Abstract
A total of 119 lactic acid bacteria (LAB) were isolated, by culture-dependant method, from rhizosphere samples of olive trees and desert truffles and evaluated for different biotechnological properties. Using the variability of the intergenic spacer 16S-23S and 16S rRNA gene sequences, the isolates were identified as the genera Lactococcus, Pediococcus, Lactobacillus, Weissella, and Enterococcus. All the strains showed proteolytic activity with variable rates 42% were EPS producers, while only 10% showed the ability to grow in 9% NaCl. In addition, a low rate of antibiotic resistance was detected among rhizospheric enterococci. Furthermore, a strong antibacterial activity against plant and/or pathogenic bacteria of Stenotrophomonas maltophilia, Pantoea agglomerans, Pseudomonas savastanoi, the food-borne Staphylococcus aureus, and Listeria monocytogenes was recorded. Antifungal activity evaluation showed that Botrytis cinerea was the most inhibited fungus followed by Penicillium expansum, Verticillium dahliae, and Aspergillus niger. Most of the active strains belonged to the genera Enterococcus and Weissella. This study led to suggest that environmental-derived LAB strains could be selected for technological application to control pathogenic bacteria and to protect food safety from postharvest deleterious microbiota.
1. Introduction
Given the world's growing demand for food, more attention is needed for food preservation, postharvest, and agricultural product preservation from different harmful factors such as the contamination caused by microbial spoilage and toxic metabolites produced by yeast, mold, and/or bacteria [1, 2], as well as the extensive use of synthetic chemicals and pesticides in food and agriculture. These factors may pose a health risk for human and animals and affect the ecological equilibrium of the environment [3].
Therefore, there is growing interest to establish alternative bioproducts to replace chemicals and toxic pesticides. For this purpose, using bacteria or natural compounds which exhibit the same inhibitory effect on phytopathogenic and spoilage microbes was not only shown to be efficient in storage life extension and nutritive and safety value retention, of food products but also the environment safeguarding [4, 5]. Such bacteria are known by “biological control agents” [6].
Lactic acid bacteria form an ecologically heterogeneous group of Gram-positive bacteria, nonspore forming, immobile, and catalase negative, that excretes lactic acid as major end product and generally recognized as safe (GRAS) organisms [7]. They are also selected as probiotic, which are able to promote health and prevent infections against enteropathogenic bacteria [8, 9]. LAB are usually harbor carbohydrate-rich environments and found in various food products such as milk, plant, meat, intestinal mucosa of human, and animals [8, 10] but especially proliferate in different fermented foods [11]. Owing to particular physiological and biochemical traits, such as exopolysaccharide production, organic acids, aromatic compounds, tolerance to low water activity, and antimicrobial production [5, 12, 13], LAB found different industrial applications, either by their biopreservatives or techno-functional properties [14]. In fact, many authors reported that some LAB strains are able to inhibit food-borne pathogens such as Staphylococcus aureus, Salmonella typhimurium, Escherichia, coli and Listeria monocytogenes [15, 16]. In addition, LAB are efficient to inhibit mycotoxicogenic fungi (Penicillium expansum, Botrytis cinerea, Aspergillus niger, Aspergillus flavus, and Fusarium graminarum) [17, 18] as well as phytopathogenic bacteria (such as Xanthomonas campestris and Erwinia carotovora) [17].
Data reporting LAB isolation from soils and plants remain scarce. However, environmental and wild LAB strains are theoretically good competitors for different growth factors and production of antagonistic compounds but often undervalued. Olive tree is one of the most important crops in Tunisia, from North to the South but also accompanied all the Mediterranean civilizations. It is recognized for its beneficial effects on human health, even by olive oil or by different derived products [19]. Moreover, truffles are ectomycorrhizal consumable tuber, which are typical of semiarid land and are known by their important economical income for local population and their good taste [20, 21]. The specificity of rhizospheric samples for bacterial isolation is their direct contact with both plant and soil, but especially because the associated bacteria have coevolved with plant pathogenic bacteria and fungi. This study aimed to isolate LAB from olive tree and desert-truffle rhizospheric soils and to evaluate their biotechnological properties.
2. Materials and Methods
2.1. Samples and Microbial Strains Origin
LAB strains were obtained from rhizospheric samples (49) which were collected from 8 sites located in the following regions in Tunisia: Jendouba, Ben Arous, Tunis, Kairouan, Gafsa, kebeli, Gabes, and Mednine. Samples were collected in sterilized bags, kept in cool box (<10°C) containing ice packs during the transport to laboratory, and processed within 7 days. Different other microbial species were used in antimicrobial testing. Pseudomonas savastanoi knW2, Pantoea agglomerans kn45, and Stenotrophomonas maltophilia KnT2 were previously isolated from olive knots [22]. Listeria monocytogenes L15 and Botrytis cinerea were obtained from the Laboratory of Microorganisms and Active Biomolecules (LMBA), Faculty of Sciences of Tunis. Penicillium expansum and Aspergillus niger from Laboratory of Microbial Ecology and Biotechnology, University of Paul Cézanne, France and Verticillium dahliae from the National Institute of Agronomic Research of Tunis (INRAT). Reference strains from the American Type Culture Collection (ATCC) were also used including Enterococcus faecium ATCC 19434, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 25923, and S. aureus ATCC 6538.
2.2. Lactic Acid Bacteria Isolation Procedure
The LAB from rhizospheres were isolated by the accumulation method as described by Chen et al. [23], with some modifications. Samples of 1 g were aseptically transferred into tubes of 15 mL containing 5 mL of MRS broth (Biolife) and incubated in anaerobic candle jars at 30°C for 3 days. After incubation, samples were serially diluted in 0.75% NaCl solution. Fractions of 0.1 mL of the dilutions ranging between 10−5 and 10−8 were plated in duplicate on the surface of MRS agar (Biolife) [24] supplemented with 0.0025% of bromocresol green (MP Biomedicals) and 0.01% cycloheximide (MP Biomedicals) to inhibit fungal growth. The plates were incubated in the same conditions. The different colonies of acid-producing bacteria, determined by a yellow zone in the media around each colony, were picked and purified on MRS agar. Gram-positive and catalase-negative isolates were selected and maintained in broth with 25% glycerol at −80°C for further identification. The isolates were also tested for gas production from D-glucose (Bio Basic) (using inverted Durham tubes in MRS broth), growth at different temperatures (10 and 45°C), different pH (4.0 and 9.6), and different concentration of NaCl (3, 6.5, 8, and 9%) in MRS broth.
2.3. DNA Extraction and PCR Amplification of 16S-23S rDNA Internal Transcribed Spacer and the 16S rDNA Gene
DNA was extracted by using a CTAB/NaCl method described by Wilson [25] and modified by using 1 mg/mL lysozyme (BIOMATIK) for cell wall digestion. DNA electrophoresis was performed on a 0.8% agarose gel and visualized under UV light according to the standard procedure of [26]. The PCR amplifications were performed using a thermal cycler (Thermal Cycler; Bio-Rad).
The bacterial collection was dereplicated by fingerprinting analysis of the rRNA 16S-23S intergenic transcribed spacer (ITS) region, using universal primers, s-d-bact-1494-a-20 and s-d-bact-0035-a-15 [27]. The ITS-PCR amplification consisted of 1X PCR reaction buffer, 1.5 mM MgCl2, 0.2 mM of dNTPs mixture, 0.5 μM of each primer, 1 U Taq polymerase (Fermentas), and 150 ng of total DNA, using the following program: 94°C for 3 min, followed by 35 cycles of 94°C for 45°C, 55°C for 1 min and 72°C for 2 min, and a final extension step at 72°C for 7 min. Strains exhibiting the same band patterns were grouped in the same ITS-haplotype. One or two representative strains from each group have been selected for subsequent identification using 16S rRNA genes sequencing. The 16S rRNA amplification was performed using the describe primers s-d-bact-0008-a-S-20 and s-d-bact-1495-a-A20 [27] and the thermal profile as mentioned previously. The ITS-PCR amplification and 16S products were migrated, respectively, on 2 and 1.5% agarose gels in 0.5 × Tris-borate-EDTA buffer (reagents from Fluka-Biochemika) and stained with ethidium bromide (Sigma-Aldrich).
2.4. 16S rDNA Gene Sequencing and Phylogenetic Analysis
The 16S rDNA PCR amplicons were purified with Exonuclease-I and Shrimp Alkaline Phosphatase (Exo-Sap, Fermentas, Life Sciences) following the manufacturer's standard protocol. Sequence analyses of the purified DNAs were performed using a Big Dye Terminator cycle sequencing kit V3.1 (Applied Biosystems) and an Applied Biosystems 3130XL Capillary DNA Sequencer machine. Sequence similarities were found by BLAST analysis [28] using the GenBank DNA databases (http://www.ncbi.nih.gov) and the Ribosomal Database Project (RDP). Phylogenetic analysis of the 16S rRNA gene sequences were conducted with Molecular Evolutionary Genetics Analysis (MEGA) software, version 5 [29]. Trees were constructed by using neighbor-joining method [30].
2.5. Nucleotide Sequence Accession Numbers
The sequences of the 16S rDNA gene of rhizospheric LAB isolates samples have been submitted to the GenBank databases under accession numbers KC568531 to KC568560.
2.6. Antibacterial Activity of LAB against Pathogen, Food-Borne, and Phytopathogenic Bacteria
The antibacterial activity test was performed using the agar-well-diffusion method described by Tagg and McGiven [31]. Five bacterial strains were used as indicators to evaluate the antibacterial activity of LAB, involving S. aureus ATCC6538, L. monocytogenes L15, St. maltophilia, Ps. savastanoi, and Pa. agglomerans. The cell-free supernatants (CFS) of LAB culture (48 h) in MRS broth were tested. All indicator strains were grown in BHI broth at 37°C. Trypticase soy agar plates were overlaid with 5 mL of soft agar (0.75%) containing 50 μL of freshly grown culture. The wells were made in agar and filled with 100 μL of the tested strain CFS. After incubation at 37°C for 18 h, the diameter of the inhibition zones was measured. The spectrum of inhibitory effect of LAB was than evaluated on indicator bacteria: E. faecium ATCC19434, E. faecalis ATCC 29212, S. aureus ATCC 6538, and S. aureus ATCC 25923. All antibacterial tests were performed in triplicate.
2.7. Antifungal Activity of LAB
The LAB isolates were tested against four phytopathogenic fungi of Aspergillus niger, Penicillium expansum, Botrytis cinerea, and Verticillium dahliae using the method described by Whipps [32] with some modifications. A dual culture of the tested pathogen fungi and the presumed antagonist LAB was established in MRS agar without sodium acetate (MRS-SA). A mycelium plug of 5 mm was taken from the peripheral edge of old cultures (5 days) on PDA plates of fungal pathogens and each plug was placed at the centre of three replicate MRS-SA plates. Bacteria were inoculated at 2 cm line from the edge of plates and allowed to grow at 30°C for 48 h. Untreated control plates were plated with pathogen plugs only. Particularly for V. dahliae, the fungus was placed on MRS-SA five days in advance, due to its relatively slower mycelium growth. All plates were incubated on adequate growth temperature of the fungi, and the percentage of growth inhibition was calculated by using the formula of Whipps [32]: [(R1 − R2)/R1]∗100, where R1 is the radial distance (mm) grown by phytopathogenic fungi in direction of the antagonist and R2 is the radial distance grown by phytopathogenic fungi.
2.8. Exopolysaccharide Production and Proteolytic Activity
The exopolysaccharide production (EPS) was evaluated by streaking fresh culture of LAB isolates on MRS agar supplemented with 2% (w/v) of sucrose (Sigma, Life science). After incubation at 30°C under anaerobic condition for 72 h, development of a mucoid colony on agar medium or long filaments (when the colony is extended with an inoculation loop) indicated the production of exopolysaccharides [33]. As well, LAB were assessed for proteolytic activity by agar-well-diffusion test in MRS containing 4% of skimmed milk (Scharlau). The diameter of the proteolysis zone was determined after incubation under anaerobic conditions at 30°C for 72 h and examined for clear zone around the wells.
2.9. Antibiotic Susceptibility
The antibiotic susceptibility was tested by disk diffusion method on BHIA as recommended by the standard criteria (CLSI, 2010). The antibiotics used (Bio-Rad Laboratoires, Hercules, CA, USA) for susceptibility of enterococci were ampicillin (AM; 10 μg), ciprofloxacin (CIP; 5 μg), chloroamphenicol (C; 30 μg), erythromycin (E; 15 μg), gentamycin (GM; 120 μg), streptomycin (S; 300 μg), tetracycline (TE; 30 μg), teicoplanin (TEI; 30 μg), and vancomycin (VAN; 30 μg). Antibiotic discs were placed on solid media and incubated at 37°C for 24 h. Based on the inhibition zone size, the results were interpreted as resistant (R), intermediate resistant (IR), or susceptible to the antimicrobial agents (S).
3. Results and Discussion
3.1. Lactic Acid Bacteria Isolation
LAB isolates were initially selected based on their ability to produce lactic acid by the presence of yellow halo surrounding the colonies on MRS-bromocresol green plates. Only Gram-positive strains exhibiting the absence of catalase and oxidase activity were kept on MRS agar for further identification. In total, 119 LAB strains were isolated from rhizospheric samples of desert truffles (4) and olive trees (49) from diverse geographic regions in Tunisia (Table 1). LAB are usually isolated from fermented products of animal and vegetable origin. However, low rate of “somnicells” of LAB [34] are naturally found in different environments which are close to these biota, such as floor of henhouse, rhizosphere of fruit trees, and around horse barn [23, 35]. Although LAB isolation from soil and water remains scarce [23, 36], their presence in rhizospheric samples seems to be more supported by the abundance of root exudates [37].
Table 1.
Origin and identification of rhizospheric LAB isolates.
| Geographical position | Sampling point | Source/number of soil samples | Number of isolates | Strains | Closest 16S rDNA sequence | Strains (access number) | % of sequence similarity |
|---|---|---|---|---|---|---|---|
| Northeast Tunisia | Ben Arous | R. of olive tree/03 | 4 | FS01 | Enterococcus hirae | ||
| FS02 | Lactococcus lactis | ||||||
| FS03 | Enterococcus faecalis | ||||||
| SSR of olive tree 01 | F S04 | Weissella confusa | FS04 ( KC568542) | 99 | |||
| Tunis | R. of olive tree/03 | 11 | FS07 | Lactococcus lactis | |||
| FS08 | Weissella halotolerans | FS08 (KC568554) | 99 | ||||
| FS11 | Enterococcus faecalis | FS11 (KC568559) | 99 | ||||
| FS13 | Leuconostoc mesenteroides | FS13 (KC568533) | 97 | ||||
| FS14 | Enterococcus durans | ||||||
| FS09, FS10, FS12, FS15 | Enterococcus faecium Enterococcus faecium | FS10 (KC568539) | 99 | ||||
| SSR of olive tree/01 | FS05, FS06 | ||||||
|
| |||||||
| Northwest Tunisia | Jendouba | R. of olive tree/21 | 50 | FS16, FS17, FS19, FS21, FS25, FS26, FS30, FS40, FS48, FS50, FS51, FS55, FS56, FS57, FS65 | Enterococcus faecium | FS25 (KC568541), FS19 (KC568549) FS26 (KC568553), FS65 (KC568552) |
99 |
| FS18 | Enterococcus faecalis | ||||||
| FS20 | Lactococcus lactis | ||||||
| FS24, FS37, FS38, FS41 | Pediococcus pentosaceus | FS24 (KC568551), FS37 (KC568550) | 99 | ||||
| FS46 | Pediococcus acidilactici | FS46 (KC568555) | 98 | ||||
| FS29, FS32, FS42, FS49 | Enterococuus durans | FS29 (KC568547) | 98 | ||||
| FS22, FS31 | Lactococcus garviae | FS31 (KC568548) | 99 | ||||
| FS33, FS34, FS62 | Lactobacillus sakei | FS33 (KC568535) | 100 | ||||
| FS35, FS59 | Lactobacillus plantarum | FS35 (KC568557) | 99 | ||||
| FS39, FS43, | Enterococcus hirae | FS39 (KC568531) | 99 | ||||
| FS45, FS60, FS64 | Weissella paramesenteroides | FS64 (KC568556) | 99 | ||||
| FS58 | Weissella halotolerans | FS58 (KC568532) | 99 | ||||
| FS36, FS44, FS52, FS53, FS54, FS61, FS63 | Weissella confusa | FS61 (KC568543), FS53 (KC568544), FS52 (KC568545), | 99 | ||||
| SSR of olive tree/04 | FS23, FS27, FS28, | Weissella confusa | FS 27 (KC568537) | 99 | |||
| FS47 | Enterococcus faecium | ||||||
|
| |||||||
| The middle Tunisia | Kairouan | R. of olive tree/03 | 9 | FS69 | Lactobacillus sakei | ||
| FS66 | Weissella confusa | FS66 (KC568540) | 99 | ||||
| FS73 | Pediococcus pentosaceus | ||||||
| FS67, FS68, FS70, FS71, FS72, FS74 | Enterococcus faecium | ||||||
|
| |||||||
| South Tunisia | Gafsa | R. of olive tree/02 | 3 | FS76 | Weissella confusa | ||
| FS75 | Lactococcus lactis | FS75 (KC568534) | 99 | ||||
| SSR/01 | FS77 | Weissella confusa | |||||
| Kebili | R. of olive tree/06 | 23 | FS81, FS86, FS100 | Enterococcus hirae | |||
| FS78, FS79, FS80, FS82, FS83, FS84, FS85, FS87, FS88 | Enterococcus faecium | FS88 (KC568538) | 99 | ||||
| FS89, FS90, FS91, FS92, FS93, FS94, FS95, FS98, FS99 | |||||||
| SSR/01 | FS96, FS97 | Enterococcus faecium | |||||
| Mednine | R. of olive tree/03 | 7 | FS101, FS102, FS103, FS105, FS106, FS107 | Enterococcus faecium | |||
| FS104 | Pediococcus pentosaceus | ||||||
| R. of truffle/04 | 12 | FS111 | Pediococcus pentosaeus | FS111 (KC568560) | 98 | ||
| (Terfezia boudieri/Pichoa) | FS119 | Lactobacillus plantarum | FS119 (KC568558) | 99 | |||
| FS110 | Enterococcus durans | ||||||
| FS112 | Enterococccus hirae | ||||||
| FS108, FS109, FS113, FS114, FS115, FS116, FS117, FS118 | Enterococcus faecium | FS118 ( KC568536) | 99 | ||||
R: Rhizosphere samples; SSR: soil surrounding rhizosphere; the underlined strains refer to LAB isolated from the SSR.
3.2. Ribotyping and Identification of Isolates by 16S rRNA Gene Sequence Analysis
Length polymorphism analysis of amplified 16S-23S internal transcribed spacer (ITS) was used to select representative strains of the different taxonomic units issued from the LAB collection. In fact, different studies have previously reported the usefulness of ITS dereplication for inter- and intradifferentiation at the genus/species level [38, 39] due to the high variability of these internal spacers. Based on this method, 16 different ITS-haplotypes designated from A to L were distinguished. ITS-PCR patterns showed 1 to 4 reproducible bands ranging from 275 to about 600 bp (Figure 1). The representative isolates of each ITS-type (30 isolates) were identified at species level by 16S rDNA gene sequence analysis and compared to the known sequences in GenBank. Phylogenetic relationship between LAB was constructed based on the 16S rDNA sequences from evolutionary distances by the neighbor-joining method (Figure 2). Phylogenetic analysis revealed the differentiation of 5 clusters (I–V) and 11 subclusters that include members of the genera Enterococcus, Lactobacillus, Pediococcus, Lactococcus, Weissella, and Leuconostoc. The cluster I formed by the strains of Enterococcus genus was divided into 2 groups. The first group included three species of E. faecium, corresponding to the ITS-types (A, B, and C), E. durans (ITS-type D), and E. hirae (ITS-type E). The second group was only represented by the strain (FS11) of E. faecalis being the most closely related species in 100% of bootstrap analyses. The cluster II grouped strains of the genera Lactobacillus and Pediococcus and presented four subclusters (3 to 6) of Lb. sakei, Lb. plantarum, Pc. acidilactici, and Pc. pentosaceus based on the ITS-types G, H, I, and J, respectively. The cluster III formed by the strains of the genus Lactococcus was divided into two subclusters (7 and 8) of L. lactis (ITS-type K) and L. garvieae (ITS-type L). Furthermore, strains of the genus Weissella were grouped in the cluster IV, including three subclusters (9 to 11) of W. halotolerans (ITS-types M), W. paramesenteroides (ITS-types N), and W. confusa (ITS-types O). The cluster V was represented by one strain of Ln. mesenteroides (ITS-type P). The used typing method showed that almost all the identified species were represented by one ITS-type, except for E. faecium, which showed an intraspecies heterogeneity with three major ITS-types (A, B, and C). In fact, E. faecium genome is extremely diverse [40] showing a high plasticity, due to the abundance of mobile genetic elements [41]. This result is in accordance with data published by Naïmi et al. [42], Park et al. [43], and Brtkova et al. [44], which reports the ITS region variability for E. faecium species. Together with W. confusa, this species was found to be the most isolated bacterium from rhizospheric samples (Table 1). Although enterococci are normal inhabitants of the human and animal gastrointestinal tract [44, 45], they are widely distributed in nature due to their high adaptation to various environmental conditions such as food, plants, water, and soil [46]. Moreover, the isolation of bacteria belonging to the genus Weissella was already reported either from soil [19] or plants [17]. With regard to others studies on LAB recovery from soil [19, 35, 47, 48] a higher number of species diversity is recorded in olive tree and truffle rhizospheric samples such as Lb. sakei, Pc. acidilactici, and W. halotolerans. These species are naturally found on several raw fermented food products of plant and animal origin [49–51]; moreover, Pc. acidilactici is emerging as a potential probiotic in animal and human [52].
Figure 1.
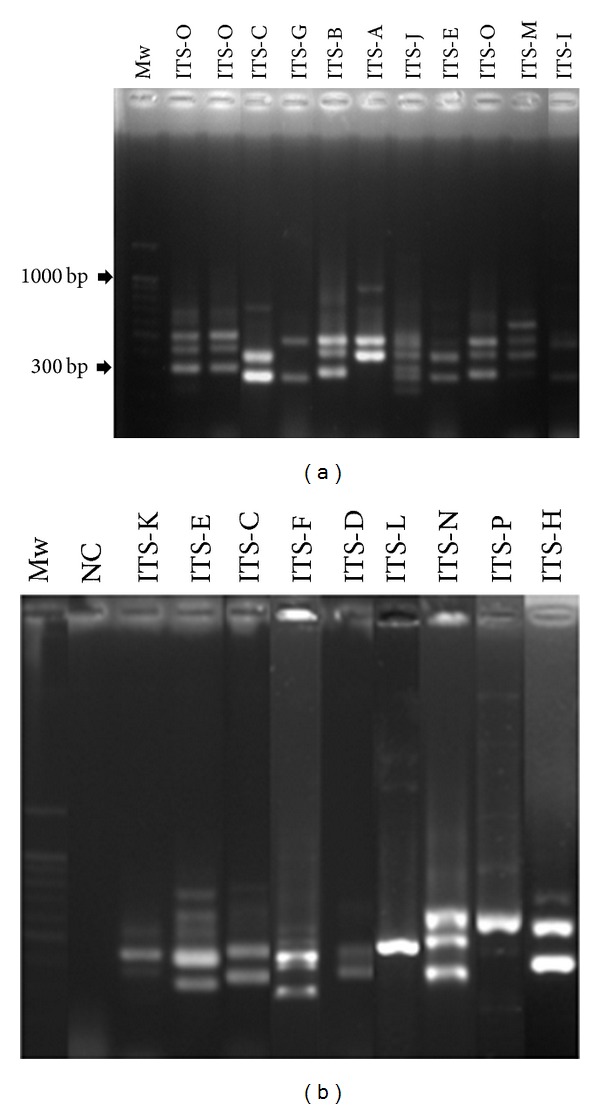
Different ITS-haplotypes (A–P) of representative rhizospheric lactic acid bacteria. Mw, Molecular weight (100 bp); NC, negative control.
Figure 2.
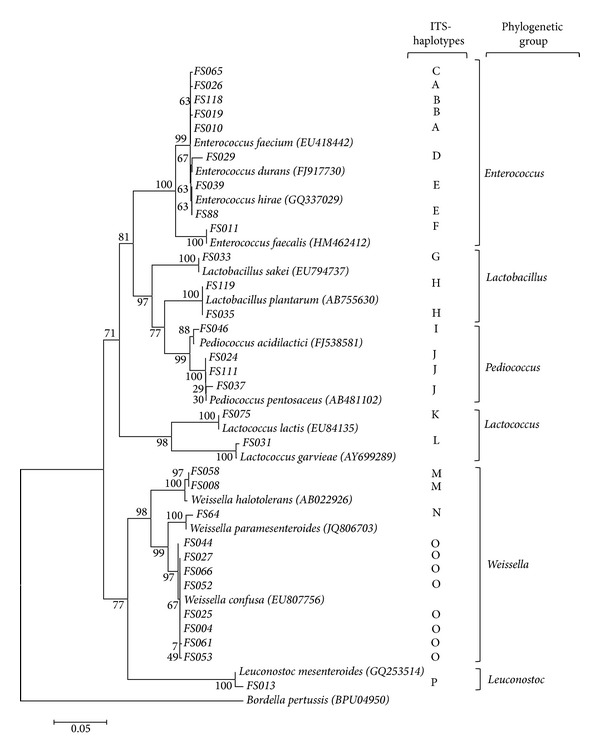
Phylogenetic tree showing the relative position of lactic acid bacteria isolates based on 16S rDNA partial sequences, using the neighbor-joining method. Bordetella pertussis was used as an out group. Bootstrap values for a total of 1000 replicates are shown at the nodes of the tree, using MEGA-5. The scale bar corresponds to 0.05 units of the number of base substitutions per site.
3.3. Physiological and Technological Properties of LAB
The physiological and biochemical characteristics including salt tolerance, growth at different temperatures, and gas production from glucose of all the strains are presented in Table 2. The majority of isolated strains were coccoid and coccoid-rods and only 5.9% showed rod shape. The majority of isolates were homofermentative, and only 13 (10.9%) were heterofermentative. From the total isolates (n = 79) 66.4% and (n = 10) 8.4% of bacterial isolates grew well in low activity water, 8 and 9% NaCl, respectively. The LAB strains with high tolerance to 9% NaCl belonged to W. halotolerans (FS58), W. confusa (FS66, FS44, FS53, FS54, and FS63), L. lactis (LFS20), Lb. sakei (FS62), and E. faecium (FS77 and FS103). All LAB isolates grew well in pH 4.0 and 10°C. A total of 28 (23%) and 14 (11.5%) of isolates were not able to grow in pH 9.6 and 45°C, respectively. Besides, LAB were screened for proteolytic activity and EPS production on MRS medium containing sucrose and skim milk, respectively. Results showed that all LAB isolates exhibited proteolytic activities in the cell-free supernatants as revealed by a clear halo surrounding the wells. However, the proteolytic activity varied among the strains according to the halo diameters. In fact, the more proteolytic strains (58.8%) exhibited a diameter greater than 15 mm. The most proteolytic activity (19 mm of diameter) was recorded for the strain Pc. acidilactici FS46. The exopolysaccharide production was detected in 42.8% of the isolates (Table 2). The good EPS-LAB producers belonged mainly to the species W. confusa (12 strains), W. paramesenteroides (FS60 and FS45), and Ln. mesenteroides FS13. The recorded physiochemical properties of the isolated rhizospheric LAB, for instance proteolytic activity, tolerance to high NaCl concentration, and the EPS production could explain their survival in such oligotrophic environments. In particular, EPSs are typically correlated with bacterial resistance and protection against different stress conditions such as desiccation, salt stress, and UV radiations [53, 54]. But it also generally implicated in their adherence to biological surface and sodium toxicity reduction [55].
Table 2.
Phenotypic characteristics of representative Gram-positive rhizospheric-LAB isolates.
| E. faecium | E. durans | E. hirae | E. faecalis | Lb. sakei | Lb. plantarum | Pc. acidilactici | Pc. pentosaceus | Lc. lactis | Lc. garvieae | W. halotolerans | W. paramesenteroides | W. confuse | Ln.mesenteroides | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS types | A, B, C | D | E | F | G | H | I | J | K | L | M | N | O | P |
| Number of strains | 64 | 06 | 07 | 03 | 04 | 03 | 01 | 07 | 03 | 03 | 02 | 03 | 12 | 01 |
| Shape | cocci | cocci | cocci | cocci | rods | rods | cocci | cocci | cocci | cocci | coccobacilli | coccobacilli | coccobacilli | cocci |
| Fermentation type | Homo | Homo | Homo | Homo | Homo | Homo | Homo | Homo | Homo | Homo | Hetero | Hetero | Hetero | Hetero |
| Catalase | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Growth at pH | ||||||||||||||
| 4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 9.6 | + | + | + | + | + | + | − | − | − | − | + | + | + | + |
| Growth in NaCl | ||||||||||||||
| 3% | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 6.5% | + | +(03) | +(04) | +(02) | +(03) | + | + | +(06) | +(03) | + | + | + | + | + |
| 8% | +(42)* | +(03) | +(01) | +(01) | +(03) | + | + | +(04) | +(02) | +(01) | + | + | + | + |
| 9% | +(02) | − | − | − | +(01) | − | − | − | +(01) | − | +(01) | − | +(05) | − |
| Growth at temperature | ||||||||||||||
| 10°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 45°C | + | + | + | + | − | + | + | + | − | − | − | − | − | − |
| EPS production | +(28) | − | − | − | − | + | + | − | +(02) | +(02) | − | +(02) | + | + |
| Proteolytic activity | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
(x): number of strains; +: positive; −: negative; Homo: homofermentative; Hetero: heterofermentative.
3.4. Antibiotic Susceptibility Testing
The antibiotic susceptibility of the isolated was mainly checked for enterococcal species, since they are the predominant isolates in the collection (Table 3) and could present a risk for antibiotic resistance gene dissemination. Rhizospheric enterococci showed low percentage of resistance to chloramphenicol (3.75%), erythromycin (3.75%), streptomycin (7.5%), and tetracycline (8.75%). Nevertheless, all the strains were susceptible to teicoplanin, ampicillin, and gentamicin. Furthermore, some strains (3.7%) exhibited intermediate resistance to vancomycin and a high frequency of resistance to ciprofloxacin (36.2%). In summary, the rhizospheric enterococci showed a low frequency of resistance to the Gram-positive target antibiotics compared to food, clinical, and animal isolates [56, 57]. This result indicates the safety of these bacteria for a potential technological application.
Table 3.
Antimicrobial susceptibility of the enterococci isolated from the rhizosphere soils.
| Antibiotics | E. faecium | E. faecalis | E. durans | E. hirae | % resistance | |
|---|---|---|---|---|---|---|
| (n = 64) | (n = 3) | (n = 6) | (n = 7) | |||
| Penicillins | AM | 0 | 0 | 0 | 0 | 0 |
| Aminoglycosides | GM | 0 | 0 | 0 | 0 | 0 |
| TE | 6 | 0 | 1 | 0 | 8,75 | |
| S | 6 | 0 | 0 | 0 | 7,5 | |
| Chloramphenicols | CH | 3 | 0 | 0 | 0 | 3,75 |
| Macrolides | E | 2 | 1 | 0 | 0 | 3,75 |
| Glycopeptides | VA | 3 | 0 | 0 | 0 | 3,75 |
| TEI | 0 | 0 | 0 | 0 | 0 | |
| Fluoroquinolones | CIP | 28 | 1 | 0 | 0 | 36,25 |
AM: ampicillin, GM: gentamicine, TE: teteracyclin, S: streptomycin, E: erythromycin, C: chloramphenicol, VA: vancomycin, TEI: teicoplanin, and CIP: ciprofloxacin. n: total number of strains; numbers indicated resistant strains within species.
3.5. In Vitro Screening of the Antagonistic Activity of LAB against Human, Plant, and Food-Borne Pathogenic Bacteria
LAB isolates were screened for antibacterial activity against human and plant pathogens, including St. maltophilia, Pa. agglomerans, and Ps. savastanoi and food-borne bacteria of S. aureus and L. monocytogenes (Figure 3(a)). According to their inhibitory effects on pathogens, LAB were differentiated into three classes: strong inhibitor (with growth inhibition diameter (d) ≥ 19 mm), medium (14 ≤ d < 19 mm), and with no significant inhibitory effect for a diameter less than 14 mm (Figure 4(a)). The results showed that 64 strains (53.8%) have significant inhibition against St. maltophilia, among them 12 strains (10%) with strong inhibitory activity. This activity was recorded for the species of Lb. plantarum, Lb. sakei, Lc. garvieae, Ln. mesenteroides, and Pc. pentosaceus and mostly for the genera Enterococcus and Weissella. Five isolates (4%) including three E. faecium (FS70, FS01, and FS03) and two Pc. pentosaceus (FS73 and FS24) showed strong inhibitory activity against Pa. agglomerans. Eleven isolates belonging to species W. confusa, Lc. Lactis, Lb. plantarum, Ln. mesenteroides, E. durans, and E. faecium showed also strong inhibitory activity against Ps. savastanoi (diameter varied between 19 to 28 mm). The recorded high level of inhibition highlights the biotechnological potential of rhizospheric-LAB to control phytopathogens, particularly for Pa. agglomerans and St. maltophilia, which are also increasingly identified as important cause of nosocomial human infections [58, 59] and among the emergent multidrug resistant Gram-negative bacteria [59]. With regard to the food-borne pathogen, efficient inhibition was recorded for W. confusa FS054 strain (28 mm) against S. aureus, for E. faecium FS106 against L. monocytogenes (20 mm) and for the strain W. confusa FS036 against both S. aureus (20 mm), and L. monocytogenes (24 mm). It is of interest to note that the genera of Enterococcus and Weissella may become potential biopreservation agents of food-poisoning and plant-borne species.
Figure 3.
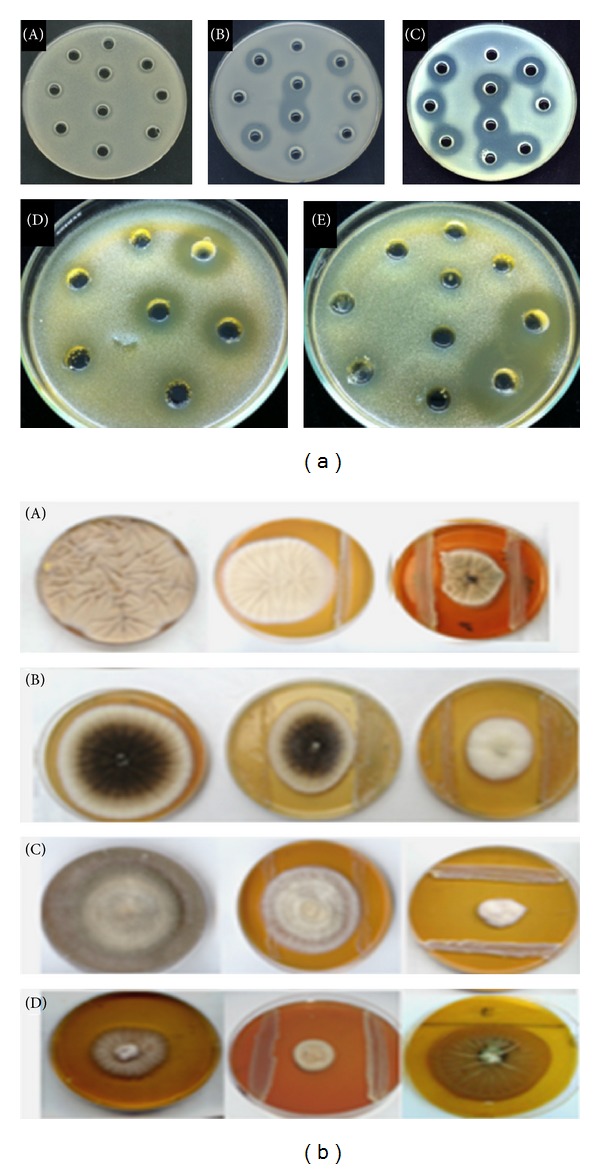
Antimicrobial activity of some rhizospheric LAB against pathogenic bacteria (a) Pa. agglomerans (A), St. maltophilia (B), Ps. savastanoi (C), L. monocytogenes (D), and S. aureus (E) by-agar well-diffusion method [31] and phytopathogen fungi (b) P. expansium (A), A. niger (B), B. cinerea (C), and V. dahliae (D) by dual culture [32].
Figure 4.
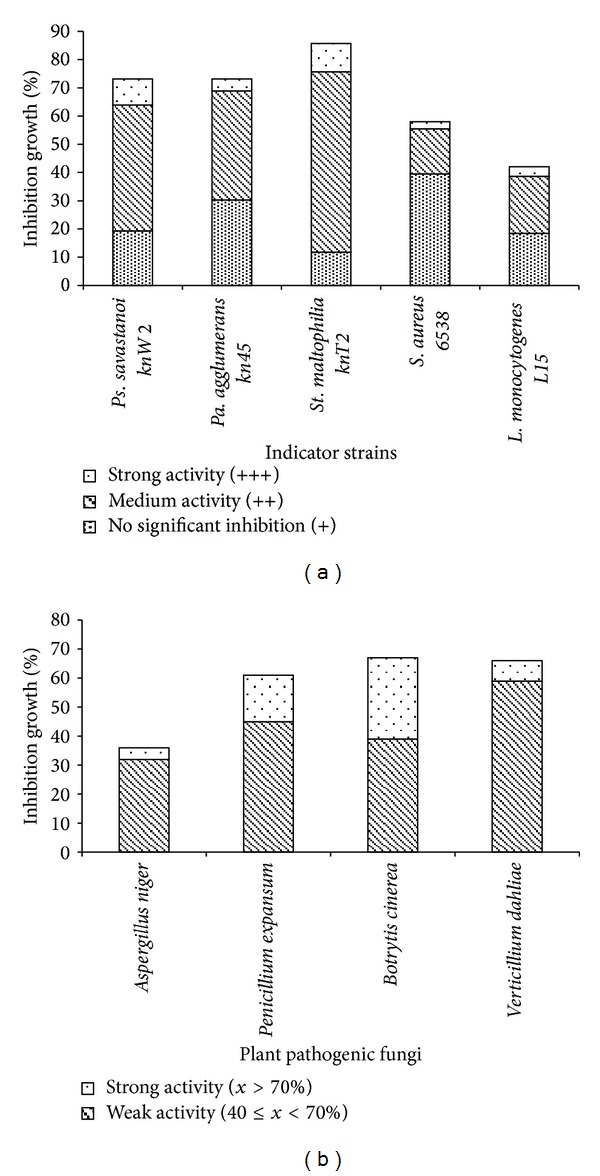
Histograms showing percentage of LAB having in vitro inhibitory effect on pathogenic and spoilage bacterial species (a) and plant pathogenic fungi (b). The experiments were repeated at least three times.
This antibacterial activity exhibited by the majority of strains especially toward Gram-negative bacteria may be due to the organic acid effect or to other compounds active in acidic conditions. For this purpose, the inhibitory effect was checked after supernatant neutralization. By this way, only three strains of E. faecium FS071, Ln. mesenteroides FS013, and W. halotolerans FS008 retained the inhibition ability against the tested pathogens (Table 4), leading to suggest the presence of bacteriocin-like substances. This result was also supported by the broad spectrum known for the majority of the identified enterocins [60, 61]. Further studies should be conducted to elucidate the nature of the antibacterial metabolites produced by selected LAB, especially by W. halotolerans FS008 strain. Moreover, different studies proposed that enterococci may have a prospectively useful role in some dairy products, due to their proteolytic and lipolytic activities, and may then contribute to the development of the organoleptic properties of fermented foods, and also due to the production of enterocins with anti-Listeria activity [62, 63].
Table 4.
Antibacterial activity spectrum of neutralized cell-free supernatant of three LAB rhizospheric isolates.
| Strains | L. monocytogenes L15 | S. aureus ATCC 6538 | S. aureus ATCC 25923 | E. faecium ATCC 19129 | E. faecalis ATCC 29212 | |
|---|---|---|---|---|---|---|
| Leuconostoc mesenteroides | FS013 | 21 ± 1.00 | 14 ± 1.00 | 12 ± 1.00 | 13 ± 1.00 | 10 ± 0.00 |
| Weissella halotolerans | FS008 | 17 ± 1.00 | 15 ± 1.00 | 20 ± 1.00 | 13.5 ± 1.00 | 11 ± 1.00 |
| Enterococcus faecium | FS071 | 16 ± 1.00 | 15 ± 1.05 | 19 ± 1.73 | 14.5 ± 0.80 | 15.5 ± 1.32 |
Numbers indicated the diameter of the inhibition zone in mm; each value represents the mean value standard deviation (SD) from three trials; values in the same column differ significantly (P < 0.05).
3.6. Antifungal Activity of LAB
LAB isolates were screened for antifungal activity against soil-borne fungi of B. cinerea and V. dahliae and postharvest contaminants of A. niger and P. expansum on MRS-SA agar medium (Figure 3(b)). According to their degree of mycelium growth reduction, active LAB were classified into two main groups: low (reduction of mycelium growth between 40 and 70%) and high antifungal activity (>70% of inhibition) (Figure 4(b)). The results showed that the maximum growth inhibition rate (28% of the strains) was registered for Botrytis cinerea (Figure 4(b)). The group of LAB strains with strong inhibition activity (75.3 to 92.6% inhibition) belonged to the species of W. paramesenteroides, W. confusa, E. durans, E. faecium and E. hirae. Besides, the most inhibitor strains toward the fungus A. niger were E. durans FS29 and four E. faecium, (FS50, FS06, FS48, and FS87) exhibiting an inhibition rate between 76.7 and 90%. Furthermore, we noted that 16% of the strains belonging to the species E. faecium, E. durans, W. halotolerans, and Lb. plantarum showed a strong inhibition rate (75.3 to 87.8%) toward P. expansum.
It is worth mentioning that strains of Enterococcus genus confirmed their antimicrobial efficacy by strong inhibition of most of the tested postharvest fungi (P. expansum, B. cinerea, and A. niger) and highlight the potential use of rhizospheric-LAB as biocontrol agents to prevent postharvest deterioration caused by these fungi. In fact, most of these fungi produce allergenic spores and mycotoxins which are responsible of the spoilage and poisoning of foods leading to serious potential health hazards [64]. It is also the case of Lb. plantarum FS119 which was isolated from desert truffle rhizosphere, and that could be considered as a potential candidate inhibitor of P. expansum, the agent of blue mold in apples. In addition strains of the genus Weissella have showed an efficient inhibition toward either pathogenic bacteria, or the different tested fungi. This result is in accordance with Valerio et al. [65] and Lee et al. [66] reporting the emergence/selection of these bacteria as biocontrol agent and potential probiotic. Regarding the vascular wilt fungi V. dahlia, Enterococcal strains were also shown to be the most efficient inhibitor strains. The highest activity was observed for E. faecium FS82 with 75% of mycelium reduction (Table 5). This result constitute a first report on the strong inhibition of this soil-born-fungus, which is responsible of Verticillium wilt, a serious worldwide disease that affects many crops including fruits, vegetables, and oilseed rape and leads to dramatically yield losses [67]. Biological compounds investigation to control this pathogen is of great significance [68], as it persists in the soil and resists different chemical treatments. The present study showed that selected environmental LAB could offer an excellent source for active metabolite to control different pathogenic bacteria and fungi. As it was reported by many authors [5, 13, 69], different substances, such as organic acids, hydrogen peroxide, cyclic dipeptides, and phenolic and proteinaceus compounds could be responsible for the detected antifungal activity. Indeed, identification of the issued rhizospheric-LAB metabolites is needed for a more target application.
Table 5.
Mycelium growth inhibition of four pathogenic fungi by selected potent antifungal rizospheric-LAB isolates using confrontation assay.
| Strains | Species | A. niger | P. expansum | B. cinerea | V. dahlia |
|---|---|---|---|---|---|
| FS29 | E. durans | ++ (76.7)* | ++ 80.2* | ++ (85.1)* | ++ (70.3)* |
| FS50 | E. faecium | ++ (79.1)* | + | ++ (82.7)* | + |
| FS06 | E. faecium | ++ (79)* | ++ (75.3)* | + | + |
| FS87 | E. faecium | ++ (79.1)* | + | ++ (80.2)* | + |
| FS48 | E. faecium | ++ (90)* | ++ (72.8)* | +++ (82.7)* | + |
| FS82 | E. faecium | + | + | +++ (85.2)* | +++ (75)* |
| FS68 | E. faecium | + | + | ++ (75.3)* | + |
| FS05 | E. faecium | + | ++ (80.2)* | ++ (72.8)* | + |
| FS14 | E. durans | + | ++ (77.7)* | ++ (82.7)* | − |
| FS21 | E. faecium | + | ++ (77.7)* | + | ++ (70.9)* |
| FS101 | E. faecium | + | ++ (75.3)* | ++ (79)* | + |
| FS32 | E. durans | + | ++ (72.8)* | ++ (82.7)* | + |
| FS107 | E. faecium | + | − | ++ (80.2)* | + |
| FS45 | W. paramesenteroides | − | + | ++ (81.4)* | + |
| FS61 | W. confusa. | − | + | ++ (85)* | − |
| FS19 | E. faecium | + | + | ++ (77.7)* | + |
| FS94 | E. faecium | + | ++ (87.6)* | ++ (82.7)* | + |
| FS119 | Lb. plantarum | − | ++ (83.8)* | ++ (70)* | + |
| FS49 | E. durans | − | ++ (75.3)* | + | + |
| FS58 | W. halotolerans | + | ++ (80.2)* | + | − |
| FS74 | E. faecium | − | ++ (72.8)* | ++ (85.2)* | ++ (74.5)* |
| FS51 | E. faecium | + | ++ (72.8)* | ++ (82.7)* | ++ (70.9)* |
| FS102 | E. faecium | − | + | ++ (81.2)* | + |
| FS16 | E. faecium | − | − | ++ (92.6)* | − |
| FS99 | E. faecium | − | + | ++ (91.3)* | + |
| FS42 | E. durans | + | + | ++ (85.1)* | + |
| FS12 | E. faecium | + | + | ++ (75.3)* | + |
| FS65 | E. faecium | + | + | ++ (77.7)* | + |
| FS15 | E. faecium | + | + | + | ++ (70.3)* |
| FS106 | E. faecium | + | ++ (70.3)* | + | + |
| FS53 | W. confusa | − | ++ (71)* | + | + |
| FS77 | E. faecium | − | ++ (78.4)* | + | + |
A. niger: Aspergillus niger, P. expansum: Penicillium expansum, B. cinerea: Botrytis cinerea, V. dahlia: Vercticillium dahliae. (+): weak antifungal activity having an inhibition rate between 40 and 70%; (++): strong activity with an inhibition rate ≥ 70%; the strains characterized with a broad range against different fungi appear in bold. Data were obtained at least three replicates. *Means within column show statistically significant difference (P < 0.05) with a control (nonexposed to the bacteria).
4. Conclusion
In this study, we reported for the first time the isolation and characterization of LAB from rhizosphere samples of olive trees and desert truffles. The results showed a high rate of antimicrobial activity among the isolates, indicating that rhizosphere may be a common source for the selection of LAB with important technological potential, which are useful for the biocontrol of food-, plant-, and soil-borne pathogenic bacteria and fungi. Further investigations to elucidate the nature of inhibiting compounds should be considered.
Acknowledgments
The authors thank financial support of the European Union in the ambit of Project 20 BIODESERT (EU FP7-CSA-SA REGPOT-2008-2, Grant agreement no. 245746) and the 21 Tunisian Ministry of Higher Education and Scientific research in the ambit of the laboratory 22 projects LR MBA206 and LR11ES31.
References
- 1.Guynes GJ, Bennett EO. Bacterial deterioration of emulsion oils. I. Relationship between aerobes and sulfate-reducing bacteria in deterioration. Applied Microbiology. 1959;7(2):117–121. doi: 10.1128/am.7.2.117-121.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pitt JI, Hocking AD. Fungi and Food Spoilage. 2nd edition. Gaithersburg, Md, USA: Aspen; 1999. (A Chapman and Hall Food Science Book). [Google Scholar]
- 3.Huang JK, Qiao FB, Zhang LX, Rozelle S. EEPSEA Working Paper. Singapore: EEPSEA; 2000. Farm pesticide, rice production, and human health. [Google Scholar]
- 4.Huang J, Hu R, Pray C, Qiao F, Rozelle S. Biotechnology as an alternative to chemical pesticides: a case study of Bt cotton in China. Agricultural Economics. 2003;29(1):55–67. [Google Scholar]
- 5.Stiles ME. Biopreservation by lactic acid bacteria. Antonie van Leeuwenhoek. 1996;70(4):331–345. doi: 10.1007/BF00395940. [DOI] [PubMed] [Google Scholar]
- 6.Pastor N, Carlier E, Andrés J, Rosas SB, Rovera M. Characterization of rhizosphere bacteria for control of phytopathogenic fungi of tomato. Journal of Environmental Management. 2012;95:S332–S337. doi: 10.1016/j.jenvman.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 7.Konings WN, Kok J, Kuipers OP, Poolman B. Lactic acid bacteria: the bugs of the new millennium. Current Opinion in Microbiology. 2000;3(3):276–282. doi: 10.1016/s1369-5274(00)00089-8. [DOI] [PubMed] [Google Scholar]
- 8.Fernández MF, Boris S, Barbés C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. Journal of Applied Microbiology. 2003;94(3):449–455. doi: 10.1046/j.1365-2672.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- 9.Skjermo J, Vadstein O. Techniques for microbial control in the intensive rearing of marine larvae. Aquaculture. 1999;177(1–4):333–343. [Google Scholar]
- 10.Amin M, Jorfi M, Khosravi AD, Samarbafzadeh AR, Sheikh AF. Isolation and identification of Lactobacillus casei and Lactobacillus plantarum from plants by PCR and detection of their antibacterial activity. Journal of Biological Sciences. 2009;9(8):810–814. [Google Scholar]
- 11.Soomro AH, Masud T, Anwaar K. Role of lactic acid bacteria in food preservation and human health—a review. Pakistan Journal of Nutrition. 2002;1(1):20–24. [Google Scholar]
- 12.Giraffa G. Microbial polysaccharides produced by lactic acid bacteria in the dairy industry. Industrie Alimentari. 1994;33(324):295–298. [Google Scholar]
- 13.Mataragas M, Drosinos EH, Metaxopoulos J. Antagonistic activity of lactic acid bacteria against listeria monocytogenes in sliced cooked cured pork shoulder stored under vacuum or modified atmosphere at 4 ± 2°C. Food Microbiology. 2003;20(2):259–265. [Google Scholar]
- 14.Bizzarro R, Tarelli GT, Giraffa G, Neviani E. Phenotypic and genotypic characterization of lactic acid bacteria isolated from Pecorino Toscano cheese. International Journal of Food Science. 2000;12(3):303–316. [Google Scholar]
- 15.Darsanaki RK, Rokhi ML, Aliabadi MA, Issazadeh K. Antimicrobial activities of Lactobacillus strains isolated from fresh vegetables. Middle-East Journal of Scientific Research. 2012;11(9):1216–1219. [Google Scholar]
- 16.Jamuna M, Jeevaratnam K. Isolation and partial characterization of bacteriocins from Pediococcus species. Applied Microbiology and Biotechnology. 2004;65(4):433–439. doi: 10.1007/s00253-004-1576-8. [DOI] [PubMed] [Google Scholar]
- 17.Trias R, Bañeras L, Montesinos E, Badosa E. Lactic acid bacteria from fresh fruit and vegetables as biocontrol agents of phytopathogenic bacteria and fungi. International Microbiology. 2008;11(4):231–236. doi: 10.2436/20.1501.01.66. [DOI] [PubMed] [Google Scholar]
- 18.Lavermicocca P, Valerio F, Evidente A, Lazzaroni S, Corsetti A, Gobbetti M. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Applied and Environmental Microbiology. 2000;66(9):4084–4090. doi: 10.1128/aem.66.9.4084-4090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laitman L. Le marché et la production de l'huile d'olive en Tunisie. Annales de Géographie. 1953;62(332):271–286. [Google Scholar]
- 20.Morte A, Zamora M, Gutiérrez A, Honrubia M. Desert truffle cultivation in semiarid Mediterranean areas. In: Azcón-Aguilar C, et al., editors. Mycorrhizas Functional Processes and Ecological Impact. chapter 15. Berlin: Springer; 2009. pp. 221–233. [Google Scholar]
- 21.Slama A, Fortas Z, Neffati M, Khabar L, Boudabous A. Etude taxinomique de quelques Ascomycota hypogés (Terfeziaceae) de la Tunisie méridionale. Bulletin de la Société Mycologique de France. 2006;122(2-3):187–195. [Google Scholar]
- 22.Ouzari H, Khsairi A, Raddadi N, et al. Diversity of auxin-producing bacteria associated to Pseudomonas savastanoi-induced olive knots. Journal of Basic Microbiology. 2008;48(5):370–377. doi: 10.1002/jobm.200800036. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y-S, Yanagida F, Shinohara I. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Letters in Applied Microbiology. 2005;40(3):195–200. doi: 10.1111/j.1472-765X.2005.01653.x. [DOI] [PubMed] [Google Scholar]
- 24.de Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. Journal of Applied Microbiology. 1960;23(1):130–135. [Google Scholar]
- 25.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, et al., editors. Current Protocols in Molecular Biology. 1987. pp. 2. 4. 1–2. 4. 5. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Daffonchio D, Borin S, Frova G, Manachini PL, Sorlini C. PCR fingerprinting of whole genomes: the spacers between the 16s and 23S rRNA genes and of intergenic tRNA gene regions reveal a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis . International Journal of Systematic Bacteriology. 1998;48(1):107–116. doi: 10.1099/00207713-48-1-107. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Tagg JR, McGiven AR. Assay system for bacteriocins. Applied Microbiology. 1971;21(5):p. 943. doi: 10.1128/am.21.5.943-943.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whipps JM. Effect of media on growth and interactions between a range of soil-borne glasshouse pathogens and antagonistic fungi. New Phytology. 1987;107(1):127–142. [Google Scholar]
- 33.Ruas-Madiedo P, de los Reyes-Gavilán CG. Invited review: methods for the screening, isolation, and characterization of exopolysaccharides produced by lactic acid bacteria. Journal of Dairy Science. 2005;88(3):843–856. doi: 10.3168/jds.S0022-0302(05)72750-8. [DOI] [PubMed] [Google Scholar]
- 34.Behrendt U, Müller T, Seyfarth W. The influence of extensification in grassland management on the populations of micro-organisms in the phyllosphere of grasses. Microbiological Research. 1997;152(1):75–85. [Google Scholar]
- 35.Yanagida F, Chen Y, Shinohara T. Isolation and characterization of lactic acid bacteria from soils in vineyards. The Journal of General and Applied Microbiology. 2005;51(5):313–318. doi: 10.2323/jgam.51.313. [DOI] [PubMed] [Google Scholar]
- 36.Yanagida F, Chen Y, Yasaki M. Isolation and characterization of lactic acid bacteria from lakes. Journal of Basic Microbiology. 2007;47(2):184–190. doi: 10.1002/jobm.200610237. [DOI] [PubMed] [Google Scholar]
- 37.Kiely PD, Haynes JM, Higgins CH, et al. Exploiting new systems-based strategies to elucidate plant-bacterial interactions in the rhizosphere. Microbial Ecology. 2006;51(3):257–266. doi: 10.1007/s00248-006-9019-y. [DOI] [PubMed] [Google Scholar]
- 38.Gürtler V, Stanisich VA. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142(1):3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 39.Daffonchio D, Cherif A, Borin S. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the ‘Bacillus cereus group’. Applied and Environmental Microbiology. 2000;66(12):5460–5468. doi: 10.1128/aem.66.12.5460-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leavis HL, Willems RJL, van Wamel WJB, Schuren FH, Caspers MPM, Bonten MJM. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium . PLoS Pathogens. 2007;3(10):p. 37. doi: 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Schaik W, Top J, Riley DR, et al. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics. 2010;11(1, article R239):18 pages. doi: 10.1186/1471-2164-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naïmi A, Beck G, Branlant C. Primary and secondary structures of rRNA spacer regions in enterococci. Microbiology. 1997;143(3):823–834. doi: 10.1099/00221287-143-3-823. [DOI] [PubMed] [Google Scholar]
- 43.Park Y, Oh E, Kim BK, Kim SM, Shim SI. Phenotypic characteristics of Enterococcus faecium variants confirmed by intergenic ribosomal polymerase chain reaction and E. faecium polymerase chain reaction. Diagnostic Microbiology and Infectious Disease. 1999;34(4):269–273. doi: 10.1016/s0732-8893(99)00041-3. [DOI] [PubMed] [Google Scholar]
- 44.Brtkova A, Filipova M, Drahovska H, Bujdakova H. Characterization of enterococci of animal and environmental origin using phenotypic methods and comparison with PCR based methods. Veterinarni Medicina. 2010;55(3):97–105. [Google Scholar]
- 45.Giraffa G. Enterococci from foods. FEMS Microbiology Reviews. 2002;26(2):163–171. doi: 10.1111/j.1574-6976.2002.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 46.Mundt JO. Occurrence of enterococci on plants in a wild environment. Applied Microbiology. 1963;11:141–144. doi: 10.1128/am.11.2.141-144.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamudio-Maya M, Narváez-Zapata J, Rojas-Herrera R. Isolation and identification of lactic acid bacteria from sediments of a coastal marsh using a differential selective medium. Letters in Applied Microbiology. 2008;46(3):402–407. doi: 10.1111/j.1472-765X.2008.02329.x. [DOI] [PubMed] [Google Scholar]
- 48.Lutz MP, Michel V, Martinez C, Camps C. Lactic acid bacteria as biocontrol agents of soil-borne pathogens. Biological Control of Fungal and Bacterial Plant Pathogens. 2012;78:285–288. [Google Scholar]
- 49.Hammes WP, Bantleon A, Min S. Lactic acid bacteria in meat fermentation. FEMS Microbiology Reviews. 1990;87(1-2):165–173. [Google Scholar]
- 50.Kandler O, Schillinger U, Weiss N. Lactobacillus halotolerans sp. nov., nom. rev. and Lactobacillus minor sp. nov., nom. rev. Systematic and Applied Microbiology. 1983;4(2):280–285. doi: 10.1016/S0723-2020(83)80056-3. [DOI] [PubMed] [Google Scholar]
- 51.Leroy F, de Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends in Food Science and Technology. 2004;15(2):67–78. [Google Scholar]
- 52.Guerra NP, Agrasar AT, Macías CL, Bernárdez PF, Castro LP. Dynamic mathematical models to describe the growth and nisin production by Lactococcus lactis subsp. lactis CECT 539 in both batch and re-alkalized fed-batch cultures. Journal of Food Engineering. 2007;82(2):103–113. [Google Scholar]
- 53.Chen L, Wang G, Hong S, Liu A, Li C, Liu Y. UV-B-induced oxidative damage and protective role of exopolysaccharides in desert cyanobacterium Microcoleus vaginatus . Journal of Integrative Plant Biology. 2009;51(2):194–200. doi: 10.1111/j.1744-7909.2008.00784.x. [DOI] [PubMed] [Google Scholar]
- 54.Ferreira AS, Silva IN, Oliveira VH, Cunha R, Moreira LM. Insights into the role of extracellular polysaccharides in Burkholderia adaptation to different environments. Frontiers in Cellular and Infection Microbiology. 2011;1:p. 16. doi: 10.3389/fcimb.2011.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qurashi AW, Sabri AN. Osmoadaptation and plant growth promotion by salt tolerant bacteria under salt stress. African Journal of Microbiology Research. 2011;5(21):3546–3554. [Google Scholar]
- 56.Abriouel H, Omar NB, Molinos AC, et al. Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. International Journal of Food Microbiology. 2008;123(1-2):38–49. doi: 10.1016/j.ijfoodmicro.2007.11.067. [DOI] [PubMed] [Google Scholar]
- 57.Poeta P, Costa D, Rodrigues J, Torres C. Antimicrobial resistance and the mechanisms implicated in faecal enterococci from healthy humans, poultry and pets in Portugal. International Journal of Antimicrobial Agents. 2006;27(2):131–137. doi: 10.1016/j.ijantimicag.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 58.Cruz AT, Cazacu AC, Allen CH. Pantoea agglomerans, a plant pathogen causing human disease. Journal of Clinical Microbiology. 2007;45(6):1989–1992. doi: 10.1128/JCM.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wjohn L, Masashi N, Kathrin M. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. The Lancet Infectious Diseases. 2009;9(5):312–323. doi: 10.1016/S1473-3099(09)70083-0. [DOI] [PubMed] [Google Scholar]
- 60.Giraffa G, Carminati D, Tarelli GT. Inhibition of listeria innocua in milk by bacteriocin-producing Enterococcus faecium 7C5. Journal of Food Protection. 1995;58(6):621–623. doi: 10.4315/0362-028X-58.6.621. [DOI] [PubMed] [Google Scholar]
- 61.Kumar RS, Kanmani P, Yuvaraj N, Paari KA, Pattukumar V, Arul V. Purification and characterization of enterocin MC13 produced by a potential aquaculture probiont Enterococcus faecium MC13 isolated from the gut of Mugil cephalus . Canadian Journal of Microbiology. 2011;57(12):993–1001. doi: 10.1139/w11-092. [DOI] [PubMed] [Google Scholar]
- 62.Ahmadova A, Dimov S, Ivanova I, et al. Proteolytic activities and safety of use of Enterococci strains isolated from traditional Azerbaijani dairy products. European Food Research and Technology. 2011;233(1):131–140. [Google Scholar]
- 63.Maisnier-Patin S, Forni E, Richard J. Purification, partial characterisation and mode of action of enterococcin EFS2, an antilisterial bacteriocin, produced by a strain of Enterococcus faecalis isolated from a cheese. International Journal of Food Microbiology. 1996;30(3):255–270. doi: 10.1016/0168-1605(96)00950-6. [DOI] [PubMed] [Google Scholar]
- 64.Nickelsen L, Jakobsen M. Quantitative risk analysis of aflatoxin toxicity for the consumers of “kenkey”—a fermented maize product. Food Control. 1997;8(3):149–159. [Google Scholar]
- 65.Valerio F, Favilla M, de Bellis P, Sisto A, de Candia S, Lavermicocca P. Antifungal activity of strains of lactic acid bacteria isolated from a semolina ecosystem against Penicillium roqueforti, Aspergillus niger and Endomyces fibuliger contaminating bakery products. Systematic and Applied Microbiology. 2009;32(6):438–448. doi: 10.1016/j.syapm.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 66.Lee KW, Park JY, Jeong HR, Heo HJ, Han NS, Kim JH. Probiotic properties of Weissella strains isolated from human faeces. Anaerobe. 2012;18(1):96–102. doi: 10.1016/j.anaerobe.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 67.Berg G, Fritze A, Roskot N, Smalla K. Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahliae Kleb. Journal of Applied Microbiology. 2001;91(3):963–971. doi: 10.1046/j.1365-2672.2001.01462.x. [DOI] [PubMed] [Google Scholar]
- 68.Mandal V, Sen SK, Mandal NC. Detection, isolation and partial characterization of antifungal compound(s) produced by Pediococcus acidilactici LAB 5. Natural Product Communications. 2007;2:671–674. [Google Scholar]
- 69.Magnusson J, Ström K, Roos S, Sjögren J, Schnürer J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiology Letters. 2003;219(1):129–135. doi: 10.1016/S0378-1097(02)01207-7. [DOI] [PubMed] [Google Scholar]


