Abstract
Mitochondria are intracellular organelles where oxidative phosphorylation is carried out to complete ATP synthesis. Mitochondria have their own genome; in metazoans, this is a small, circular molecule encoding 13 electron transport proteins, 22 tRNAs, and 2 rRNAs. In invertebrates, mitochondrial gene rearrangement is common, and it is correlated with increased substitution rates. In vertebrates, mitochondrial gene rearrangement is rare, and its relationship to substitution rate remains unexplored. Mitochondrial genes can also show spatial variation in substitution rates around the genome due to the mechanism of mtDNA replication, which produces a mutation gradient. To date, however, the strength of the mutation gradient and whether movement along the gradient in rearranged (or otherwise modified) genomes impacts genic substitution rates remain unexplored in the majority of vertebrates. Salamanders include both normal mitochondrial genomes and independently derived rearrangements and expansions, providing a rare opportunity to test the effects of large-scale changes to genome architecture on vertebrate mitochondrial gene sequence evolution. We show that: 1) rearranged/expanded genomes have higher substitution rates; 2) most genes in rearranged/expanded genomes maintain their position along the mutation gradient, substitution rates of the genes that do move are unaffected by their new position, and the gradient in salamanders is weak; and 3) genomic rearrangements/expansions occur independent of levels of selective constraint on genes. Together, our results demonstrate that large-scale changes to genome architecture impact mitochondrial gene evolution in predictable ways; however, despite these impacts, the same functional constraints act on mitochondrial protein-coding genes in both modified and normal genomes.
Keywords: gene rearrangement, genome expansion, substitution rates, selective constraint
Introduction
Mitochondria are intracellular organelles where oxidative phosphorylation (OXPHOS) is carried out to complete the process of ATP synthesis via cellular respiration. These organelles have their own genome (retained from their alpha-proteobacterial ancestor), which over evolutionary time has experienced extensive gene transfer to the nucleus (Andersson and Kurland 1998; Gray et al. 2001; Adams and Palmer 2003). In metazoans, this streamlining process has resulted in a small, circular DNA molecule encoding 13 peptides essential for electron transport, 22 tRNAs, and 2 rRNAs (Scheffler 2008). In addition, the mitochondrial genome also contains a control region (CR), a noncoding region that contains a replication origin and transcriptional promoter (Boore 1999; Saccone et al. 1999; Gissi et al. 2008; Scheffler 2008). Because of its small size, the mitochondrial genome has served as an important tractable model for examining the relationships among the different forces shaping the evolution of genes and genomes. Such forces include point mutations, larger mutations impacting genome size and architecture, selection on protein function, selection on transcriptional and translational efficiency, and genetic drift (Moritz et al. 1987; Rand 1993; Ballard and Dean 2001; Fernandez-Silva et al. 2003; Rand et al. 2004; Lynch et al. 2006; Detmer and Chan 2007; Scheffler 2008; Galtier et al. 2009; Boussau et al. 2011). Despite much progress, many important questions remain unanswered about how these forces interact to drive molecular evolution in mitochondrial genomes.
Although they have evolutionarily conserved genomic content, metazoan mitochondrial genomes show a diversity of gene orders (Boore and Brown 1998; Boore 1999; Saccone et al. 1999; Xu et al. 2006). Rearrangement of gene order, often accompanied by expansion of noncoding regions, results from gene duplications followed by random loss of one of the paralogs (Moritz and Brown 1987; Boore 1999; Mueller and Boore 2005; San Mauro et al. 2006) or by intramolecular recombination (Stanton et al. 1994). Variation in gene order is substantial across invertebrates, but is far less common in vertebrates (Boore 1999; Scheffler 2008). The reasons why some clades have more dynamic genomes than others remain unknown. The high rates of gene rearrangement in invertebrate mitochondrial genomes have been positively correlated with rates of nucleotide substitution within mitochondrial genes in mollusks, insects, crustaceans, and other arthropods (Hoffmann et al. 1992; Hoeh et al. 1996; Shao et al. 2003; Xu et al. 2006). The relationship between gene rearrangement and substitution rate is unexplored in vertebrates, and the mechanisms underlying this correlation remain unknown across all taxa.
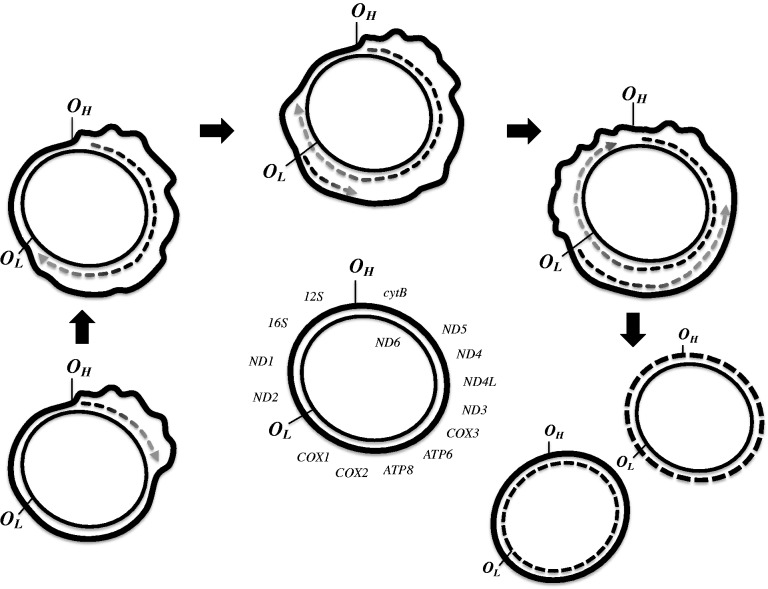
In addition to variation in substitution rates across taxa associated with gene rearrangements, mitochondrial genes in some taxa also show spatial variation in substitution rates around the genome due to the mechanism underlying mitochondrial DNA (mtDNA) replication. Vertebrate mitochondrial replication initiates from two replication origins that are offset from each other, causing asynchronous replication across both strands (fig. 1). Specifically, mtDNA replication begins at the H-strand replication origin (OH), displacing one strand and leaving it single-stranded, until the newly synthesized strand reaches the L-stand replication origin (OL) and synthesis begins in the reverse direction along the displaced strand. Because single-stranded DNA is prone to mutation, mtDNA replication results in a gradient of mutation accumulation where regions that persist in the single-stranded state for longer experience more AT-biased mutations (Reyes et al. 1998; Faith and Pollock 2003; Krishnan et al. 2004; Broughton and Reneau 2006; Scheffler 2008). The mitochondrial environment is mutagenic, in part, because of the presence of reactive oxygen species (ROS), byproducts of oxidative phosphorylation (Scheffler 2008). Metabolic rates vary dramatically across vertebrates, which may, in turn, yield different levels of mutagenicity, impacting the strength of the mutation gradient (Gatten et al. 1992; Santos 2012). To date, however, 1) the strength of the mutation gradient, 2) whether the mutation gradient produces a substitution rate gradient, and 3) whether genes in rearranged and/or expanded genomes move to more or less vulnerable positions along the gradient remain unexplored in the majority of vertebrates.
Fig. 1.—
Schematic representation of vertebrate mitochondrial genome replication. Replication begins at the OH and proceeds along the heavy strand. Once the replication fork passes the OL, replication begins along the light strand.
Substitution rates in mitochondrial genes are also shaped by functional constraints on mitochondrial proteins, which differ across both genes and lineages; these differences are associated with variation in organismal traits (Martin and Palumbi 1993; Rand 1994; Galtier et al. 2009; Sun et al. 2011). For example, high metabolic energy demands in birds and mammals are positively correlated with strong selective constraint (i.e., low levels of nonsynonymous relative to synonymous substitutions) on mitochondrial genes, whereas low metabolic demand in salamanders is correlated with weaker selective constraint (i.e., higher levels of nonsynonymous substitutions) (Shen et al. 2009; Chong and Mueller 2013). It remains unclear whether organisms experiencing weaker selection on mitochondrial gene sequences, which underlie OXPHOS protein function, also experience weaker selection on genome architecture (i.e., genome size and order), which impacts OXPHOS protein transcription and translation.
Salamanders, an amphibian clade of 648 species, include both normal mitochondrial genomes that have the vertebrate consensus mitochondrial gene order and a range of independently derived, modified genomes that have both gene order rearrangements and genome expansions. Because this genomic diversity is unusual among vertebrates, salamanders provide a rare opportunity to test the effects of genomic modification on vertebrate mitochondrial gene sequence evolution. In this study, we show that: 1) mitochondrial protein-coding genes within modified salamander genomes have significantly higher synonymous and nonsynonymous substitution rates than genes within normal salamander genomes; 2) despite expansions of up to 6 kb, most genes in modified salamander genomes maintain their position along the mutation gradient; the genes that do move are not substantially impacted by their new position; and the mutation gradient in salamanders is weak; and 3) gene rearrangements and genomic expansion events occur independent of levels of selective constraint acting on mitochondrial genes. Taken together, our results demonstrate that large-scale changes to genome architecture impact mitochondrial gene sequence evolution in predictable ways within salamanders; however, despite these impacts, the same functional constraints are acting on mitochondrial protein-coding genes in both modified and normal salamander genomes.
Materials and Methods
Sequence Data and Genome Characteristics
We obtained 62 complete salamander mitochondrial genome sequences, each representing a unique species, from the GenBank RefSeq database (release 53). Our data set includes representatives from 6 of the 10 salamander families, with predominant taxonomic sampling representing the largest family, Plethodontidae. We extracted all 13 protein-coding genes from each mitochondrial genome based on GenBank genome annotations.
We characterized mitochondrial genomes based on gene order and genome size, where genomes with gene arrangements that deviated from the consensus vertebrate mitochondrial gene order and genomes that were expanded (>17.5 kb) were labeled as modified (in contrast to normal). Gene order was described relative to the CR (the initiation site for mtDNA replication and transcription), beginning with tRNA-Phe, using a purpose-built perl script (Minxiao et al. 2011). We extracted mitochondrial genome size data from the GenBank files. These results are summarized in supplementary file S1, Supplementary Material online.
Compositional analyses were carried out for all genomes in the data set using custom perl scripts (available by request from author). For each of the 13 protein-coding genes, we estimated base composition for the whole gene and for the third position of 4-fold degenerate codons (P4FD): alanine-GCN, glycine-GGN, leucine-CTN, proline-CCN, arginine-CGN, serine-TCN, threonine-CAN, and valine-GTN. Base frequencies, as well as AT skew and GC skew, were also calculated for the whole genome. These results are summarized in supplementary file S1, Supplementary Material online. We tested for differences in AT skew and GC skew values between normal and modified genomes using a two-way Mann–Whitney test implemented in R.
Initial Phylogenetic Analysis
Multiple sequence alignments were performed based on amino acid sequences for each mitochondrial protein-coding gene using MUSCLE v.3.8 (Edgar 2004). We then estimated a maximum likelihood tree for the concatenated mitochondrial gene data set using RAxML v.7.2 (Stamatakis 2006). The data were partitioned by gene and codon position and analyzed using the GTR + Γ model of nucleotide substitution for each partition.
Nonsynonsymous and Synonymous Substitution Analysis
For each mitochondrial gene, we estimated nonsynonymous (dN) and synonymous (dS) substitution rates. We first estimated dN and dS using a single value for each parameter across all branches (Model 0 in Codeml, implemented in PAML v4.4 [Yang 2007]) using the fixed topology of the maximum likelihood tree estimated from the concatenated mitochondrial sequences. Subsequently, we reestimated these parameters under Model 1 (in Codeml), which allows these parameter values to vary for each branch. These results are summarized in supplementary file S1, Supplementary Material online. For each gene, we tested for differences in dN and dS values between normal and modified genomes using a two-way Mann–Whitney test implemented in R.
Mutation Gradient Analysis
The site-specific mutation rate around the mitochondrial genome is associated with the duration of time spent single stranded, which is determined by the position of a site relative to the origins of replication. The duration of single-stranded state of the parental H strand (DssH) is defined by the duration between the displacement of the heavy strand by the replication fork and synthesis of its complement; for a given gene, it is estimated using the formula DssH = (L − 2 [x − OL])/L for the two genes located upstream of the origins of replication, ND1 and ND2, and DssH = (2 [x − OL])/L for the remaining genes, where L is the total length of the genome, OL is the position of the light strand origin of replication, and x is the midpoint of a gene (Tanaka and Ozawa 1994; Reyes et al. 1998). We used a modification of these formulas to estimate absolute mutation position, which is not standardized by genome size, for each mitochondrial gene for all genomes because our goals include comparing estimates of mutation position for a given gene across genomes of different sizes. Specifically, we hypothesize that rearrangements and expansions may alter the mutation position of genes in modified genomes. Mutation position is estimated using (L − 2 [x − OL]) for the two genes located upstream of the origins of replication, ND1 and ND2, and (2 [x − OL]) for the remaining genes.
Functional Constraint on Mitochondrial Genes
To test whether mitochondrial genome expansions and rearrangements reflect an overall relaxation of selective constraint on mitochondrial function, we estimated ω, which is the ratio of nonsynonymous to synonymous substitutions (ω = dN/dS), for each mitochondrial gene. The ratio ω is used to measure the strength of selection, where for values of ω between 0 and 1, a smaller ω indicates stronger purifying selection and a larger ω indicates weaker purifying selection. We first estimated ω using a single value across all branches (Model 0 in Codeml, implemented in PAML v4.4 [Yang 2007]) using the fixed topology of the concatenated mitochondrial maximum likelihood tree. We then reestimated ω under Model 1 where parameter values can vary for all branches. These results are summarized in supplementary file S1, Supplementary Material online. For each gene, we tested whether ω was greater in modified genomes using a one-way Mann–Whitney test implemented in R.
Results and Discussion
Mitochondrial Genome Characteristics
For mitochondrial genomes from 62 salamander species, we estimated genome composition and organization to identify and characterize the differences between normal and modified genomes. We identified 14 mitochondrial genomes as modified based on evidence of gene rearrangement, a large increase in genome size (>17.5 kb total length), or a combination of the two. Eight of these modified genomes show extensive gene rearrangements that include one or more of the origins of replication and the regions flanking them, including both protein-coding genes and tRNAs. The remaining six modified genomes maintain the ancestral vertebrate gene order, but are larger than 19 kb in size. This size increase reflects the accumulation of tandem repeats of noncoding sequence in the CR and/or in the IGS, an intergenic spacer region between tRNA-Thr and tRNA-Pro present in diverse salamander clades (Wallis 1987; McKnight and Shaffer 1997; Mueller and Boore 2005). Similar patterns of noncoding sequence accumulation exist in other taxonomic groups (e.g., mammals, caecilians, fish, and invertebrates) (Stewart and Baker 1994; Prager et al. 1996; Delarbre et al. 2001; San Mauro et al. 2006; Minxiao et al. 2011). Tandem repetitive noncoding sequences are also present in seven of the eight genomes with rearranged gene order. Overall, genome size is significantly larger in modified salamander genomes (19,775 ± 1,809 bp) than in normal salamander genomes (16,475 ± 209 bp). Both pseudogenes and additional copies of duplicate genes are present in at least six of the modified salamander mitochondrial genomes, suggesting that these rearrangements were mediated by duplication of a portion of the genome (Mueller and Boore 2005). The localization of genomic modification to regions containing and flanking the two origins of replication, as well as the presence of tandem repetitive sequences, suggests that these genomic regions are particularly susceptible to slipped-strand mispairing, intramolecular recombination, and imprecise replication.
We estimated several measures of genome composition for normal and modified genomes and compared the two groups. Average base composition did not differ between normal (A: 0.3352, T: 0.3104, C: 0.1390, G: 0.2152) and modified genomes (A: 0.3414, T: 0.3029, C: 0.1307, G: 0.2250). Average GC content also did not differ between the two groups (35.4% and 35.5%, respectively). In contrast, normal and modified genomes did differ slightly in both average AT skew (0.039 and 0.046, respectively; P = 0.087) and GC skew (−0.214 and −0.226, respectively; P < 0.001).
Substitution Rates in Normal and Modified Genomes
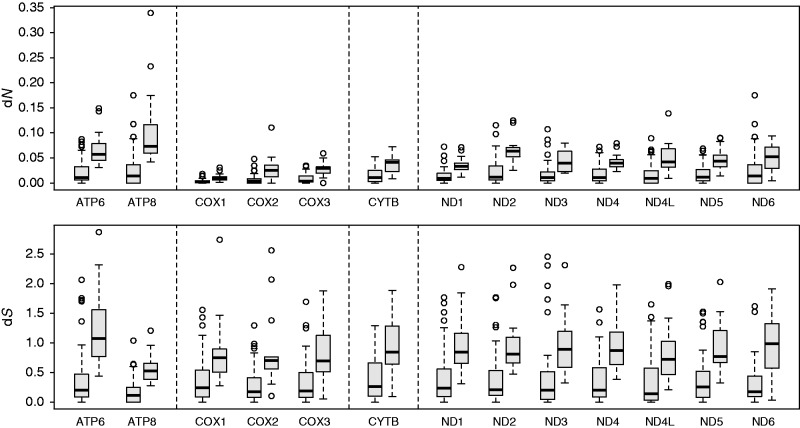
To test for differences in rates of evolution between normal and modified genomes, we estimated rates of synonymous (dS) and nonsynonymous (dN) substitution across a maximum likelihood tree for each of the 13 mitochondrial protein-coding genes. Our results show that mitochondrial genes in modified genomes have a significantly elevated dS (P < 0.001 for all genes) and dN (P < 0.006 for all genes). Modified genomes show, on average, a 2.66-fold increase in dS compared with normal genomes; COX1 shows the smallest increase (2.31-fold), whereas ATP6 shows the greatest increase (3.17-fold). Modified genomes show, on average, a 3.07-fold increase in dN compared with normal genomes; ND4 shows the smallest increase (2.42-fold) and COX2 shows the largest increase (4.50-fold). These results demonstrate that mitochondrial genome modification is correlated with an absolute increase in both synonymous and nonsynonymous substitution rates for mitochondrial protein-coding genes, independent of phylogeny (fig. 2).
Fig. 2.—
Comparison of nonsynonymous and synonymous substitution rates for the 13 mitochondrial protein-coding genes. Normal genomes are on the left; modified genomes are on the right. The dashed lines separate mitochondrial genes that belong to different OXPHOS function complexes. For all comparisons, substitution rates of modified genomes are significantly higher than substitution rates of normal genomes.
Although the mechanisms that give rise to the correlation between substitution rate and mitochondrial rearrangement remain unknown in salamanders as well as other taxa, a number of hypotheses have been proposed. Some such hypotheses state that an increase in substitution rates causes elevated rates of gene rearrangement. Specifically, increased substitutions accumulating in the sequences regulating replication initiation and termination can decrease replication fidelity, resulting in duplications, deletions, and rearrangements (Shao et al. 2003). The high rates of substitution may also cause high rates of DNA damage and double-strand breaks, which can facilitate intramolecular recombination (Dowton and Campbell 2001). Alternatively, the presence of gene rearrangements may cause an increase in substitution rates by an undetermined mechanism. Differences in population biology have also been proposed to explain the correlation between elevated substitution rates and genomic modifications; relatively strong genetic drift would cause fixation of both point mutations and large-scale genomic modifications (Lynch et al. 2006; Boussau et al. 2011). Comparative studies that test for the signatures of these putative molecular and demographic mechanisms are required to determine the cause of this correlation in salamanders as well as other taxa.
Mutation Gradient in Normal and Modified Genomes
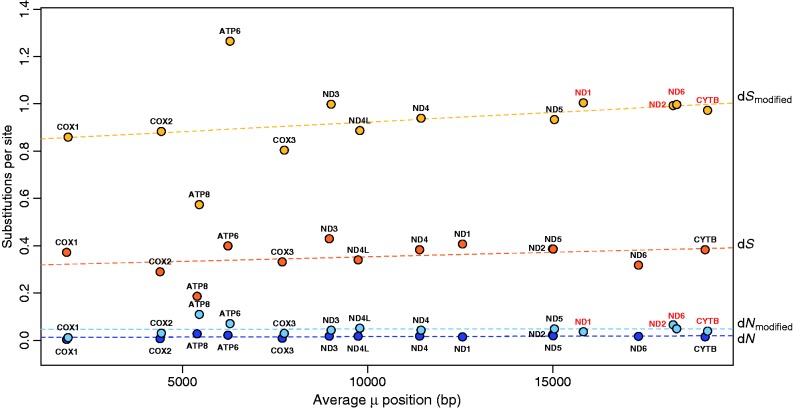
To test for differences in gene placement along the mutation gradient between normal and modified mitochondrial genomes, we first estimated the average mutation position for each gene in each species. We define average mutation position as the duration spent single stranded, measured in base pairs. This measurement reflects the probability of mutation for a given position in the mitochondrial genome due to its exposure, in the single-stranded state, to the mutagenic mitochondrial environment during replication. We do not standardize for total genome size, as done in previous studies on mitochondrial mutational gradients (Tanaka and Ozawa 1994; Faith and Pollock 2003; Krishnan et al. 2004), because our goals include comparing estimates of mutation position for a given gene across genomes of different sizes. Specifically, we hypothesize that rearrangements and expansions may alter the mutation position of genes in modified genomes. Our results show that alterations to mutation position are restricted to a subset of genes in modified genomes. Specifically, genes located upstream of both the OL and the OH—ND1 and ND2 (fig. 1)—experience a large increase in average mutation position in modified genomes (3.3 ± 1.9 kb each) (fig. 3). This pattern reflects the fact that the majority of genomic expansions occur near the OH. During replication, such expansions are not encountered until replication has begun from the OL (fig. 1) and completed the majority of both strands. Therefore, only genes located upstream of both the OL and the OH—ND1 and ND2—spend a longer duration in the single-stranded state in expanded genomes. Of the remaining 11 genes positioned downstream of both replication origins, 9 are never involved in gene rearrangements and have very similar mutation positions in both modified and normal genomes; on average, these genes shift less than 70 bp (fig. 3). In contrast, ND6 and CYTB have been involved in four independent gene rearrangements (Mueller and Boore 2005). Mutation position of ND6 either decreases by 4.0 kb or increases by up to 8.5 kb, depending on lineage-specific gene rearrangements. Mutation position of CYTB either decreases by 1.2 kb or increases by up to 2.5 kb, depending on lineage-specific gene rearrangements.
Fig. 3.—
Plot of average mutation position (x axis) and average nonsynonymous (dN) and synonymous (dS) substitution rates (y axis) for each mitochondrial gene for both normal and modified genomes. Average dN for normal mitochondrial genomes are dark blue, average dN for modified mitochondrial genomes are light blue, average dS for normal mitochondrial genomes are dark orange, and average dS for modified mitochondrial genomes are light orange. The mutation position for a given gene is the average time spent single stranded measured in base pairs. Genes within modified genomes do not significantly differ in mutation position from genes within normal genomes, with the exception of four genes (labeled red): average mutation positions of ND1 and ND2 increase by 3.3 kb within modified genomes. Mutation position of ND6 either decreases by 4.0 kb or increases by up to 8.5 kb within rearranged genomes, depending on lineage-specific gene rearrangements. Mutation position of CYTB either decreases by 1.2 kb or increases by up to 2.5 kb, depending on lineage-specific gene rearrangements. Despite the presence of a weak mutational gradient, no significant relationship exists between average mutation position and substitution rates (dN or dS) in salamanders. Thus, change in mutation position has no significant impact on rates of molecular evolution.
We then tested for a gradient generated by mutational bias during replication in normal and modified salamander genomes. We estimated base compositional asymmetry (i.e., AT skew) of the third position of 4-fold degenerate codons (P4FD) for each gene. A linear regression of AT skew on mutation position indicates significant positive correlation for normal genomes, though only a small proportion of compositional variation can be explained by mutation position (y = 0.000006874x + 0.271, r2 = 0.056, P < 0.001). The positive relationship between AT skew and mutation position is not significant in modified genomes (y = 0.000003913x + 0.318, r2 = 0.018, P = 0.068), suggesting that modified genomes are not at the same base compositional equilibrium as normal salamander genomes. However, this mutational gradient does not translate into a gradient in either dN or dS (fig. 3). Thus, although the mutation positions of ND1, ND2, ND6, and CYTB are altered during genomic expansion and/or rearrangement, their movement to this new position has no significant impact on their substitution rates. Salamanders have the lowest aerobic metabolic demands of any tetrapod vertebrates, suggesting that their levels of ROS (a mutagenic byproduct of oxidative phosphorylation) may be low. This may underlie their weak mitochondrial mutational gradient, although comparative studies of this gradient across taxa with different metabolic rates are required to test this hypothesis.
Functional Constraints on Mitochondrial Genes in Normal and Modified Genomes
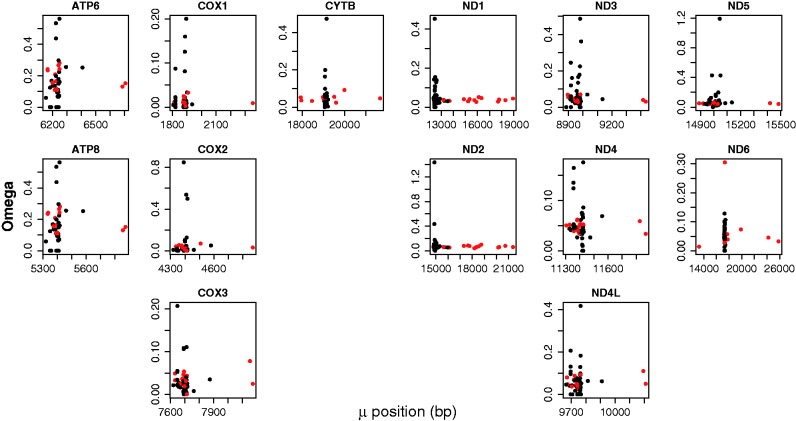
To test whether mitochondrial genome expansions and rearrangements reflect an overall relaxation of selective constraint on mitochondrial function, we compared estimates of ω (dN/dS, a measure of the strength of selection) for all 13 mitochondrial genes between normal and modified genomes. Specifically, we tested whether ω is greater in modified genomes, indicative of weaker selective constraint. Although both dN and dS are elevated for all genes in modified genomes, ω values for genes in modified genomes are not significantly greater than ω values in normal genomes (fig. 4; P > 0.189 for all genes). Thus, genes within modified genomes are not experiencing weaker selective constraint than genes within normal genomes. This result demonstrates that mitochondrial genome expansions and rearrangements likely do not reflect an overall relaxation of selective constraint on mitochondrial function. Such an overall relaxation would affect protein sequence evolution as well as transcriptional and translational efficiency, which are impacted by changes in genome size and gene order (Fernandez-Silva et al. 2003; Bonawitz et al. 2006; Satoh et al. 2010; Chong and Mueller 2013). In contrast, our results show that functional constraints on protein-coding sequences are not weaker in lineages with modified genomes than in lineages with normal genomes.
Fig. 4.—
Plots of levels of selective constraint, represented by ω (dN/dS), acting on the 13 mitochondrial genes for both normal (black) and modified (red) genomes. Omega plots for each gene are grouped to reflect protein functional complexes (i.e., COX1-3, ATP6&8, CYTB, ND1-6, and 4L). ω values for modified genomes are not larger than those for normal genomes, showing that genes in modified genomes are not experiencing weaker selective constraint.
Supplementary Material
Supplementary file S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Science Foundation (NSF-DEB 1021489 to R.L.M.) and by Colorado State University. Analyses were performed on the CSU ISTeC HPC System (NSF CNS-0923886). Publication of this work was supported by the Colorado State University Libraries Open Access Research and Scholarship Fund. Members of the Mueller lab, W. C. Funk, C. T. Webb, and two anonymous reviewers provided helpful discussion and comments.
Literature Cited
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Ballard JW, Dean MD. The mitochondrial genome: mutation, selection and recombination. Curr Opin Genet Dev. 2001;11:667–672. doi: 10.1016/s0959-437x(00)00251-3. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 1998;8:668–674. doi: 10.1016/s0959-437x(98)80035-x. [DOI] [PubMed] [Google Scholar]
- Boussau B, Brown JM, Fujita MK. Nonadaptive evolution of mitochondrial genome size. Evolution. 2011;65:2706–2711. doi: 10.1111/j.1558-5646.2011.01322.x. [DOI] [PubMed] [Google Scholar]
- Broughton RE, Reneau PC. Spatial covariation of mutation and nonsynonymous substitution rates in vertebrate mitochondrial genomes. Mol Biol Evol. 2006;23:1516–1524. doi: 10.1093/molbev/msl013. [DOI] [PubMed] [Google Scholar]
- Chong RA, Mueller RL. Low metabolic rates in salamanders are correlated with weak selective constraints on mitochondrial genes. Evolution. 2013;67:894–899. doi: 10.1111/j.1558-5646.2012.01830.x. [DOI] [PubMed] [Google Scholar]
- Delarbre C, Rasmussen A-S, Arnason U, Gachelin G. The complete mitochondrial genome of the hagfish Myxine glutinosa: unique features of the control region. J Mol Evol. 2001;53:634–641. doi: 10.1007/s002390010250. [DOI] [PubMed] [Google Scholar]
- Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Dowton M, Campbell NJH. Intramitochondrial recombination—is it why some mitochondrial genes sleep around? Trends Ecol Evol. 2001;16:269–271. doi: 10.1016/s0169-5347(01)02182-6. [DOI] [PubMed] [Google Scholar]
- Edgar R. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Pollock DD. Likelihood analysis of asymmetrical mutation bias gradients in vertebrate mitochondrial genomes. Genetics. 2003;165:735–745. doi: 10.1093/genetics/165.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- Galtier N, Jobson RW, Nabholz B, Glemin S, Blier PU. Mitochondrial whims: metabolic rate, longevity and the rate of molecular evolution. Biol Lett. 2009;5:413–416. doi: 10.1098/rsbl.2008.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatten REJ, Miller K, Full RJ. Energetics at rest and during locomotion. In: Feder ME, editor. Environmental physiology of the amphibians. Chicago (IL) University of Chicago Press; 1992. pp. 314–377. [Google Scholar]
- Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews1018. 1018.1011–1018.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeh WR, Stewart DT, Sutherland BW, Zouros E. Cytochrome c oxidase sequence comparisons suggest an unusually high rate of mitochondrial DNA evolution in Mytilus (Mollusca: Bivalvia) Mol Biol Evol. 1996;13:418–421. doi: 10.1093/oxfordjournals.molbev.a025600. [DOI] [PubMed] [Google Scholar]
- Hoffmann RJ, Boore JL, Brown WM. A novel mitochondrial genome organization for the blue mussel, Mytilus edulis. Genetics. 1992;131:397–412. doi: 10.1093/genetics/131.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan NM, Seligmann H, Raina SZ, Pollock DD. Detecting gradients of asymmetry in site-specific substitutions in mitochondrial genomes. DNA Cell Biol. 2004;23:707–714. doi: 10.1089/1044549042476901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- Martin AP, Palumbi SR. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci U S A. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight ML, Shaffer HB. Large, rapidly evolving intergenic spacers in the mitochondrial DNA of the salamander family Ambystomatidae (Amphibia: Caudata) Mol Biol Evol. 1997;14:1167–1176. doi: 10.1093/oxfordjournals.molbev.a025726. [DOI] [PubMed] [Google Scholar]
- Minxiao W, Song S, Chaolun L, Xin S. Distinctive mitochondrial genome of Calanoid copepod Calanus sinicus with multiple large non-coding regions and reshuffled gene order: useful molecular markers for phylogenetic and population studies. BMC Genomics. 2011;12:73. doi: 10.1186/1471-2164-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Brown WM. Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc Natl Acad Sci U S A. 1987;84:7183–7187. doi: 10.1073/pnas.84.20.7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Dowling TE, Brown WM. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu Rev Ecol Evol Sys. 1987;18:269–292. [Google Scholar]
- Mueller RL, Boore JL. Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol Biol Evol. 2005;22:2104–2112. doi: 10.1093/molbev/msi204. [DOI] [PubMed] [Google Scholar]
- Prager EM, Tichy H, Sage RD. Mitochondrial DNA sequence variation in the eastern house mouse, Mus musculus: comparison with other house mice and report of a 75-bp tandem repeat. Genetics. 1996;143:427–446. doi: 10.1093/genetics/143.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM. Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol. 1993;37:281–295. doi: 10.1007/BF00175505. [DOI] [PubMed] [Google Scholar]
- Rand DM. Thermal habit, metabolic rate and the evolution of mitochondrial DNA. Trends Ecol Evol. 1994;9:125–131. doi: 10.1016/0169-5347(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Reyes A, Gissi C, Pesole G, Saccone C. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 1998;15:957–966. doi: 10.1093/oxfordjournals.molbev.a026011. [DOI] [PubMed] [Google Scholar]
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 1999;238:195–209. doi: 10.1016/s0378-1119(99)00270-x. [DOI] [PubMed] [Google Scholar]
- San Mauro D, Gower DJ, Zardoya R, Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol Biol Evol. 2006;23:227–234. doi: 10.1093/molbev/msj025. [DOI] [PubMed] [Google Scholar]
- Santos JC. Fast molecular evolution associated with high active metabolic rates in poison frogs. Mol Biol Evol. 2012;29:2001–2018. doi: 10.1093/molbev/mss069. [DOI] [PubMed] [Google Scholar]
- Satoh T, Sato Y, Masuyama N, Miya M, Nishida M. Transfer RNA gene arrangement and codon usage in vertebrate mitochondrial genomes: a new insight into gene order conservation. BMC Genomics. 2010;11:479. doi: 10.1186/1471-2164-11-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler IE. Mitochondria. Hoboken (NJ): John Wiley & Sons, Inc; 2008. [Google Scholar]
- Shao R, Dowton M, Murrell A, Barker SC. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol Evol. 2003;20:1612–1619. doi: 10.1093/molbev/msg176. [DOI] [PubMed] [Google Scholar]
- Shen Y, Shi P, Sun Y, Zhang Y. Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Res. 2009;19:1760–1765. doi: 10.1101/gr.093138.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stanton DJ, Daehler LL, Moritz CC, Brown WM. Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics. 1994;137:233–241. doi: 10.1093/genetics/137.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DT, Baker AJ. Patterns of sequence variation in the mitochondrial D-loop region of shrews. Mol Biol Evol. 1994;11:9–21. doi: 10.1093/oxfordjournals.molbev.a040096. [DOI] [PubMed] [Google Scholar]
- Sun YB, Shen YY, Irwin DM, Zhang YP. Evaluating the roles of energetic functional constraints on teleost mitochondrial-encoded protein evolution. Mol Biol Evol. 2011;28:39–44. doi: 10.1093/molbev/msq256. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Ozawa T. Strand asymmetry in human mitochondrial DNA mutations. Genomics. 1994;22:327–335. doi: 10.1006/geno.1994.1391. [DOI] [PubMed] [Google Scholar]
- Wallis GP. Mitochondrial DNA insertion polymorphism and germ line heteroplasmy in the Triturus cristatus complex. Heredity. 1987;58:229–238. doi: 10.1038/hdy.1987.37. [DOI] [PubMed] [Google Scholar]
- Xu W, Jameson D, Tang B, Higgs PG. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J Mol Evol. 2006;63:375–392. doi: 10.1007/s00239-005-0246-5. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.