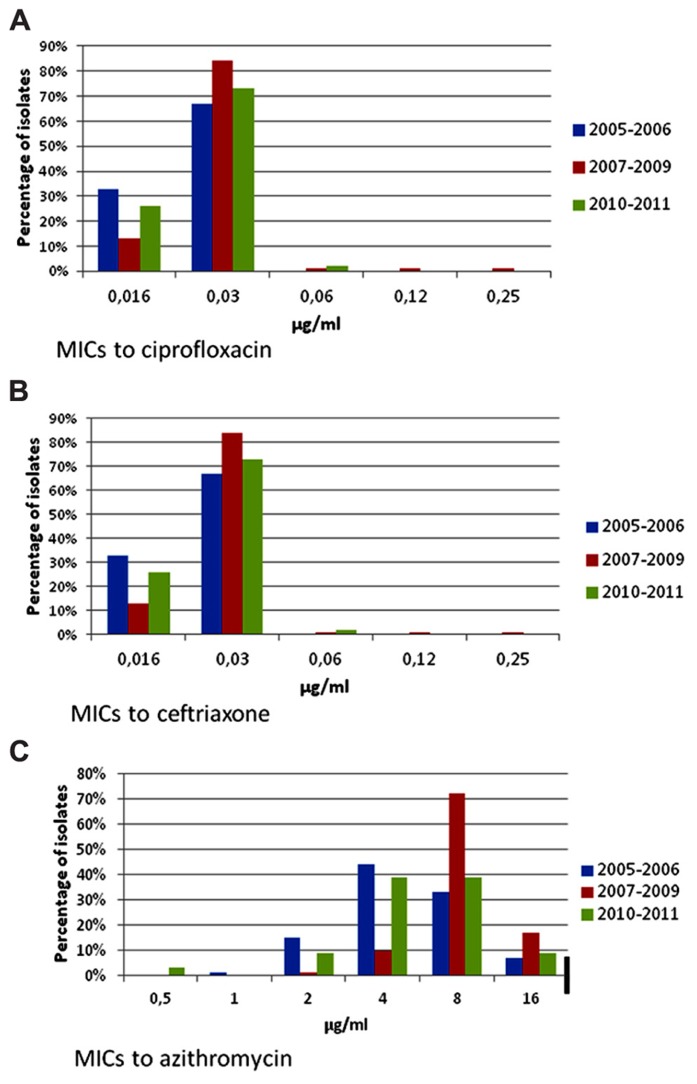
Shigella isolates were frequently resistant to tetracycline, ampicillin, furazolidone, and trimethoprim–sulfamethoxazole (Table 3); all isolates were susceptible to ciprofloxacin, ceftriaxone, and azithromycin. Only one isolate was resistant to nalidixic acid. Resistance to furazolidone decreased from 56.1% in 2005 to 27.1% in 2011 (p < 0.001). There were few changes in the MIC distributions for azithromycin, ceftriaxone, and ciprofloxacin (Figures 3A–C), and no significant resistance trends during the different time periods (Table 3). MIC50 and MIC90 values for the whole study period were, respectively, 0.03 and 0.03 μg/ml for ciprofloxacin, 0.06 and 0.06 μg/ml for ceftriaxone, and 8 and 16 μg/ml for azithromycin.
Table 3.
Antimicrobial susceptibility in Shigella isolates from ill children inYucatan, Mexico, 2005–2011. Percentage of resistance*.
| Year of isolation | AMP | CRO | AZT | CIP | NAL | CHL | FZD† | STX | TET |
|---|---|---|---|---|---|---|---|---|---|
| 2005–2006 (n = 73) | 39.8 | 0 | 0 | 0 | 0 | 16.4 | 57.5 | 57.5 | 83.6 |
| 2007–2009 (n = 79) | 44.3 | 0 | 0 | 0 | 1.3 | 24.1 | 25.4 | 59.5 | 92.4 |
| 2010–2011 (n = 66) | 34.8 | 0 | 0 | 0 | 0 | 12.1 | 25.8 | 59.0 | 93.9 |
AMP, ampicillin; AZT, azithromycin; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; CRO, ceftriaxone; FZD, furazolidone; GEN, gentamicin; NAL, nalidixic acid, STX, trimethoprim–sulfamethoxazole; TET, tetracycline.
Includes resistant and intermediate strains.
Difference from 2005 to 2011: p < 0.001.
FIGURE 3.
Minimum inhibitory concentrations to ciprofloxacin, ceftriaxone, and azithromycin in Shigella isolates from children with gastroenteritis at a state referral hospital inYucatan, Mexico, 2005–2011. Thick black lines indicate breakpoints for resistance. Number of isolates for each period was as follows: 2005–2006, n = 73; 2007–2009, n = 79 and 2010–2011, n = 66. (A) Ciprofloxacin MICs. Isolates were fully susceptible to ciprofloxacin throughout the study. Two isolates had ciprofloxacin MICs ≥ 0.12 μg/ml, only one of which was resistant to nalidixic acid. CLSI breakpoints for susceptibility and resistance are 1 and 4 μg/ml, respectively. (B) Ceftriaxone MICs. All isolates remained fully susceptible to ceftriaxone with no changes over the study period. CLSI breakpoints for susceptibility and resistance are 1 and 4 μg/ml, respectively. (C) Azithromycin MICs. Suggested breakpoints are the same as those proposed for Salmonella. Although isolates were apparently susceptible to azithromycin throughout the study period, 7–17% of these had an MIC = 16.
PREVALENCE OF qnr AND BETA-LACTAMASE GENES
Of the 117 Salmonella isolates with a ciprofloxacin MIC between 0.125 and 2 μg/ml, 42 (36%) harbored qnr genes. Four (3%) carried qnrA genes and 38 (32%) carried qnrB genes. The number of isolates with qnr genes peaked in 2007 (29%) and decreased to 10% by 2011 (p < 0.001). The qnrA genes, found in two serovars, were 99% identical to qnrA1 gene variants. qnrB genes, 99% identical to qnrB19 variants, were distributed in 11 different serovars (Table 4). Both Qnr determinants presented 100% identity to the originally reported amino acid sequences. All 61 ESC-resistant Salmonella isolates carried a blaCMY gene. The sequences of all PCR products were 99% identical to the prototype blaCMY-2 gene and had 100% identity to the those obtained for blaCMY-2 genes found in S. Typhimurium strains from Yucatan and three other states in Mexico (Wiesner et al., 2009). Only one isolate also harbored a TEM-1 gene; none harbored SHV, CTX, or OXA genes.
Table 4.
Serotype distribution and antimicrobial susceptibility of Salmonella isolates positive for the qnr gene, Yucatan, Mexico, 2005–2011.
| Qnr variant | ID number | Serotype | Year of isolation | Resistance phenotypes to non-quinolone antibiotics | Ciprofloxacin MIC (μg/ml) | Nalidixic acid MIC (μg/ml) |
|---|---|---|---|---|---|---|
| A1 | yuhs10-35 | Enteritidis | 2010 | Nal, Fzd | 1 | 128 |
| yuhs05-13 | Havana | 2005 | Amp, Sxt, Tet | 0.5 | 32 | |
| yuhs05-87 | Havana | 2005 | Amp, Sxt, Tet, Nal | 0.5 | 16 | |
| yuhs05-106 | Havana | 2005 | Amp, Sxt, Tet, Cro | 0.5 | 16 | |
| B19 | yuhs07-5 | Adelaide | 2007 | Nal, Fzd | 0.12 | 32 |
| yuhs07-44 | Adelaide | 2007 | Nal | 0.5 | 64 | |
| yuhs07-71 | Adelaide | 2007 | Nal | 1 | 64 | |
| yuhs08-31 | Adelaide | 2008 | Nal | 1 | 64 | |
| yuhs07-40 | Agona | 2007 | Nal | 0.5 | 32 | |
| yuhs08-34 | Agona | 2008 | Nal | 1 | 64 | |
| yuhs07-38 | Anatum | 2007 | Amp, Sxt, Tet, Nal, Cro | 1 | 64 | |
| yuhs07-53 | Anatum | 2007 | Amp, Sxt, Tet, Nal | 2 | 128 | |
| yuhs09-10 | Derby | 2009 | Tet, Nal | 0.5 | 64 | |
| yuhs07-61 | Enteritidis | 2007 | Nal, Fzd | 1 | 64 | |
| yuhs08-57 | Enteritidis | 2008 | Nal, Fzd | 1 | 64 | |
| yuhs08-59 | Enteritidis | 2008 | Nal, Fzd | 1 | 64 | |
| yuhs09-1 | Enteritidis | 2009 | Nal, Fzd | 2 | 64 | |
| yuhs09-7 | Enteritidis | 2009 | Nal, Fzd | 1 | 64 | |
| yuhs11-43 | Enteritidis | 2011 | Nal, Fzd | 1 | 64 | |
| yuhs07-45 | Havana | 2007 | Sxt, Tet, Chl | 0.5 | 16 | |
| yuhs07-28 | Muenchen | 2007 | Nal | 1 | 32 | |
| B19 | yuhs07-41 | Muenchen | 2007 | Nal | 1 | 32 |
| yuhs08-8-1 | Muenchen | 2008 | Nal | 0.5 | 32 | |
| yuhs08-10 | Muenchen | 2008 | Sxt, Nal | 0.5 | 32 | |
| yuhs08-30-2 | Muenchen | 2008 | Nal | 0.5 | 32 | |
| yuhs11-33 | Muenchen | 2011 | Nal | 0.5 | 32 | |
| yuhs05-97 | Muenster | 2005 | Amp, Sxt, Tet, Nal, Chl, Fzd | 0.12 | 32 | |
| yuhs07-74-2 | Muenster | 2007 | Sxt, Tet, Nal, Chl | 1 | 32 | |
| yuhs07-75 | Muenster | 2007 | Sxt, Tet, Nal, Chl | 1 | 32 | |
| yuhs10-7 | Muenster | 2010 | Sxt, Tet, Chl, Nal | 1 | 32 | |
| yuhs11-10 | Muenster | 2011 | Sxt, Tet, Chl, Nal | 1 | 32 | |
| yuhs06-59 | Reading | 2006 | Tet, Nal | 1 | 32 | |
| yuhs06-60 | Reading | 2006 | Tet, Nal | 1 | 32 | |
| yuhs06-65 | Reading | 2006 | Tet, Nal | 1 | 32 | |
| yuhs07-78 | Typhimurium | 2007 | Amp, Tet, Nal, Cro, Chl | 1 | 32 | |
| yuhs07-79 | Typhimurium | 2007 | Amp, Tet, Nal, Cro, Chl | 1 | 32 | |
| yuhs07-9 | Typhimurium | 2007 | Amp, Sxt, Tet, Nal, Cro, Chl | 1 | 32 | |
| yuhs08-60 | Typhimurium | 2008 | Amp, Sxt, Tet, Nal, Cro, Chl, Fzd, Gen | 2 | 64 | |
| yuhs10-16 | Typhimurium | 2010 | Amp, Sxt, Tet, Nal, Cro, Chl | 0.25 | 32 | |
| yuhs11-28 | Typhimurium | 2011 | Amp, Sxt, Tet, Nal, Cro, Chl | 2 | 64 | |
| yuhs11-47 | Typhimurium | 2011 | Tet, Chl, Nal | 1 | 32 | |
| yuhs07-20 | Untypable | 2007 | Nal | 1 | 32 |
AMP, ampicillin; CHL, chloramphenicol; CRO, ceftriaxone; FZD, furazolidone; GEN, gentamicin; NAL, nalidixic acid; STX, trimethoprim–sulfamethoxazole; TET, tetracycline.
DISCUSSION
Illness from Salmonella has decreased in the Yucatan Peninsula while severe Shigella infections have increased among children with gastroenteritis requiring hospitalization. There is a high frequency of resistance to ampicillin and trimethoprim–sulfamethoxazole in Shigella isolates, but these remain uniformly susceptible to ESC, ciprofloxacin, and azithromycin. While Shigella resistance to clinically important antibiotics over the last 7 years remained basically unchanged, resistance to most of these, with the exception of ciprofloxacin, declined in Salmonella. Resistance to ESC continues to be prevalent among Salmonella isolates, and is mediated by the blaCMY-2 gene. qnrB genes are widely distributed among our Salmonella population, but their overall frequency appear to be decreasing in recent years.
The increase in shigellosis in our pediatric population is a major public health concern, as a high proportion of our patients presented bloody diarrhea, high fever, and dehydration. The frequency of seizures (11%), moreover, is much higher than that reported in a recent study in Asia (3%; von Seidlein et al., 2006). This could be explained by the higher proportion of S. flexneri infections at our center and by the fact that our study was confined to children with diarrhea sufficiently severe to seek medical care at a hospital emergency room.
Our Shigella strains were frequently resistant to trimethoprim–sulfamethoxazole and ampicillin, the first line empirical therapy for bloody diarrhea at our public hospitals. Although all strains were susceptible to azithromycin, ESCs, and ciprofloxacin, none of these antimicrobials are provided in oral form by our government healthcare system, which services 70% of the population. Consequently, patients with therapeutic failure ultimately required hospitalization with parenteral ceftriaxone or out-of-pocket purchase of oral azithromycin or cefixime, escalating costs for both the public health sector and families.
Although the increase in Shigella gastroenteritis was not statistically significant, we believe, nonetheless, that our findings are epidemiologically and clinically significant, as the prevalence of Shigella gastroenteritis had never, during the last decade, reached 12%. It has remained at this level during 2012 and 2013 – a worrisome trend that warrants close scrutiny by public health authorities.
Our results concur with those of other investigators who challenge the widely accepted notion of a worldwide decline in shigellosis. Recent studies using active or passive surveillance have detected high incidence rates of shigellosis in Peru, Thailand, and China (Chompook et al., 2005; Kosek et al., 2008; Xia et al., 2011). Unlike Southeast Asia (von Seidlein et al., 2006; Vinh et al., 2009), where the dominant species has shifted from S. flexneri to S. sonnei, the former continues to be the major culprit of severe shigellosis in Yucatan, as is the case in Peru and China.
The troubling recent rise in shigellosis could well be linked to the current economy. Between 2008 and 2010, the number of people below the poverty line in Yucatan increased by 35,000 (CONEVAL, 2012). Several independent studies have shown that the risk factors for S. flexneri infection include poverty and overcrowded living conditions (Admoni et al., 1995; Kosek et al., 2008). Interestingly, a massive spike in shigellosis has recently been reported in the USA (Andrews, 2012).
In addition to a significant decrease in Salmonella gastroenteritis, isolates showed a downward trend in resistance to almost all antibiotics, which was unrelated to changes in serovar distribution. The spread of ESC resistance to other serovars suggests that the blaCMY-2 gene previously found in MDR S. Typhimurium isolates in Yucatan (Zaidi et al., 2007) may have spread, as noted in other reports (Arlet et al., 2006), through horizontal transfer. There was remarkably little diversity of extended spectrum beta-lactamase genes as only blaCMY-2 was present in our Salmonella strains. blaCMY-2 also predominates in Salmonella isolates from humans, retail meat and animals in the USA (Zhao et al., 2009; Folster et al., 2012), although blaCTX has been found in almost 7% of clinical turkey isolates (Wittum et al., 2012). The near absence of blaSHV, blaOXA, and blaCTX in the Salmonella population of USA and Mexico strongly contrasts with reports from Europe and Asia. Recent studies from Spain (de Toro et al., 2011; Perez-Moreno et al., 2013) found an assortment of bla genes in human Salmonella isolates, including blaTEM-1, blaSHV, blaCTX-M, blaPSE-1, blaOXA, and blaDHA-1, while blaCTX-M, blaTEM, and blaSHV were more frequently found in poultry isolates from the Netherlands (Dierikx et al., 2010). Likewise, blaTEM, blaSHV, blaDHA, blaOXA, and blaCTX-M are common in non-typhoidal salmonellae from humans and food-animals in India, China, and Korea (Menezes et al., 2010; Tamang et al., 2011a; Yu et al., 2011a). Diversification of beta-lactamase genes depends on a myriad of factors which include, among others, selective pressures in the environment as well as enzyme efficiency (Galan et al., 2013). Unraveling the complexity of the evolutionary processes that lead to the development of resistance and transfer of resistance genes may allow us to better explain their different distributions around the world.
Reduced susceptibility to ciprofloxacin and the presence of qnr genes was frequent in our Salmonella isolates; 32% presented MICs between 0.125 and 2 μg/ml, and 12% harbored a qnrA or qnrB gene. These findings are comparable with those of other reports from Latin America in which qnrB19 was the predominant variant. It has been detected in Salmonella isolates from food in Colombia (Karczmarczyk et al., 2010), and was highly prevalent in commensal enterobacteria isolated from healthy children in Peru and Bolivia (Pallecchi et al., 2009). Notably, it was also found in 32% of screened individuals living in a remote village in the Amazonas (Pallecchi et al., 2011). In contrast, a recent study conducted on Salmonella isolates from humans, retail meat and animals in the USA detected qnr genes in only 0.3% of human isolates, and none from animal sources (Sjolund-Karlsson et al., 2010). qnr genes are also reportedly low in China, Korea, and India (on the order of 0–3%), where aac(6′)-lb-cr is more commonly present (Menezes et al., 2010; Tamang et al., 2011b; Yu et al., 2011b; Chen et al., 2012; Kim et al., 2013). The origin and the mechanisms for widespread dissemination of qnrB genes is currently unfolding. Recent evidence points to the chromosome of Citrobacter spp. as the likely origin of plasmid-mediated qnrB (Jacoby et al., 2011). The presence of qnrB19 genes on small colE-type plasmids (Karczmarczyk et al., 2010; Pallecchi et al., 2010), as well as their insertion on transposons located on larger plasmids (Cattoir et al., 2008; Dionisi et al., 2009) are believed to contribute to their dissemination and persistence, even in the absence of selective pressure. This could explain its ubiquity in commensal fecal flora and horizontal transfer and spread among unrelated species.
It is likely that many of our isolates with decreased susceptibility to ciprofloxacin have mutations in the quinolone resistance determining region, which was not assessed in this study. Furthermore, several of our nalidixic acid-susceptible isolates with reduced susceptibility to ciprofloxacin are likely to harbor other resistance genes such as aac(6′)-lb-cr, qep, qnrC, and qnrD that have been reported by other investigators. A more thorough search for plasmid-mediated genes, nucleotide mutations in the DNA gyrase and topoisomerase IV genes, as well as plasmid characterization will be conducted in the future.
It is noteworthy that the prevalence of blaCMY-2 and qnr genes decreased during the study period, possibly due to a reduced selective pressure in the regional Salmonella population. Although detected in several other serovars, the blaCMY-2 gene is mainly confined to our S. Typhimurium isolates for which swine is the major reservoir (Zaidi et al., 2007). Our qnr genes, on the other hand, have dispersed more widely. In the Yucatan Peninsula, all of the serovars harboring qnr genes are mainly associated with swine except for S. Enteritidis, which is poultry-associated, and S. Muenchen, which is more prevalent in cattle.
The encouraging decline in both the incidence and the antimicrobial resistance of Salmonella is likely due to interventions at the farm level. During recent years, swine producers in Yucatan have decreased the overall usage of antimicrobial compounds by the implementation of stricter biosecurity measures, greater use of vaccines, and stringent hygienic measures. Licensed veterinarians are solely responsible for determining the antibiotic regimens on farms (Jose Cervera, Asociacion Ganadera Local de Porcicultores de Merida, personal communication). Our previous molecular studies have shown that Salmonella strains in humans closely reflect those of locally produced food-animals, particularly swine (Zaidi et al., 2007). Improved farm management practices would thus have a direct impact on zoonotic pathogens such as Salmonella, but would be unlikely to have any effect on strictly human pathogens such as Shigella, an assumption supported by our study results. A comprehensive study in Denmark (World Health Organization [WHO], 2003) showed that an overall reduction in usage of antimicrobial compounds in food-animals was associated with decreased resistance in foodborne pathogens.
The epidemiology of Salmonella and Shigella in the Yucatan Peninsula, as in other regions of the world, is evolving. Typhoid fever and Shigella dysenteriae type 1infections, the major scourges four decades ago, are now rare. The new challenges, as evidenced from this study, are MDR, ESC-resistant Salmonella with rising resistance to ciprofloxacin and severe S. flexneri gastroenteritis. Our findings underscore the need for greater efforts to preserve critically important antibiotic classes such as ESC and fluoroquinolones. The agricultural sector in Mexico has taken important steps to reduce the burden of salmonellosis. Comparable action to ameliorate the impact of shigellosis is urgently required. If worsening socioeconomic conditions are a major determinant of this increase, action by health authorities alone will not suffice. Policy makers must take account of the full consequences of social inequality and act accordingly (Stiglitz, 2012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jose Cervera, President of the Swine Producer Association for the State of Yucatan (Asociacion Ganadera Local de Porcicultores de Merida) for providing information on farm management practices during the study period. This study was supported by NIH grant U01A/0082110, grant no. FD-U-001934 and funding from Fundacion Mexicana para la Salud, Capitulo Peninsular.
REFERENCES
- Admoni O., Yagupsky P., Golan A., Kenes Y., Schifroni G., Horowitz I. (1995). Epidemiological, clinical and microbiological features of shigellosis among hospitalized children in Northern Israel. Scand. J. Infect. Dis. 27 139–144 10.3109/00365549509018994 [DOI] [PubMed] [Google Scholar]
- Andrews J. (2012). Ohio Sees Increase in Shigella Cases. Available at: http://www.foodsafetynews.com/2012/11/ohio-sees-massive-spike-in-shigella-cases/#.UXlBpUrNmSo (accessed November 20, 2012) [Google Scholar]
- Arlet G., Barrett T. J., Butaye P., Cloeckaert A., Mulvey M. R., White D. G. (2006). Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8 1945–1954 10.1016/j.micinf.2005.12.029 [DOI] [PubMed] [Google Scholar]
- Bauernfeind A., Stemplinger I., Jungwirth R., Giamarellou H. (1996). Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin (resistance). Antimicrob. Agents Chemother. 40 221–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briñas L., Zarazaga M., Sáenz Y., Ruiz-Larrea F., Torres C. (2002). Beta-lactamases in ampicillin-resistant Escherichia coli isolated from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 46 3156–3163 10.1128/AAC.46.10.3156-3163.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir V., Nordmann P., Silva-Sanchez J., Espinal P., Poirel L. (2008). ISEcp-1-mediated transposition of qnrB-like gene in Escherichia coli. Antimicrob. Agents Chemother. 52 2929–2932 10.1128/AAC.00349-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang W., Pan W., Yin J., Pan Z., Gao S., et al. (2012). Prevalence of qnr, aac(6′)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob. Agents Chemother. 56 3423–3427 10.1128/AAC.06191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chompook P., Samosonsuk S., von Seidlein L., Jitsanguansuk S., Sirima N., Sudjai S., et al. (2005). Estimating the burden of shigellosis in Thailand: 36-month population-based surveillance study. Bull. World Health Organ. 83 739–746 [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control (CDC). (1988). Shigella dysenteriae type 1 in tourists to Cancun, Mexico. MMWR Morb. Mortal. Wkly. Rep. 37 465 [PubMed] [Google Scholar]
- CLSI. (2012a). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-second Informational Supplement. CLSI Document M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute [Google Scholar]
- CLSI. (2012b). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 9th Edn. CLSI Document M07-A9. Wayne, PA: Clinical and Laboratory Standards Institute [Google Scholar]
- CONEVAL (2012). Medición de la pobreza, Estados Unidos Mexicanos, 2012. Evolución de la pobreza y pobreza extrema nacional en entidades federativas. 2008–2012 Available at: http://coneval.gob.mx/Medición/Paginas/Medición/Pobreza2012/Anexo-estadístico-pobreza-2012.aspx [Google Scholar]
- Datta N., Olarte J. (1974). R factors in strains of Salmonella typhi and Shigella dysenteriae 1 isolated during epidemics in Mexico: classification by compatibility. Antimicrob. Agents Chemother. 5 310–317 10.1128/AAC.5.3.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Toro M., Sáenz Y., Cercenado E., Rojo-Bezares B., García-Campello M., et al. (2011). Genetic characterization of the mechanisms of resistance to amoxicillin/clavulanate and third-generation cephalosporins in Salmonella enterica from three Spanish hospitals. Int. Microbiol. 14 173–181 [DOI] [PubMed] [Google Scholar]
- Dierikx C., van Essen-Zandbergen A., Veldman K., Smith H., Mevius D. (2010). Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 26 273–278 10.1016/j.vetmic.2010.03.019 [DOI] [PubMed] [Google Scholar]
- Dionisi A. M., Lucarelli C., Owczarek S., Luzzi I., Villa L. (2009). Characterization of the plasmid-borne quinolone resistance gene qnrB19 in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 53 4019–4021 10.1128/AAC.00294-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein M., Pimkin M., Palagin I., Edelstein I., Stratchounski L. (2003). Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47 3724–3732 10.1128/AAC.47.12.3724-3732.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folster J. P., Pecic G., Singh A., Duval B., Rickert R., Ayers S., et al. (2012). Characterization of cephalosporin-resistant Salmonella enterica serovar Heidelberg isolated from food animals, retail meat, and humans in the United States 2009. Foodborne Pathog. Dis. 9 638–645 10.1089/fpd.2012.1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. C., Gonzalez-Candelas F., Rolain J. M., Canton R. (2013). Antibiotics as selectors and accelerators of diversity in the mechanisms of resistance: from the resistome to genetic plasticity in the β-lactamases world. Front. Microbiol. 4:9 10.3389/fmicb.2013.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G., Cattoir V., Hooper D., Martinez-Martinez L., Nordmann P., Pascual A., et al. (2008). qnr gene nomenclature. Antimicrob. Agents Chemother. 52 2297–2299 10.1128/AAC.00147-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Griffin C. M., Hooper D. C. (2011). Citrobacter spp. as a source of qnrB alleles. Antimicrob. Agents Chemother. 55 4979–4984 10.1128/AAC.05187-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Walsh K. E., Walker V. J. (2006). Identification of extended-spectrum, AmpC, and Carbapenem-hydrolyzing β-lactamases in Escherichia coli and Klebsiella pneumoniae by disk tests. J. Clin. Microbiol. 44 1971–1976 10.1128/JCM.00062-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczmarczyk M., Martins M., McCusker M., Mattar S., Amaral L., Leonard N., et al. (2010). Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol. Lett. 313 10–19 10.1111/j.1574-6968.2010.02119.x [DOI] [PubMed] [Google Scholar]
- Kim J. H., Cho J. K., Kim K. S. (2013). Prevalence and characterization of plasmid-mediated quinolone resistance genes in Salmonella isolated from poultry in Korea. Avian Pathol. 42 221–229 10.1080/03079457.2013.779636 [DOI] [PubMed] [Google Scholar]
- Kosek M., Yori P. P., Pan W. K., Olortegui M. P., Gilman R. H., Perez J., et al. (2008). Epidemiology of highly endemic multiply antibiotic-resistant shigellosis in children in the Peruvian Amazon. Pediatrics 122 541–549 10.1542/peds.2008-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K. L., Nataro J. P., Blackwelder W. C., Nasrin D., Farag T. H., Panchalingam S., et al. (2013). Burden and etiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet 382 209–222 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- Menezes G. A., Khan M. A., Harish B. N., Parija S. C., Goessens W., Vidyalakshmi K., et al. (2010). Molecular characterization of antimicrobial resistance in non-typhoidal Salmonellae associated with systemic manifestations from India. J. Med. Microbiol. 59 1477–1483 10.1099/jmm.0.022319-0 [DOI] [PubMed] [Google Scholar]
- Niyogi S. K. (2005). Shigellosis. J. Microbiol. 43 133–143 [PubMed] [Google Scholar]
- Pallecchi L., Riccobono E., Mantella A., Bartalesi F., Sennati S., Gamboa H., et al. (2009). High prevalence of qnr genes in commensal enterobacteria from healthy children in Peru and Bolivia. Antimicrob. Agents Chemother. 53 2632–2635 10.1128/AAC.01722-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallecchi L., Riccobono E., Mantella A., Fernandez C., Bartalesi F., Rodriguez H., et al. (2011). Small qnrB-harbouring ColE-like plasmids widespread in commensal enterobacteria from a remote Amazonas population not exposed to antibiotics. J. Antimicrob. Chemother. 66 1176–1178 10.1093/jac/dkr026 [DOI] [PubMed] [Google Scholar]
- Pallecchi L., Riccobono E., Sennati S., Mantella A., Bartalesi F., Trigoso C., et al. (2010). Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob. Agents Chemother. 54 678–682 10.1128/AAC.01160-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry C. M., Threlfall E. J. (2008). Antimicrobial resistance in typhoidal and nontyphoidal Salmonellae. Curr. Opin. Infect. Dis. 21 531–538 10.1097/QCO.0b013e32830f453a [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M. O., Picó-Plana E., de Toro M., Grande-Armas J., Quiles-Fortuny V., Pons M. J., et al. (2013). β-lactamases, transferable quinolone resistance determinants, and class 1 integron-mediated antimicrobial resistance in human clinical Salmonella enterica isolates of non-Typhimurium serotypes. Int. J. Med. Microbiol. 303 25–31 10.1016/j.ijmm.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Rampling A., Upson R, Brown D. F. J. (1990). Nitrofurantoin resistance in isolates of Salmonella enteritidis phage type 4 from poultry and humans. J. Antimicrob. Chemother. 25 285–290 10.1093/jac/25.2.285 [DOI] [PubMed] [Google Scholar]
- Robicsek A., Strahilevitz J., Sahm D. F., Jacoby G. A., Hooper D. C. (2006). qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50 2872–2874 10.1128/AAC.01647-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti P. J. (2006). Shigellosis: an old disease in new clothes? PLoS Med. 3:e354 10.1371/journal.pmed.0030354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund-Karlsson M., Howie R., Rickert R., Krueger A., Tran T. T., Zhao S., et al. (2010). Plasmid-mediated quinolone resistance among non-Typhi Salmonella enterica isolates, USA. Emerg. Infect. Dis. 16 1789–1791 10.3201/eid1611.100464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund-Karlsson M., Joyce K., Blickenstaff K., Ball T., Haro J., Medalla F. M., et al. (2011). Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrob. Agents Chemother. 55 3985–3989 10.1128/AAC.00590-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiglitz J. E. (2012). The Price of Inequality: How Today’s Divided Society Endangers Our Future. New York: W.W. Norton & Company [Google Scholar]
- Sutcliffe J. G. (1979). Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb. Symp. Quant. Biol. 43 77–90 10.1101/SQB.1979.043.01.013 [DOI] [PubMed] [Google Scholar]
- Tamang M. D., Nam H. M., Kim T. S., Jang G. C., Jung S. C., Lim S. K. (2011a). Emergence of extended-spectrum β-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J. Clin. Microbiol. 49 2671–2675 10.1128/JCM.00754-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang M. D., Nam H. M., Kim A., Lee H. S., Kim T. S., Kim M. J., et al. (2011b). Prevalence and mechanisms of quinolone resistance among selected nontyphoid Salmonella isolated from food animals and humans in Korea. Foodborne Pathog. Dis. 8 1199–1206 10.1089/fpd.2011.0899 [DOI] [PubMed] [Google Scholar]
- Vinh H., Nhu N. T., Nga T. V., Duy P. T., Campbell J. I., Hoang N. V., et al. (2009). A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect. Dis. 9:204 10.1186/1471-2334-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Seidlein L., Kim D. R., Ali M., Lee H., Wang X. Y., Thiem V. D., et al. (2006). A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 3:e353 10.1371/journal.pmed.0030353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner M., Zaidi M. B., Calva E., Fernandez-Mora M., Calva J. J., Silva C. (2009). Association of virulence plasmid and antibiotic resistance determinants with chromosomal multilocus genotypes in Mexican Salmonella enterica serovar Typhimurium strains. BMC Microbiol. 9:131 10.1186/1471-2180-9-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittum T. E., Mollenkopf D. F., Erdman M. M. (2012). Detection of Salmonella enterica isolates producing CTX-M cephalosporinase in U. S. livestock populations. Appl. Environ. Microbiol. 78 7487–7491 10.1128/AEM.01682-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woc-Colburn L., Bobak D. A. (2009). The expanding spectrum of disease due to Salmonella: an international perspective. Curr. Infect. Dis. Rep. 11 120–124 10.1007/s11908-009-0018-2 [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) (2003). Impacts of Antimicrobial Growth Promoter Termination in Denmark. The WHO International Review Panel’s Evaluation of the Termination of the Use of Antimicrobial Growth Promoters in Denmark. Geneva: WHO [Google Scholar]
- Xia S., Xu B., Huang L., Zhao J. Y., Ran L., Zhang J., et al. (2011). Prevalence and characterization of human Shigella infections in Henan Province, China, in 2006. J. Clin. Microbiol. 49 232–242 10.1128/JCM.01508-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Chen Q., Yu X., Li Q., Ding B., Yang L., et al. (2011a). High prevalence of extended-spectrum beta lactamases among Salmonella enterica Typhimurium isolates from pediatric patients with diarrhea in China. PLoS ONE 6:e16801 10.1371/journal.pone.0016801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Chen Q., Yu X., Pan J., Li Q., Yang L., et al. (2011b). High prevalence of plasmid-mediated quinolone resistance determinant aac(6′)-lb-cr amongst Salmonella enterica serotype Typhimurium isolates from hospitalized paediatric patients with diarrhea in China. Int. J. Antimicrob. Agents 37 152–155 10.1016/j.ijantimicag.2010.10.021 [DOI] [PubMed] [Google Scholar]
- Zaidi M. B., Leon V., Canche C., Perez C., Zhao S., Hubert S. K., et al. (2007). Rapid and widespread dissemination of multidrug-resistant blaCMY-2 Salmonella Typhimurium in Mexico. J. Antimicrob. Chemother. 60 398–401 10.1093/jac/dkm168 [DOI] [PubMed] [Google Scholar]
- Zaidi M. B., McDermott P. F., Fedorka-Cray P., Leon V., Canche C., Hubert S. K., et al. (2006). Nontyphoidal Salmonella from human clinical cases, asymptomatic children, and raw retail meats in Yucatan, Mexico. Clin. Infect. Dis. 42 21–28 10.1086/498508 [DOI] [PubMed] [Google Scholar]
- Zhao S., Blickenstaff K., Glenn A., Ayers L., Friedman S. L., Abbott J. W., et al. (2009). β-lactam resistance in Salmonella strains isolated from retail meats in the United States by the National Antimicrobial Resistance Monitoring System between 2002 and 2006. Appl. Environ. Microbiol. 75 7624–7630 10.1128/AEM.01158-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., White D. G., McDermott P. F., Friedman S., English L., Ayers S., et al. (2001). Identification and expression of cephamycinase bla (CMY) genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45 3647–3650 10.1128/AAC.45.12.3647-3650.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]