Abstract
Comparative mitochondrial genomics of arbuscular mycorrhizal fungi (AMF) provide new avenues to overcome long-lasting obstacles that have hampered studies aimed at understanding the community structure, diversity, and evolution of these multinucleated and genetically polymorphic organisms. AMF mitochondrial (mt) genomes are homogeneous within isolates, and their intergenic regions harbor numerous mobile elements that have rapidly diverged, including homing endonuclease genes, small inverted repeats, and plasmid-related DNA polymerase genes (dpo), making them suitable targets for the development of reliable strain-specific markers. However, these elements may also lead to genome rearrangements through homologous recombination, although this has never previously been reported in this group of obligate symbiotic fungi. To investigate whether such rearrangements are present and caused by mobile elements in AMF, the mitochondrial genomes from two Glomeraceae members (i.e., Glomus cerebriforme and Glomus sp.) with substantial mtDNA synteny divergence, were sequenced and compared with available glomeromycotan mitochondrial genomes. We used an extensive nucleotide/protein similarity network-based approach to investigate dpo diversity in AMF as well as in other organisms for which sequences are publicly available. We provide strong evidence of dpo-induced inter-haplotype recombination, leading to a reshuffled mitochondrial genome in Glomus sp. These findings raise questions as to whether AMF single spore cultivations artificially underestimate mtDNA genetic diversity. We assessed potential dpo dispersal mechanisms in AMF and inferred a robust phylogenetic relationship with plant mitochondrial plasmids. Along with other indirect evidence, our analyses indicate that members of the Glomeromycota phylum are potential donors of mitochondrial plasmids to plants.
Keywords: arbuscular mycorrhizal fungi, mitochondrial genome rearrangements, homologous recombination, plasmid-related DNA polymerase genes, short inverted repeats, nucleotide/protein similarity network
Introduction
The association between arbuscular mycorrhizal fungi (AMF) and plant roots is one of the most widespread symbioses involving plants, and thus has an important role in terrestrial ecosystems (Smith and Read 2008). In exchange for carbohydrates, AMF improve plant fitness by enhancing mineral nutrient and water uptake (Ruiz-Lozano et al. 1995). AMF have also been shown to alleviate salt stress (Evelin et al. 2009) and to provide protection against root pathogens (St-Arnaud and Vujanovik 2007; Ismail and Hijri 2012). Despite the fact that these symbioses contribute to important services in natural, agricultural and reclaimed ecosystems (Gianinazzi et al. 2010), the species richness, community structure and functional diversity of AMF is not well understood (Wehner et al. 2010) due to a lack of reliable molecular tools. The intra-isolate genetic diversity of nuclear DNA and RNA sequences observed in AMF (Sanders et al. 1995; Kuhn et al. 2001; Pawlowska and Taylor 2004; Mathimaran et al. 2008; Stockinger et al. 2009; Boon et al. 2010), has made it difficult to determine evolutionary relatedness at high levels of resolution (i.e., between genetically similar species and/or isolates).
In contrast, the first mitochondrial (mt) genome sequenced from Glomus irregulare (synonym Rhizophagus irregularis; formerly G. intraradices) was shown to be homogeneous (Lee and Young 2009), a state often referred to as homoplasmic. Since then, the mitochondrial large subunit rRNA gene has been explored for its usefulness as a marker (Raab et al. 2005; Börstler et al. 2008; Thiéry et al. 2010), but determination of its specificity at the isolate level is challenging due to a lack of genetic variability. Recently, the mitochondrial genomes of six G. irregulare isolates (Formey et al. 2012) and a closely related species (Beaudet et al. 2013) were compared. Striking features of these data sets were the presence and variability among isolates of numerous mobile elements such as plasmid-related DNA polymerase genes (dpo), short inverted repeats (SIRs), and homing endonuclease genes, which have given rise to mitochondrial gene hybrids through horizontal gene transfer (HGT). Intra-specific divergence in the eroded portions of these elements revealed the high value of designing much-needed isolate-specific molecular markers (Corradi and Bonen 2012).
Mitochondrial DNA has long been considered a valuable tool in evolutionary biology because of its high mutation rate and the assumption that individuals only possessed one mtDNA haplotype, directly maternally inherited, without recombination (Birky et al. 1978; Birky 2001). However, reports of mitochondrial recombination events and heteroplasmy in animals, but especially in plants and fungi, led to re-evaluate this assertion (Barr et al. 2005; White et al. 2008). Molecular recombination is an important evolutionary mechanism that allows deleterious mutations removal and generates allelic diversity in a population. The main causes of heteroplasmy are bi-parental inheritance and/or mutations, which result in the coexistence of mitochondrial genomes that differ in either nucleotide composition (site heteroplasmy) or length (length heteroplasmy). The latter can involve large-scale rearrangements or insertion/loss of coding regions (Boursot et al. 1987; Volz-Lingenhöhl et al. 1992). In AMF, transient mitochondrial length heteroplasmy through anastomosis has recently been demonstrated for geographically distant G. irregulare in vitro isolates (de la Providencia et al. 2013). The divergence in length between the two mtDNA haplotypes was caused by a variation in plasmid-related dpo insertions. The integration of elements such as dpo and SIRs has been shown to produce genomic rearrangements through recombination in other organisms (Schofield et al. 1992; Bi and Liu 1996; Paquin et al. 2000; Ferandon et al. 2008; Tanaka et al. 2012).
Integrated plasmid-related dpo genes have been reported in all AMF mitochondrial genomes that have been sequenced thus far. Plasmids are self-replicating extra-chromosomal DNA molecules that were originally identified in bacteria. Analogous molecules have now been reported in eukaryotes, notably plants and numerous fungal species. The majority of plasmids described in filamentous fungi are strictly mitochondrial, but cases of nuclear localization have been reported in yeasts (reviewed in Griffiths [1995]) and a Mucor-like fungus (Hänfler et al. 1992). Plasmid structure varies widely, from circular molecules to linear invertrons with terminal inverted repeats (TIRs). Both types harbor one or two open reading frames (ORFs) encoding DNA and/or RNA polymerase genes (Nargang et al. 1984; Akins et al. 1988; Pande et al. 1989; Sakaguchi 1990; Shiu-Shing Chan et al. 1991; Court and Bertrand 1992; Li and Nargang 1993). Plasmids have also been shown to integrate into fungal mtDNA through recombination and can either be cryptic or elicit changes in phenotypic expression (Akins et al. 1986; Robison et al. 1991; Griffiths 1992; Hänfler et al. 1992; Oeser et al. 1993; Hermanns et al. 1994). Such insertions have been reported to be acquired vertically from a common ancestor in plants (Robison and Wolyn 2005) and also in fungi (Robison et al. 1991). However, numerous cases of HGT of those elements have been reported in fungi, offering an explanation for their ubiquitous distribution in this group (Collins and Saville 1990; Griffiths et al. 1990; Kempken et al. 1992; Arganoza et al. 1994; Debets et al. 1994). Furthermore, plant mitochondrial plasmids are suspected to have been acquired from a fungal donor based on their similar structure and low GC content, which is a common feature of fungal mitochondrial genomes (reviewed in Rosewitch and Kistler [2000]).
Another type of mobile element found in the mtDNA of Glomeromycota, SIRs (Formey et al. 2012), were observed in the mitochondrial genome of the green algae Chlamydomonas reinhardtii (Boer and Gray 1991), but their structure is reminiscent of GC clusters in yeast (de Zamaroczy and Bernardi 1986) and the Pst I palindromes of Neurospora crassa (Yin et al. 1981). SIRs can be folded into hairpin secondary structures, and have been suspected to have created genome rearrangements in Chlamydomonas spp. by intramolecular recombination (Denovan-Wright and Lee 1994; Nedelcu and Lee 1998) through a model that is analogous to what has been observed in fungi (Almasan and Mishra 1991; Weiller et al. 1991), plants (André et al. 1992) and bacteria (Schofield et al. 1992; Bi and Liu 1996). They are putatively mobile, although their transposition mechanism is not well understood (Grindley and Reed 1985; Nakazono et al. 1994). Nevertheless, mobility has been demonstrated in similar repeats that fold into double hairpin structures in Allomyces spp. mtDNA (Paquin et al. 2000).
No information about the underlying mechanisms and consequences of such integrations into the mtDNA of Glomeromycota is currently available. Understanding whether mitochondrial recombination occurs in AMF is important because it can modify the interpretation of several aspects of research, including phylogenetic and population genetic analyses (Ballard and Whitlock 2004). To investigate this issue, we compared two novel mitochondrial genomes from the Glomeraceae family with a G. irregulare isolate. As all AMF mitochondrial genomes from the Glomeraceae family that have been sequenced so far share the same mitochondrial synteny, we used this as a comparative basis for our study. We assessed mitochondrial genome evolution with regards to two other species of this ubiquitous AMF family, which showed divergence in mitochondrial gene organization. We wanted to know whether the new mtDNA reported here, from the species Glomus sp. DAOM-240422 and G. cerebriforme DAOM-227022, could shed light on the impact of mobile element insertions with regards to genome organization, and thus assessed the diversity, distribution, and propagation of mitochondrial dpo sequences within the Glomeromycota phylum. We also explored their evolutionary relatedness to other taxa, with respect to their global positioning within all known plasmid-related dpo sequences in the public databases.
Materials and Methods
Fungal Material and DNA Extraction
Spores and mycelium of G. irregulare (DAOM-234179), Glomus sp. (DAOM-240422), and G. cerebriforme (DAOM-229022) were cultivated in vitro on a minimal (M) medium solidified with 0.4% (w/v) gellan gum (Gelzantm, Gelrite) in association with Ri-T transformed carrot roots following the protocol described by Bécard and Fortin (1988). Plates were incubated in the dark in an inverted position at 25 °C, and after several weeks, abundant mycelia and spores were produced. Spores and mycelia were extracted from M medium by solubilization of the gellan gum in 10 mM sodium acetate-citrate buffer (pH 6) (Doner and Becard 1991) and washed in sterile water. The resulting fungal material was further purified by hand under a binocular microscope to remove root fragments.
Spores and mycelium were suspended in 400 µL of the DNeasy Plant Mini Kit AP1 buffer (Qiagen) and crushed with a pestle in 1.5 ml micro tubes, and the DNA was purified according to the manufacturer’s recommendations. Purified DNA in a final elution volume of 40 µL was stored at −20 °C until use.
Long Polymerase Chain Reactions
The long polymerase chain reaction (PCR) protocol was performed using Takara LA Taq (Takara Bio, Canada) following the manufacturer’s recommendations in a volume of 50 µl containing 0.2 mM deoxynucleotide (dNTP) solution mix, 1.5 mM MgCl2, and 0.5 µM of each primer and 1 µl of template DNA. The primer pairs summarized in table 1 were used to validate the synteny in Glomus sp. DAOM-240422. Cycling parameters were 94 °C/3 min, followed by 38 cycles of: 94 °C/30 s, 54 °C/25 s, 68 °C/12 min, and a final elongation at 72 °C for 8 min. PCR products were separated by electrophoresis in a 1% (w/v) agarose gel and visualized with GelRed under ultraviolet light.
Table 1.
Long PCR Primers Used to Validate the Glomus sp. DAOM-240422 Mitochondrial Synteny
| mtDNA region | Primers | Sequences (5′–3′) | Size (bp) |
|---|---|---|---|
| atp9-cox1 | F | CTTGGCTCTATTCGCCTTAATGA | 5,029 |
| R | ACCAGGAAGAAGATCATAACGA | ||
| nad4L-rnl | F | TGATAGGATTGATGGGTTTCATAG | 13,094 |
| R | GAACATACTTAGCTTTGATGATGGT | ||
| cox3-rns | F | TACGGGTGTACAGCTCTATGAGT | 13,610 |
| R | TGGACTACGAGGGTATCTAATCCT | ||
| cox1 control | F | TGCTAAAGATATTGGGACTCTCT | 8,785 |
| R | GTAATCTGGTATTCTTCGAGGCA | ||
Sequencing, Assembly, and Gene Annotation
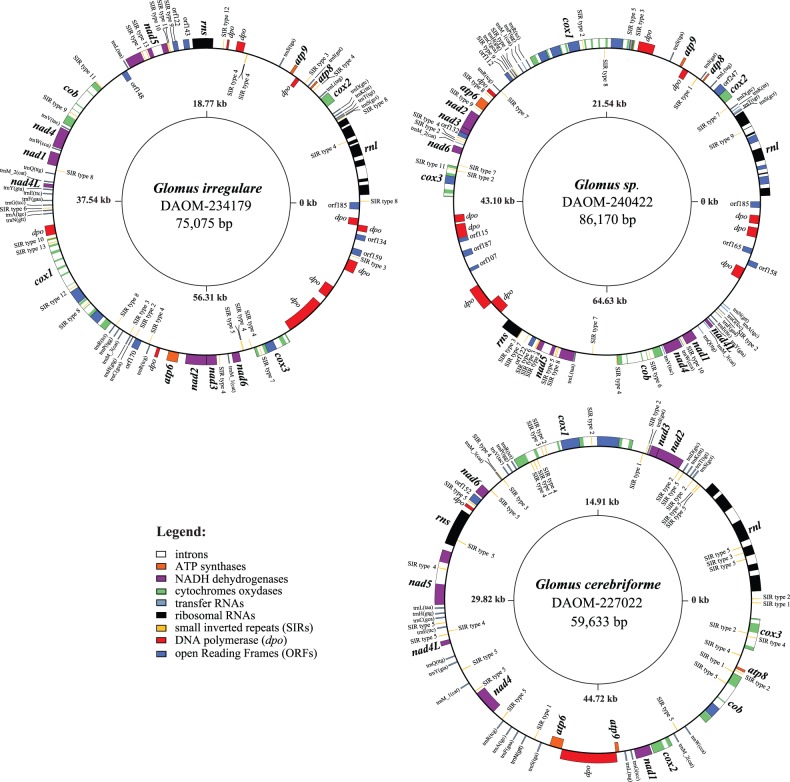
Glomus irregulare DAOM-234179, Glomus sp. DAOM-240422, and G. cerebriforme total DNA was sequenced using 454 Titanium FLX shotgun technology and the resulting reads were assembled in one mitochondrial contig using Newbler (Genome Quebec Innovation Center, McGill University, Montreal, Canada). Gene annotation was performed with the automated organellar annotation software MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl, last accessed August 23, 2013) (for more information on MFannot see Burger et al. [2013]), followed by manual inspection for the presence of frame shifts and the introduction of missing gene features. SIRs were also manually annotated using NCBI BLAST, and their secondary structure was predicted using the RNAfold web server (Gruber et al. 2008). The annotated sequences of the three new AMF mitochondrial genomes were deposited in GenBank under the accession numbers KC164354, KC164355, and KC164356 for G. irregulare DAOM-234179, Glomus sp. DAOM-240422, and G. cerebriforme, respectively. Circular and linear maps of the mitochondrial genomes (figs. 1 and 2) were created using OGDraw v.1.1 (Lohse et al. 2007).
Fig. 1.—
Comparison of Glomus irregulare DAOM-234179, Glomus sp. DAOM-240422, and G. cerebriforme mitochondrial genomes. The circular-mapping genomes were opened upstream of rnl to allow for easier comparisons. Genes on the outer and inner circumference are transcribed in a clockwise and counterclockwise direction, respectively. Gene and corresponding product names are atp6, 8, 9, ATP synthase subunit 6; cob, apocytochrome b; cox1–3, cytochrome c oxidase subunits; nad1–4, 4L, 5–6, NADH dehydrogenase subunits; rnl, rns, large and small subunit rRNAs; A-W, tRNAs, the letter corresponding to the amino acid specified by the particular tRNA followed by their anticodon. ORFs smaller than 100 amino acids are not shown.
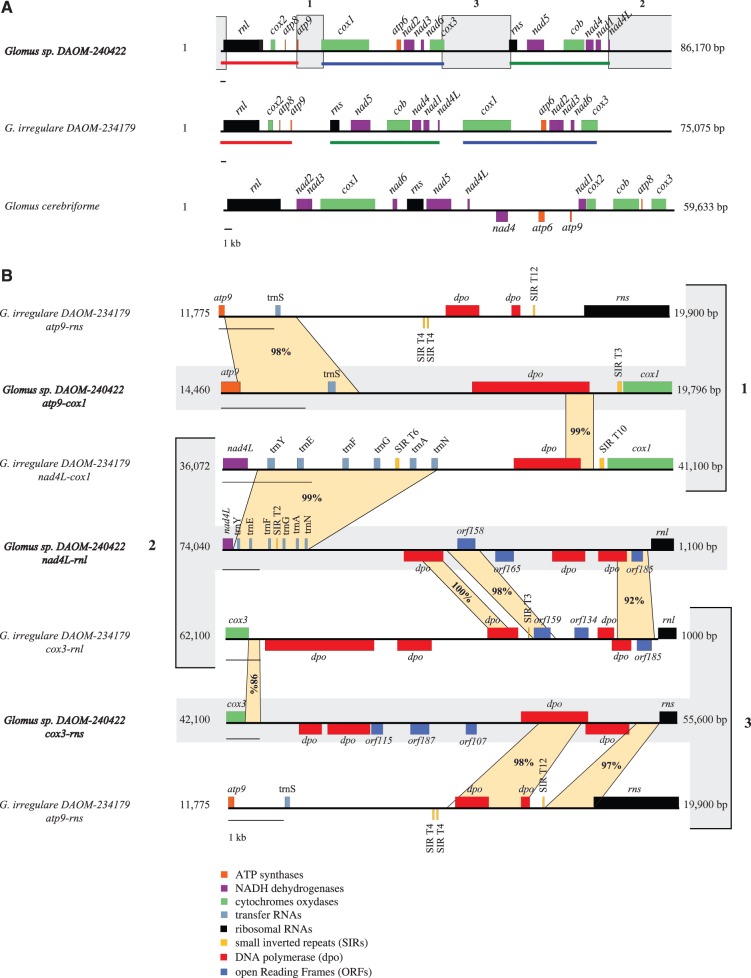
Fig. 2.—
(A) Linear genome representation to compare the mitochondrial synteny between Glomus irregulare DAOM-234179, Glomus sp. DAOM-240422, and G. cerebriforme. The linear-mapping genomes were opened upstream of rnl to allow for easier comparisons. Corresponding gene clusters between G. irregulare DAOM-234179 and Glomus sp. DAOM-240422 are underlined in red, green, and blue. The newly formed intergenic regions of Glomus sp. DAOM-240422 are boxed in gray and tagged with a number. (B) Nucleotide identity comparison with BLASTn between the reshuffled Glomus sp. DAOM-240422 intergenic regions (atp9-cox1 [box 1], nad4L-rnl [box 2], and cox3-rns [box 3]) with their putative homologous sequence in G. irregulare DAOM-234179. Homologous regions are indicated by the projections with their corresponding percent identity.
Network Analyses
Two similarity network analyses, where each node represents a dpo sequence and an edge represents a sequence similarity/identity were performed using EGN (Halary et al. 2013). The first network was constructed from a data set of 91 Glomeromycotan dpo sequences (from our collection and retrieved from GenBank). All of the sequences were compared with each other using BLASTn (Altschul et al. 1990) according to the following parameters: reciprocal best hit with a minimum of 1E−10 e-value, and 30% minimum identity covering at least 30% of the smallest sequence. The second and larger network includes an extensive sampling of all DNA polymerases from a broad range of taxa. This data set was built using translated Glomeromycota dpo sequences as queries for BLASTp searches on the Genbank nr database. After retrieving hits that showed a minimal e-value of 1E−05, we performed a second BLASTp search to be confident that we had covered the largest possible diversity of DNA polymerase gene sequences. The protein similarity network was calculated using BLASTp with a minimal e-value threshold of 1E−40, 20% minimal similarity covering at least 20% of the smallest sequence. The network layouts were further produced by Cytoscape software, using an edge-weighted force-directed model, meaning that genes sharing more DNA identity/protein similarity appear closer in the display.
Phylogenetic Analyses
Maximum likelihood phylogenetic analyses of AMF mitochondrial genes were performed with a data set of seven mitochondrial gene nucleotide sequences (atp6, atp9, cox2, nad1, nad2, nad4, and nad6) that were concatenated using DAMBE software version 5.2.13 (Xia and Xie 2001). The alignment was done using MUSCLE version 3.8.31. The analyses were performed using the GTR + G model (with five distinct gamma categories), with 1,000 bootstrap replicates. Bayesian analysis was performed using MrBayes version 3.2 with the GTR + G model (with five distinct gamma categories), four independent chains, one million cycles, tree sampling every 10 generations and a burn-in value of 40% (supplementary fig. S1, Supplementary Material online), which was adequate for Markov chain convergence.
The global dpo protein phylogeny (fig. 4B) was performed with a data set extracted from the connected component of the protein similarity network containing fungal and plant sequences. The MCODE Cytoscape plugin (Bader and Hogue 2003) was used to extract the most connected cluster, that is, the cluster of nodes which connect the most sequences to each other. This process allows a sampling in which sequences will share a maximum similarity, improving the downstream phylogenetic inference. The selected protein sequences were locally aligned with COBALT version 2.01 (Papadopoulos and Agarwala 2007). The maximum likelihood phylogenetic analysis was performed with RaxML using the rtREV model with the CAT option (PROTRTREVCAT) and 1,000 bootstrap replicates. This phylogenetic model is an amino acid substitution matrix that has been shown to be appropriate for the analyses of polymerase genes (Dimmic et al. 2002). Further, Bayesian inference was performed with MrBayes using the rtREV + CAT model, four independent chains, one million cycles, tree sampling every 10 generations, and a burn-in value of 40%, which was adequate for Markov chain convergence.
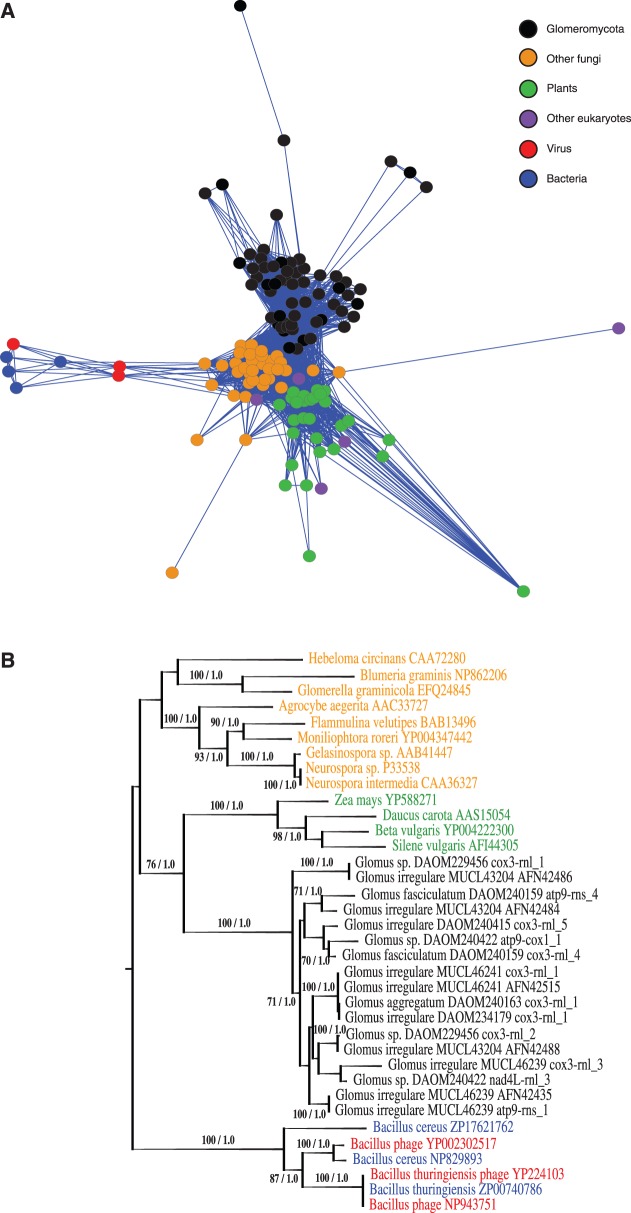
Fig. 4.—
(A) Sub-cluster of the global network of shared amino acid similarities between the Glomeromycota DPO proteins and all homologous sequences found on GenBank. Building this data set was performed using translated Glomeromycota sequences as queries for a BLASTp search on the GenBank nr database (minimal e-value threshold of 1E−40, 20% minimal similarity covering at least 20% of the smallest sequence). The network layouts were further produced by Cytoscape software, using an edge-weighted force-directed model, meaning that genes sharing more protein similarity appear closer in the display. There are 135 nodes in that network with 2,520 edges. (B) DPO protein maximum likelihood tree obtained with the rtREV + CAT phylogenetic model. The bacterial/viral cluster was used to root the tree. Number on branches indicates bootstrap support values (<60% cut-off) and Bayesian inference values, respectively. The Bayesian analyses gave a similar topology (data not shown). The tree includes AMF (black), other fungi (orange), plants (green), virus (red), and bacteria (blue).
The Glomeromycota dpo phylogeny (supplementary fig. S6, Supplementary Material online) was performed on a conserved domain of 75 amino acids that was present in 60 out of the 91 AMF dpo sequences. The sequence alignment was done with COBALT version 2.01. The maximum likelihood analysis was conducted with the rtREV + G model (with five distinct gamma categories), with 1,000 bootstrap replicates. Bayesian inference was performed with MrBayes using the rtREV + G model, four independent chains, one million cycles, tree sampling every 10 generations, and a burn-in value of 40%, which was adequate for Markov chain convergence.
All three phylogenetic analyses were done using the integrative software TOPALi version 2.5 (Milne et al. 2004). The best phylogenetic model was determined using TOPALi version 2.5 model selection test based on the Akaike information criterion. The tree figures were completed using TreeGraph version 2.0.47 (Stover and Muller 2010).
Recombination Analyses
The recombination analyses were performed on the Glomeromycota nucleotide identity network group 1, 2, and 8 (where the Glomus sp. 240422 dpo inserted in the reshuffled intergenic regions clustered). To easily detect the presence of distinct recombination signatures, a distance-based recombination analyses (supplementary fig. S7, Supplementary Material online) was achieved with the recombination analyses tool (RAT) (Etherington et al. 2005), with the low threshold set to 70% nucleotide identity and the high threshold set to over 90% identity. To further confirm the occurrence of the putative recombination events, we tested their statistical significance with a phylogenetic method using the Hidden Markov Model (HMM) with the F84 + gaps nucleotide substitution model (Husmeier and Wright 2001), performed with TOPALi v2.5 (default parameters).
Results
Mitochondrial Genome Description and Comparison
The complete mtDNA sequences of G. irregulare (DAOM-234179), Glomus sp. (DAOM-240422), and G. cerebriforme (DAOM-229022) are double-stranded circular DNA molecules, are homogeneous within isolate with no DNA sequence polymorphisms, that is, genetic segregation of mtDNA in these strains appears to be as effective as in other published AMF genome. These mtDNAs harbor the typical set of 41 mitochondrial genes reported so far in AMF (Lee and Young 2009): two rRNAs, 14 protein-coding genes (PCGs) and 25 tRNAs. The PCGs include three ATP synthetase (atp), one cytochrome b (cob), three cytochrome c oxydase (cox), and seven NADH dehydrogenase (nad) genes (fig. 1). The G. irregulare isolate DAOM-234179 mtDNA has a genome size of 75,075 bp with a GC content of 36.7%, which is within range of the sizes of other sequences reported for this species (68,995–87,754 bp with a ≈37% average GC content) (Lee and Young 2009; Formey et al. 2012; Nadimi et al. 2012). The only true outlying G. irregulare isolate with respect to its mitochondrial genome was reported by Formey et al. (2012) (i.e., MUCL43204) and has a size of 87,754 bp, whereas its conserved mitochondrial genes show only 65.8% identity (including introns and ORFs) with G. irregulare isolate 494. The Glomus sp. DAOM-240422 and G. cerebriforme mtDNAs were 86,170 and 59,633 bp, respectively, with GC contents of 36.7% and 46.7%. The latter is the smallest AMF mitochondrial genome yet reported, and is highly divergent when compared with the available mtDNAs of Glomus spp. and Gigaspora spp. The inflated GC% observed in G. cerebriforme is mostly due to reduced intergenic region size and the lack of mobile elements, which are mostly AT-rich regions, compared with the other strains. The difference in the mtDNA sequences observed between these taxa is caused by variation in intron content, dpo insertions and the presence of ORFs (table 2). Glomus cerebriforme genes are encoded on both strands, their order is completely reshuffled compared with the other Glomus spp., and they are also phylogenetically distant from those of G. irregulare (supplementary fig. S1, Supplementary Material online). On the other hand, Glomus sp. DAOM-240422 showed high similarity to G. irregulare with regards to coding sequence identity, but surprisingly, the synteny of these two isolates differed substantially.
Table 2.
AMF Mitochondrial Genome Features
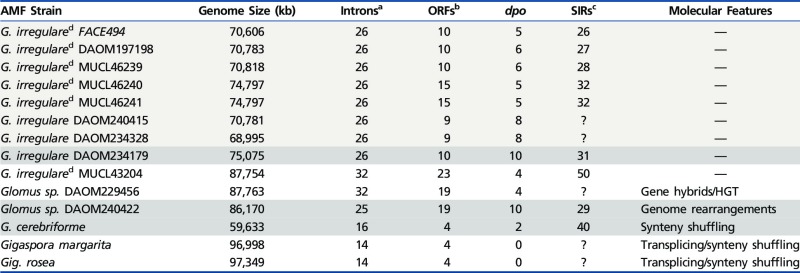 |
aAll types including group I and II introns.
bORFs greater than 100 amino acids encoding unknown protein, LAGLIDADG and GIYYIG endonuclease are listed, including intronic ORFs but not dpo fragments.
cSIRs reported so far in AMF mitochondrial genomes.
dBased on data reported in Formey et al. (2012), but eroded ORFs were excluded.
Plasmid-Related DNA Polymerase and Sequence Diversity of Small Inverted Repeats
We found 10 dpo insertions in G. irregulare DAOM-234179 and Glomus sp. DAOM-240422, making them the most dpo-rich AMF isolate reported so far, but only two copies were observed in G. cerebriforme. The three isolates harbor a huge bona fide dpo with ORF extensions, rendering a structure similar to the mitochondrial plasmid found in other fungi (Kim et al. 2000). However, we did not find TIRs, which are usually a characteristic of recent plasmid integration. We also found numerous SIRs in the three genomes. The 31 SIRs in G. irregulare DAOM-234179, 29 in Glomus sp. DAOM-240422, and 40 in G. cerebriforme were divided into 13, 11, and 5 distinct types, respectively, based on their hairpin secondary structure (supplementary figs. S3–S5, Supplementary Material online). The high number of SIR types, compared with the five previously described by Formey et al., result from the characterization of SIRs found in single copy, homologous to those previously described that could not have been detected otherwise. Some types are present in all G. irregulare isolates and closely related species such as Glomus sp. DAOM-240422, whereas others are endogenous to each strain, giving rise to a high SIRs sequence diversity. These elements were always present in intergenic regions, introns, or at the edge of endonuclease-encoding ORFs (supplementary tables S1–S3, Supplementary Material online), and suggest a close relationship with the homing mechanism for endonuclease integrations, as proposed by Formey et al. (2012). However, the diversity of possible cleaving sites seems to be greater than previously expected with the here-reported novel mtDNA sequences.
Divergent Synteny in Two Novel Glomeraceae Species mtDNA
The G. cerebriforme mtDNA shows a completely different gene order compared with the other AMF mitochondrial genomes sequenced so far, with many type 5 SIR insertions in intergenic regions that could potentially be involved in synteny rearrangements. Nevertheless, as mentioned earlier, only two dpo insertions are present, in the cox3-rnl and nad6-rns intergenic regions. As G. cerebriforme is phylogenetically distant from the publicly available mtDNAs of Glomus spp. and Gigaspora spp., no reference scaffold is available to confirm any gene order reorganization. In contrast, the comparison of Glomus sp. DAOM-240422 mitochondrial genome with its close relative G. irregulare DAOM-234179, revealed a reshuffling of two large gene clusters (fig. 2A). One gene cluster encompasses cox1, atp6, nad2, nad3, nad6, and cox3, whereas the other contains rns, nad5, cob, nad4, nad1, and nad4L. This rearrangement creates three novel intergenic regions in Glomus sp. DAOM-240422, which are atp9-cox1, cox3-rns, and nad4L-rnl. This result was confirmed by long PCR performed with primers spanning those regions (supplementary fig. S2, Supplementary Material online). We then assessed the sequence identity between those intergenes and their corresponding regions in G. irregulare DAOM-234179 (fig. 2B). We compared the Glomus sp. DAOM-240422 atp9-cox1 with G. irregulare DAOM-234179 atp9-rns and nad4L-cox1 intergene; the cox3-rns with G. irregulare DAOM-234179 cox3-rnl and atp9-rns regions; and finally the nad4L-rnl was compared with nad4L-cox1 and cox3-rnl. Each Glomus sp. DAOM-240422 intergenic region showed high sequence identity to its putative former counterpart in G. irregulare DAOM-234179, but covered only a small fraction of the intergene length, because many more ORFs (mostly dpo insertions) are present in Glomus sp. DAOM-240422. Local alignment using BLASTn, between the newly formed intergenic regions, did not reveal high similarity matches. We also constructed a sequence identity matrix of all the dpo sequences in its mtDNA and the highest match was only 53.1%.
Glomeromycota dpo Nucleotide Identity Network and Phylogenetic Analyses
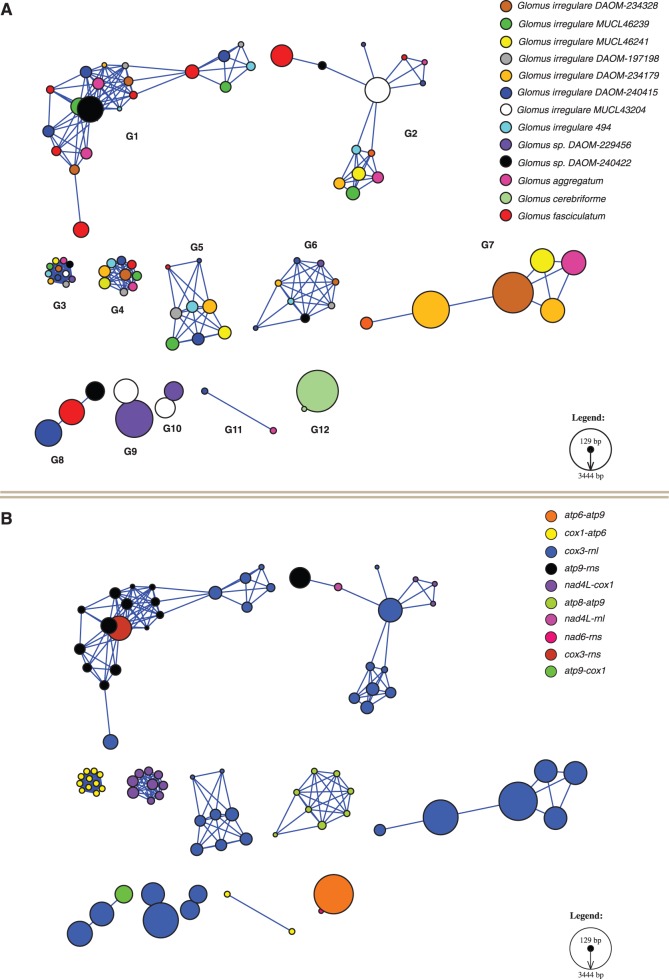
To test whether the dpo sequences in Glomus sp. DAOM-240422 were orthologous with the sequences found in other AMF and to assess evolution of dpo within the Glomeromycota, we constructed a nucleotide similarity network of all dpo genes reported so far in AMF mitochondrial genomes (fig. 3A). In total, 12 similarity groups were formed and seven singletons were left aside. We observed that five Glomus sp. DAOM-240422 dpo sequences (black circles) clustered in distinct identity groups (G1, G2, G3, G6, and G8), whereas the rest were singletons inserted in reshuffled intergenic regions (cox3-rnl and nad4L-rnl), with low sequence similarity to each other or to known AMF dpo sequences. The Gig. rosea putative mitochondrial plasmid that was previously described (Nadimi et al. 2012) was also one of the singletons. The two G. cerebriforme dpo copies (light green circles) were the sole representatives of similarity group 12. Also, the mtDNA-integrated dpo showed a clustering tendency toward their intergenic region localization (fig. 3B). Interestingly, three Glomus sp. DAOM-240422 dpo sequences which are inserted in the reshuffled intergenic regions atp9-cox1, cox3-rns, and nad4L-rnl (fig. 2) clustered, respectively, with the AMF dpo group 8 (cox3-rnl), group 1 (atp9-rns), and group 2 (cox3-rnl and the Glomus fasciculatum atp9-rns_3 sequence).
Fig. 3.—
Similarity network of dpo sequences inserted in AMF mitochondrial genomes. (A) Each node represents a dpo insert colored by mitochondrial genome. The node size is proportional to the sequence length. Two nodes are connected by an edge when they share significant similarity (reciprocal best BLAST hit with a minimum of 1E−10 score, and 30% minimum identity covering at least 30% of the smallest sequence). The layout was produced by Cytoscape, using an edge-weighted force-directed model, which brings closer sequences sharing more similarity. Twelve homology groups were formed (labeled G1–G12). There are 97 nodes on that network with 336 edges. (B) Same network but nodes are colored by their mitochondrial intergenic region localization. The legend shows a linear size relationship between the smallest dpo sequence (Glomus irregulare DAOM-240415_cox3-rnl_4, 129 bp) and the largest one (G. cerebriforme atp6-atp9, 3444 bp).
To compare the identity network and the phylogenetic approach, and to assess the relationship of the dpo present within each respective mtDNA (i.e., paralogy vs. orthology), we constructed a phylogeny based on a conserved protein domain. The alignment of all 91 Glomeromycota dpo protein sequences pointed out to a 75 amino acid region that was common in 60 of the sequences (supplementary fig. S6A, Supplementary Material online). Interestingly, most of the dpo were truncated either upstream and/or downstream of that conserved domain. The resulting maximum likelihood phylogenetic tree showed robust bootstrap values (>75%) in the distal branches that support clades corresponding to the similarity groups observed in the gene network, whereas the basal branches displayed were weakly supported (supplementary fig. S6B, Supplementary Material online). All similarity network groups were represented in the tree with strong statistical support in bootstrap and Bayesian inference values, except for the identity group 11 that was not represented, because the sequences did not harbor the conserved gene core. Surprisingly, this tree revealed only one occurrence of paralogy, in the group 12, for the two dpo sequences inserted in G. cerebriforme mtDNA (like it was also observed in the nucleotide network). Every other dpo phylogenetic clusters represent orthology groups. Finally, only one of the three Glomus sp. DAOM-240422 dpo sequences found in reshuffled intergenic regions was present in the phylogenetic analyses, in the group 8 cluster.
Glomus sp. DAOM-240422 dpo Recombination Analyses
Three Glomus sp. dpo genes present in the reshuffled intergenic regions clustered with dpo of other AMF taxa inserted in different mtDNA localization, rather than with paralogous mtDNA sequences (within DAOM-240422 isolate) in the nucleotide network. To easily screen for homologous recombination, we performed a distance-based recombination analyses in which these Glomus sp. dpo sequences were compared with their three closest relatives in the nucleotide identity network. For the Glomus sp. atp9-cox1 and cox3-rns (supplementary fig. S7A and B, Supplementary Material online) comparison, we observed a disruption in the identity of the dpo sequences going from 70% to 80% identity and spiking abruptly to a near perfect 100% nucleotide identity. When compared together, the Glomus sp. nad4L-rnl_1 dpo sequence started off with less than 80% identity to the G. fasciculatum atp9-rns_3 dpo and then rose sharply to almost 95% identity. Conversely, the G. irregulare MUCL43204_cox3-rnl_1 dpo started at more than 99% similar to the Glomus sp. nad4L-rnl_1 dpo sequence, but then dropped to less than 80% identity (supplementary fig. S7C, Supplementary Material online). These dpo sequences showed typical signatures of homologous recombinants regarding their sequence identity, highly reminiscent to what was found for a recombinant HIV-1 strain (Liitsola et al. 2000).
To further confirm these findings, we performed a statistical phylogenetic recombination analysis using the HMM. The Glomus sp. DAOM-240422 atp9-cox1 showed a perfect Bayesian inference value of 1.0 for each three topologies in a given portion of the alignment (supplementary fig. S7A, Supplementary Material online), which allowed the finding of putative recombination break points (between two given topologies where one reach a Bayesian inference value >0.95 and the other <0.05) at nucleotide position 357 and 613 bp of the sequence, respectively. The Glomus sp. DAOM-240422 nad4L-rnl also showed a perfect Bayesian inference value of 1.0, but for only two topologies (I and II), thus allowing for the determination of one putative break point at position 161 bp (supplementary fig. 7C, Supplementary Material online). Conversely, the Glomus sp. DAOM-240422 cox3-rns analyses did not provide statistical support for any given topologies tested with the HMM phylogenetic method (supplementary fig. 7B, Supplementary Material online).
Global Protein Similarity Network with All Known DNA Polymerase Genes in the Public Database
To assess a possible origin and transmission mode of AMF mitochondrial dpo sequences, we constructed a protein similarity network using a broad spectrum of dpo sequences. Interestingly, most of the glomeromycotan plasmid-related DNA polymerase proteins, 60 in total, were found clustered with 35 other fungi, 28 plants (including two red algae of the genus Porphyra), five other eukaryotes (Physarum polycephalum, Ochromonas danica, two Placozoan sp. and a Cnidaria species), four bacteria (Bacillus thuringiensis, two Bacillus cereus, and Brevibacillus sp.), and three bacteriophages (fig. 4A). All dpo sequences are present within their respective mtDNA or are located in a mitochondrial plasmid. The patchy distribution observed among phylogenetically distant taxa such as plants and fungi is similar to that of a cox1 intron that invaded plant mitochondria and was thought to have been acquired horizontally from a fungal donor (Cho et al. 1998). To assess the evolutionary relationship of AMF DPO proteins with the other taxa present in the cluster, we selected the group gathering the most connected nodes, that is, the sequences sharing the most similarity all together. The corresponding sequences (including plant, Glomeromycotan and other fungal sequences), were used to perform a maximum likelihood phylogenetic analyses (fig. 4B). We used the viral and bacterial proteins present in the sub-network to root the tree, because they present a distal position in the network. The resulting tree showed robust support (100% bootstrap and 1.0 Bayesian inference value) for the viral-bacterial group, and surprisingly, the plant DPO proteins clustered with those of the Glomeromycota as a sister group, supported by a 76% bootstrap and 1.0 Bayesian inference value. Except for the other fungi group clade, the phylogeny was robustly supported either for the basal and the distal branches.
Discussion
Interhaplotype Homologous Recombination in Glomus sp. DAOM-240422
The numerous type 5 SIRs present in G. cerebriforme intergenic regions may have had the potential to trigger intra-molecular gene reshuffling. The two G. cerebriforme dpo sequences were shown to be paralogs, with a nucleotide identity of 81%. However, we cannot be sure that intra-molecular recombination occurred in G. cerebriforme, because we do not have closely related taxa as comparators. In the Glomus sp. DAOM-240422 mitochondrial genome, the two most similar dpo sequences share a low nucleotide identity percentage of 53.1%. However, we observe a near perfect sequence identity for PCGs and some intergenic regions with close relative G. irregulare. Taken together, these observations suggest that the rearrangements observed in Glomus sp. DAOM-240422 are recent and do not result from intramolecular recombination from an ancient dpo duplication. Further, no recent dpo duplications were observed in any given AMF strains surveyed. The Glomus sp. DAOM-240422 dpo sequences that are inserted in the reshuffled intergenic regions clustered in the nucleotide network with dpo sequences of other AMF species that were inserted in the putatively former intergenic arrangement. Comparison of the Glomus sp. DAOM-240422 atp9-cox1 and nad4L-rnl_1 dpo, with their three closest relatives showed a typical signature of homologous recombination (supplementary fig. S7A and C, Supplementary Material online), with genes sharing a high sequence identity with different portions of the alignment, where intersections represent putative recombination break points. These recombination events were strongly statistically supported with the HMM method. Even if the Glomus sp. 240422 cox3-rns dpo recombination was not supported by HMM, it still clustered with dpo present in other intergenic regions, and the lack of statistical support might be explained by the absence of its recombination partner in the cluster. These observations strongly suggest that the mitochondrial gene cluster reorganization in Glomus sp. DAOM-240422 is the result of dpo-mediated homologous recombination between different mitochondrial haplotypes harboring highly similar dpo orthologs present in distinct genome localization.
Two types of mobile elements reported in AMF mitochondrial genomes have been previously shown to be involved in genome rearrangements through homologous recombination in other organisms: the plasmid-related dpo genes in Basidiomycota (Ferandon et al. 2008) and SIRs in bacteria and plants (Schofield et al. 1992; Bi and Liu 1996; Tanaka et al. 2012). Furthermore, the fact that the number of dpo sequences varies in different intergenic regions among isolates of the same species and closely related ones (Formey et al. 2012; Beaudet et al. 2013), combined with our findings suggest that 1) there is no evidence of intra-molecular recombination in Glomus sp. DAOM-240422; 2) recombination between distinct mtDNA haplotypes harboring different dpo insertions could occur; 3) as there is no intra-isolate mtDNA polymorphism, the reshuffled mtDNA haplotype in Glomus sp. DAOM-240422 is fixed in the population; and 4) those rearrangements are triggered by dpo insertions through homologous recombination.
Such recombination events could take place following hyphal fusion (i.e., anastomosis). This last feature not only facilitates the distribution of nutrients and signaling molecules through the entire AMF mycelium, but also plays a key role in genetic exchange (Giovannetti et al. 1999) and segregation (Angelard and Sanders 2011). Those anastomoses can occur either between genetically different AMF isolates originating from the same experimental field (Croll et al. 2009; Angelard et al. 2010) or between strains isolated from distant locations (e.g., from the same [Purin and Morton 2012], or even different continents [de la Providencia et al. 2013]). Both of these studies suggested that mitochondria might also be exchanged via anastomosis, thus creating a heteroplasmic state.
Evidence of Heteroplasmy Challenges the Concept of an AMF Individual
All published AMF mitochondrial genomes are homoplasmic, that is, homogeneous within isolates. In other fungi, mechanisms contributing to homoplasmy are often related to the recombination of parental mtDNAs or by the selection of one of the parental haplotypes due to the presence of segregation mechanisms (reviewed by Birky [2001]). In the absence of external factors, heteroplasmy should be the default state for mtDNA under a simple mutation-drift scenario (White et al. 2008). In yeasts, the presence of a heteroplasmic state was shown to be transient due to the presence of segregation mechanisms, such as nucleoid formation, and mtDNA molecular repair pathways which maintain mitochondrial genome integrity (Zimmer et al. 1991; Hu et al. 1995; Yasuhira and Yasui 2000). The presence of nucleoids, similar mtDNAs linked by Holliday junctions, induces a genetic bottleneck responsible for a fast segregation (Lockshon et al. 1995; White and Lilley 1997; MacAlpine et al. 1998). The mitochondrial genome homogeneity observed for in vitro and in vivo AMF cultures could be the result of such effective mtDNA segregation and repair mechanisms that would take place during the subcultivation processes.
The AMF colonies are characterized by the interconnectedness of its different parts by means of hyphal fusion (i.e., anastomosis) (Giovannetti et al. 1999; de la Providencia et al. 2005; Voets et al. 2006; Purin and Morton 2012), which can lead to genetic exchange, even between genetically distinct individuals. This hyphal fusion dynamic in natural populations can also prevent the lost of genetic diversity (Bever and Wang 2005). Recently, anastomoses between geographically distant G. irregulare isolates cultivated in vitro were shown to induce transient mitochondrial length heteroplasmy (with variation in dpo insertions between the two mitochondrial haplotypes) (de la Providencia et al. 2013). Although in situ heteroplasmy have never been demonstrated, these observations, in addition to the interhaplotype dpo homologous recombination evidences presented here, suggest that transient heteroplasmy could occur in the field.
The coexistence of numerous AMF mitochondrial haplotypes in the same cytoplasm and the occurrence of homologous mitochondrial recombination might be commonplace in nature, as it was shown in natural populations of the basidiomycete fungus Armillaria gallica (Saville et al. 1998). The heteroplasmy stability in natural populations, may provide useful additional information for defining haplotypes, and resolving further the relationships among individuals at a population level (White et al. 2008). Given the increasing use of mitochondrial markers in AMF research, the demonstration that genetically distinct mtDNAs can be exchanged through anastomosis, coexist in a common cytoplasm and give rise to a recombinant haplotype, should lead to a thorough discussion on the AMF individuality concept. If heteroplasmy is confirmed in natural populations, the use of mtDNA as a criterion to define a reliable AMF taxonomic unit would certainly facilitate our understanding of the processes occurring in natural populations, although caution should still be applied because it would be artificial, and would not reflect the genotypic diversity of AMF nuclear DNA, because mitochondrial and nuclear DNA segregation are mostly stochastic mechanisms.
Vertical Versus Horizontal Inheritance of the AMF Mitochondrial dpo
Given the high genetic diversity of dpo and their variable truncation patterns, a classical phylogenetic approach requiring multiple alignment of highly similar sequences, is not suitable to analyze their overall evolutionary relationships, especially for recombinant ones (Bapteste et al. 2012). Indeed, the classical approach only allowed us to analyze 60 of 91 Glomeromycota dpo sequences, on a limited 75 amino acids conserved domain, leading to a tree with poorly supported basal branches (supplementary fig. 6B, Supplementary Material online). As an alternative, gene similarity network analyses have already been proven to be useful in the study of highly diverse gene families and introgressive descent events, such as recombination and HGT (Bapteste et al. 2012).
The AMF dpo nucleotide network highlights the existence of a clustering tendency for dpo sequences located in the same intergenic regions (fig. 3B). The high similarity (short edges) observed between the sequences of each group and the phylogenetic topology (supplementary fig. S6B, Supplementary Material online) suggest that they are orthologs that could have recently been acquired vertically from a common ancestor. This is not surprising for the G. irregulare isolates, but implies that the other species that shared those sequences have also diverged recently (i.e., G. fasciculatum and G. aggregatum) given the high within-group sequence similarity. However, the seven singletons left aside by the network nucleotide similarity clustering (five in the closely related species Glomus sp. DAOM-240422) suggest that numerous independent plasmid-mediated dpo insertions, from unknown origin, and/or followed by a fast divergence might also have occurred. The putative Gig. rosea plasmid had low sequence similarity with the other dpo insertions in AMF mtDNA and no extra chromosomal dpo were found within any of the surveyed AMF genomes. This dpo sequence diversity could indicate that AMF mitochondria harbor a broad range of plasmid types, as described in Neurospora species, where seven circular and four linear homology groups of mitochondrial plasmids have been reported (Schulte and Lambowitz 1991; Yang and Griffiths 1993a, 1993b; Arganoza et al. 1994; Marcinko-Kuehn et al. 1994).
It has been found that mitochondrial plasmids in plants could be horizontally acquired through pollen (Handa 2007). HGT has also been hypothesized to be an important factor in the dispersal of mitochondrial plasmids between fungal strains and species. That assumption is based on 1) discordant phylogenetic relationships between plasmids and host genomes (Kempken et al. 1992), 2) the geographic distribution of plasmids within and among host species (Arganoza et al. 1994), and 3) direct experimental demonstration of HGT (Collins and Saville 1990; Griffiths et al. 1990; Debets et al. 1994). Until now, no direct experimental evidence of HGT of dpo sequences was provided, and no mitochondrial plasmid has yet been found in AMF (except a putative linear plasmid in Gig. rosea). However, the distribution of the plasmid-related dpo sequences is uneven between isolates of the same AMF species (Formey et al. 2012) and we showed that there is a large diversity of dpo sequences within AMF (12 orthology groups, along with seven singletons). Another intriguing observation is that the dpo sequences are also present in all AMF family surveyed including the Glomeraceae, Gigasporaceae, Ambisporaceae, Diversisporaceae, and Archaeosporaceae (data not shown, based on a preliminary 454 assembly). A mechanism known as post-fusion incompatibility might be the most plausible explanation for the ubiquitous distribution of AMF mtDNA dpo. This mechanism implies a brief hyphal fusion between incompatible AMF species and the formation of septa. This process has already been shown to allow exchange of nuclear genetic material (Croll et al. 2009), and also offers an explanation for the mobile endonuclease-mediated HGT, leading to the formation of gene hybrids in the mitochondrial genome of Glomus diaphanum-like species (Beaudet et al. 2013). Keeping in perspective that divergent AMF taxa can simultaneously colonize roots of the same plant species, it is likely that a mitochondrial plasmid could spread rapidly through the Glomeromycota phylum even between phylogenetically distant taxa.
Glomeromycota dpo Are Closely Related to Plant Mitochondrial Plasmids
More than 50 linear plasmids have been reported in the mitochondria of 20 fungal species, in contrast to the 14 found in eight plant species. These have only been observed twice in animal mtDNA, in the placozoan Trichoplax adhaerens and in the moon jelly Aurelia aurita (Shao et al. 2006; Signorovitch et al. 2007). This uneven distribution of mitochondrial plasmids within the Eukaryota raises questions about their origin and their mode of transmission.
The structure of the plant mitochondrial plasmids consists of an invertron structure, long inverted repeats with proteins covalently bound to their 5’-ends. This structure is remarkably similar to fungal plasmids and to some DNA viruses (Rosewich and Kistler 2000). Additional evidence, such as low GC content in plant plasmids (30% in the Brassica 11.6 kb plasmid, 38.9% in the 10.4 kb sugar beet plasmid, and 37–39% in the maize S plasmids) compared with their respective mtDNA (Brassica napus 45.2% [Handa 2003], Beta vulgaris 43.9% [Kubo et al. 2000]) and the maize 44.0% (GenBank accession DQ490951), supports HGT from a fungal donor as the source of linear mitochondrial plasmids in plants, because low GC content is typically found in fungal mtDNA (Handa 2008) (with an average of 37% in G. irregulare isolates [Handa 2008; Formey et al. 2012]). Moreover, our phylogenetic analyses robustly support the DPO proteins of plants and Glomeromycota as a sister group. Taken together, these evidences suggest that AMF could be potential donors of plasmid-related dpo to plants. However, species of Glomeromycota colonize the same ecological niche and often co-occur with many pathogenic or endophytic fungi in plant roots, and one of these co-occurring fungi could be the common donor to both plants and AMF, also plant plasmids could have different fungal origins. If this is the case, the organism in question has yet to be sequenced.
To further study the AMF mitochondrial plasmid-related dpo genes, the mitochondrial genome of Geosiphon pyriformis, an ancestral member of Glomeromycota that lives in endocytobiotic association with the cyanobacterium Nostoc punctiforme (Gehrig et al. 1996), could provide insights into the origin and evolution of those elements in that phylum. Also, some DNA polymerase genes were present in the mtDNA of the ascomycete fungal partner of the lichen Peltigera malacea (Xavier et al. 2012). Those sequences clustered within the other fungi group in the global DPO protein network. Another interesting avenue would be to assess the horizontal transmission of mitochondrial plasmids in different fungi–plant symbiotic systems.
Conclusions and Outlook
The mitochondrial genome comparison of the three Glomus species showed the extent of mtDNA plasticity with regards to their synteny. It also offers a basis to develop molecular tool kits aimed specifically at identifying and quantifying taxa of the ecologically and agriculturally important Glomeraceae family. However, finding evidence of interhaplotype homologous recombination suggests the occurrence of mtDNA heteroplasmy in natural populations. Although this has yet to be demonstrated, it raises questions about the artificiality of single spore cultivations of these fungi, which probably induces an underestimation of the mtDNA diversity in situ. Approaches like genotyping-by-sequencing and ecotilling could be useful to assess this particular hypothesis. Mitochondrial genome comparisons can also provide insights into molecular processes related to the biology and community structure of AMF. The close phylogenetic relationship between the plasmid-related dpo of plants and Glomeromycota suggests that HGT events could have occurred between plants and their fungal symbionts. The intimate relationship between plants and AMF in nature may unravel other transfer events that have taken place through evolution. Also, the role of viruses and bacteria and their interactions with fungi and plants represents an interesting research avenue to generate knowledge about the evolution and origin of mitochondrial plasmids in those groups.
Supplementary Material
Supplementary tables S1–S3 and figures S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Biopterre centre du développement des bioproduits and CRBM. They thank Dr B.F. Lang for bioinformatics assistance and access to an automated organelle genome annotation software, Dr Simon Joly for recommendations on the phylogenetic analyses, Stéphanie Berthiaume for her assistance in long PCRs, and Dr Terrence Bell and Karen Fisher Favret for English editing and comments on the manuscript. They also thank two anonymous reviewers for helpful comments. This work is a part of a research project organized and coordinated by Premier Tech. This work was supported by the NSERC Cooperative Research and Development grant (RDCPJ 395241-09), Premier Tech, and CRIBIQ.
Literature Cited
- Akins RA, et al. Nucleotide sequence of the Varkud mitochondrial plasmid of Neurospora and synthesis of a hybrid transcript with a 5′ leader derived from mitochondrial RNA. J Mol Biol. 1988;204:1–25. doi: 10.1016/0022-2836(88)90594-3. [DOI] [PubMed] [Google Scholar]
- Akins RA, Kelley RL, Lambowitz AM. Mitochondrial plasmids of Neurospora: integration into mitochondrial DNA and evidence for reverse transcription in mitochondria. Cell. 1986;47:505–516. doi: 10.1016/0092-8674(86)90615-x. [DOI] [PubMed] [Google Scholar]
- Almasan A, Mishra NC. Recombination by sequence repeats with formation of suppressive or residual mitochondrial DNA in Neurospora. Proc Natl Acad Sci U S A. 1991;88:7684–7688. doi: 10.1073/pnas.88.17.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- André C, Levy A, Walbot V. Small repeated sequences and the structure of plant mitochondrial genomes. Trends Genet. 1992;8:128–132. doi: 10.1016/0168-9525(92)90370-J. [DOI] [PubMed] [Google Scholar]
- Angelard C, Colard A, Niculita-Hirzel H, Croll D, Sanders IR. Segregation in a mycorrhizal fungus alters rice growth and symbiosis-specific gene transcription. Curr Biol. 2010;20:1216–1221. doi: 10.1016/j.cub.2010.05.031. [DOI] [PubMed] [Google Scholar]
- Angelard C, Sanders IR. Effect of segregation and genetic exchange on arbuscular mycorrhizal fungi in colonization of roots. New Phytol. 2011;189:652–657. doi: 10.1111/j.1469-8137.2010.03602.x. [DOI] [PubMed] [Google Scholar]
- Arganoza MT, Min J, Hu Z, Akins RA. Distribution of seven homology groups of mitochondrial plasmids in Neurospora: evidence for widespread mobility between species in nature. Curr Genet. 1994;26:62–73. doi: 10.1007/BF00326306. [DOI] [PubMed] [Google Scholar]
- Bader G, Hogue C. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- Bapteste E, et al. Evolutionary analyses of non-genealogical bonds produced by introgressive descent. Proc Natl Acad Sci U S A. 2012;109:18266–18272. doi: 10.1073/pnas.1206541109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr CM, Neiman M, Taylor DR. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005;168:39–50. doi: 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- Beaudet D, Nadimi M, Iffis B, Hijri M. Rapid mitochondrial genome evolution through invasion of mobile elements in two closely related species of arbuscular mycorrhizal fungi. PLoS One. 2013;8:e60768. doi: 10.1371/journal.pone.0060768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard G, Fortin JA. Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 1988;108:211–218. doi: 10.1111/j.1469-8137.1988.tb03698.x. [DOI] [PubMed] [Google Scholar]
- Bever JD, Wang M. Arbuscular mycorrhizal fungi: hyphal fusion and multigenomic structure. Nature. 2005;433:E3–E4. doi: 10.1038/nature03294. [DOI] [PubMed] [Google Scholar]
- Bi X, Liu LF. DNA rearrangement mediated by inverted repeats. Proc Natl Acad Sci U S A. 1996;93:819–823. doi: 10.1073/pnas.93.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- Birky CW, Strausberg RL, Forster JL, Perlman PS. Vegetative segregation of mitochondria in yeast: estimating parameters using a random model. Mol Gen Genet. 1978;158:251–261. [Google Scholar]
- Boer PH, Gray MW. Short dispersed repeats localized in spacer regions of Chlamydomonas reinhardtii mitochondrial DNA. Curr Genet. 1991;19:309–312. doi: 10.1007/BF00355060. [DOI] [PubMed] [Google Scholar]
- Boon E, Zimmerman E, Lang BF, Hijri M. Intra-isolate genome variation in arbuscular mycorrhizal fungi persists in the transcriptome. J Evol Biol. 2010;23:1519–1527. doi: 10.1111/j.1420-9101.2010.02019.x. [DOI] [PubMed] [Google Scholar]
- Börstler B, Raab PA, Thiéry O, Morton JB, Redecker D. Genetic diversity of the arbuscular mycorrhizal fungus Glomus intraradices as determined by mitochondrial large subunit rRNA gene sequences is considerably higher than previously expected. New Phytol. 2008;180:452–465. doi: 10.1111/j.1469-8137.2008.02574.x. [DOI] [PubMed] [Google Scholar]
- Boursot P, Yonekawa H, Bonhomme F. Heteroplasmy in mice with deletion of a large coding region of mitochondrial DNA. Mol Biol Evol. 1987;4:46–55. doi: 10.1093/oxfordjournals.molbev.a040421. [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout Jakobid protists. Genome Biol Evol. 2013;5:418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Qiu Y-L, Kuhlman P, Palmer JD. Explosive invasion of plant mitochondria by a group I intron. Proc Natl Acad Sci U S A. 1998;95:14244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RA, Saville BJ. Independent transfer of mitochondrial chromosomes and plasmids during unstable vegetative fusion in Neurospora. Nature. 1990;345:177–179. doi: 10.1038/345177a0. [DOI] [PubMed] [Google Scholar]
- Corradi N, Bonen L. Mitochondrial genome invaders: an unselfish role as molecular markers. New Phytol. 2012;196:963–965. doi: 10.1111/j.1469-8137.2012.04354.x. [DOI] [PubMed] [Google Scholar]
- Court DA, Bertrand H. Genetic organization and structural features of maranhar; a senescence-inducing linear mitochondrial plasmid of Neurospora crassa. Curr Genet. 1992;22:385–397. doi: 10.1007/BF00352440. [DOI] [PubMed] [Google Scholar]
- Croll D, et al. Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2009;181:924–937. doi: 10.1111/j.1469-8137.2008.02726.x. [DOI] [PubMed] [Google Scholar]
- de la Providencia IE, Nadimi M, Beaudet D, Rodriguez Morales G, Hijri M. Detection of a transient mitochondrial DNA heteroplasmy in the progeny of crossed genetically divergent isolates of arbuscular mycorrhizal fungi. New Phytol. 2013 doi: 10.1111/nph.12372. Advance Access published June 24, 2013, doi: 10.1111/nph.12372. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M, Bernardi G. The GC clusters of the mitochondrial genome of yeast and their evolutionary origin. Gene. 1986;41:1–22. doi: 10.1016/0378-1119(86)90262-3. [DOI] [PubMed] [Google Scholar]
- Debets F, Yang X, Griffiths AF. Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet. 1994;26:113–119. doi: 10.1007/BF00313797. [DOI] [PubMed] [Google Scholar]
- Denovan-Wright EM, Lee RW. Comparative structure and genomic organization of the discontinuous mitochondrial ribosomal RNA genes of Chlamydomonas eugametos and Chlamydomonas reinhardtii. J Mol Biol. 1994;241:298–311. doi: 10.1006/jmbi.1994.1505. [DOI] [PubMed] [Google Scholar]
- Dimmic MW, Rest JS, Mindell DP, Goldstein RA. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J Mol Evol. 2002;55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- Doner L, Becard G. Solubilization of gellan gels by chelation of cations. Biotechnol Tech. 1991;5:25–28. [Google Scholar]
- Etherington GJ, Dicks J, Roberts IN. Recombination analysis tool (RAT): a program for the high-throughput detection of recombination. Bioinformatics. 2005;21:278–281. doi: 10.1093/bioinformatics/bth500. [DOI] [PubMed] [Google Scholar]
- Evelin H, Kapoor R, Giri B. Arbuscular mycorrhizal fungi in alleviation of salt stress: a review. Ann Bot. 2009;104:1263–1280. doi: 10.1093/aob/mcp251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferandon C, Chatel SEK, Castandet B, Castroviejo M, Barroso G. The Agrocybe aegerita mitochondrial genome contains two inverted repeats of the nad4 gene arisen by duplication on both sides of a linear plasmid integration site. Fungal Genet Biol. 2008;45:292–301. doi: 10.1016/j.fgb.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Formey D, et al. Comparative analysis of mitochondrial genomes of Rhizophagus irregularis—syn. Glomus irregulare—reveals a polymorphism induced by variability generating elements. New Phytol. 2012;196:1217–1227. doi: 10.1111/j.1469-8137.2012.04283.x. [DOI] [PubMed] [Google Scholar]
- Gehrig H, Schüßler A, Kluge M. Geosiphon pyriforme, a fungus forming endocytobiosis withNostoc (Cyanobacteria), is an ancestral member of the glomales: evidence by SSU rRNAnalysis A. J Mol Evol. 1996;43:71–81. doi: 10.1007/BF02352301. [DOI] [PubMed] [Google Scholar]
- Gianinazzi S, et al. Agroecology: the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza. 2010;20:519–530. doi: 10.1007/s00572-010-0333-3. [DOI] [PubMed] [Google Scholar]
- Giovannetti M, Azzolini D, Citernesi AS. Anastomosis formation and nuclear and protoplasmic exchange in arbuscular mycorrhizal fungi. Appl Environ Microbiol. 1999;65:5571–5575. doi: 10.1128/aem.65.12.5571-5575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths AF, et al. Heterokaryotic transmission of senescence plasmid DNA in Neurospora. Curr Genet. 1990;17:139–145. [Google Scholar]
- Griffiths AJ. Natural plasmids of filamentous fungi. Microbiol Rev. 1995;59:673–685. doi: 10.1128/mr.59.4.673-685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths AJF. Fungal senescence. Annu Rev Genet. 1992;26:351–372. doi: 10.1146/annurev.ge.26.120192.002031. [DOI] [PubMed] [Google Scholar]
- Grindley NDF, Reed RR. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNWebsuite A. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halary S, Mclnerney JO, Lopez P, Bapteste E. EGN: a wizard for construction of gene and genome similarity networks. BMC Evol Biol. 2013;13:146. doi: 10.1186/1471-2148-13-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa H. Investigation of the origin and transmission of linear mitochondrial plasmid based on phylogenetic analysis in Japanese rapeseed varieties. Genome. 2007;50:234–240. doi: 10.1139/g06-150. [DOI] [PubMed] [Google Scholar]
- Handa H. Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions? Mitochondrion. 2008;8:15–25. doi: 10.1016/j.mito.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Hänfler J, et al. Circular extrachromosomal DNA codes for a surface protein in the (+) mating type of the zygomycete Absidia glauca. Curr Genet. 1992;22:319–325. doi: 10.1007/BF00317929. [DOI] [PubMed] [Google Scholar]
- Hermanns J, Asseburg A, Osiewacz HD. Evidence for a life span-prolonging effect of a linear plasmid in a longevity mutant of Podospora anserina. Mol Gen Genet. 1994;243:297–307. doi: 10.1007/BF00301065. [DOI] [PubMed] [Google Scholar]
- Hu J, Vanderstraeten S, Foury F. Isolation and characterization of ten mutator alleles of the mitochondrial DNA polymerase-encoding MIP1 gene from Saccharomyces cerevisiae. Gene. 1995;160:105–110. doi: 10.1016/0378-1119(95)00215-r. [DOI] [PubMed] [Google Scholar]
- Husmeier D, Wright F. Detection of recombination in DNA multiple alignments with hidden Markov models. J Comput Biol. 2001;8:401–427. doi: 10.1089/106652701752236214. [DOI] [PubMed] [Google Scholar]
- Ismail Y, Hijri M. Arbuscular mycorrhisation with Glomus irregulare induces expression of potato PR homologues genes in response to infection by Fusarium sambucinum. Funct Plant Biol. 2012;39:236–245. doi: 10.1071/FP11218. [DOI] [PubMed] [Google Scholar]
- Kempken F, Hermanns J, Osiewacz HD. Evolution of linear plasmids. J Mol Evol. 1992;35:502–513. doi: 10.1007/BF00160211. [DOI] [PubMed] [Google Scholar]
- Kim E-K, Jeong J-H, Youn HS, Koo YB, Roe J-H. The terminal protein of a linear mitochondrial plasmid is encoded in the N-terminus of the DNA polymerase gene in white-rot fungus Pleurotus ostreatus. Curr Genet. 2000;38:283–290. doi: 10.1007/s002940000157. [DOI] [PubMed] [Google Scholar]
- Kubo T, et al. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNACys(GCA) Nucleic Acids Res. 2000;28:2571–2576. doi: 10.1093/nar/28.13.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn G, Hijri M, Sanders IR. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature. 2001;414:745–748. doi: 10.1038/414745a. [DOI] [PubMed] [Google Scholar]
- Lee J, Young JP. The mitochondrial genome sequence of the arbuscular mycorrhizal fungus Glomus intraradices isolate 494 and implications for the phylogenetic placement of Glomus. New Phytol. 2009;183:200–211. doi: 10.1111/j.1469-8137.2009.02834.x. [DOI] [PubMed] [Google Scholar]
- Li Q, Nargang FE. Two Neurospora mitochondrial plasmids encode DNA polymerases containing motifs characteristic of family B DNA polymerases but lack the sequence Asp-Thr-Asp. Proc Natl Acad Sci U S A. 1993;90:4299–4303. doi: 10.1073/pnas.90.9.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liitsola K, et al. An AB recombinant and its parental HIV type 1 strains in the area of the former Soviet Union: low requirements for sequence identity in recombination. AIDS Res Hum Retroviruses. 2000;16:1047–1053. doi: 10.1089/08892220050075309. [DOI] [PubMed] [Google Scholar]
- Lockshon D, et al. A role for recombination junctions in the segregation of mitochondrial DNA in yeast. Cell. 1995;81:947–955. doi: 10.1016/0092-8674(95)90014-4. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Perlman PS, Butow RA. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc Natl Acad Sci U S A. 1998;95:6739–6743. doi: 10.1073/pnas.95.12.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinko-Kuehn M, Yang X, Debets F, Jacobson DJ, Griffiths AJF. A kalilo-like linear plasmid in Louisiana field isolates of the pseudohomothallic fungus Neurospora tetrasperma. Curr Genet. 1994;26:336–343. doi: 10.1007/BF00310498. [DOI] [PubMed] [Google Scholar]
- Mathimaran N, et al. Microsatellites for disentangling underground networks: Strain-specific identification of Glomus intraradices, an arbuscular mycorrhizal fungus. Fungal Genet Biol. 2008;45:812–817. doi: 10.1016/j.fgb.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Milne I, et al. TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics. 2004;20:1806–1807. doi: 10.1093/bioinformatics/bth155. [DOI] [PubMed] [Google Scholar]
- Nadimi M, Beaudet D, Forget L, Hijri M, Lang BF. Group I intron-mediated trans-splicing in mitochondria of Gigaspora rosea, and a robust phylogenetic affiliation of arbuscular mycorrhizal fungi with Mortierellales. Mol Biol Evol. 2012;29:2199–2210. doi: 10.1093/molbev/mss088. [DOI] [PubMed] [Google Scholar]
- Nakazono M, Kanno A, Tsutsumi N, Hirai A. Palindromic repeated sequences (PRSs) in the mitochondrial genome of rice: evidence for their insertion after divergence of the genus Oryza from the other Gramineae. Plant Mol Biol. 1994;24:273–281. doi: 10.1007/BF00020167. [DOI] [PubMed] [Google Scholar]
- Nargang FE, Bell JB, Stohl LL, Lambowitz AM. The DNA sequence and genetic organization of a Neurospora mitochondrial plasmid suggest a relationship to introns and mobile elements. Cell. 1984;38:441–453. doi: 10.1016/0092-8674(84)90499-9. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Lee RW. Short repetitive sequences in green algal mitochondrial genomes: potential roles in mitochondrial genome evolution. Mol Biol Evol. 1998;15:690–701. doi: 10.1093/oxfordjournals.molbev.a025972. [DOI] [PubMed] [Google Scholar]
- Oeser B, Rogmann-Backwinkel P, Tudzynski P. Interaction between mitochondrial DNA and mitochondrial plasmids in Claviceps purpurea: analysis of plasmid-homologous sequences upstream of the IrRNA-gene. Curr Genet. 1993;23:315–322. doi: 10.1007/BF00310892. [DOI] [PubMed] [Google Scholar]
- Pande S, Lemire EG, Nargang FE. The mitochondrial plasmid from Neurospora intermedia strain Labelle-1b contains a long open reading frame with blocks of amino acids characteristic of reverse transcriptases and related proteins. Nucleic Acids Res. 1989;17:2023–2042. doi: 10.1093/nar/17.5.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Paquin B, Laforest M-J, Lang BF. Double-hairpin elements in the mitochondrial DNA of Allomyces: evidence for mobility. Mol Biol Evol. 2000;17:1760–1768. doi: 10.1093/oxfordjournals.molbev.a026274. [DOI] [PubMed] [Google Scholar]
- Pawlowska TE, Taylor JW. Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature. 2004;427:733–737. doi: 10.1038/nature02290. [DOI] [PubMed] [Google Scholar]
- Purin S, Morton J. Anastomosis behavior differs between asymbiotic and symbiotic hyphae of Rhizophagus clarus. Mycologia. 2012;105:589–602. doi: 10.3852/12-135. [DOI] [PubMed] [Google Scholar]
- Raab PA, Brennwald A, Redecker D. Mitochondrial large ribosomal subunit sequences are homogeneous within isolates of Glomus (arbuscular mycorrhizal fungi, Glomeromycota) Mycol Res. 2005;109:1315–1322. doi: 10.1017/s0953756205003977. [DOI] [PubMed] [Google Scholar]
- Robison MM, Royer JC, Horgen PA. Homology between mitochondrial DNA of Agaricus bisporus and an internal portion of a linear mitochondrial plasmid of Agaricus bitorquis. Curr Genet. 1991;19:495–502. doi: 10.1007/BF00312742. [DOI] [PubMed] [Google Scholar]
- Robison MM, Wolyn DJ. A mitochondrial plasmid and plasmid-like RNA and DNA polymerases encoded within the mitochondrial genome of carrot (Daucus carota) Curr Genet. 2005;47:57–66. doi: 10.1007/s00294-004-0549-x. [DOI] [PubMed] [Google Scholar]
- Rosewich UL, Kistler HC. Role of horizontal gene transfer in the evolution of fungi. Annu Rev Phytopathol. 2000;38:325–363. doi: 10.1146/annurev.phyto.38.1.325. [DOI] [PubMed] [Google Scholar]
- Ruiz-Lozano JM, Azcon R, Gomez M. Effects of arbuscular-mycorrhizal Glomus species on drought tolerance: physiological and nutritional plant responses. Appl Environ Microbiol. 1995;61:456–460. doi: 10.1128/aem.61.2.456-460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements, and genomes of adeno-type viruses. Microbiol Rev. 1990;54:66–74. doi: 10.1128/mr.54.1.66-74.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders IR, Alt M, Groppe K, Boller T, Wiemken A. Identification of ribosomal DNA polymorphisms in spores of the Glomales: application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol. 1995;130:419–427. [Google Scholar]
- Saville BJ, Kohli Y, Anderson JB. mtDNA recombination in a natural population. Proc Natl Acad Sci U S A. 1998;95:1331–1335. doi: 10.1073/pnas.95.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield MA, Agbunag R, Miller JH. DNA inversions between short inverted repeats in Escherichia coli. Genetics. 1992;132:295–302. doi: 10.1093/genetics/132.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte U, Lambowitz AM. The LaBelle mitochondrial plasmid of Neurospora intermedia encodes a novel DNA polymerase that may be derived from a reverse transcriptase. Mol Cell Biol. 1991;11:1696–1706. doi: 10.1128/mcb.11.3.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Graf S, Chaga OY, Lavrov DV. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene. 2006;381:92–101. doi: 10.1016/j.gene.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Shiu-Shing Chan B, Court DA, Vierula PJ, Bertrand H. The kalilo linear senescence-inducing plasmid of Neurospora is an invertron and encodes DNA and RNA polymerases. Curr Genet. 1991;20:225–237. doi: 10.1007/BF00326237. [DOI] [PubMed] [Google Scholar]
- Signorovitch AY, Buss LW, Dellaporta SL. Comparative genomics of large mitochondria in Placozoans. PLoS Genet. 2007;3:e13. doi: 10.1371/journal.pgen.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Read D, editors. Mycorrhizal symbiosis. Cambridge: Academic Press; 2008. [Google Scholar]
- St-Arnaud M, Vujanovik V. Effects of arbuscular mycorrhizal fungi on plant diseases and pests mycorrhizae in crop production: applying knowledge. Binghampton (NY): Haworth Press; 2007. [Google Scholar]
- Stockinger H, Walker C, Schussler A. ‘Glomus intraradices DAOM197198’, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 2009;183:1176–1187. doi: 10.1111/j.1469-8137.2009.02874.x. [DOI] [PubMed] [Google Scholar]
- Stover B, Muller K. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Tsuda M, Yasumoto K, Yamagishi H, Terachi T. A complete mitochondrial genome sequence of Ogura-type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.) BMC Genomics. 2012;13:352. doi: 10.1186/1471-2164-13-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiéry O, Börstler B, Ineichen K, Redecker D. Evolutionary dynamics of introns and homing endonuclease ORFs in a region of the large subunit of the mitochondrial rRNA in Glomus species (arbuscular mycorrhizal fungi, Glomeromycota) Mol Phylogenet Evol. 2010;55:599–610. doi: 10.1016/j.ympev.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Voets L, De La Providencia IE, Declerck S. Glomeraceae and Gigasporaceae differ in their ability to form hyphal networks. New Phytologist. 2006;172:185–188. doi: 10.1111/j.1469-8137.2006.01873.x. [DOI] [PubMed] [Google Scholar]
- Volz-Lingenhöhl A, Solignac M, Sperlich D. Stable heteroplasmy for a large-scale deletion in the coding region of Drosophila subobscura mitochondrial DNA. Proc Natl Acad Sci U S A. 1992;89:11528–11532. doi: 10.1073/pnas.89.23.11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner J, Antunes PM, Powell JR, Mazukatow J, Rillig MC. Plant pathogen protection by arbuscular mycorrhizas: a role for fungal diversity? Pedobiologia. 2010;53:197–201. [Google Scholar]
- Weiller GF, Bruckner H, Kim SH, Pratje E, Schweyen RJ. A GC cluster repeat is a hotspot for mit—macro-deletions in yeast mitochondrial DNA. Mol Gen Genet. 1991;226:233–240. doi: 10.1007/BF00273608. [DOI] [PubMed] [Google Scholar]
- White DJ, Wolff JN, Pierson M, Gemmell NJ. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol. 2008;17:4925–4942. doi: 10.1111/j.1365-294X.2008.03982.x. [DOI] [PubMed] [Google Scholar]
- White MF, Lilley DM. Characterization of a Holliday junction—resolving enzyme from Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:6465–6471. doi: 10.1128/mcb.17.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier BB, Miao VPW, Jónsson ZO, Andrésson ÓS. Mitochondrial genomes from the lichenized fungi Peltigera membranacea and Peltigera malacea: features and phylogeny. Fungal Biol. 2012;116:802–814. doi: 10.1016/j.funbio.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Yang X, Griffiths AJ. Male transmission of linear plasmids and mitochondrial DNA in the fungus Neurospora. Genetics. 1993a;134:1055–1062. doi: 10.1093/genetics/134.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Griffiths AJF. Plasmid diversity in senescent and nonsenescent strains of Neurospora. Mol Gen Genet. 1993b;237:177–186. doi: 10.1007/BF00282799. [DOI] [PubMed] [Google Scholar]
- Yasuhira S, Yasui A. Alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe operates both in nucleus and in mitochondria. J Biol Chem. 2000;275:11824–11828. doi: 10.1074/jbc.275.16.11824. [DOI] [PubMed] [Google Scholar]
- Yin S, Heckman J, RajBhandary UL. Highly conserved GC-rich palindromic DNA sequences flank tRNA genes in Neurospora crassa mitochondria. Cell. 1981;26:325–332. doi: 10.1016/0092-8674(81)90201-4. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Krabusch M, Wolf K. Characterization of a novel open reading frame, urf a, in the mitochondrial genome of fission yeast: correlation of urf a; mutations with a mitochondrial mutator phenotype and a possible role of frameshifting in urf a; expression. Curr Genet. 1991;19:95–102. doi: 10.1007/BF00326289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.