Abstract
Environmental or geological changes can create new niches that drive ecological species divergence without the immediate cessation of gene flow. However, few such cases have been characterized. On a recently formed volcano, Mt. Etna, Senecio aethnensis and S. chrysanthemifolius inhabit contrasting environments of high and low altitude, respectively. They have very distinct phenotypes, despite hybridizing promiscuously, and thus may represent an important example of ecological speciation “in action,” possibly as a response to the rapid geological changes that Mt. Etna has recently undergone. To elucidate the species’ evolutionary history, and help establish the species as a study system for speciation genomics, we sequenced the transcriptomes of the two Etnean species, and the outgroup, S. vernalis, using Illumina sequencing. Despite the species’ substantial phenotypic divergence, synonymous divergence between the high- and low-altitude species was low (dS = 0.016 ± 0.017 [SD]). A comparison of species divergence models with and without gene flow provided unequivocal support in favor of the former and demonstrated a recent time of species divergence (153,080 ya ± 11,470 [SE]) that coincides with the growth of Mt. Etna to the altitudes that separate the species today. Analysis of dN/dS revealed wide variation in selective constraint between genes, and evidence that highly expressed genes, more “multifunctional” genes, and those with more paralogs were under elevated purifying selection. Taken together, these results are consistent with a model of ecological speciation, potentially as a response to the emergence of a new, high-altitude niche as the volcano grew.
Keywords: speciation genomics, altitude adaptation, ecological speciation, hybrid zone, gene flow, RNA-seq
Introduction
Despite concerted recent efforts, many major questions regarding the process of speciation remain unanswered. For example, it is unclear to what extent new ecological opportunities can drive speciation by way of divergent selection in the face of gene flow, and what proportion of the genome is typically involved in the formation of species differences (Nosil et al. 2009; Pinho and Hey 2010). Recent technological advances are revolutionizing the study of speciation genomics by allowing nonmodel organisms to be studied on a genome-wide scale (Rokas and Abbot 2009; Ekblom and Galindo 2011; Heliconius Genome Consortium 2012; Jones et al. 2012). It is becoming feasible to choose study species on the basis of their ecological suitability for answering specific biological questions, rather than, as before, the existence of genomic resources. Furthermore, a wider range of study systems makes the discrimination of system-specific phenomena from those that are more general much less challenging, so the need for a greater number of evolutionary study systems with sufficient genomic resources is significant. Specific to the field of speciation research, examples of “speciation in action” can now be chosen, in which the speciation process took place very recently or is ongoing.
In plants, one classic example of speciation in action can be found in Sicilian Senecio (Ragworts). Senecio aethnensis Jan ex DC. grows at high altitudes on Mt. Etna (>1,600 m), whereas S. chrysanthemifolius Poir. is largely confined to the volcano’s lower slopes (<1,000 m). At intermediate altitudes, where the ranges of these two species approach one another, there is a stable hybrid zone (Chapman et al. 2005; James and Abbott 2005). Although “purer” forms of S. aethnensis and S. chrysanthemifolius differ in several phenotypic characteristics, such as leaf shape, capitulum (inflorescence) size, and ray flower size, and so are regarded as “good species,” plants in the hybrid zone show a range of intermediate phenotypes that track an altitudinal cline (James and Abbott 2005; Brennan et al. 2009). Ecological factors that are likely to differ between their high- and low-altitude habitats include UV light exposure, temperature, and water availability (James and Abbott 2005; Brennan et al. 2009), and germination temperature in the greenhouse has been shown to correlate with the altitude of their source population, likely as a result of selection (Ross et al. 2012). Given the high interfertility of S. aethnensis and S. chrysanthemifolius (Chapman et al. 2005), this system provides an excellent platform to examine the selective forces that maintain these species’ considerable phenotypic divergence in the face of what may be substantial gene flow, and to identify the loci responsible. Furthermore, material collected from this hybrid zone in the late 17th century and cultivated at Oxford Botanic Garden gave rise to the invasive British homoploid species Senecio squalidus (Oxford ragwort) (James and Abbott 2005), adding importance of this system for studies of the speciation process in general.
Clinal analysis of the Senecio hybrid zone using quantitative traits and a small (n = 13) cohort of molecular markers showed that the hybrid zone is shaped by gene flow and selection against hybrids (Brennan et al. 2009). Differences between the clines for certain quantitative traits suggested that environmental selection is largely responsible for structuring trait differentiation across the hybrid zone. In contrast, at finer spatial scales, gene flow and intrinsic selection against hybrids may be more important than environmental selection. To date, however, all studies of genetic divergence in the system have been limited by the small number of loci that they have used (Brennan et al. 2009) and by biased selection of loci (Muir et al. 2013).
In the current study, we aimed to elucidate the speciation process in these species using a massive and unbiased data set with the aims of 1) producing the first reference transcriptomes for these species, 2) elucidating the mode and estimating the demographic parameters of their speciation (to be compared with the timescale of Mt. Etna’s growth), and 3) estimating the strength and direction of selective pressures acting across the transcriptome and how this varies among loci.
For this purpose, we used Illumina RNA-seq to sequence the transcriptomes of one individual each, of high-altitude S. aethnensis and low-altitude S. chrysanthemifolius, as well as an outgroup, S. vernalis. These were used for a comprehensive analysis of molecular divergence in the system. In addition to our evolutionary analyses, we used these data to further develop a public database (http://www.seneciodb.org/, last accessed September 16, 2013), which represents an important resource for further investigations into the evolutionary genomics of Senecio.
Materials and Methods
Sequencing
Single plants of S. aethnensis, S. chrysanthemifolius, and S. vernalis were grown in the glasshouse from seeds collected in the wild. S. aethnensis was collected from 37.42 °N, 14.59 °E, 2,097 m above sea level; S. chrysanthemifolius was collected from 57.57 °N, 14.57 °E, 763 m above sea level (both locations are on Mt. Etna, Sicily); S. vernalis seeds were obtained from the Millennium Seed Bank and were originally collected in Cyprus. Total RNA was extracted from actively growing shoots with a single capitulum bud to maximize the number of genes represented in the transcriptome. Extractions were conducted using the Qiagen Plant RNeasy kit. Poly-A selection, reverse transcription, library construction, and sequencing were performed according to Illumina RNA-seq protocol at the genomic sequencing facility at the Wellcome Trust Centre for Human Genetics (WTCHG) in Oxford. S. chrysanthemifolius and S. aethnensis transcriptomes were multiplexed, and 100-bp paired-end reads were produced by running the multiplexed samples twice over two independent Illumina HiSeq 2000 runs to maximize the sequencing depth. S. vernalis 100-bp paired-end reads were generated in a single Illumina HiSeq 2000 run (part of another multiplexed run for which not all data are used here). Raw data were uploaded to the Sequence Read Archive (SRA) database (Study Accession Number: SRP028289).
Data Set Preparation
Basecalling, adaptor trimming, and sorting of reads by multiplex tags were undertaken as part of the WTCHG bioinformatics pipeline. This uses Illumina’s native basecalling pipeline (Bustard 1.9) with default parameters, and demultiplexing is performed using an in-house Perl script at the WTCHG. Reads received from the WTCHG were quality trimmed using the modified Mott trimming algorithm in CLC Genomics Workbench v5 (CLC bio, Aarhus, Denmark). Default settings of two unknown nucleotides (Ns) allowed per read and an error probability cutoff of 0.05 were used (see CLC Genomics Workbench v5 manual for details). Amplification artifacts that occur during high-throughput library preparation can make de novo assemblies inaccurate (Kozarewa et al. 2009), so we used the CLC Genomics Workbench Duplicate Reads Removal Plugin to remove them. This uses an algorithm that distinguishes duplicate reads arising from amplification artifacts (as opposed to identical reads in high-coverage regions) by first identifying “neighbors” (reads that share most of their sequence but with an offset). It then identifies read pairs that share identical sequence at high copy numbers compared with neighbors, and in which all, or nearly all, duplicates are on the same strand. Identical read pairs with such a signature are extremely unlikely to occur by chance so these were reduced to one copy before assembly.
De novo assembly of the transcriptomes from each individual was conducted using CLC Genomics Workbench v5 with a k-mer length (termed word size in the CLC Genomics documentation) of 28 nt. Other settings used were a minimal contig length of 300 bp, automatically determined maximum bubble size, and use of the scaffolding algorithm, which uses paired-end information to scaffold the contigs (using the following settings: mismatch cost = 3; insertion cost = 3; deletion cost = 3; similarity fraction = 0.95; length fraction = 0.5). We also produced assemblies using a range of alternative k-mer sizes, which we assessed by BlastX comparison to the Arabidopsis thaliana proteome (supplementary table S1, Supplementary Material online), but the parameter combination described above provided the best assembly (i.e., the assembly in which the highest proportion of contigs produced a full-length coding sequence relative to their top Arabidopsis BlastX hit; the analysis is described below in the Transcriptome Annotation and Validation section).
To investigate divergence between the species, we used a reference-guided mapping approach using the assembled transcriptome of S. vernalis, which yielded the greatest number of reads and contigs, as a reference. Quality-trimmed reads (discussed earlier) from each of the three species were mapped to the reference using CLC Genomics Workbench v5 with strict settings (mismatch cost = 3; insertion cost = 3; deletion cost = 3; similarity fraction = 0.95; length fraction = 0.9). BAM files for each mapping were then exported and input into the samtools package (Li et al. 2009) for local phasing, variant calling, filtering, and consensus calling. Several of our downstream analyses require haploid sequences, so samtools phase function was used first to locally phase alleles. The samtools phase algorithm is conservative because it phases heterozygous bases only when phase information is available within individual reads. When phase cannot be determined, it outputs IUPAC ambiguity characters. BAM files for both phases of each species were then used to create a pileup file using samtools’ “mpileup” function with a strict PHRED-scaled base quality cutoff of Q = 30 (equivalent to a 1/1,000 probability of incorrect base assignment). Indels were not called, as they were not relevant to any of our downstream analyses, although the base alignment quality (BAQ) algorithm in samtools was used to decrease the probability of false-positive single nucleotide polymorphisms (SNPs) arising from proximity to indels. Outputted binary call format (BCF) files were then converted to variant call format (VCF) with BCFtools’ “view” function. VCF files, in conjunction with the consensus sequence, were used to create consensus fastq files for each species using the samtools helper script, vcfutils.pl (Li et al. 2009). Conversion to fastq format included an additional filtering step that required all bases called to be represented by at least three independent reads. Finally, fastq files were converted to fasta format using seqtk (part of the samtools package), and bases that failed the depth and quality filters (represented as lowercase bases in the vcfutils.pl output) were converted to ambiguity characters (Ns).
All filtered consensus fasta files were then imported into Proseq3 (Filatov 2009) to create alignments for all reference contigs. As only a single sequence for each species was required for downstream analyses, one phase of each species was randomly removed for each contig. The resulting tripartite alignments were then further filtered in Proseq3, with only contigs with at least 300 bp of aligned sequence for all three species retained for further analysis.
Transcriptome Annotation and Validation
To annotate the reference de novo transcriptome assembly, to filter out sequences of dubious origin (i.e., sequences from contaminating organisms and chloroplast genomic sequence), and to assess the completeness of the transcriptome, we conducted a series of Blast-based analyses. First, sequences for each locus were used as queries in BlastX searches (Altschul et al. 1990) against the NCBI nonredundant (nr) protein database, using BLAST2GO (Conesa et al. 2005) with default settings. Gene Ontology terms (GO Consortium) were then assigned using BLAST2GO based on the results of the BlastX. Results were manually filtered by species, and those with top hits from nonplants were removed from further analysis. The resulting functional annotations were then imported and assigned to the aligned data sets using Proseq3. To remove chloroplast genome sequence from the data set, we performed a BlastN search against the closely related Jacobea vulgaris (formerly Senecio jacobea) chloroplast genome sequence (Doorduin et al. 2011). Sequences with more than 95% identity over 100 bp with J. vulgaris cpDNA were removed from further analysis. To assess the quality of the transcriptomes, a second BlastX search was performed against the TAIR10 Arabidopsis thaliana proteins (ftp://ftp.arabidopsis.org/home/tair/Proteins/TAIR10_protein_lists/TAIR10_pep_20101214, last accessed January 5, 2013; Swarbreck et al. 2008) using the BLAST+ suite (version: BLAST2.2.25+; Camacho et al. 2009) with default length and e-value settings. The length of the BlastX alignment and that of the top-hit sequences were then compared to evaluate transcript completeness. A final filtering step based on ORF analysis was then conducted in Proseq3. To minimize errors arising from incorrect coding region assignment, loci that included premature stop codons or more than one overlapping putative coding sequence (CDS) in a different reading frame or orientation were removed. The inferred CDS were used to make a coding region-only data set for subsequent divergence analysis.
Divergence Analysis
To compare the divergence between the two Etnean species from their outgroup S. vernalis, genes were concatenated and subjected to Tajima’s relative rate tests (Tajima 1993) implemented in MEGA 5 (Tamura et al. 2011). Branch-specific dN, dS, and dN/dS were estimated with the codeml program in PAML 4 (Yang 2007) using the M1, free-ratios model. We determined whether the median dN/dS from the outgroup to S. aethnensis and from the outgroup to S. chrysanthemifolius were significantly different using a Wilcoxon signed-rank test. To detect positive selection, several pairs of nested models in codeml were compared using likelihood ratio tests (LRTs). The PAML branch site test of positive selection (Yang and Nielsen 2002; Yang et al. 2005; Zhang et al. 2005) detects positive selection at a subset of sites in a specific lineage. Two branch-site models, one specifying positive selection at a subset of sites on each of the S. aethnensis and S. chrysanthemifolius lineages (model = 2, NSsites = 2), were compared with the null model with dN/dS in tested branches set to 1. Two pairs of site models, which detect selection at a subset of sites, were also compared with LRTs (M7 vs. M8 and M1a vs. M2a; Nielsen and Yang 1998; Yang et al. 2000). Correction for multiple tests was implemented using the false discovery rate method (FDR; Benjamini and Hochberg 1995) using an alpha level of 0.05 to determine significance. All PAML analyses were automated using in-house Perl scripts.
Factors Affecting the Genome-Wide Selective Landscape
To investigate which factors may affect the differences in selective constraint among loci in the species, the dS and dN estimates for the whole tree (i.e., the sum of the estimates from all three branches estimated in codeml, discussed earlier) were used to calculate dN/dS for each of the contigs. Those for which dN/dS could not be calculated (i.e., those in which dN or dS was zero) were excluded from this analysis.
Following gene duplication, selection pressures may be altered relative to nonduplicated genes, and those that tend to be retained as duplicates may preferentially belong to functional groups that are under different selection pressures than those that are not. Therefore, we aimed to determine the relationship between dN/dS and the presence/absence of paralogs by predicting duplicate genes within our reference transcriptome using the method of McKain et al. (2012). Genes were then divided into two groups, those with at least one paralog in the transcriptome versus those without, and the distributions of dN/dS in two groups of genes were compared using a Mann–Whitney U test.
To determine the level of correlation between dN/dS and the number of paralogs present, and whether this significantly differed from zero, we conducted a Spearman’s rank correlation test between dN/dS for each locus and the number of paralogs detected. To investigate the possibility that any difference in dN/dS was due to inflated dS (either an artificial increase because of incorrect alignment of paralogs or a real one, due to selection on synonymous sites), a further Spearman’s rank test was applied to dS and number of paralogs.
In other species (Koonin and Wolf 2006; Slotte et al. 2011), expression level has been found to be correlated with the level of selective constraint, so we examined the relationship between expression level and dN/dS in Senecio. Reads per kilobase per million mapped reads (RPKM) was determined using CLC Genomics Workbench v5. This was tested for correlation with dN/dS using a Spearman’s rank correlation test.
To determine whether there was enrichment of any specific GO terms in genes with high dN/dS, one-tailed Fisher’s exact tests were performed for each GO term in BLAST2GO (Conesa et al. 2005) with FDR correction for multiple tests. These were performed comparing loci in the top 5% and 10% for dN/dS with all other loci.
Genes with more distinct functions may be expected to be under stronger selective constraint. To assess the association between the level of purifying selection on a locus and the “multifunctionality” (sensu Salathé et al. 2006) of the protein it encodes, total numbers of GO terms assigned to each locus, as well as the number from each of the three main GO ontologies, were compared with dN/dS using Spearman’s rank correlation coefficient.
Tests for enrichment of GO terms in loci with high dN/dS and the comparisons of the distribution of GO term number with that of dN/dS could potentially be biased by some genes (e.g., especially those with low expression) being fragmented into several contigs in our data set (and GO terms being counted multiply for such genes). To control for this, we used the STM scaffolding method (Surget-Groba and Montoya-Burgos 2010) to scaffold contigs that may be derived from single transcripts by comparison to the Arabidopsis thaliana proteome (TAIR 10; discussed earlier). Contigs were subjected to a BlastX search, using the suggested e-value cutoff of 10−5, and xml outputs were run through the STM program using default settings. Contigs that were scaffolded using this process were then merged into single entries (and their GO terms were combined), and the tests were re-performed. The STM scaffolding method was not used for the main data set because it introduces an increased risk of producing chimeric sequences, and is thus less conservative for most analyses.
Testing the Mode of Speciation
To determine whether the species had diverged in the presence of gene flow, and to estimate their divergence times and population sizes, we used the likelihood ratio test and maximum likelihood models implemented in 3s (version 2.0a; Yang 2010). In this analysis we used only 4-fold degenerate sites to minimize the influence of selection and to allow the mutation rate to be estimated more accurately. Filtering of 4-fold degenerate sites from our “CDS-only” data sets was undertaken using Proseq3 (Filatov 2009). 3s was run with default settings with the exception that 32 points (parameter k in 3s) were used in the Gaussian quadrature, which produces a more accurate, but more computationally expensive, result (Yang 2010). We also used the 3s program’s implementation of the model of Yang (2002) to estimate four demographic parameters of the species’ divergence, namely, θacv = 4Nacvµ, θac = 4Nacvµ, τacv = Tacvµ, and τac = Tacµ, where N is the effective population size, T is the divergence time (in years because generation time was assumed to be 1 per year), µ is the mutation rate, and subscript letters denote the common ancestors of all three species (acv) and of just the two focal species (ac). These were converted into more biologically meaningful units as follows: Ne = θ/(4µ) for the effective population sizes of each ancestral population; and T = τ/µ for the divergence times to each ancestral node. The mutation rate is not known in Senecio, although a neutral mutation rate of 1 × 10−8 has been reported within the Asteraceae, of which Senecio is a member (used in Strasburg and Rieseberg 2008). The Asteraceae-specific rate was used for conversions, but because the average plant mutation rate (5 × 10−9) estimated by Wolfe et al. (1987) has been used in a previous demographic analysis of Senecio (Muir et al. 2013), parameter estimates were also scaled using this mutation rate for enhanced comparability between the studies. The Asteraceae-specific rate is likely to be the most accurate for Senecio. 3s analyses were run twice to ensure that the results were stable.
Results
Transcriptome Sequencing, Validation, and Characterization
In total, we generated ca. 1.48, 1.23, and 3.64 Gbp of sequence data for S. aethnensis, S. chrysanthemifolius, and S. vernalis transcriptomes, respectively. De novo assemblies of these sequences yielded 30,560, 26,536, and 35,770 contigs over 300 bp, respectively (with total lengths of S. aethnensis 26897354; S. chrysanthemifolius: 21356997; and S. vernalis: 27343943; supplementary table S2, Supplementary Material online). De novo assembled contigs for these transcriptomes were uploaded to our SenecioDB database (http://www.seneciodb.org/, last accessed September 16, 2013).
As an approximate estimate of transcriptome completeness and quality, we performed a further BlastX search against the Arabidopsis thaliana proteome and determined the proportion of the top hit that was covered by each contig in the BlastX alignment. This revealed that (after filtering described in methods) 23.42% of Senecio contigs covered at least 90% of their top hit (supplementary fig. S1, Supplementary Material online), suggesting that these sequences may represent approximately full-length transcripts. In addition, 57.09% of contigs covered at least 50% of their top hit (supplementary fig. S1, Supplementary Material online). Our contigs were annotated with a wide range of GO terms, suggesting that we captured a diverse array of functional gene categories (supplementary table S3, Supplementary Material online). After filtering, 12,679 contigs were successfully assigned coding regions longer than 100 codons, and these made up our CDS-only data set for subsequent dN/dS-based analyses.
To estimate the number of contigs that may have been unscaffolded regions of the same transcript, we used an STM scaffolding approach that takes advantage of homology and functional annotation to scaffold cDNA contigs likely to come from the same gene (Surget-Groba and Montoya-Burgos 2010). This analysis identified only 498 (3.93%) contigs that were putatively fragments of a larger transcript and were assembled by STM into 214 scaffolds. Although this approach can increase the contiguity of a transcriptome, it also risks creating chimeric sequences from fragments of related but distinct genes. Because it relies on a high quality, closely related reference proteome, which is not available for Senecio or close relatives (we used Arabidopsis thaliana as the reference), this risk is greatly increased and could severely affect our dN/dS and demographic analyses. For this reason, we deemed it more conservative to use the non-STM scaffolded reference for the majority of our analyses. The scaffolded transcriptome was only used as a control for our GO enrichment analyses.
In our analysis of paralog evolution, 2,598 genes were identified as having a putative paralog among the whole reference assembly (20.49% of the transcriptome). The distribution of the number of paralogs present (supplementary fig. S2, Supplementary Material online) revealed that more than 75% of identified genes with paralogs had only one transcribed paralog. Median synonymous divergence (dS) between paralogous pairs was 0.90, and paralogs were significantly enriched for 190 GO terms (when reduced to most specific terms; supplementary table S4, Supplementary Material online). Using the STM data set for the same analysis, the results were broadly similar (187 GO terms were overrepresented in the STM data set all together, and 175 GO terms were significantly overrepresented in both data sets). These results suggest that there is a high proportion of retained paralogs in S. aethnensis and S. chrysanthemifolius and that many paralogs are expressed simultaneously.
Transcriptome-Wide Analyses of dN/dS
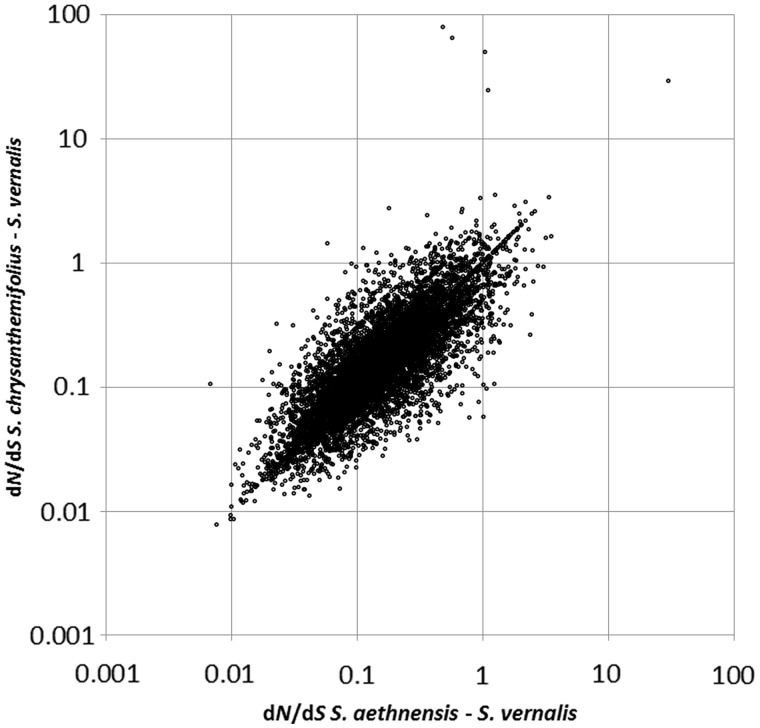
We then attempted to characterize dN/dS, an indicator of the strength and type of selection, across the transcriptome, and to determine its relationship to several important parameters: expression level, presence of paralogs, and gene function. Median dN/dS was 0.153 between S. vernalis and S. aethnensis and 0.148 between S. vernalis and S. chrysanthemifolius, suggesting that purifying selection is the dominant selective force acting on the loci. This ratio varied widely among the genes (fig. 1) and there was only a modest significant correlation between dN/dS on the S. aethnensis and S. chrysanthemifolius branches (Spearman’s rank: rs = 0.241, P = 2.0 × 10−21). This correlation increases to 0.81 (Spearman’s rank: rs = 0.810, P < 1 × 10−100) when the whole distance between each of the Etnean species and S. vernalis is considered, but most SNPs in the data set are between S. vernalis and the Etnean species, and are invariant between S. aethnensis and S. chrysanthemifolius, so this figure is misleadingly inflated. Despite the fact that the correlation between dN/dS in the two Etnean lineages was fairly weak, there was no significant difference between median dN/dS on the S. aethnensis and S. chrysanthemifolius branches (Wilcoxon test: W = 5.79 × 105; P = 0.189), indicating that the overall level of selective constraint in the two lineages is comparable.
Fig. 1.—
A scatterplot of dN/dS for each locus between S. aethnensis and S. vernalis versus dN/dS between S. chrysanthemifolius and S. vernalis.
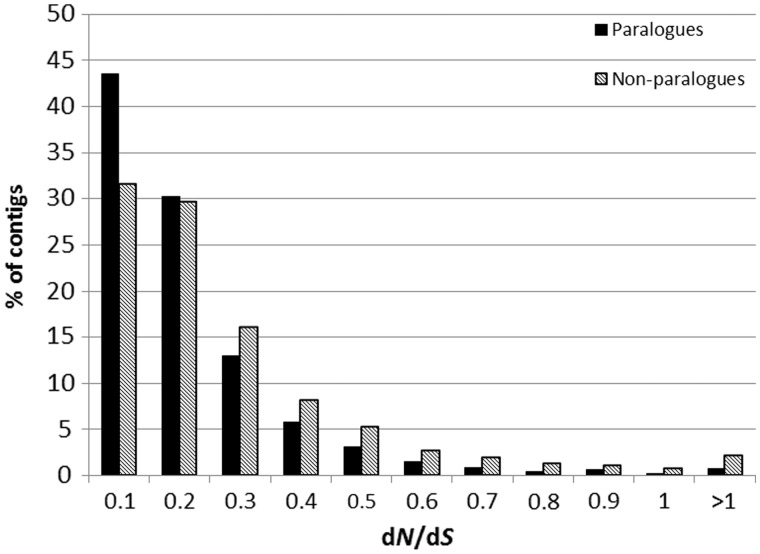
We then attempted to identify transcriptome-wide trends that may influence dN/dS. Interestingly, across all loci, there was a weak but significant negative correlation between total level of expression and dN/dS (Spearman’s rank: rs = −0.222, P = 1.9 × 10−119), suggesting that more highly expressed genes are under stronger purifying selection. dN/dS was significantly lower in genes with putative paralogs (median = 0.169 ± 0.188 [SD]) than those without (median = 0.237 ± 0.415 [SD]; fig. 2; Mann–Whitney U = 1.18 × 106; P = 2.97 × 10−8; genes for which dN/dS could not be calculated were excluded from the test). Furthermore, there was a weak but highly significant negative correlation between the number of paralogs for a gene and its dN/dS ratio compared with orthologs (Spearman’s rank: rs = −0.131, P = 4.33 × 10−42), suggesting that genes that are retained as paralogs tend to be under stronger purifying selection. It is possible that a correlation between number of paralogs and dN/dS could arise because, in genes that are part of high copy gene families, there could be an increase in incorrect mapping of reads from their many different paralogs. This would also produce an increase in dS in high copy number gene families. There was no significant correlation, however, between dS and number of paralogs, suggesting that this was not the case.
Fig. 2.—
Genes with expressed paralogs (black) in the transcriptome have significantly lower dN/dS than those without (gray). The x axis values are the maximum value of dN/dS in each bin range. The percentage of contigs (in each of the two classes, i.e., with or without paralogs) in each bin are displayed on the y axis.
Finally, we examined the relationship between predicted function and dN/dS. Fisher’s exact tests with FDR correction showed that genes with the highest dN/dS were not significantly enriched for any GO terms (neither genes in the top 5% nor 10% tails of the dN/dS distribution). This was true when dN/dS in the S. aethnensis or S. chrysanthemifolius branches as well as across the whole tree was considered, and was also true of the STM-scaffolded, control data set (see Materials and Methods). However, the number of GO terms annotated to a gene (multifunctionality; sensu Salathé et al. 2006) showed a highly significant but weak negative correlation with dN/dS (Spearman’s rank: rs = −0.191, P = 1.31 × 10−88). This was also true when GO terms were divided into their three ontology categories: molecular function (Spearman’s rank: rs = −0.127, P = 7.02 × 10−40), cellular component (Spearman’s rank: rs = −0.169, P = 2.17 × 10−69), and biological process (Spearman’s rank: rs = −0.152, P = 1.58 × 10−56). This may be evidence that more multifunctional genes, those that are involved in a greater number of distinct processes at molecular or organismal levels, or that function in more cellular locations, are more likely to experience a high level of purifying selection.
To attempt to identify specific loci under positive selection, we used several dN/dS-based tests of positive selection (Nielsen and Yang 1998; Yang et al. 2000; Yang and Nielsen 2002; Yang et al. 2005; Zhang et al. 2005; Yang 2007). Using an FDR cutoff of 0.05, however, no locus showed evidence for positive selection in the branches leading to S. aethnensis, S. chrysanthemifolius, or across the whole tree. However, with only three species and a very large number of loci, the test may have lacked sufficient power.
Testing the Mode of Speciation
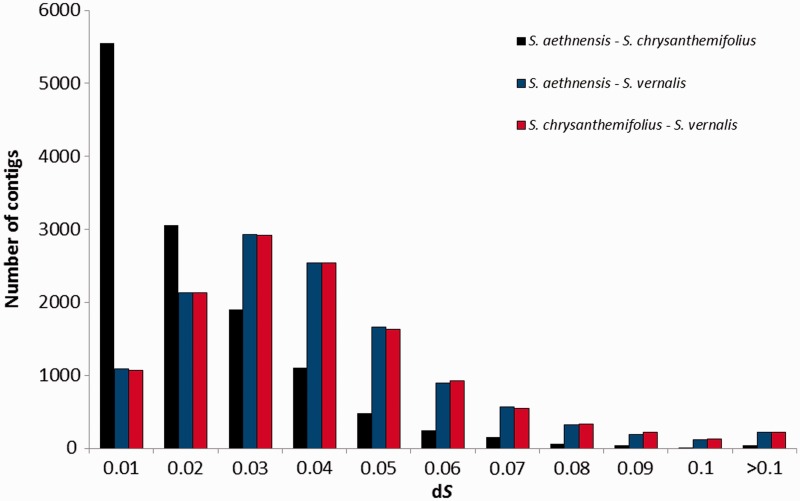
S. aethnensis and S. chrysanthemifolius might be assumed to represent a good example of ecological speciation (Chapman et al. 2005; James and Abbott 2005; Brennan et al. 2009), as these species are closely related but adapted to contrasting environments. Indeed, mean synonymous (dS) and nonsynonymous (dN) divergence between the orthologous genes of the two Mt. Etna species was low (dS = 0.016 ± 0.017 [SD]; dN = 0.002 ± 0.003 [SD]), which is consistent with a recent origin of these species. To determine whether the substitution rate differed between the species, a Tajima’s relative rate test was performed (Tajima 1993) on concatenated alignments; however, there was no significant difference between the rates of S. aethnensis and S. chrysanthemifolius (χ2 = 1.98, P = 0.16). The divergence of the Mt. Etna species from the outgroup S. vernalis was about 2-fold higher (S. aethnensis: dS = 0.035 ± 0.023 [SD]; dN = 0.005 ± 0.005 [SD]; S. chrysanthemifolius: dS = 0.035 ± 0.023 [SD]; dN = 0.005 ± 0.005 [SD]), confirming previous findings (Comes and Abbott 2001) that S. aethnensis and S. chrysanthemifolius form a monophyletic clade with regard to S. vernalis and thus validating its choice as an appropriate outgroup for the study (fig. 3).
Fig. 3.—
Distribution of contig-specific synonymous divergence between S. aethnensis and S. chrysanthemifolius (black), between S. aethnensis and S. vernalis (red), and between S. chrysanthemifolius and S. vernalis (blue). The x axis values are the maximum value in each bin range.
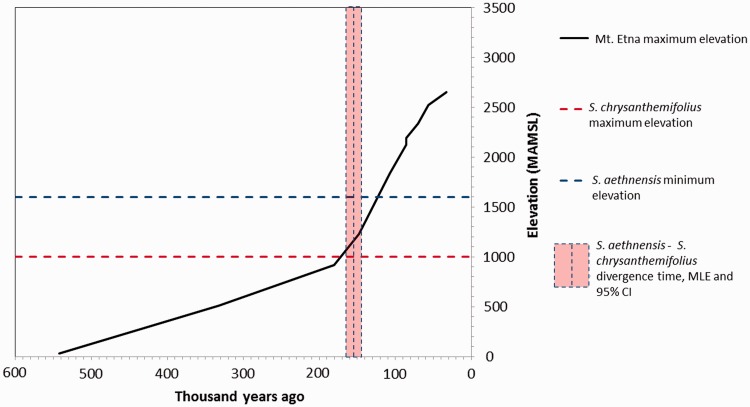
Little is known about the demographic history of S. aethnensis and S. chrysanthemifolius’ speciation, and how it relates to the geological evolution of Mt. Etna. To estimate the demographic parameters of the species split, we used the model developed by Yang (2002). This model provided raw maximum likelihood estimates for all four parameters (θacv = 0.0097 ± 0.0003 [SE], θac = 0.0012 ± 0.0004 [SE]; τacv = 0.0108 ± 0.0001 [SE]; and τac = 0.0015 ± 0.0001 [SE]; see Materials and Methods for parameter details). The conversion of these into more intuitively obvious demographic units requires knowledge of the mutation rate, which is not known in Senecio. Therefore, we used an estimate from the Asteraceae (1 × 10−8; used in Strasburg and Rieseberg 2008), which we expect to be fairly accurate for Senecio, as well as the estimated average plant rate (5 × 10−9; Wolfe et al. 1987) for comparability with a previous study that used it (Muir et al. 2013), which we nevertheless expect to be less accurate because of its phylogenetic generality. This was translated into ancestral population sizes of Nacv = 243,238 ± 6,338 (SE) for the ancestor of all three species and Nac = 312,023 ± 9,833 (SE) for the ancestor of the two Etnean species using the Asteraceae-specific mutation rate. Using the generic plant mutation rate, population size estimates were higher (Nacv = 486,475 ± 12,675 [SE] and Nac = 624,045 ± 19,665 [SE]). Divergence times were estimated to be Tacv = 1,077,760 ± 12,240 (SE) for the split between the ancestors of S. vernalis and the Etnean species and Tac = 153,080 ± 11,470 (SE) for the split between S. aethnensis and S. chrysanthemifolius, using the Asteraceae-specific mutation rate. These estimates of the divergence time between S. aethnensis and S. chrysanthemifolius occur around the period in its geological history during which Mt. Etna began to exceed the altitude above which S. aethnensis is now found and S. chrysanthemifolius is not (De Beni et al. 2011; fig. 4), suggesting that the creation of a new niche as the mountain grew may have been a catalyst for the plant’s speciation. Divergence time estimates increased moderately when the plant mutation rate was used (Tacv = 2,155,520 ± 24,480 [SE] and Tac = 306,160 ± 22,940 [SE]).
Fig. 4.—
The estimated divergence time of the S. aethnensis and S. chrysanthemifolius occurs shortly after Mt. Etna exceeded the elevations that partition the species today (converted into years using an Asteraceae-specific mutation rate of 1 × 10−8 and a generation time of 1 year). Age of volcanic layers of different elevations estimated from 40Ar/39Ar isotopic dating in De Beni et al. (2011).
To test a null hypothesis of allopatric speciation with no significant subsequent gene flow, compared with a model of divergence with gene flow, we used the likelihood ratio test developed by Yang (2010). The test was highly significant (χ2 = 129.43; P = 2.73 × 10−30), indicating that there has been significant gene flow between the species since their divergence. This is consistent with an ecological speciation model, but is also compatible with a scenario of secondary contact following a period of allopatric divergence.
Discussion
Characterization of the Senecio Transcriptomes
Genome or transcriptome-wide data sets for closely related species are essential for evolutionary genomic analyses, but they have historically been restricted to a few model species. However, with the advent of next-generation sequencing, the situation has begun to change, with genomic resources for nonmodel organisms becoming available (Rokas and Abbot 2009). De novo sequencing and assembly of nonmodel organisms, however, presents significant bioinformatic challenges, particularly if, as in Senecio, no high-quality reference genome or transcriptome is available from a closely related species. Transcriptome sequences for three Senecio species generated in our study represent the first step toward the development of comprehensive genomic resources for this fascinating system, which promises to be a rich resource for studies of adaptation and speciation in plants. De novo assembly and annotation of cDNA contigs, more than 20% of which covered the full length of their nearest Arabidopsis homolog, indicate that Illumina sequencing of transcriptomes is an effective strategy for producing large and high-quality comparative transcriptomic data sets, even in the absence of a reference genome. The annotated transcriptomes we present here are the first published transcriptomes of the species and greatly increase the amount of data available to the community. SenecioDB (http://www.seneciodb.org/, last accessed September 9, 2013) now contains sequences from six Senecio species, complementing the existing Compositae Genome Project database (http://compgenomics.ucdavis.edu/, last accessed September 9, 2013). The data will thus further facilitate large-scale comparative genomic analyses of divergence in one of the largest families of flowering plants (Panero and Funk 2008).
Recent Speciation May Have Been Driven by the Growth of Mt. Etna
We used the data to gain a better understanding of the demographic history of the species. Species adapting to altitude are attractive systems for the study of ecological speciation for several reasons. First, four distinct environmental variables that can exert strong selective pressures (atmospheric pressure, temperature, total solar radiation, and fraction of UV-B radiation; Körner 2007) co-vary with altitude universally. Second, several other factors such as precipitation, seasonality, and biotic interactions are often linked to elevation on a case-by-case basis (Körner 2007; Defossez et al. 2011). Thus, altitudinal gradients can exert strong divergent selection over a short geographical distance, and a simple measure of altitude can be a proxy for many of these. Furthermore, Etnean Senecio are unusual among cases of altitude-related speciation in that the mountain on which the species are found is a volcano, and arose relatively recently and rapidly (Branca et al. 2011; De Beni et al. 2011; fig. 4). Although volcanic activity in the area began over 1.5 Ma, the oldest dated rock samples over 1,000 m above mean sea level (MAMSL) were formed only 147.7 kya, and the oldest over 1,600 MAMSL only 107.2 kya, with the current mountain standing at over 3,000 MAMSL (fig. 4; De Beni et al. 2011). Our finding that the species split between ∼141 and ∼164 kya fits strikingly well with the time that the volcano attained the elevations that partition the species today. This suggests that the species may have diverged in response to a new high-altitude niche rapidly becoming available as the volcano grew. Although the estimate is dependent on the mutation rate used for conversion (Yang 2010), we used two different mutation rates, one a general estimate for plants and the other an Asteraceae-specific estimate, and both give estimates of divergence time within the volcano’s period of rapid recent growth. Because mutation rate is taxonomically correlated in plants (Eyre-Walker and Gaut 1997), we expect the Asteraceae-specific estimate to be more accurate for Senecio.
Our recent study into signatures of selection on differentially expressed versus nondifferentially expressed genes in S. aethnensis and S. chrysanthemifolius (Muir et al. 2013) used an isolation–migration model to estimate divergence times, population size, and migration rate between the species. This found an even more recent estimate of divergence time (15–75 kya). However, in that article, only a small number of loci were used (15 nuclear genes and 5 microsatellites) without distinguishing between nonsynonymous, synonymous, and noncoding sites, and used only one mutation rate (5 × 10−9) in conversions. Thus, their estimates are likely to be fairly approximate, as was acknowledged in that work. Our use, in the current study, of only 4-fold degenerate sites means substitution rates within our sample are likely to be far more homogenous, and any influence from selection is likely to be minimized. Our far larger data set and use of two different mutation rates in conversions resulted in more precise estimates with narrow confidence intervals.
Our test of a model of divergence with gene flow versus a null hypothesis of allopatric speciation was extremely significant, suggesting that gene flow has occurred since the species split. Therefore, in addition to gene flow from the parent species into the hybrid zone, there has also been significant exchange of genes between the native ranges of each species where the parents appear “phenotypically pure” (James and Abbott 2005). The significant result cannot, however, be taken as confirmation of speciation with gene flow sensu stricto. This is because the test does not distinguish between divergence in the presence of gene flow and allopatric speciation followed by secondary contact and gene flow. However, the high significance of the test rejecting a no gene flow model in our study suggests that gene flow has occurred between these two species over sufficient time to have had a considerable impact on the pattern of species divergence across the genome. Moreover, there are several pieces of “circumstantial evidence” that the species diverged with gene flow. The species are wind dispersed; they display no intrinsic barriers to interspecific hybridization (Chapman et al. 2005) and occur in proximity to each other with no geographic barriers between them; and, presuming they diverged in situ, this is likely to have been the case throughout their history (Branca et al. 2011). Taking these factors into account, speciation with gene flow, in the absence of a significant allopatric phase, seems the most parsimonious account of their history. Interspecific gene flow can play contrasting roles in evolution. Although it can act in opposition to local adaptation, by homogenizing genomic regions under diversifying selection between two species, it can also allow globally adaptive alleles to be shared between interfertile species, aiding adaptation (Seehausen 2004; Abbott et al. 2013). Genomes of such hybridizing species may be expected to exhibit a mosaic structure, with high levels of divergence restricted to “speciation islands,” which contain the loci responsible for species differences (Wu 2001). Thus, our finding of divergence with gene flow between the species indicates significant gene sharing between Etnean Senecio species; loci containing fixed differences that may be identified in future DNA polymorphism analyses are likely to be under diversifying selection.
Taking our demographic analyses together, in addition to previous work on the system (Chapman et al. 2005; Brennan et al. 2009; Muir et al. 2013) and on the geological evolution of Mt. Etna (Branca et al. 2011; De Beni et al. 2011), the most plausible scenario of the species divergence is as follows. Several hundred thousand years ago, Mt. Etna rapidly began growing in altitude, with the oldest dated sections above which pure S. chrysanthemifolius does not grow today being formed around 148 kya and those above which S. aethnensis is found today around 108 kya (De Beni et al. 2011). This led to the creation of a new high-altitude niche, and the very different environments of the mountain’s upper and lower slopes created strong divergent selection between the plants growing in the two habitats and they subsequently diverged, although gene flow persisted. Thus, the species appear to be a classic example of ecological speciation in response to rapid geological upheaval.
The Genome-Wide Landscape of Selection in Senecio
Because selection is likely to have played a major role in the species’ divergence, we then attempted to elucidate the selective landscape across the genomes of the two species. dN/dS varied widely across the genome but was correlated in the two Etnean lineages, although several genes had much higher dN/dS in one species than the other on visual inspection of the data (fig. 1). The fact that no genes had a significant signature of selection at an FDR cutoff of 0.05 is most likely due to a lack of power. First, PAML likelihood ratio tests (Yang 2007) lack power with few species and second, the FDR over several thousand tests requires an extremely high level of significance.
More revealing was our investigation of dN/dS in relation to other genomic parameters. Recent work in the emerging field of evolutionary systems biology has begun to uncover several “laws of genome evolution” (Koonin and Wolf 2006; Koonin 2011), such as genome-wide correlations with evolutionary rate. Several studies have shown that gene expression level is negatively correlated with dN/dS (Koonin and Wolf 2006; Slotte et al. 2011) potentially due to increased selection for translational robustness against protein misfolding in highly expressed genes (Drummond et al. 2005). Our results are consistent with these previous studies, finding a negative correlation between dN/dS and expression level. A significantly lower dN/dS in genes with paralogs, compared with those without paralogs, was perhaps more surprising.
Gene duplication and subsequent functional divergence is thought to be one of the most important mechanisms for the generation of evolutionary novelty in plants (Ohno 1970; Moore and Purugganan 2005), and recently duplicated genes are expected to have higher dN/dS ratios. This is based on the prediction that immediately following gene duplication, functional constraint is reduced and therefore dN/dS might be higher as one or both paralogs evolve complementary (or novel) functions, or that one paralog becomes pseudogenized. However, in contrast to this theory, genes in our transcriptomes that had at least one paralog had a significantly lower dN/dS than those genes with no paralogs, suggesting they were under stronger purifying selection. One explanation for this finding comes from the observation that the major peak in the dS distribution (between paralogs) was around 0.9–1.0 (supplementary fig. S3, Supplementary Material online), likely corresponding to the whole genome duplication (WGD) detected at the base of the Asteraceae ∼50 Ma by Barker et al. (2008). Thus, any relaxation in purifying selection immediately following WGD may long have passed. Indeed, it is possible that, following a WGD, many of the genes that are retained as pairs of paralogs (as opposed to one copy undergoing pseudogenization) may preferentially be from functional categories that are particularly highly constrained. Several studies have shown evidence for two such opposing forces affecting the value of dN/dS in duplicate genes (Davis and Petrov 2004; Jordan et al. 2004; Yang and Gaut 2011). Although there is a decrease in purifying selection immediately following duplication, which may lead to a higher dN/dS, there is also a general tendency for genes that are retained in duplicate to be more conserved over longer timescales, leading to a decreased dN/dS (Jordan et al. 2004). Hence, although a higher than average dN/dS ratio (caused by relaxed purifying selection) may be expected in recent duplicates, over much longer timescales after whole genome duplications, the opposite may be observed, and the trends may be different for different functional categories of genes.
Although there were no significantly overrepresented GO terms in those loci with high values of dN/dS, the overall number of GO terms was negatively correlated with dN/dS. Genes with a greater number of distinct functions may be more constrained because they are more pleiotropic (Salathé et al. 2006), and are thus more likely to disrupt a phenotype deleteriously. However, it should be noted that these GO annotations were ascertained by sequence homology rather than direct experimentation so should be taken with caution. Nevertheless, the fact that a negative correlation still exists even with the poor functional annotation available for Senecio may indicate that, with more accurate annotation, multifunctionality may be a reliable predictor of the strength of purifying selection on a locus.
The correlations of dN/dS with duplication status, expression level, and multifunctionality, which we have uncovered, add to the growing body of evidence that the evolutionary rates of genes and proteins are influenced by surprisingly constant relationships with several aspects of gene function (Koonin 2011; Yang and Gaut 2011). Here we have shown that, even in species that appear to be undergoing strong diversifying selection, and over the relatively short time since the species diverged, many of these “laws” appear to hold. Thus, although a subset of the genome may be involved in adaptive responses to the species’ radically different habitats, the majority of loci may continue to evolve at a rate overwhelmingly determined by variation in the strength of long-term purifying selection.
Conclusions
Taken together, our results suggest that, despite their considerable phenotypic divergence, the two focal species, S. aethnensis and S. chrysanthemifolius, are extremely closely related genetically and that there has been a substantial amount of gene flow since their divergence. Intriguingly, our estimated time of divergence is strikingly close to estimates of the time when the height of Mt. Etna was first approaching the altitude above which S. aethnensis occurs today, alluding to the possibility that the volcano’s rapid emergence as a mountain of over 3000 MAMSL could have been responsible for the species’ divergence.
The current study was not primarily designed to detect specific loci under selection, but we were able to undertake the first characterization of selective constraint across the genome. Identification of specific loci under positive selection is a key focus of our ongoing work and, based on our findings here, we aim to characterize the genomic basis for such dramatic phenotypic divergence over such a short space of time.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank two anonymous reviewers for their helpful suggestions; Mark Chapman for useful discussion and helpful comments on the manuscript and for providing the S. vernalis RNA; Xin-Sheng Hu for statistical assistance; Michael McKain for running the paralog identification analysis; and Gary Barker for assistance with database construction. This work was funded by the Natural Environment Research Council (NERC; Grant code: NE/G017646/1 to D.A.F. and S.J.H.) and a PhD studentship grant to O.G.O. from the Gatsby Charitable Foundation.
Literature Cited
- Abbott RJ, et al. Hybridization and speciation. J Evol Biol. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barker MS, et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Mol Biol Evol. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Branca S, Coltelli M, Groppelli G. Geological evolution of a complex basaltic stratovolcano: Mount Etna, Italy. Ital J Geosci (Boll Soc Geol It). 2011;130:306–317. [Google Scholar]
- Brennan AC, Bridle JR, Wang A-L, Hiscock SJ, Abbott RJ. Adaptation and selection in the Senecio (Asteraceae) hybrid zone on Mount Etna, Sicily. New Phytol. 2009;183:702–717. doi: 10.1111/j.1469-8137.2009.02944.x. [DOI] [PubMed] [Google Scholar]
- Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Forbes DG, Abbott RJ. Pollen competition among two species of Senecio (Asteraceae) that form a hybrid zone on Mt. Etna, Sicily. Am J Bot. 2005;92:730–735. doi: 10.3732/ajb.92.4.730. [DOI] [PubMed] [Google Scholar]
- Comes HP, Abbott RJ. Molecular phylogeography, reticulation, and lineage sorting in Mediterranean Senecio sect. Senecio (Asteraceae) Evolution. 2001;55:1943–1962. [PubMed] [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beni E, Branca S, Coltelli M, Groppelli G, Wijbrans JR. Ar-40/Ar-39 isotopic dating of Etna volcanic succession. Ital J Geosci (Boll Soc Geol It). 2011;130:292–305. [Google Scholar]
- Defossez E, et al. Do interactions between plant and soil biota change with elevation? A study on Fagus sylvatica. Biol Lett. 2011;7:699–701. doi: 10.1098/rsbl.2011.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduin L, et al. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SKIPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 2011;18:93–105. doi: 10.1093/dnares/dsr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom R, Galindo J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity. 2011;107:1–15. doi: 10.1038/hdy.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut BS. Correlated rates of synonymous site evolution across plant genomes. Mol Biol Evol. 1997;14:455–460. doi: 10.1093/oxfordjournals.molbev.a025781. [DOI] [PubMed] [Google Scholar]
- Filatov DA. Processing and population genetic analysis of multigenic datasets with ProSeq3 software. Bioinformatics. 2009;25:3189–3190. doi: 10.1093/bioinformatics/btp572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JK, Abbott RJ. Recent, allopatric, homoploid hybrid speciation: the origin of Senecio squalidus (Asteraceae) in the British Isles from a hybrid zone on Mount Etna, Sicily. Evolution. 2005;59:2533–2547. [PubMed] [Google Scholar]
- Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Wolf YI, Koonin EV. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Are there laws of genome evolution? PLoS Comput Biol. 2011;7:e1002173. doi: 10.1371/journal.pcbi.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI. Evolutionary systems biology: links between gene evolution and function. Curr Opin Biotech. 2006;17:481–487. doi: 10.1016/j.copbio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Körner C. The use of “altitude” in ecological research. Trends Ecol Evol. 2007;22:569–574. doi: 10.1016/j.tree.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kozarewa I, et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G plus C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKain MR, et al. Phylogenomic analysis of transcriptome data elucidates co-occurrence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae) Am J Bot. 2012;99:397–406. doi: 10.3732/ajb.1100537. [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Muir G, Osborne OG, Sarasa J, Hiscock SJ, Filatov DA. Recent ecological selection on regulatory divergence is shaping clinal variation in Senecio on Mount Etna. Evolution. Advance Access published June 21, 2013. Forthcoming 2013 doi: 10.1111/evo.12157. doi: 10.1111/evo.12157. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang ZH. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Panero JL, Funk VA. The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol Phylogenet Evol. 2008;47:757–782. doi: 10.1016/j.ympev.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Pinho C, Hey J. Divergence with gene flow: models and data. Annu Rev Ecol Evol Syst. 2010;41:215–230. [Google Scholar]
- Rokas A, Abbot P. Harnessing genomics for evolutionary insights. Trends Ecol Evol. 2009;24:192–200. doi: 10.1016/j.tree.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Ross RIC, Agren JA, Pannell JR. Exogenous selection shapes germination behaviour and seedling traits of populations at different altitudes in a Senecio hybrid zone. Ann Bot. 2012;110:1439–1447. doi: 10.1093/aob/mcs211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathé M, Ackermann M, Bonhoeffer S. The effect of multifunctionality on the rate of evolution in yeast. Mol Biol Evol. 2006;23:721–722. doi: 10.1093/molbev/msj086. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Slotte T, et al. Genomic determinants of protein evolution and polymorphism in Arabidopsis. Genome Biol Evol. 2011;3:1210–1219. doi: 10.1093/gbe/evr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasburg JL, Rieseberg LH. Molecular demographic history of the annual sunflowers Helianthus annuus and H. petiolaris—large effective population sizes and rates of long-term gene flow. Evolution. 2008;62:1936–1950. doi: 10.1111/j.1558-5646.2008.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surget-Groba Y, Montoya-Burgos JI. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 2010;20:1432–1440. doi: 10.1101/gr.103846.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:1009–1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI. The genic view of the process of speciation. J Evol Biol. 2001;14:851–865. [Google Scholar]
- Yang ZH. Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics. 2002;162:1811–1823. doi: 10.1093/genetics/162.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang ZH. A likelihood ratio test of speciation with gene flow using genomic sequence data. Genome Biol Evol. 2010;2:200–211. doi: 10.1093/gbe/evq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Gaut BS. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol. 2011;28:2359–2369. doi: 10.1093/molbev/msr058. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang ZH. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.