Abstract
Nuclear distribution element 1 (NDE1, also known as NudE) and NDE-like 1 (NDEL1, also known as Nudel) are paralogous proteins essential for mitosis and neurodevelopment that have been implicated in psychiatric and neurodevelopmental disorders. The two proteins possess high sequence similarity and have been shown to physically interact with one another. Numerous lines of experimental evidence in vivo and in cell culture have demonstrated that these proteins share common functions, although instances of differing functions between the two have recently emerged. We review the key aspects of NDE1 and NDEL1 in terms of recent advances in structure elucidation and cellular function, with an emphasis on their differing mechanisms of post-translational modification. Based on a review of the literature and bioinformatics assessment, we advance the concept that the twin proteins NDE1 and NDEL1, while sharing a similar ‘nature’ in terms of their structure and basic functions, appear to be different in their ‘nurture’, the manner in which they are regulated both in terms of expression and of post-translational modification within the cell. These differences are likely to be of significant importance in understanding the specific roles of NDE1 and NDEL1 in neurodevelopment and disease.
Keywords: NDE1/NudE, NDEL1/Nudel, neurodevelopment, phosphorylation, post-translational modification
Introduction
Nuclear distribution element 1 (NDE1, also known as NudE) and NDE-like 1 (NDEL1, also known as Nudel) are a pair of highly similar coiled-coil-containing proteins (Figure 1), believed to have evolved from a common ancestral gene, such as the NudE gene of Aspergillus nidulans, after which they were named (1-5). Both of these proteins are known to be critical for neurodevelopment and have been implicated in a range of mental health measures and neurodevelopmental conditions. Copy number variations (CNVs) at the 16p13.11 chromosomal locus, that contains the NDE1 gene among others, have been associated with intellectual disability, autism, attention deficit hyperactivity disorder, schizophrenia and epilepsy (6-18). In contrast, the region of the 17p13.1 locus where NDEL1 resides has not been directly implicated in brain disorders through CNV analysis. However, it should be noted that genomic structural variation at the 16p13.11 locus is more common than at 17p13.1, even among healthy individuals [database of genomic variants; ref. (19)]. Of the genes disrupted by CNVs at 16p13.11, disruption of normal NDE1 function stands out as the most likely cause of the associated mental health problems due to the known functional roles of the NDE1 and NDEL1 proteins through interaction with Disrupted In Schizophrenia 1 (DISC1), a molecule strongly implicated in risk of mental illness (reviewed in refs. 20-23).
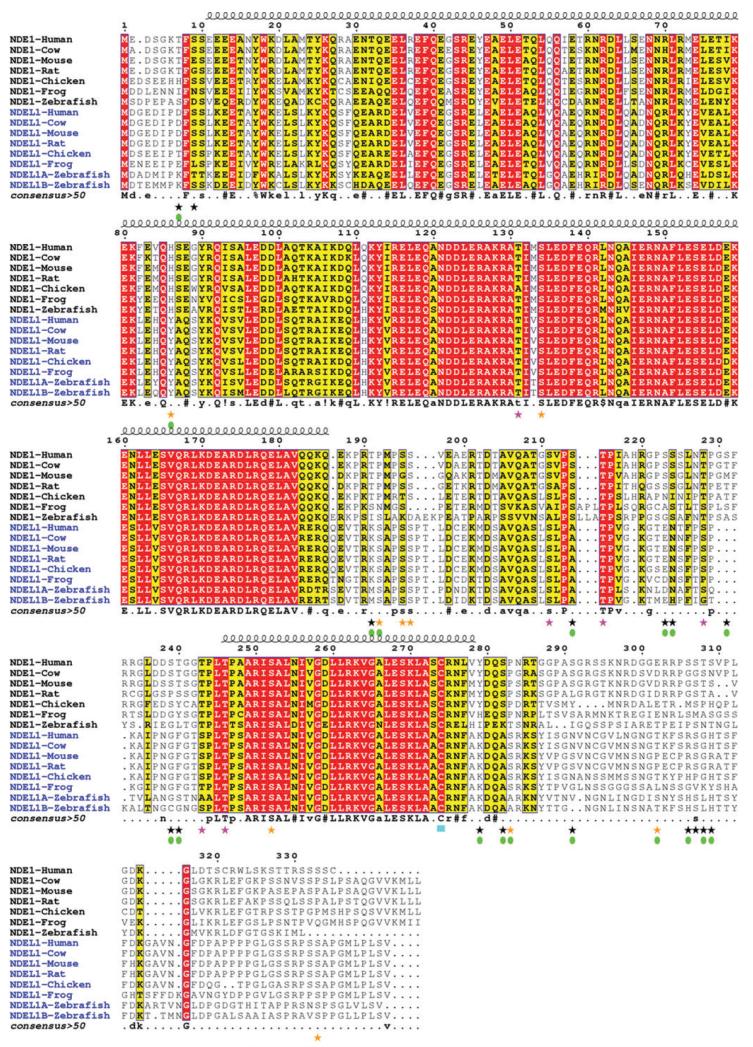
Figure 1.
Multiple sequence alignment of NDE1 and NDEL1 orthologs.
Orthologs of NDE1 and NDEL1 across vertebrate species were identified using the UCSC Genome Browser (http://genome-euro.ucsc.edu). These were aligned initially using Clustal Omega (108, 109) with further manual editing for optimum alignment. The sequence numbering at the top of each alignment block corresponds to the sequence of human NDE1. Conserved residues are colored with a red background; conservatively substituted residues are shown with a yellow background. The location of the N-terminal coiled-coil α-helical domain (residue ~10–185) and the predicted C-terminal α-helix (residue ~247–278) are shown above the alignment. Consensus amino acids are shown at the bottom of the each alignment block (uppercase is strictly conserved; lowercase is consensus level > 0.5;! is I or V; $ is L or M; % is F or Y; = is N, D, Q or E). Despite 60% sequence identity there are at least 18 specific phosphorylation sites for each human protein – i.e., phosphorylation possible only in one protein. Some of these phosphorylation sites have been derived from high-throughput proteomic screening using mass spectrometry (90), while others have been experimentally verified in the cell. See the main text and Table 2 for more information. Those sites known to be specific to either NDE1 or NDEL1 are highlighted with a green filled oval under the corresponding star; black star=phosphorylation site in NDE1; orange star=phosphorylation site in NDEL1; pink star=phosphorylation site in both NDE1 and NDEL1; cyan block=palmitoylation site in both proteins. The figure was generated with ESPript v2.2 (111).
The DISC1 gene was discovered because it is directly disrupted by a balanced translocation that segregates with major mental illness in a single large Scottish pedigree (24, 25). Initial efforts to characterize DISC1 function revolved around attempts to discover its protein interaction partners, on the assumption that such knowledge would suggest biological functions in which the novel and unique DISC1 protein might partake. This led to the formation of the DISC1 pathway hypothesis, which hypothesized that disruption of components of the DISC1 pathway could influence susceptibility to schizophrenia and related disorders (26, 27). NDE1 and NDEL1 were both found to directly interact with DISC1 (28-33) and have since become central to this hypothesis (20-23).
In genetic association studies of NDE1 and NDEL1 using single-nucleotide polymorphisms and haplotypes, both genes have provided evidence for involvement in mental illness phenotypes, with significant association ascertained independently in a Finnish family cohort for schizophrenia (26, 34), and in a Caucasian-American schizophrenia cohort where the association depended on the status of a common variant in DISC1 (C704) (29). A variant at the NDEL1 locus has also been demonstrated to associate with schizophrenia in interaction with an independently significant variant from the CIT gene, encoding another DISC1 interacting protein (35). However, such significant associations have not been uniformly observed (35-39), nor have they yet been found to be significant in genome-wide association studies (40).
Deletions of the 16p13.11 locus are also associated with mild microcephaly (7). Again, NDE1 stands out as the most likely gene to be responsible for the greater part of the effect, as extreme microcephaly with lissencephaly was reported in multiple patients with biallelic frameshift mutations leading to truncation of the NDE1 protein (41, 42). Additionally, a biallelic mutant predicted to abolish NDE1 expression has been reported in patients with microhydranencephaly (43). A recent study investigating association of the 16p13.11 region with microcephaly and a fetal brain disruption-like phenotype also found a similar phenotype to occur in two patients with deletion of one copy of 16p13.11, combined with a frameshift mutation in the remaining NDE1 gene (44).
Therefore, genetic association evidence exists implicating both NDE1 and NDEL1 in psychiatric illness, with additional evidence from CNV and gene-disrupting mutations implicating NDE1 specifically in a wider array of neurodevelopmental conditions. In this review, we assess both proteins, with a focus on what properties they share in common, but also the manner in which their regulation and functions differ – differences that may help explain their specific roles in neurodevelopment and disease and why two highly similar proteins have been retained in vertebrate evolution.
NDE1 and NDEL1 play overlapping roles in the cell and developing brain
NDE1 and NDEL1 were initially identified as part of a highly evolutionarily conserved pathway consisting of the motor protein dynein and its various ancillary and regulatory proteins (1-5). These included the developmentally critical Lissencephaly 1 protein [LIS1, encoded by the PAFAH1B1 gene; ref. (45)]. NDE1 is known to recruit LIS1 to dynein, where they together induce a force-state, facilitating the movement of dynein along microtubules. In contrast, on its own NDE1 inhibits dynein motility by causing it to dissociate from the microtubules (46). NDEL1 appears to act in the same way, with its LIS1-binding N-terminus stimulating the movement of dynein, while its C-terminus, expressed alone, causes dynein-microtubule dissociation (47). Interestingly, while both proteins can interact with the intermediate chain of the dynein complex (2, 48), NDEL1, but not NDE1, can also bind the motor domain of dynein, in its heavy-chain (1, 2, 46). Additionally, NDE1 is known to compete with dynactin (dynein-activator) for binding to the dynein intermediate chain, and possesses a distinct site for interaction with dynein light chain 8 (LC8) (49). NDEL1 is not known to share this latter feature, and the LC8 binding region of NDE1 (residues 200–203) is only partially conserved in NDEL1 (Figure 1).
NDEL1 is known to affect many aspects of dynein function, including transport along the microtubule network of vesicles (50, 51) and lysosomes (52), with effects on the correct structure of the Golgi apparatus, as well as on the transport of intermediate filament proteins (53), short microtubules and viral glycoproteins (54). Of these known roles of NDEL1, NDE1 has also been shown to affect the size of the Golgi apparatus, with knockdown of NDE1 in HeLaM cells having a smaller effect than NDEL1, but with the double-knockdown having a greater effect still, implying that they can partially compensate for each other’s loss (55). Peptides derived from each of NDE1 and NDEL1 also affect actin transport in a squid axonal model (56).
NDE1 and NDEL1 have also been demonstrated to be of significant importance in mitosis, with Nde1 knockout mice displaying frequent instances of misaligned spindles in their cortical progenitor cells (57). Use of an antibody against both NDE1 and NDEL1 to disrupt their function induces similar defects, and disrupts the dynein-related removal of proteins from the kinetochore essential for mitotic progression at metaphase (48). Interestingly, this effect may be mediated by NDE1 alone, as suppression of NDE1, but not NDEL1, stopped dynein from localizing to the kinetochore and the cells from passing this mitotic checkpoint (58). This NDE1-specific role has, however, been contradicted by another study (59), while expression of Xenopus Ndel1 alone was sufficient to rescue spindle alignment defects caused by depletion of endogenous Nde1 and Ndel1 from egg extracts (60). NDEL1 has also been shown to stabilize dynein at the kinetochore (61) as well as having additional mitotic roles in the breakdown of the nuclear envelope (62) and the assembly of the lamin B spindle matrix (63). NDE1 also has a dynein LC8-related effect on ciliogenesis during mitosis (64).
Nde1 knockout mice display thinning of the superficial layers of the cerebral cortex leading to a reduction in brain volume (57) reminiscent, but not as severe as, the microcephaly displayed in human patients with non-functional NDE1 genes (41, 42). This appears to occur due to defects in neuronal migration and neurogenesis amongst neuronal progenitors, potentially as a result of incorrect orientation of the spindle during cell division (57). Additional effects of mouse Nde1 on cortical neurodevelopment in conjunction with Lis1 via radial glial cell function and spatial co-ordination of MAPK signaling pathways have also been described (65, 66).
In contrast Ndel1 knockout mice are not viable, with blastocysts perishing within days of fertilization (51). Studies depleting the level of Ndel1 in the developing mouse by various means, have also demonstrated defects in cortical neuronal migration (51, 67-69) leading to a thinning of the cortex (69) and implying an overlapping role with NDE1. This seemingly occurs due to uncoupling of the centrosome from the nucleus (68), an effect which is exaggerated when Lis1 expression is also knocked-out (51, 69). Aberrant positioning of both embryonic and adult-born hippocampal neurons (51, 69, 70) and of neurite extension in dorsal root ganglia (71) have also been reported following NDEL1 depletion.
A number of additional functions for NDEL1 have also been reported including its facilitation of neurofilament polymerization (72), promotion of axonal regeneration (73), regulation of Cdc42 at the leading edge of migrating neurons (74), regulating the GTPase activity of the microtubule remodeling protein Dynamin 2 (75), facilitating the actin polymerization-promoting activity of the WAVE complex (76) and a peptidase activity (77). It is not currently known whether NDE1 shares any of these functions. For more detail on the cytoskeletal functions of NDEL1 and its role in signaling pathways, readers are referred to the comprehensive review by Chansard et al. (78).
It is therefore apparent that, in terms of the regulation of dynein, effects on mitotic progression and regulation of cortical neuronal development and migration, NDE1 and NDEL1 possess functions which largely overlap and that the two proteins are able, to some extent, to compensate for the loss of one another. Several lines of evidence suggest that the two proteins are not fully redundant, however, notably the potential differences in their dynein-related regulation of mitotic checkpoints (58), overlapping but distinct regions of binding to the dynein protein complex (1, 2, 46, 49), the difference in viability of Nde1 and Ndel1 knockout mice (51, 57) and the dramatic microcephaly phenotypes seen in patients with loss of functional NDE1, but presumably not functional NDEL1 (41-44). There also remain a large number of functions of one of the paralogs, for which the importance of the other is undetermined. There are also notable differences in the expression of the two proteins over development, at the mRNA level (Figure 2). According to data from the Human Brain Transcriptome project [hbatlas.org; ref. (79)], while expression of NDEL1 across the brain is relatively steady throughout the lifetime, NDE1 is considerably higher during the first couple of months after fertilization, before dropping dramatically at birth. A smaller increase in NDE1 expression is also notable in some brain regions in early childhood. Given the high degree of amino acid conservation between NDE1 and NDEL1 (Figure 1; Table 1), we will now discuss the potential basis of some of these similarities and differences: how do their structures compare? Do the NDE1 and NDEL1 proteins function differently, or are they proteins with a common function, but with distinct regulation?
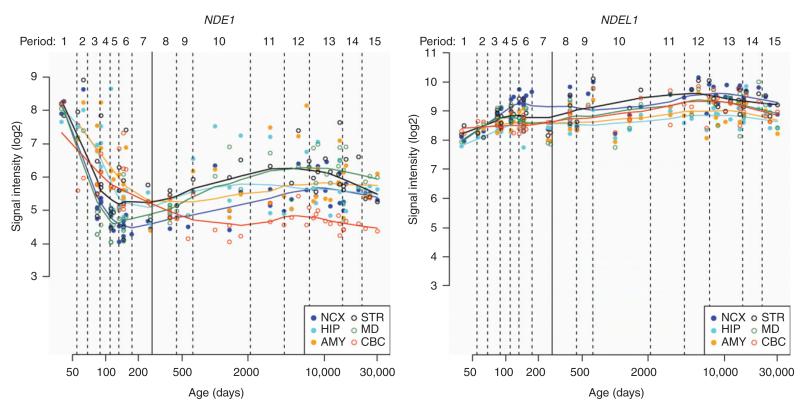
Figure 2.
Differential expression of NDE1 and NDEL1 transcripts in the human brain across the lifetime.
Change in NDE1 mRNA expression with time is shown in the left panel and NDEL1 in the right panel; developmental age shown on the x-axis in days; mRNA expression signal intensity (Log2) shown on the y-axis. While, expression of NDEL1 across the brain remains relatively steady, NDE1 shows a far more specialized pattern of expression. Abbreviations for brain regions used in figure: NCX, neocortex; STR, striatum; HIP, hippocampus; MD, mediodorsal nucleus of the thalamus; AMY, amygdala; CBC, cerebellar cortex. Data and figure accessed from the Human Brain Transcriptome project (http://hbatlas.org/pages/hbtd; used with permission) (79).
Table 1.
Sequence comparison of NDE1 and NDEL1 shows greater conservation of NDEL1 across vertebrate species.
| Cow | 88.66 | Amino acid conservation in NDE1 (% identity) | ||||
| (Bos | 99.13 | Amino acid conservation in NDEL1 (% identity) | ||||
| taurus) | -10.47 | Difference in conservation (% identity) | ||||
| Mouse | 85.89 | 87.50 | ||||
| (Mus | 95.94 | 96.52 | ||||
| musculus) | -10.05 | -9.02 | ||||
| Rat | 83.13 | 84.59 | 91.57 | |||
| (Rattus | 95.94 | 96.52 | 100.00 | |||
| norvegicus) | -12.81 | -11.93 | -8.43 | |||
| Chicken | 73.33 | 73.90 | 73.45 | 72.27 | ||
| (Gallus | 92.13 | 92.71 | 90.67 | 90.67 | ||
| gallus) | -18.80 | -18.81 | -17.22 | -18.40 | ||
| Frog | 68.56 | 66.96 | 67.06 | 65.60 | 67.74 | |
| (Xenopus | 81.23 | 81.52 | 80.35 | 80.35 | 83.48 | |
| tropicalis) | -12.67 | -13.56 | -13.29 | -14.75 | -15.74 | |
| Zebrafish | 58.54 | 57.93 | 57.98 | 58.28 | 58.95 | 56.50 |
| (Danio | 72.19/76.33 | 71.89/76.04 | 70.41/74.56 | 70.71/74.56 | 71.73/75.30 | 69.03/71.98 |
| rerio) | -13.65/-17.79 | -13.96/-18.11 | -12.43/-16.58 | -12.13/-16.28 | -12.78/-16.35 | -12.53/-15.48 |
| Human | Cow | Mouse | Rat | Chicken | Frog | |
| (Homo sapiens) |
(Bos taurus) |
(Mus musculus) |
(Rattus norvegicus) |
(Gallus gallus) |
(Xenopus tropicalis) |
The percentage amino acid sequence identity was compared using Clustal Omega (108, 109) after alignment shown in Figure 1. All-against-all pairwise inter-species percentage identity for NDE1 is shown on the top row on a lighter gray background, and for NDEL1 on the middle row on a white background. The difference between these values is shown on the bottom row in bold type on a darker gray background. As there are two NDEL1 genes in zebrafish (110), values for both encoded proteins are displayed, ndel1a on the left and ndel1b on the right. It can be seen that in every instance, between all species examined, NDEL1 is more highly conserved than NDE1.
NDE1 and NDEL1 have a similar nature: sequence, structure and oligomeric state
At the sequence level, NDE1 and NDEL1 share approximately 60% amino acid sequence identity (~80% sequence similarity) and are thought to have evolved from a common ancestral gene (1-5). This duplication and divergence into two genes appears to have occurred around the time of the emergence of vertebrate species (54), with NDEL1 orthologs showing greater inter-species conservation compared to NDE1 orthologs (Table 1). Multiple isoforms for both proteins have been confirmed, mainly consisting of a variety of alternative extreme C-terminal regions (33). Both evolutionarily conserved proteins have a highly conserved N-terminal half (residue~1–185), while the C-terminal sequence is comparatively more variable (Figure 1). Sequence analysis revealed predominance of the classical ‘heptad’ repeat that could confer the coiled-coil motif to the N-terminal ~190 amino acids (80).
Early structure-based work focused exclusively on NDEL1. Sasaki et al. showed that full-length murine Ndel1 interacted strongly with itself, and by means of yeast two-hybrid based assays using NDEL1 truncation constructs, showed that this potential ‘homodimerization’ resided within the putative coiled-coil domain (amino acids 56–166) (1). The emergence of high-resolution crystal structures and biophysical characterization of an N-terminal coiled-coil domain fragment for rat Ndel1 (identical in amino acid sequence to human NDEL1 for the region solved) (81) revealed a highly extended, continuous, tightly associating coiled-coil protein that facilitated both parallel dimerization and anti-parallel tetramerization (Figure 3); the latter composed of two parallel dimers associating in anti-parallel fashion. Initial chemical cross-linking analysis coupled with mass spectrometry (MS) supported the parallel dimer arrangement of NDEL1 in solution (82). High molecular weight-only species (~400 kDa) for His6-tagged recombinant NDEL1 were also noted (83), while predominantly tetrameric species of GB1-tagged NDEL1 were described by Narayanan et al. using size-exclusion chromatography and analytical ultracentrifugation (84). Tarricone et al. (85) showed that a dimer of NDEL1 was important for interaction with LIS1, and Narayanan et al. (84) suggested that a stable tetramer was responsible for interaction with DISC1. Sequence-analysis and homology-based modeling suggested that NDE1 could indeed form both the dimeric and tetrameric species, and coiled-coil orientation (86), as observed in the equivalent NDEL1 crystal structures (81). Monomeric (~40 kDa) species of NDEL1 with a cysteine peptidase activity and composed of ~40% β-sheet by circular dichroism have also been reported (77), but are inconsistent with the remainder of the biophysical literature on NDEL1.
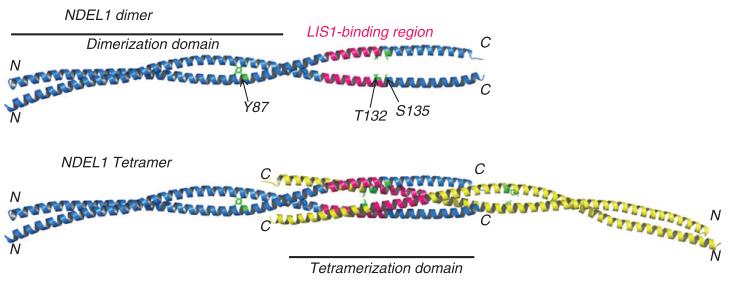
Figure 3.
Coiled-coil crystal structure of NDEL1 with known post-translational modifications mapped.
Only three known PTMs map onto the crystal structure of the NDEL1 coiled-coil domain fragment (residues 8–167; PDB ID: 2V71); these side-chains are shown in green and labeled on cartoon representations of the NDEL1 crystal structure, parallel dimer (top panel) and anti-parallel tetramer (bottom panel) (81); N- and C-termini for each chain and regions that facilitate dimerization and tetramerization are indicated. The essential LIS1-binding region on NDEL1 114–133 (96) is also shown (pink). Phosphorylation of NDE1 at T131 (directly equivalent to T132 of NDEL1) reduces its ability to interact with LIS1 (86). It can therefore be hypothesized that phosphorylation of T132 and/or S135 of NDEL1 may have similar effects on LIS1-binding. The Y87 site is highly solvent-exposed on the dimerization domain of coiled-coil structure and is thus readily accessible to kinases.
Our more recent biophysical characterization using size-exclusion chromatography, circular dichroism, negative-stain electron microscopy, and chemical crosslinking coupled with MS analysis and bioinformatics confirmed that both N-terminally His6-tagged NDE1 and NDEL1 exhibit similar secondary structure, tertiary structure and oligomeric state (87). Both proteins form dimers, tetramers and high molecular weight species in solution thus unifying disparate observations in the literature. The application of chemical cross-linking/MS methodology allowed investigation of the C-terminal regions and thus provided insight into the conformation and overall architecture of the native proteins and was the first biophysical study that investigated the structure of NDE1. The C-terminal portion of the proteins was shown to ‘bend-back’, potentially using the proline-rich region in the C-terminal half of the protein that is predicted to be unstructured (88), and interact with the N-terminal coiled-coil domain (87). This result was consistent with functional observations of two disparate dynein binding sites on NDEL1 at its N- and C-termini (60, 89). Dynamic change in conformation for NDEL1 was also previously observed (90), in keeping with the flexible role of the C-terminus. We also showed via a novel application of stable-isotope labeling in cell culture (SILAC) chemical cross-linking and MS that NDE1 can directly and physically interact with NDEL1 through the formation of heterotetramers and higher oligomeric species (87). This is in agreement with the observation that the two proteins exist within the same protein complex in the cell, as determined by co-immunoprecipitation experiments (29, 33).
NDE1 and NDEL1 have different nurture: post-translational modification
Early work revealed that NDEL1 (1, 2) was a phospho-protein and NDE1 was predicted to contain similar posttranslational modifications (PTMs) (3, 4). Since then a number of phosphorylation sites, along with one palmitoylation site, have been experimentally demonstrated to exist in each protein, while a number of others have been suggested with high-throughput proteomic (HTP) experiments using MS (91) (Figure 1; Table 2).
Table 2.
Post-translational modification on NDE1 and NDEL1.
| NDE1 residue |
Known or predicted kinase/ enzyme |
NDEL1 residue |
Known or predicted kinase/enzyme |
|---|---|---|---|
| T7-p | CKI | ||
| S9-p | CKII | ||
| Y87-p | SRC | ||
| T131-p | PKA (86) | T132-p | PKA |
| S135-p | – | ||
| T191-p | p38MAPK, GSK3 | ||
| S194-p | CKI, PKC, PKA | ||
| S197-p | CKI | ||
| S198-p | Cdk1, Cdk5 (2, 97) | ||
| S211-p | PKC | S215-p | PKC |
| S214-p | – | ||
| T215-p | Cdk1 (41) | T219-p a |
Cdk1, Cdk5, Erk2 (2, 96, 97) |
| S223-p | RSK, PKC, PKA | ||
| S224-p | – | ||
| T228-p | p38MAPK, Cdk5 | S231-pa | Cdk1, Cdk5 (2, 97) |
| S231-p | PKC, Cdkl | ||
| S239-p | – | ||
| T240-p | – | ||
| T243-p | Cdkl, Cdk5 | S242-p | Cdk1 (96) |
| T246-p | Cdk1 (41) | T245-p | Erk2 (96) |
| S251-p | Aurora-A kinase (97) | ||
| C274-s | DHHC2, 3, 7 (54) | C273-s | DHHC2, 3, 7 (54) |
| Y279-p | – | ||
| S282-p | p38MAPK, Cdk5 | ||
| S282-p | PKC | ||
| S291-p | PKC | ||
| T301-p | PKC | ||
| S306-p | PKA (88) | ||
| S307-p | – | ||
| T308-p | – | ||
| S309-p | Cdk1 | ||
| S336-p | Aurora-A and B | ||
| kinases (101) |
Experimentally determined phosphorylation sites (indicated with ‘-p’) from the literature and HTP/MS screens and palmitoylation sites (indicated with ‘-s’) are shown for each protein along with corresponding known kinase/enzyme or NetPhosK v1.0 (94) predicted kinase (stringency prediction threshold=0.5). Where PTM occurs at the equivalent amino acid position in NDE1 and NDEL1 following alignment (Figure 1), these are shown side-by-side. Where the kinase/enzyme is known, these are shown in bold type and their corresponding reference provided; these PTMs are discussed in more detail in the text. All other phosphorylation sites were obtained from the PhosphoSitePlus database (91).
The vast majority of experimentally determined phosphorylation sites reside within unstructured regions of the proteins, predominantly in their C-terminal regions, in particular within the ‘flexible’ proline-rich linker region (87, 88) that connects the coiled-coil domain to the C-terminal α-helix, and others located towards their extreme C-termini. These unstructured regions are likely to be highly solvent-exposed and thus accessible to enzymatic action from protein kinases. Phosphorylation and other PTM sites are usually short motifs that occur within rapidly evolving unstructured regions (92). A number of these phosphorylation sites are not conserved between NDE1 and NDEL1; there are at least 18 sites specific to only one of the proteins (Figure 1). This is in keeping with the notion that rapid evolution of these sites may be linked to gene retention among highly similar duplicated genes such as NDE1 and NDEL1. These differences in PTM present a ready means of effecting functional rewiring (93).
A total of 32 sites have been shown to be posttranslationally modified in NDE1 and/or NDEL1 (Table 2; where the equivalent residue is putatively modified in both proteins, this is counted only once). For some of the phosphorylation sites on NDE1 and NDEL1 the kinase or kinases responsible have been identified, while others can be speculated by means of a kinase prediction search of their respective sequences using Net-PhosK (94). These results taken together show NDE1 and NDEL1 are substrates for a wide array of kinases (Table 2). It is well known that substrate specificity of protein kinases is highly dependent on the primary amino acid sequence immediately flanking the site of phosphorylation (95); these are classified into basophilic (preference for positively charged amino acids, e.g., PKA and PKC), acidophilic (preference for negatively charged amino acids, e.g., CKI, CKII), proline-directed kinases (preference for proline, e.g., Cdk1, Cdk5, Erk2, GSK-3, MAPK) and other kinases. Both NDE1 and NDEL1 have proline-rich linker regions between their N-terminal coiled-coil domain and predicted C-terminal α-helix, and are significantly enriched for phosphorylation sites therein; it is reasonable to assume that prolinedirected kinases are likely to act here. Indeed, both T215 and T246 in NDE1 that possess adjacent proline residues are phosphorylated by proline-directed kinase Cdk1 (41). Likewise, known sites in NDEL1 – S198, T219, S231, S242 and T245 – are also located adjacent to proline residues and are phosphorylated by Cdk1, Cdk5 and/or Erk2 (2, 96, 97). The C-terminal regions of NDE1 and NDEL1 are highly basic in overall charge (87, 89) and are thus more amenable to basophilic kinases. In support of this, many of the C-terminal sites are suggested to be targets for PKA or PKC, with S306 in NDE1 experimentally verified as a PKA site (88). Future work should aim to experimentally verify the existence of the HTP/MS phosphorylation sites using additional techniques such as amino acid sequencing, phospho-specific antibodies, site-directed mutagenesis, dominant-negative constructs etc. and confirm the remaining kinases that act on NDE1 and NDEL1 based upon this exercise.
Structural landscape of post-translational modification: implications for NDE1 and NDEL1 function
In order to discuss in detail the potential and known effects of NDE1 and NDEL1 PTM, each structural domain or region of the proteins will now be considered in turn. Schematics displaying the overall PTM landscape on NDE1 and NDEL1 structure along with known protein-protein interaction sites are shown in Figure 4.
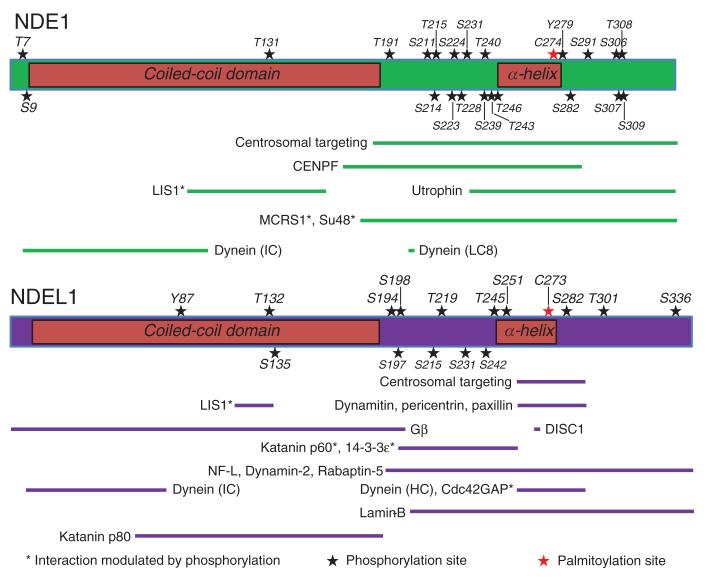
Figure 4.
Domain architecture, post-translation modification and binding/functional regions of NDE1 and NDEL1.
Schematics of NDE1 (top panel) and NDEL1 (bottom panel) drawn to scale depicting the N-terminal coiled-coil domain and the predicted C-terminal α-helix (labeled); the central flexible linker region connects these two structured regions, while another unstructured region lies at the extreme C-terminus of the proteins. Numerous PTMs (stars) reside within delineated protein-protein interaction sites, sub-cellular localization regions, or oligomerization domains with the vast majority located within predicted unstructured regions of the proteins. Minimal essential protein-binding sites from the literature are shown drawn to scale (NDE1: green bars; NDEL1: purple bars). References for each NDE1 interaction site are as follows: Centrosomal targeting (98); Centromere protein F (CENPF)/Mitosin/LEK1 (112); LIS1 (4); Utrophin (66); MCRS1/p78 (99); Su48/DBZ/ZNF365 (98); Dynein IC (89); Dynein (LC8) (49). References for each NDEL1 interaction site are as follows: Centrosomal targeting (113); LIS1 (96); Dynamitin (50); Pericentrin/Kendrin (113); Paxillin (114); G-protein β (Gβ) (115); DISC1 (28); Katanin p60 and p80 (68); 14-3-3ε (100); Neurofilament (NF)-L (72); Dynamin-2 (75); Rabaptin-5 (78); Dynein intermediate chain (IC) (89); Dynein heavy chain (HC) (1); Cdc42GAP (74); Lamin-B (63). Note: the palmitoylation site C273 (red star) on NDEL1 is also the site of endooligopeptidase activity (77). In the majority of cases, these sites have only been investigated for one of NDE1 or NDEL1; it is likely that some of them will be conserved between the two proteins. The effect of PTMs should be tested on protein conformation, stability, oligomerization, modulation of protein-protein interaction and cellular trafficking/targeting.
N-terminal coiled-coil domain
The N-terminal region of NDEL1 that is involved in the formation of oligomers through coiled-coil interactions remains the only region of the two proteins to be experimentally determined by crystallography (81), allowing the locations of the known phosphorylation sites in this region to be mapped on its three-dimensional structure (Figure 3).
Only three phosphorylation sites are known to reside within the N-terminal 190-amino acid coiled-coil-containing region for each of NDE1 and NDEL1 to date. NDE1 was shown to be phosphorylated by PKA at T131 with both DISC1 and phosphodiesterase 4 (PDE4), interaction partners of NDE1, able to modulate this post-translational event (86). Phosphorylation of this site causes increased interaction of NDE1 with NDEL1 in COS7 cells, but decreased interaction with LIS1 (86). Given that this region of NDE1 and NDEL1 governs their oligomerization/tetramerization state (81, 87) and that NDE1 and NDEL1 are capable of interacting in the same manner as NDE1 and NDEL1 homomeric species (87), one possibility is that this site may modulate the dimeric vs. tetrameric state of NDE1 generally, with the observed effect on hetero-oligomerization of NDE1-NDEL1 representing a special case of this. The HTP/MS-derived phosphorylation site at S135 of NDEL1 would likewise be predicted to impact on similar processes given its similar characteristics to T131, such as its location within an incomplete heptad repeat within the tetramerization domain (81, 86) and proximity to the LIS1-binding region (Figure 3). Potentially as a consequence of this, expression of NDE1 with a T131 phosphomimic mutant leads to decreased neurite outgrowth in a NS-1 cell assay (86). Based on amino acid sequence identity, NDEL1 would be predicted to be phosphorylated at its equivalent site, T132, however two studies have been unsuccessful in demonstrating this (86, 90), although it has been detected in a single HTP/MS screen (91). An antibody which could recognize both T131-phosphorylated NDE1 and putative T132-phosphorylated NDEL1 species, showed such species to be present at post-synaptic densities and cortical primary axons of mice, and to remain at the spindle poles of COS7 cells throughout cell mitosis, in contrast to non-phosphorylated species (86).
Central flexible linker region
As previously noted, the unstructured region between the N-terminal coiled-coil and predicted C-terminal α-helix of both NDE1 and NDEL1 are highly phosphorylated, with NDEL1 being a substrate for Cdk5 at S198, T219 and S231 (2). Cdk1 has been variously reported to either phosphorylate these sites or T219, S242 and T245 (96, 97), while Erk2 acts at T219 and T245 (96). Equivalent phosphorylation of NDE1 is less well studied; however, it is known to be a substrate for Cdk1 (98), which acts on T215, T246 and at least one additional undefined site (41). Cross-talk between these kinases also occurs, with use of a dominant-negative version of Cdk5 preventing Cdk1 from phosphorylating S231 of NDEL1, while the reverse was not true when a dominant-negative form of Cdk1 was used (62).
Several studies have looked at the expression and function of NDEL1 species in which five putative phosphorylation sites in this region (S198, T219, S231, S242 and T245), including all known Cdk1, Cdk5 and Erk2 sites were substituted to make either phosphodead alanine or valine residues, or to mimic phosphorylation with glutamic acid residues. NDEL1 carrying phosphodead mutations of all five of these residues are found prominently at the centrosome during interphase, as was wild-type NDEL1; in contrast, NDEL1 with five phosphomimic residues was only weakly detectable there (96). Similarly, the phosphomimic failed to transfer to the spindle poles during telophase in the manner of wild-type NDEL1; by contrast, the phosphodead mutant was prominent at the spindle from metaphase (96). Phosphorylation of NDEL1 therefore appears to block its spindle localization until the appropriate part of the cell cycle. Similar results were also seen when five putative phosphorylation sites in this region of NDE1 (T191, T215, T228, T243 and T246), plus one immediately C-terminal of its predicted α-helix (S282), were all mutated to phosphodead alanine or valine residues (98). Dramatically, knock-out of one such site on NDE1, T246, arrested mitotic HEK293T cells at the G2/M phase boundary (41).
Phosphorylation of residues in the central flexible linker region have been shown to modulate the interactions with other centrosomal proteins, specifically of NDE1 with the centrosomal protein Su48 (also known as DBZ or ZNF365) (98) and mitotic protein Microspherule protein 1 (MCRS1; also known as p78) (99), as well as the interactions of NDEL1 with the microtubule severing protein katanin (68) and 14-3-3ε (100) (Figure 4); although other phosphorylation events have been reported to have a greater impact upon 14-3-3ε interaction (101). Mock phosphorylation of NDEL1 at these residues also leads to increased activation of Cdc42GAP (74). Of particular interest though is the effect of these phosphorylation events on LIS1 and dynein. Application of either Cdk1 or Cdk5 to NDEL1 led to an increase in its interaction with LIS1, while causing dissociation of LIS1 to dynein (62). Successful dissociation of LIS1 is vital in the process by which dynein moves along microtubules with its cargo (102) and the importance of NDEL1 phosphorylation in this is seemingly demonstrated by the fact that expression of mutant NDEL1 lacking the five putative phosphorylation sites, or expression of a dominant-negative Cdk5 mutant, each effectively abolished dynein-related organelle transport along the axons of rat neurons (103). It is therefore possible that Cdk5 as well as potentially Cdk1 and Erk2 phosphorylation of NDEL1 may modulate dynein activity through altering LIS1-dynein interaction. Mutation of NDEL1 residues T219 and T245 was also shown to block NDEL1-dynein interactions (89). Other dynein-related effects of phosphorylation sites in this region are known; they are required to increase prophase nuclear envelope invagination in response to Cdk1 or Cdk5 (62), while their mutation inhibits cell migration during scratch-wound assays (74).
It has recently been shown that the C-terminal α-helix of NDE1 and NDEL1 can interact directly with its N-terminal coiled-coil domain (87) facilitated by this central proline-rich unstructured region that permits a bent-back conformation. It is therefore reasonable to suggest that many of the effects attributed to phosphorylation of residues in this region could be a consequence of regulation of this conformational process. For example, the effects on dynein function may occur because altered phosphorylation of this region affects the ability of NDE1 and NDEL1 to bring their distinct N- and C-terminal dynein binding domains (60, 89) into close physical proximity.
At least two separate phosphatases are involved in the removal of phosphate groups from this region of NDEL1, with protein phosphatase 4 catalytic subunit (PP4c) acting on T219 (104) and protein phosphatase 2 (PP2) acting on S231 (100) (Table 2). Interestingly, the former phosphatase is present at the centrosome at all times except during mitosis, suggesting that it may be involved in temporal regulation of NDEL1 function there (104).
Predicted C-terminal α-helix
The predicted α-helix of NDEL1 (87, 88) contains the DISC1 binding site (28) and overlaps with the ‘minimally’ defined binding regions of the dynein heavy chain (1) (Figure 4). The most investigated PTM in this region, is phosphorylation of NDEL1 by Aurora-A kinase at S251 during prophase, with expression of a NDEL1-S251E phosphomimic mutant being sufficient to rescue mitotic entry defects caused by Aurora-A knockdown (97). At the end of prophase, these phosphorylated NDEL1 species rapidly disappear, seemingly due to ubiquitination followed by degradation by the proteasome (97). This phosphorylation event also occurs in post-mitotic neurons, where phosphorylation of NDEL1 at S251 occurs as a result of Aurora-A-kinase activation by PKC-ζ at connections of the neuronal soma with neurites (71). Disruption of the function of any of PKC-ζ, Aurora-A kinase or NDEL1, variously by use of pseudo-substrates, kinase-deficient mutants or RNAi, led to decreased neurite extension in dorsal root ganglia neurons (71). Expression of a NDEL1-S251E phosphomimic mutant was also able to rescue migration defects in granular neurons caused by either conditional knockdown of endogenous NDEL1 or inhibition of Aurora-A kinase (105). The mechanism by which S251 phosphorylation causes these effects is unclear, although one possibility emerges from the fact that, unlike wild-type NDEL1, an S251A phosphodead mutant was not able to interact with dynein (89).
Additionally, NDEL1 is known to interact with 14-3-3ε as well as potentially 14-3-3α/β, γ, τ and ζ (100). S251 of NDEL1 is a good match for the canonical phosphorylation-dependent binding site of 14-3-3 proteins and indeed mutation of S251 to alanine reduces NDEL1 interaction with 14-3-3 (101). Additionally, interaction between recombinant forms of the two proteins is increased following Aurora-A kinase treatment (101).
NDEL1 can also be palmitoylated within this predicted helix at C273 by three enzymes, DHHC2, 3 and 7, causing a reduction in interaction between NDEL1 and dynein and an increase in NDEL1 at neurite tips, indicating reduced retrograde transport by dynein (54). Other evidence that NDEL1 palmitoylation limits dynein function can be seen through its impaired ability to transport proteins, short microtubules and Golgi vesicles following co-transfection of DHHC and wild-type NDEL1, but not when a DHHC and a NDEL1-C273S palmitoylation-resistant mutant or NDEL1 alone were used (54). It has also been reported that expression of NDEL1 with a C273A mutation inhibits the growth of neurites of PC12, in contrast to wild-type NDEL1 which increases it, although a specific role of palmitoylation in this process has not been established (106). Similarly, wild-type, but not C273A, NDEL1 can rescue neurite outgrowth defects caused by knockdown of endogenous NDEL1 (106). NDE1 can also be palmitoylated at its equivalent site, C274, by the same DHHC enzymes as NDEL1; however, strikingly, palmitoylation of NDE1 had no effect on its interaction with dynein or any of the other functional tests of NDEL1 palmitoylation, indicating a fundamental difference in the function of NDE1 and NDEL1 (54).
C273 is also the catalytic residue for the oligopeptidase activity of NDEL1 (77), although it remains unclear whether this is blocked by palmitoylation, or whether the effect on neurite outgrowth ascribed to NDEL1 species in which C273 has been mutated is due to altered enzymatic activity, altered interaction with dynein or a combination of the two. The possibility exists that this palmitoylation event may act as a switch between these two, seemingly distinct, functions of the NDEL1 protein. Although the putative catalytic cysteine residue is also present in NDE1, no study to date has investigated whether it also possesses a similar oligopeptidase activity.
Extreme C-terminal unstructured region
The extreme C-terminal region of NDE1 and NDEL1 is known to interact with a number of different protein binding partners, and it is therefore likely that the PTMs within this region influence NDE1 and NDEL1 function at least in part through regulation of these interactions. Of these, NDE1 is known to be phosphorylated by PKA at S306 (88), in a DISC1- and PDE4-modulated process (86), while the equivalent residue in NDEL1 is non-phosphorylatable (Figure 1). The function of this site remains unknown; however, NS-1 cells expressing NDE1-S306D phosphomimic mutants display an increase in neurite number compared to those expressing phosphodead S306A mutants (86). It is notable that proteomics screening implies S306 is part of a cluster of four directly concurrent phosphorylated residues, three of which are specific to NDE1 (Figure 1). The functional significance of this cluster remains to be determined.
Additionally, NDEL1 has a serine at residue 336 which can be phosphorylated by Aurora-A and -B kinases, creating a 14-3-3 binding site (101). Mutation of this site to alanine has a greater effect on 14-3-3 interaction than similar mutation of S251 described earlier, with mutation of both abolishing the majority of interaction (101). Notably, however, this site exists only within a specific isoform of NDEL1 and because both NDE1 and NDEL1 are known to exist as multiple splice variants, many of which vary only in their extreme C-termini (33) it can therefore be speculated that one purpose of the multiple splice variants is to introduce alternative PTM regulation at the extreme C-terminus of a specific subset of NDE1/NDEL1 molecules in vivo, and potentially ones with unique patterns of temporal or spatial expression with the organism.
Conclusions
Since their first description, NDE1 and NDEL1 have emerged as key players in mitosis and neurodevelopment with overlapping yet distinct functional profiles. While they share high sequence similarity and have been shown to adopt similar structure and oligomeric states, they are clearly differentially regulated post-translationally. A mass of PTM data has emerged for the two proteins over the past decade and recent HTP data via tandem MS will continue to inform on regulatory mechanisms of protein function. Notwithstanding the expert manually curated effort of the PhosphoSitePlus database (91) to catalogue experimentally determined PTM sites, it should be borne in mind there could be some data quality issues for assignments; false positives and/or incorrect assignments to either NDE1 or NDEL1, given their high sequence identity, is a possibility, especially for some of the sites identified purely via MS/HTP-discovery, whose assignments are probabilistic by nature. Nonetheless, nine out of ten sites verified in the primary literature for NDE1 and NDEL1 were seen in the MS-only assignments in the database suggesting a high true positive rate. Even for the one site T131 in NDE1, not discovered by MS, the equivalent NDEL1 site (T132) was indeed observed. Additionally, for each protein >50% of sites assigned only using the MS/HTP criteria had five or more references corresponding to each site in the database, indicating a high potential for these sites to be true positives too. Conversely, it should also be noted that there could be a failure to detect some of the phosphorylation sites in a protein sample because of the inherent sensitivity of the methodology whereby the phosphate groups can be lost during sample preparation and analysis. Hence, failure to detect phosphorylation should not necessarily be considered as evidence that the site in not actually phosphorylated under certain relevant conditions (107).
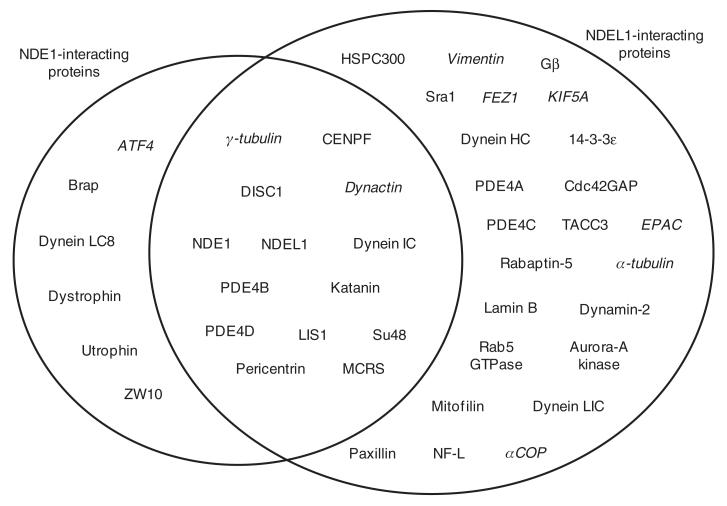
A major issue in clarifying the relationship between NDE1 and NDEL1 remains the relative lack of directly comparable information between them. For example, while there are some proteins confirmed to be interaction partners of both NDE1 and NDEL1, there are a greater number which are known to interact with NDEL1, but for which comparable interaction studies with NDE1 have not yet been performed (Figure 5). Some examples of protein interaction partners proven to bind NDE1, but not yet tested for NDEL1, also exist.
Figure 5.
Protein interaction partners of NDE1 and NDEL1.
Proteins in the left circle interact with NDE1, those in the right circle with NDEL1. Proteins in the intersecting region are known to interact with both proteins. In the majority of instances where a protein is listed as interacting with only one of NDE1/NDEL1, the reciprocal experiment to test for interaction with the other of NDEL1/NDE1 has not been published to the best of our knowledge. Proteins listed in italics are known to exist in a complex with NDE1/NDEL1 but do not, or have not yet been shown to, directly bind to it. Data are taken from the following papers (1-5, 27-29, 31-33, 48, 50, 53, 57, 58, 63, 65, 66, 68, 72, 74-76, 78, 80, 88, 90, 96-98, 100, 101, 113-121). In some cases the essential binding regions for these interaction partners have been delineated on NDE1 or NDEL1; these are shown schematically in Figure 4.
From the information currently available, however, it is possible to begin to speculate on the fundamental differences between the NDE1 and NDEL1 proteins which have warranted their conserved existence throughout vertebrate evolution. Functionally, the proteins appear to be highly similar, but with potential differences in dynein binding (49) and mitotic function (58). By contrast, their post-translational regulation is distinct; this suggests that, due to the fundamental roles the two proteins play in neuronal function, the existence of two distinct ‘NudE-derived’ proteins allows a tighter level of regulation of their roles. It is also noteworthy that while Ndel1 knock-out mice are not viable (51), Nde1 knock-out mice survive to birth (57). We therefore hypothesize that while NDEL1 plays the “core” role, conserved across evolution of NDE1/NDEL1 orthologs, NDE1 can act as an ancillary version, providing additional or perhaps slightly modified versions of the common NDE1/NDEL1 functions where necessary under a different system of regulation. Such differences are likely to be both temporal, with NDE1 having a more specialized pattern of expression during early neurodevelopment, and spatial, with NDEL1 known to have both cortical and hippocampal developmental functions (51), while NDE1 has mainly been described as affecting the cortex (57). An alternative theory to explain the mouse viability, i.e., Ndel1 has a specific essential function in the early blastocyst, must also be considered. The former model would, however, be consistent with the relatively higher conservation of NDEL1 throughout evolution, compared to the more recent, faster evolution of NDE1 (54) (Figure 1; Table 1). This would also be consistent with the apparent role of NDE1 CNVs in mental health and other neurodevelopmental conditions: while gross disruption of the more ‘essential’ NDEL1 protein would be critical and perhaps incompatible with life, disruption of NDE1 causes more subtle effects by comparison, notwithstanding the gross microcephaly phenotypes associated with complete loss of NDE1 function. The issue of exactly when and where NDE1 and NDEL1 perform their functions is further complicated by their ability to form hetero-oligomers (29, 33, 87), leading to the possibility of unique functions of a mixed NDE1-NDEL1 species, in addition to those performed by each protein alone. Teasing apart this relationship remains an important avenue for future research; research that will be of significant importance in understanding their shared and specific roles in psychiatric disorders and neurodevelopment.
Acknowledgments
The authors would like to thank David Porteous, Kirsty Millar, Pippa Thomson and Darragh Crummie for critical reading of the manuscript, useful discussion and comments. This work was supported by the Alexander von Humboldt Foundation to N.J.B., the Academy of Finland to W.H. and a Wellcome Trust grant (088179/A/09/Z) and Medical Research Council funding to D.C.S. The authors declare no competing interests exist.
Biographies

Nicholas Bradshaw received his BSc in Natural Sciences (Biology with Physics) from Durham University in 2005 and undertook his doctorate at the University of Edinburgh under the supervision of Dr Kirsty Millar and Prof David Porteous on the interactions of the schizophrenia-related proteins NDE1 and DISC1. After receiving his PhD in 2009, Nick continued his research into the biochemical and biophysical properties of these and related proteins pertinent to major mental illness in Edinburgh, in collaboration with Dr Dinesh Soares. In 2011 he received a postdoctoral fellowship from the Alexander von Humboldt Foundation to further develop his research in the laboratory of Prof Dr Carsten Korth at the Heinrich Heine University, Düsseldorf.

William Hennah is an Academy of Finland Research Fellow at the Institute for Molecular Medicine Finland FIMM, University of Helsinki. William received his BSc in Molecular Genetics and MSc in Neuroscience from King’s College London, and obtained his PhD in Human Genetics from the University of Helsinki. His current research focus is in the field of neuropsychiatric genetics, using the unique advantages afforded by the Finnish population registers to study the role of the DISC1 gene network in major mental illness.

Dinesh Soares is a Research Fellow at the MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh. Dinesh received his BSc in Biochemistry and Zoology from St. Xavier’s College, University of Mumbai, an M.Res. in Bioinformatics from the University of York, UK and subsequently a PhD in Structural Bioinformatics from the School of Chemistry at the University of Edinburgh, UK in 2007. Dinesh has since worked both as an experimental biochemist/biophysicist and computational structural biology specialist at the University of Edinburgh on a range of projects, but chiefly applied to the broad areas of immunology, neuroscience and psychiatry.
Contributor Information
Nicholas J. Bradshaw, Department of Neuropathology, Heinrich Heine University, Düsseldorf, University Medical School, Moorenstrasse 5, D-40225, Düsseldorf, Germany.
William Hennah, Institute for Molecular Medicine Finland FIMM, University of Helsinki, Helsinki, Finland; and National Institute for, Health and Welfare, Department of Mental Health and Substance, Abuse Services, Helsinki, Finland.
Dinesh C. Soares, MRC Institute of Genetics and Molecular Medicine (MRC IGMM), University of Edinburgh, Western General, Hospital, Crewe Road South, Edinburgh EH4 2XU, UK.
References
- 1.Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–96. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 2.Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 3.Kitagawa M, Umezu M, Aoki J, Koizumi H, Arai H, Inoue K. Direct association of LIS1, the lissencephaly gene product, with a mammalian homologue of a fungal nuclear distribution protein, rNUDE. FEBS Lett. 2000;479:57–62. doi: 10.1016/s0014-5793(00)01856-1. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Olson EC, Stukenberg PT, Flanagan LA, Kirschner MW, Walsh CA. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–79. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- 5.Sweeney KJ, Prokscha A, Eichele G. NudE-L, a novel Lis1-interacting protein, belongs to a family of vertebrate coiled-coil proteins. Mech Dev. 2001;101:21–33. doi: 10.1016/s0925-4773(00)00543-8. [DOI] [PubMed] [Google Scholar]
- 6.de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, Kluck C, Muhle H, von Spiczak S, Ostertag P, Obermeier T, Kleefuss-Lie AA, Hallmann K, Steffens M, Gaus V, Klein KM, Hamer HM, Rosenow F, Brilstra EH, Trenite DK, Swinkels ME, Weber YG, Unterberger I, Zimprich F, Urak L, Feucht M, Fuchs K, Moller RS, Hjalgrim H, De Jonghe P, Suls A, Ruckert IM, Wichmann HE, Franke A, Schreiber S, Nurnberg P, Elger CE, Lerche H, Stephani U, Koeleman BP, Lindhout D, Eichler EE, Sander T. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, Breuning MH, Fryns JP, Devriendt K, Van Buggenhout G, Vogels A, Stewart H, Hennekam RC, Cooper GM, Regan R, Knight SJ, Eichler EE, Vermeesch JR. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–32. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinzen EL, Radtke RA, Urban TJ, Cavalleri GL, Depondt C, Need AC, Walley NM, Nicoletti P, Ge D, Catarino CB, Duncan JS, Kasperaviciute D, Tate SK, Caboclo LO, Sander JW, Clayton L, Linney KN, Shianna KV, Gumbs CE, Smith J, Cronin KD, Maia JM, Doherty CP, Pandolfo M, Leppert D, Middleton LT, Gibson RA, Johnson MR, Matthews PM, Hosford D, Kalviainen R, Eriksson K, Kantanen AM, Dorn T, Hansen J, Kramer G, Steinhoff BJ, Wieser HG, Zumsteg D, Ortega M, Wood NW, Huxley-Jones J, Mikati M, Gallentine WB, Husain AM, Buckley PG, Stallings RL, Podgoreanu MV, Delanty N, Sisodiya SM, Goldstein DB. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 2010;86:707–18. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP, Buizer-Voskamp JE, Strengman E, Francks C, Muglia P, Gylfason A, Gustafsson O, Olason PI, Steinberg S, Hansen T, Jakobsen KD, Rasmussen HB, Giegling I, Moller HJ, Hartmann A, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Bramon E, Kiemeney LA, Franke B, Murray R, Vassos E, Toulopoulou T, Muhleisen TW, Tosato S, Ruggeri M, Djurovic S, Andreassen OA, Zhang Z, Werge T, Ophoff RA, Rietschel M, Nothen MM, Petursson H, Stefansson H, Peltonen L, Collier D, Stefansson K, St Clair DM. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, Franke A, Malafosse A, Genton P, Thomas P, Gurnett CA, Schreiber S, Bassuk AG, Guipponi M, Stephani U, Helbig I, Eichler EE. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramalingam A, Zhou XG, Fiedler SD, Brawner SJ, Joyce JM, Liu HY, Yu S. 16p13.11 duplication is a risk factor for a wide spectrum of neuropsychiatric disorders. J Hum Genet. 2011;56:541–4. doi: 10.1038/jhg.2011.42. [DOI] [PubMed] [Google Scholar]
- 12.Sahoo T, Theisen A, Rosenfeld JA, Lamb AN, Ravnan JB, Schultz RA, Torchia BS, Neill N, Casci I, Bejjani BA, Shaffer LG. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med. 2011;13:868–80. doi: 10.1097/GIM.0b013e3182217a06. [DOI] [PubMed] [Google Scholar]
- 13.Tropeano M, Ahn JW, Dobson RJ, Breen G, Rucker J, Dixit A, Pal DK, McGuffin P, Farmer A, White PS, Andrieux J, Vassos E, Ogilvie CM, Curran S, Collier DA. Male-biased autosomal effect of 16p13.11 copy number variation in neurodevelopmental disorders. PLoS One. 2013;8:e61365. doi: 10.1371/journal.pone.0061365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, Banna L, Brereton AV, Hill A, Bisgaard AM, Muller I, Hultschig C, Erdogan F, Wieczorek G, Ropers HH. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–82. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 15.Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R, Stefansson H, Stefansson K, Magnusson P, Gudmundsson OO, Gustafsson O, Holmans P, Owen MJ, O’Donovan M, Thapar A. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–8. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, Shianna KV, Yoon W, Kasperaviciute D, Gennarelli M, Strittmatter WJ, Bonvicini C, Rossi G, Jayathilake K, Cola PA, McEvoy JP, Keefe RS, Fisher EM, St Jean PL, Giegling I, Hartmann AM, Moller HJ, Ruppert A, Fraser G, Crombie C, Middleton LT, St Clair D, Roses AD, Muglia P, Francks C, Rujescu D, Meltzer HY, Goldstein DB. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagamani SC, Erez A, Bader P, Lalani SR, Scott DA, Scaglia F, Plon SE, Tsai CH, Reimschisel T, Roeder E, Malphrus AD, Eng PA, Hixson PM, Kang SH, Stankiewicz P, Patel A, Cheung SW. Phenotypic manifestations of copy number variation in chromosome 16p13.11. Eur J Hum Genet. 2011;19:280–6. doi: 10.1038/ejhg.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisniowiecka-Kowalnik B, Kastory-Bronowska M, Bartnik M, Derwinska K, Dymczak-Domini W, Szumbarska D, Ziemka E, Szczaluba K, Sykulski M, Gambin T, Gambin A, Shaw CA, Mazurczak T, Obersztyn E, Bocian E, Stankiewicz P. Application of custom-designed oligonucleotide array CGH in 145 patients with autistic spectrum disorders. Eur J Hum Genet. 2013;21:620–5. doi: 10.1038/ejhg.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 20.Yerabham AS, Weiergraber OH, Bradshaw NJ, Korth C. Revisiting Disrupted in Schizophrenia 1 as a scaffold protein. Biol Chem. 2013 doi: 10.1515/hsz-2013-0178. DOI: 10.1515/hsz-2013-0178. [DOI] [PubMed] [Google Scholar]
- 21.Bradshaw NJ, Porteous DJ. DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology. 2012;62:1230–41. doi: 10.1016/j.neuropharm.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12:707–22. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soares DC, Carlyle BC, Bradshaw NJ, Porteous DJ. DISC1: structure, function, and therapeutic potential for major mental illness. ACS Chem Neurosci. 2011;2:609–32. doi: 10.1021/cn200062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–23. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 25.St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–6. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 26.Hennah W, Tomppo L, Hiekkalinna T, Palo OM, Kilpinen H, Ekelund J, Tuulio-Henriksson A, Silander K, Partonen T, Paunio T, Terwilliger JD, Lonnqvist J, Peltonen L. Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum Mol Genet. 2007;16:453–62. doi: 10.1093/hmg/ddl462. [DOI] [PubMed] [Google Scholar]
- 27.Millar JK, Christie S, Porteous DJ. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun. 2003;311:1019–25. doi: 10.1016/j.bbrc.2003.10.101. [DOI] [PubMed] [Google Scholar]
- 28.Brandon NJ, Handford EJ, Schurov I, Rain JC, Pelling M, Duran- Jimeniz B, Camargo LM, Oliver KR, Beher D, Shearman MS, Whiting PJ. Disrupted in Schizophrenia 1 and Nudel form a neurodevelopmentally regulated protein complex: implications for schizophrenia and other major neurological disorders. Mol Cell Neurosci. 2004;25:42–55. doi: 10.1016/j.mcn.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, DeRosse P, Ishizuka K, Elashvili S, Arai H, Goldman D, Sawa A, Malhotra AK. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–73. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamiya A, Tomoda T, Chang J, Takaki M, Zhan C, Morita M, Cascio MB, Elashvili S, Koizumi H, Takanezawa Y, Dickerson F, Yolken R, Arai H, Sawa A. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum Mol Genet. 2006;15:3313–23. doi: 10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- 31.Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In- Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- 32.Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, Ross CA, Sawa A. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA. 2003;100:289–94. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradshaw NJ, Christie S, Soares DC, Carlyle BC, Porteous DJ, Millar JK. NDE1 and NDEL1: multimerisation, alternate splicing and DISC1 interaction. Neurosci Lett. 2009;449:228–33. doi: 10.1016/j.neulet.2008.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomppo L, Hennah W, Lahermo P, Loukola A, Tuulio-Henriksson A, Suvisaari J, Partonen T, Ekelund J, Lonnqvist J, Peltonen L. Association between genes of Disrupted in Schizophrenia 1 (DISC1) interactors and schizophrenia supports the role of the DISC1 pathway in the etiology of major mental illnesses. Biol Psychiatry. 2009;65:1055–62. doi: 10.1016/j.biopsych.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicodemus KK, Callicott JH, Higier RG, Luna A, Nixon DC, Lipska BK, Vakkalanka R, Giegling I, Rujescu D, St Clair D, Muglia P, Shugart YY, Weinberger DR. Evidence of statistical epistasis between DISC1, CIT and NDEL1 impacting risk for schizophrenia: biological validation with functional neuroimaging. Hum Genet. 2010;127:441–52. doi: 10.1007/s00439-009-0782-y. [DOI] [PubMed] [Google Scholar]
- 36.Ikeda M, Hikita T, Taya S, Uraguchi-Asaki J, Toyo-oka K, Wynshaw-Boris A, Ujike H, Inada T, Takao K, Miyakawa T, Ozaki N, Kaibuchi K, Iwata N. Identification of YWHAE, a gene encoding 14-3-3 ε, as a possible susceptibility gene for schizophrenia. Hum Mol Genet. 2008;17:3212–22. doi: 10.1093/hmg/ddn217. [DOI] [PubMed] [Google Scholar]
- 37.Kahler AK, Djurovic S, Kulle B, Jonsson EG, Agartz I, Hall H, Opjordsmoen S, Jakobsen KD, Hansen T, Melle I, Werge T, Steen VM, Andreassen OA. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1089–100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- 38.Numata S, Ueno S, Iga J, Nakataki M, Ohmori T, Tanahashi T, Itakura M, Sano A, Ohi K, Hashimoto R, Takeda M. No association between the NDE1 gene and schizophrenia in the Japanese population. Schizophr Res. 2008;99:367–9. doi: 10.1016/j.schres.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 39.Rastogi A, Zai C, Likhodi O, Kennedy JL, Wong AH. Genetic association and post-mortem brain mRNA analysis of DISC1 and related genes in schizophrenia. Schizophr Res. 2009;114:39–49. doi: 10.1016/j.schres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 40.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–76. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, Hill RS, Barry BJ, Partlow JN, Gascon GG, Kentab A, Jan M, Shaheen R, Feng Y, Walsh CA. Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am J Hum Genet. 2011;88:536–47. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakircioglu M, Carvalho OP, Khurshid M, Cox JJ, Tuysuz B, Barak T, Yilmaz S, Caglayan O, Dincer A, Nicholas AK, Quarrell O, Springell K, Karbani G, Malik S, Gannon C, Sheridan E, Crosier M, Lisgo SN, Lindsay S, Bilguvar K, Gergely F, Gunel M, Woods CG. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am J Hum Genet. 2011;88:523–35. doi: 10.1016/j.ajhg.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guven A, Gunduz A, Bozoglu TM, Yalcinkaya C, Tolun A. Novel NDE1 homozygous mutation resulting in microhydranencephaly and not microlyssencephaly. Neurogenetics. 2012;13:189–94. doi: 10.1007/s10048-012-0326-9. [DOI] [PubMed] [Google Scholar]
- 44.Paciorkowski AR, Keppler-Noreuil K, Robinson L, Sullivan C, Sajan S, Christian SL, Bukshpun P, Gabriel SB, Gleeson JG, Sherr EH, Dobyns WB. Deletion 16p13.11 uncovers NDE1 mutations on the non-deleted homolog and extends the spectrum of severe microcephaly to include fetal brain disruption. Am J Med Genet A. 2013;161:1523–30. doi: 10.1002/ajmg.a.35969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissen- cephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–21. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 46.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–14. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torisawa T, Nakayama A, Furuta K, Yamada M, Hirotsune S, Toyoshima YY. Functional dissection of LIS1 and NDEL1 towards understanding the molecular mechanisms of cytoplasmic dynein regulation. J Biol Chem. 2011;286:1959–65. doi: 10.1074/jbc.M110.169847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stehman SA, Chen Y, McKenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol. 2007(178):583–94. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem. 2011;286:39615–2. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang Y, Yu W, Li Y, Yang Z, Yan X, Huang Q, Zhu X. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J Cell Biol. 2004;164:557–66. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, Muramatsu M, Miyagawa S, Hiraiwa N, Yoshiki A, Wynshaw- Boris A, Hirotsune S. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–27. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Q, Wang F, Cao J, Shen Y, Huang Q, Bao L, Zhu X. Nudel promotes axonal lysosome clearance and endo-lysosome formation via dynein-mediated transport. Traffic. 2009;10:1337–49. doi: 10.1111/j.1600-0854.2009.00945.x. [DOI] [PubMed] [Google Scholar]
- 53.Shim SY, Samuels BA, Wang J, Neumayer G, Belzil C, Ayala R, Shi Y, Tsai LH, Nguyen MD. Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J Biol Chem. 2008;283:12232–40. doi: 10.1074/jbc.M710200200. [DOI] [PubMed] [Google Scholar]
- 54.Shmueli A, Segal M, Sapir T, Tsutsumi R, Noritake J, Bar A, Sapoznik S, Fukata Y, Orr I, Fukata M, Reiner O. Ndel1 palmitoylation: a new mean to regulate cytoplasmic dynein activity. EMBO J. 2010;29:107–19. doi: 10.1038/emboj.2009.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lam C, Vergnolle MA, Thorpe L, Woodman PG, Allan VJ. Functional interplay between LIS1, NDE1 and NDEL1 in dynein- dependent organelle positioning. J Cell Sci. 2010;123:202–12. doi: 10.1242/jcs.059337. [DOI] [PubMed] [Google Scholar]
- 56.Segal M, Soifer I, Petzold H, Howard J, Elbaum M, Reiner O. Ndel1-derived peptides modulate bidirectional transport of injected beads in the squid giant axon. Biol Open. 2012;1:220–31. doi: 10.1242/bio.2012307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–93. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 58.Vergnolle MA, Taylor SS. Cenp-F links kinetochores to Ndel1/ Nde1/Lis1/dynein microtubule motor complexes. Curr Biol. 2007;17:1173–9. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- 59.Raaijmakers JA, Tanenbaum ME, Medema RH. Systematic dissection of dynein regulators in mitosis. J Cell Biol. 2013;201:201–15. doi: 10.1083/jcb.201208098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Zheng Y. Identification of a novel dynein binding domain in nudel essential for spindle pole organization in Xenopus egg extract. J Biol Chem. 2011;286:587–93. doi: 10.1074/jbc.M110.181578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang Y, Yu W, Li Y, Yu L, Zhang Q, Wang F, Yang Z, Du J, Huang Q, Yao X, Zhu X. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol Biol Cell. 2007;18:2656–66. doi: 10.1091/mbc.E06-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hebbar S, Mesngon MT, Guillotte AM, Desai B, Ayala R, Smith DS. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol. 2008;182:1063–71. doi: 10.1083/jcb.200803071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma L, Tsai MY, Wang S, Lu B, Chen R, Iii JR, Zhu X, Zheng Y. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat Cell Biol. 2009;11:247–56. doi: 10.1038/ncb1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat Cell Biol. 2011;13:351–60. doi: 10.1038/ncb2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanctot AA, Peng CY, Pawlisz AS, Joksimovic M, Feng Y. Spatially dependent dynamic MAPK modulation by the Nde1-Lis1-Brap complex patterns mammalian CNS. Dev Cell. 2013;25:241–55. doi: 10.1016/j.devcel.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pawlisz AS, Feng Y. Three-dimensional regulation of radial glial functions by Lis1-Nde1 and dystrophin glycoprotein complexes. PLoS Biol. 2011;9:e1001172. doi: 10.1371/journal.pbio.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–77. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 68.Toyo-Oka K, Sasaki S, Yano Y, Mori D, Kobayashi T, Toyoshima YY, Tokuoka SM, Ishii S, Shimizu T, Muramatsu M, Hiraiwa N, Yoshiki A, Wynshaw-Boris A, Hirotsune S. Recruitment of katanin p60 by phosphorylated NDEL1, an LIS1 interacting protein, is essential for mitotic cell division and neuronal migration. Hum Mol Genet. 2005;14:3113–28. doi: 10.1093/hmg/ddi339. [DOI] [PubMed] [Google Scholar]
- 69.Youn YH, Pramparo T, Hirotsune S, Wynshaw-Boris A. Distinct dose-dependent cortical neuronal migration and neurite extension defects in Lis1 and Ndel1 mutant mice. J Neurosci. 2009;29:15520–30. doi: 10.1523/JNEUROSCI.4630-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, Liu CY, Ganesan S, Cheng HJ, Ming GL, Lu B, Song H. Disrupted-in-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–58. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mori D, Yamada M, Mimori-Kiyosue Y, Shirai Y, Suzuki A, Ohno S, Saya H, Wynshaw-Boris A, Hirotsune S. An essential role of the aPKC-Aurora A-NDEL1 pathway in neurite elongation by modulation of microtubule dynamics. Nat Cell Biol. 2009;11:1057–68. doi: 10.1038/ncb1919. [DOI] [PubMed] [Google Scholar]
- 72.Nguyen MD, Shu T, Sanada K, Lariviere RC, Tseng HC, Park SK, Julien JP, Tsai LH. A NUDEL-dependent mechanism of neurofilament assembly regulates the integrity of CNS neurons. Nat Cell Biol. 2004;6:595–608. doi: 10.1038/ncb1139. [DOI] [PubMed] [Google Scholar]
- 73.Toth C, Shim SY, Wang J, Jiang Y, Neumayer G, Belzil C, Liu WQ, Martinez J, Zochodne D, Nguyen MD. Ndel1 promotes axon regeneration via intermediate filaments. PLoS One. 2008;3:e2014. doi: 10.1371/journal.pone.0002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen Y, Li N, Wu S, Zhou Y, Shan Y, Zhang Q, Ding C, Yuan Q, Zhao F, Zeng R, Zhu X. Nudel binds Cdc42GAP to modulate Cdc42 activity at the leading edge of migrating cells. Dev Cell. 2008;14:342–53. doi: 10.1016/j.devcel.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 75.Chansard M, Wang J, Tran HC, Neumayer G, Shim SY, Park YU, Belzil C, Le HT, Park SK, Nguyen MD. The cytoskeletal protein Ndel1 regulates dynamin 2 GTPase activity. PLoS One. 2011;6:e14583. doi: 10.1371/journal.pone.0014583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu S, Ma L, Wu Y, Zeng R, Zhu X. Nudel is crucial for the WAVE complex assembly in vivo by selectively promoting subcomplex stability and formation through direct interactions. Cell Res. 2012;22:1270–84. doi: 10.1038/cr.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hayashi MA, Portaro FC, Bastos MF, Guerreiro JR, Oliveira V, Gorrao SS, Tambourgi DV, Sant’Anna OA, Whiting PJ, Camargo LM, Konno K, Brandon NJ, Camargo AC. Inhibition of NUDEL (nuclear distribution element-like)-oligopeptidase activity by disrupted-in-schizophrenia 1. Proc Natl Acad Sci USA. 2005;102:3828–33. doi: 10.1073/pnas.0500330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chansard M, Hong JH, Park YU, Park SK, Nguyen MD. Ndel1, Nudel (Noodle): flexible in the cell? Cytoskeleton (Hoboken) 2011;68:540–54. doi: 10.1002/cm.20532. [DOI] [PubMed] [Google Scholar]
- 79.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE, Sestan N. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150:681–8. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Derewenda U, Tarricone C, Choi WC, Cooper DR, Lukasik S, Perrina F, Tripathy A, Kim MH, Cafiso DS, Musacchio A, Derewenda ZS. The structure of the coiled-coil domain of Ndel1 and the basis of its interaction with Lis1, the causal protein of Miller-Dieker lissencephaly. Structure. 2007;15:1467–81. doi: 10.1016/j.str.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 82.Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics. 2007;6:2200–11. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- 83.Leliveld SR, Bader V, Hendriks P, Prikulis I, Sajnani G, Requena JR, Korth C. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J Neurosci. 2008;28:3839–45. doi: 10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Narayanan S, Arthanari H, Wolfe MS, Wagner G. Molecular characterization of disrupted in schizophrenia-1 risk variant S704C reveals the formation of altered oligomeric assembly. J Biol Chem. 2011;286:44266–76. doi: 10.1074/jbc.M111.271593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tarricone C, Perrina F, Monzani S, Massimiliano L, Kim MH, Derewenda ZS, Knapp S, Tsai LH, Musacchio A. Coupling PAF signaling to dynein regulation: structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 2004;44:809–21. doi: 10.1016/j.neuron.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 86.Bradshaw NJ, Soares DC, Carlyle BC, Ogawa F, Davidson-Smith H, Christie S, Mackie S, Thomson PA, Porteous DJ, Millar JK. PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1. J Neurosci. 2011;31:9043–54. doi: 10.1523/JNEUROSCI.5410-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soares DC, Bradshaw NJ, Zou J, Kennaway CK, Hamilton RS, Chen ZA, Wear MA, Blackburn EA, Bramham J, Bottcher B, Millar JK, Barlow PN, Walkinshaw MD, Rappsilber J, Porteous DJ. The mitosis and neurodevelopment proteins NDE1 and NDEL1 form dimers, tetramers, and polymers with a folded back structure in solution. J Biol Chem. 2012;287:32381–93. doi: 10.1074/jbc.M112.393439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bradshaw NJ, Ogawa F, Antolin-Fontes B, Chubb JE, Carlyle BC, Christie S, Claessens A, Porteous DJ, Millar JK. DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem Biophys Res Commun. 2008;377:1091–6. doi: 10.1016/j.bbrc.2008.10.120. [DOI] [PubMed] [Google Scholar]
- 89.Zylkiewicz E, Kijanska M, Choi WC, Derewenda U, Derewenda ZS, Stukenberg PT. The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J Cell Biol. 2011;192:433–45. doi: 10.1083/jcb.201011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Collins DM, Murdoch H, Dunlop AJ, Charych E, Baillie GS, Wang Q, Herberg FW, Brandon N, Prinz A, Houslay MD. Ndel1 alters its conformation by sequestering cAMP-specific phospho- diesterase-4D3 (PDE4D3) in a manner that is dynamically regulated through Protein Kinase A (PKA) Cell Signal. 2008;20:2356–69. doi: 10.1016/j.cellsig.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 91.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a compre- hensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–49. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Amoutzias GD, He Y, Gordon J, Mossialos D, Oliver SG, Van de Peer Y. Posttranslational regulation impacts the fate of duplicated genes. Proc Natl Acad Sci USA. 2010;107:2967–71. doi: 10.1073/pnas.0911603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phospho- rylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–49. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 95.Kreegipuu A, Blom N, Brunak S, Jarv J. Statistical analysis of protein kinase specificity determinants. FEBS Lett. 1998;430:45–50. doi: 10.1016/s0014-5793(98)00503-1. [DOI] [PubMed] [Google Scholar]
- 96.Yan X, Li F, Liang Y, Shen Y, Zhao X, Huang Q, Zhu X. Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol Cell Biol. 2003;23:1239–50. doi: 10.1128/MCB.23.4.1239-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mori D, Yano Y, Toyo-oka K, Yoshida N, Yamada M, Muramatsu M, Zhang D, Saya H, Toyoshima YY, Kinoshita K, Wynshaw- Boris A, Hirotsune S. NDEL1 phosphorylation by Aurora-A kinase is essential for centrosomal maturation, separation, and TACC3 recruitment. Mol Cell Biol. 2007;27:352–67. doi: 10.1128/MCB.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirohashi Y, Wang Q, Liu Q, Li B, Du X, Zhang H, Furuuchi K, Masuda K, Sato N, Greene MI. Centrosomal proteins Nde1 and Su48 form a complex regulated by phosphorylation. Oncogene. 2006;25:6048–55. doi: 10.1038/sj.onc.1209637. [DOI] [PubMed] [Google Scholar]
- 99.Hirohashi Y, Wang Q, Liu Q, Du X, Zhang H, Sato N, Greene MI. p78/MCRS1 forms a complex with centrosomal protein Nde1 and is essential for cell viability. Oncogene. 2006;25:4937–46. doi: 10.1038/sj.onc.1209500. [DOI] [PubMed] [Google Scholar]
- 100.Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, Hirotsune S, Wynshaw-Boris A. 14-3-3 ε is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–85. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 101.Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–86. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pandey JP, Smith DS. A Cdk5-dependent switch regulates Lis1/ Ndel1/dynein-driven organelle transport in adult axons. J Neurosci. 2011;31:17207–19. doi: 10.1523/JNEUROSCI.4108-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toyo-oka K, Mori D, Yano Y, Shiota M, Iwao H, Goto H, Inagaki M, Hiraiwa N, Muramatsu M, Wynshaw-Boris A, Yoshiki A, Hirotsune S. Protein phosphatase 4 catalytic subunit regulates Cdk1 activity and microtubule organization via NDEL1 dephosphorylation. J Cell Biol. 2008;180:1133–47. doi: 10.1083/jcb.200705148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takitoh T, Kumamoto K, Wang CC, Sato M, Toba S, Wynshaw-Boris A, Hirotsune S. Activation of Aurora-A is essential for neuronal migration via modulation of microtubule organization. J Neurosci. 2012;32:11050–66. doi: 10.1523/JNEUROSCI.5664-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hayashi MA, Guerreiro JR, Charych E, Kamiya A, Barbosa RL, Machado MF, Campeiro JD, Oliveira V, Sawa A, Camargo AC, Brandon NJ. Assessing the role of endooligopeptidase activity of Ndel1 (nuclear-distribution gene E homolog like-1) in neurite outgrowth. Mol Cell Neurosci. 2010;44:353–61. doi: 10.1016/j.mcn.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 107.Negrutskii B, Vlasenko D, El’skaya A. From global phosphoproteomics to individual proteins: the case of translation elongation factor eEF1A. Expert Rev Proteomics. 2012;9:71–83. doi: 10.1586/epr.11.71. [DOI] [PubMed] [Google Scholar]
- 108.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–9. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Drerup CM, Ahlgren SC, Morris JA. Expression profiles of ndel1a and ndel1b, two orthologs of the NudE-Like gene, in the zebrafish. Gene Expr Patterns. 2007;7:672–9. doi: 10.1016/j.modgep.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 111.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–8. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 112.Soukoulis V, Reddy S, Pooley RD, Feng Y, Walsh CA, Bader DM. Cytoplasmic LEK1 is a regulator of microtubule function through its interaction with the LIS1 pathway. Proc Natl Acad Sci USA. 2005;102:8549–54. doi: 10.1073/pnas.0502303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guo J, Yang Z, Song W, Chen Q, Wang F, Zhang Q, Zhu X. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol Biol Cell. 2006;17:680–9. doi: 10.1091/mbc.E05-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shan Y, Yu L, Li Y, Pan Y, Zhang Q, Wang F, Chen J, Zhu X. Nudel and FAK as antagonizing strength modulators of nascent adhesions through paxillin. PLoS Biol. 2009;7:e1000116. doi: 10.1371/journal.pbio.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wan Y, Yang Z, Guo J, Zhang Q, Zeng L, Song W, Xiao Y, Zhu X. Misfolded Gbeta is recruited to cytoplasmic dynein by Nudel for efficient clearance. Cell Res. 2012;22:1140–54. doi: 10.1038/cr.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Starr D, Saffery R, Li Z, Simpson A, Choo K, Yen T, Goldberg M. HZwint-1, a novel human kinetochore component that interacts with HZW10. J Cell Sci. 2000;113:1939–50. doi: 10.1242/jcs.113.11.1939. [DOI] [PubMed] [Google Scholar]
- 117.Taya S, Shinoda T, Tsuboi D, Asaki J, Nagai K, Hikita T, Kuroda S, Kuroda K, Shimizu M, Hirotsune S, Iwamatsu A, Kaibuchi K. DISC1 regulates the transport of the NUDEL/LIS1/14-3-3 ε complex through kinesin-1. J Neurosci. 2007;27:15–26. doi: 10.1523/JNEUROSCI.3826-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Park YU, Jeong J, Lee H, Mun JY, Kim JH, Lee JS, Nguyen MD, Han SS, Suh PG, Park SK. Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc Natl Acad Sci USA. 2010;107:17785–90. doi: 10.1073/pnas.1004361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Singh KK, Ge X, Mao Y, Drane L, Meletis K, Samuels BA, Tsai L-H. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hutchins JRA, Toyoda Y, Hegemann B, Poser I, Heriche J-K, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, Conrad C, Comartin D, Schleiffer A, Sarov M, Pozniakovsky A, Slabicki MM, Schloissnig S, Steinmacher I, Leuschner M, Ssykor A, Lawo S, Pelletier L, Stark H, Nasmyth K, Ellenberg J, Durbin R, Buchholz F, Mechtler K, Hyman AA, Peters J-M. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–9. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kang EC, Burdick KE, Kim JY, Duan X, Guo JU, Sailor KA, Jung DE, Ganesan S, Choi S, Pradhan D, Lu B, Avramopoulos D, Christian K, Malhotra AK, Song HJ, Ming GL. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron. 2011;72:559–71. doi: 10.1016/j.neuron.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]