Abstract
Embryonic stem cell (ESC) identity and self-renewal is maintained by extrinsic signaling pathways and intrinsic gene regulatory networks. Here, we show that three members of the Ccr4-Not complex, Cnot1, Cnot2, and Cnot3, play critical roles in maintaining mouse and human ESC identity as a protein complex and inhibit differentiation into the extraembryonic lineages. Enriched in the inner cell mass of blastocysts, these Cnot genes are highly expressed in ESC and downregulated during differentiation. In mouse ESCs, Cnot1, Cnot2, and Cnot3 are important for maintenance in both normal conditions and the 2i/LIF medium that sup ports the ground state pluripotency. Genetic analysis indicated that they do not act through known self-renewal pathways or core transcription factors. Instead, they repress the expression of early trophectoderm (TE) transcription factors such as Cdx2. Importantly, these Cnot genes are also necessary for the maintenance of human ESCs, and silencing them mainly lead to TE and primitive endoderm differ entiation. Together, our results indicate that Cnot1, Cnot2, and Cnot3 represent a novel component of the core self-renewal and pluripotency circuitry conserved in mouse and human ESCs.
Keywords: Ccr4-Not, Embryonic stem cell, Self-renewal, Pluripotency, Extraembryonic differentiation
Introduction
Embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of the blastocyst-stage embryos. They have two unique characteristics: pluripotency and self-renewal. Under standing the molecular basis of these two defining features of ESCs will provide insights to the early embryonic development in mammals and facilitate the use of pluripotent stem cells for disease modeling, drug discovery, and stem cell therapies (reviewed in [1, 2]).
For both mouse and human ESCs, a comprehensive model of the self-renewal mechanism is still being defined. It has been shown that self-renewal is controlled by a combination of extracellular signal transduction pathways, transcription factors, and epigenetic regulators [3–7]. In mouse ESCs, signaling pathways including the leukemia inhibitory factor (LIF)-signal transducer and activator of transcription 3 (Stat3) [8], bone morphogenetic protein (BMP)-Smad [9], and Wnt pathways support self-renewal [10], while the fibroblast growth factor (FGF)-extracellular signal-regulated kinase (ERK) pathway induces differentiation [11]. In addition, the self-renewal transcription factors and cofactors form a com plex transcriptional network necessary for the maintenance of the ESC state [3, 5, 7]. The core pluripotency factors, Nanog, Oct4, and Sox2, positively regulate their own expression, and they co-occupy and activate other self-renewal genes while repressing factors necessary for lineage specification [12, 13]. Many others including Dax1, Esrrb, Klf4, Nac1, Nr5a2, Tcfcp2l1, Tcf3, and Zfp281 modulate and refine the core transcription circuitry established by Nanog, Oct4, and Sox2 [14, 15]. At the same time, Myc, Cnot3, Trim28, Zfx, Ronin, and Rex1 form a separate transcription module and regulate ESC genes that are not controlled by the core factors [16]. Finally, epigenetic regulators such as histone-modifying enzymes, ATP-dependent chromatin-remodeling complexes, micro-RNAs, and noncoding RNAs also play important roles. They can facilitate or inhibit the binding of the above transcription factors, fine-tune the expression of key developmental genes, and modulate ESC proliferation [3, 5, 7].
Human ESCs differ from mouse ESCs in morphology, growth, differentiation behavior, and requirement for culture conditions. They depend on FGF-ERK and Activin/Nodal pathways for self-renewal but do not respond to LIF, BMP, or Wntpathways [17, 18]. Much less is known about the regulatory networks governing self-renewal in human ESCs. How ever, it is known that they require the same set of core self-renewal factors, Nanog, Oct4, and Sox2 [19–21].
Recent RNAi screens have identified many novel players and have greatly extended our knowledge on the self-renewal regulatory network [16, 22–25]. One member of the Ccr4-Not complex, Cnot3, was found to be important for mouse ESC self-renewal in our screen [16]. The Ccr4-Not complex is known for the regulation of mRNA stability [26–28]. Different members of the complex are associated with various functions, such as transcription initiation and elongation, deadeny-lation, and protein ubiquitination. This complex has also been implicated in various cellular activities such as DNA repair, histone methylation, spindle positioning, microtubule length regulation, and spermatogenesis. However, the role of this complex in ESC self-renewal has not been fully investigated. In this study, we present evidence that Cnot1, Cnot2, and Cnot3 maintain self-renewal in both mouse and human ESCs as a protein complex, possibly by inhibiting extraembryonic lineage differentiation. They represent a critical component in the self-renewal circuitry that has not been previously characterized.
Materials and Methods
Mouse ESC Culture, Differentiation, and Transfection
Oct4GiP and Cdx2 deletion dKO23-5 cells were kindly provided by Drs. Austin Smith and Hitoshi Niwa. E14Tg2a and J1 cells were obtained from Mutant Mouse Research Resource Centers and American Type Culture Collection. ESCs were maintained on gelatin-coated plates in the ESGRO complete plus clonal grade medium (Millipore).
For embryoid body (EB) formation, ESCs were seeded at 25-50 × 103 cells per square centimeter in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in low-attachment plates (Corning). For LIF-with-drawal, cells were seeded at 10-20 × 103 cells per square centimeter in DMEM supplemented with 10% FBS on gelatin-coated plates. For retinoic acid treatment, cells were cultured in LIF-withdrawal conditions plus 0.2 μM all-trans retinoic acid. Cells were collected at the indicated time points.
For transfections, ESCs were cultured on gelatin-coated plates in M15 medium: DMEM (Invitrogen) supplemented with 15% FBS, 10 μM 2-mercaptoethanol, 0.1 mM nonessential amino acids (Invitrogen), 1× EmbryoMax nucleosides (Millipore), 1,000 U of ESGRO (Millipore). ESCs (20-25 × 103 ) were transfected with siRNAs at 100 nM using lipofectamine 2000 (Invitrogen) in one well of a 96-well plate on day 1. Fifty to hundred percentage of the cells in each well was replated into one well of a 12-well plate on day 2 and cultured for another 3 days. For lineage marker and microarray analysis, cells were harvested for RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR). For alkaline phosphatase (AP) staining, cells were stained for AP activity with the Alkaline Phosphatase Detection Kit (Millipore). To determine the knockdown efficiency of siR NAs, 50-100 × 103 ESCs were transfected with siRNAs at 50 nM in one well of a 24-well plate. Cells were harvested 48 hours after transfection for qRT-PCR or Western blot.
Mouse Trophoblast Stem Cell Culture
Mouse trophoblast stem (TS) cells were kindly provided by Dr. Janet Rossant and were cultured in mouse embryonic fibroblast (MEF) conditioned medium supplemented with heparin and FGF4 without feeders [29].
Oct4GiP Reporter Assay
Oct4GiP reporter assay was carried out as described previously [16]. For each experimental condition, 3–6 independent transfections were carried out and data were plotted as mean ± SEM.
qRT-PCR
Total RNAs were prepared from cells with the Aurum Total RNA kit (Bio-Rad), and cDNAs were generated using the iScript kit (Bio-Rad) according to the manufacturer's instructions. qPCRs were performed using the SsoFast EvaGreen supermix (Bio-Rad) on the Bio-Rad CFX-384 or CFX-96 Real-Time PCR System. Actin was used for normalization. Two to four biological repeats were carried out for each experiment and one representative result was shown. For every biological repeat, triplicate or quadruplicate PCR reactions were performed, and data were plotted as mean ± SEM. Supporting Information Table 1 for primers used.
Western Blot
Cells were lysed in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and total protein con centration was quantitated with the RC DC protein assay kit (Bio-Rad). Alternatively, cells were lysed in SDS-PAGE sample buffer at the same cell concentration. Equal amount of total protein or equivalent proportion was loaded in each lane. Two to four biological repeats were carried out for each experiment and one representative result was shown. Primary antibodies used in this study were as follows Oct4 (Abcam, Cambridge, MA; Cat#AB19857, www.abcam.com), Nanog (Millipore, Billerica, MA; Cat#AB9220, www.millipore.com), Smad1-5-8-phospho-specific (Cell Signaling Technology, Beverly, MA; Cat#9512, www.cellsignal.com), Smad1 (Cell Signaling Technology, Cat#9511), Stat3-S727-phosphospecific (Cell Signaling Technology, Cat#9134), Stat3Y705-phospho-specific (Cell Signaling Technology, Cat#9145), Stat3 (Cell Signaling Technology, Cat#9132), b-Catenin-Ser45-phospho-specific (Cell Signaling Technology, Cat#9564), b-Catenin (Cell Signaling Technology, Cat#9562), Hemagglutinin (HA) (Covance, Princeton, NJ; Cat#MMS-101P, www.covance.com), Ran (BD Biosciences, San Jose, CA; Cat#610340, www.bdbiosciences.com), Cnot1 (Protein-tech, Chicago, IL; Cat#14276-1-AP, www.ptglab.com), Cnot2 (Proteintech, Cat#10313-1-AP). Rabbit polyclonal antibody against mouse Cnot3 was raised using mouse Cnot3 amino acid 1-232.
Overexpression Mouse ESC Lines
Mouse Cnot2 or Cnot3 coding region was PCR cloned into the pDNR223 vector and then transferred into destination expression vectors using the Gateway technology (Invitrogen). The destination expression vectors used are: pHAGE-EF-HA-Puro-DEST and pHAGE-EF-HA-Neo-DEST (see attached maps). The resulting Cnot2 and Cnot3 expression pHAGE vectors were packaged into viruses in 293T cells using standard protocols.
Oct4GiP cells were infected with the pHAGE-EF-Cnot2-HA-Neo or pHAGE-EF-Cnot3-HA-Neo virus, drug was selected, and single clones were picked and amplified. Expression of the exogenous Cnot2-HA or Cnot3-HA was verified by Western blot with the HA antibody, and the level of overexpression at the mRNA level was estimated by qRT-PCR. Three independent clones for the Cnot2-HA line were selected and tested in the rescue experiments, and all three clones rescued Cnot2 siRNA-induced differentiation in the Oct4GiP reporter assay. Similarly, three independent clones for the Cnot3-HA line were selected and tested, and all three clones rescued Cnot3 siRNA-induced differentiation. One clone from each line was then used for all the experiments. E14Tg2a cells were infected with the pHAGE-EF-Cnot2-HA-Puro virus, and a clonal line expressing exogenous Cnot2-HA was generated similarly as described above.
Immunoprecipitation
E14Tg2a cells expressing Cnot2-HA were lysed in lysis buffer (1% NP-40, 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, phenylmethanesulfonylfluoride (PMSF), and Roche EDTA-free Protease inhibitors). Lysates were cleared by sonication and centrifuged to remove insoluble materials and then precleared for 1 hour at 4°C with protein-A agarose beads (Invitrogen). Immunoprecipitations were performed using Roche anti-HA matrix for 4 hours at 4°C. Beads were washed with lysis buffer, and proteins were eluted using 2× SDS-PAGE sample loading buffer (Invitrogen) and heating at 95°C for 10 minutes.
Whole-Mount In Situ Hybridization
Whole-mount in situ hybridization was carried out using an established protocol [30]. Briefly, E3.5 blastocyst embryos from CD-1 mice were collected, fixed, permeabilized, and hybridized to digoxigenin-labeled probes (10 μg/ml). They were washed and incubated with the antidigoxigenin-AP antibody (Roche), and the staining was visualized with BM purple (Roche). Stained embryos were imaged with Leica M-165C stereomicroscope.
For the hybridization probes, Cnot1, Cnot2, and Cnot3 fragments were PCRed from mouse ESC cDNAs using the following primers: Cnot1; 5′-ATGTATTGGCTGAGCTGCATCAGG, 5′-TTG TCCTGAGCTAACTGAGCAGCAG; Cnot2: 5′-TATGCCAAAGC AACAGCCTTCTCG, 5′-AGATGTACCATTCCTGGGTCTGTC; Cnot3: 5′-AGCAAACTGCACTACGGAAAACTC, 5′-ACTGCTAC TCAGGGTCACCTCPCR. PCR products were cloned into pCR-BluntII-TOPO vector (Invitrogen) and sequenced. Probes were generated using the digoxigenin RNA labeling kit (Roche) following manufacturer's instructions.
Microarray Analysis
ESCs were transfected with siRNAs as described above in triplicates. Gene expression analysis was conducted using Agilent Whole Mouse Genome 4 × 44 multiplex format oligo arrays (Agilent Technologies) following the Agilent 1-color microarray-based gene expression analysis protocol.
Raw data files were processed using relevant R/Bioconductor packages [31] including “Agi4 × 44PreProcess” for background correction and normalization. Probe intensities were transformed to log 2 scale and an intensity filter was applied to remove probes whose intensity values were constitutively below the first quartile in each array. Probes were averaged to obtain a single-expression intensity measure per Entrez gene ID and array.
To determine differentially expressed genes, a family-wise mod erated t test [32]was performed followed by Bonferroni multiple-testing correction. Genes were considered differentially expressed if they had an adjusted p value of less than 10−4 and a fold change of greater than 1.5. Fisher's exact test was used to determine the statistical significance of the observed overlap between gene lists. Functional enrichment analysis of upregulated and downregulated genes and cluster analysis of per-gene normalized expression levels was performed using the CLEAN software package [33].
For Figure 3C, the set of 2,463 genes and corresponding expression data were obtained from Aiba et al. [34]. Additional data (Cdx2 overexpression) from Nishiyama et al. [35] and Cnot1, 2, 3 intensity data (control and knockdown samples) were also added using same set of genes. Principal component analysis (PCA) was performed using R and visualized using R package “rgl.” DC, NS, and PL samples were not shown in the PCA plot.
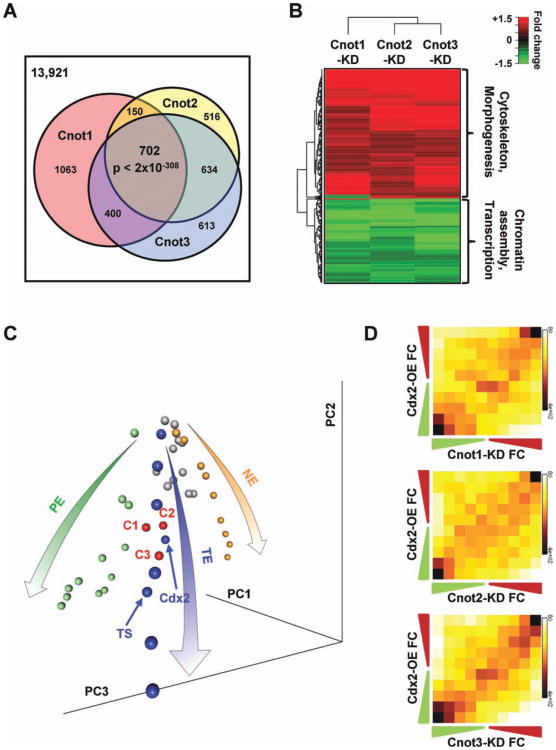
Figure 3.
Silencing Cnot1, Cnot2, or Cnot3 induce differentiation primarily into the TE lineage. (A): Cnot1, Cnot2, or Cnot3 knockdown induced similar gene expression changes. Venn diagram of genes that showed 1.5-fold changes after Cnot1, Cnot2, or Cnot3 knockdown. (B): Heatmap of expression changes for genes that are commonly affected by Cnot1, Cnot2, and Cnot3 knockdown. (C): Cnot1, Cnot2, or Cnot3 knockdown primarily led to TE differentiation. Principal component analysis (PCA) of embryonic stem cell (ESC) differentiation into three different lineages: pluripotent cells (gray spheres), TE differentiation time course and TE cells (blue spheres), PE differentiation time course (green spheres), and neural ectoderm differentiation time course (orange spheres). The three lineage-specific differentiation time courses form a tripod like structure in the PCA space (represented by the arrows). Cnot1 (C1), Cnot2 (C2), or Cnot3 (C3) silencing (red spheres) clustered closely to TE cells (blue). (D): Cdx2 overexpression and Cnot1, Cnot2, or Cnot3 knockdown resulted in similar gene expression changes. Two-dimensional matrix and heat map depicting gene expression changes in Cdx2-overexpression (Cdx2-OE) and Cnot1 (Cnot1-KD), Cnot2 (Cnot2-KD), or Cnot3 (Cnot3-KD) knockdowns, compared with control ESCs. Axes indicate degree of fold change, from nil (middle of axis) to greater than 1.5-fold (outermost square). Numbers indicate the median fold change of genes in each column or row. The intensity of each square represents the number of genes that fall in that square. The Pearson's correlation coefficients for the plots are: 0.24 for Cnot1-KD versus Cdx2-OE, 0.22 for Cnot2-KD versus Cdx2-OE, and 0.34 for Cnot3-KD versus Cdx2-OE. Abbreviations: KD, knockdown; NE, neuroectoderm; PE, primitive endoderm; TE, trophectoderm; TS, trophoblast stem.
For Figure 3D, histogram heat maps showing log fold changes after Cnot1, 2, and 3 knockdown, respectively, against log fold changes 72 hours after Cdx2 overexpression were com puted. First, datasets were mapped using only Entrez gene IDs represented in both datasets. Next, for each dataset, genes were evenly distributed among 10 bins based on their respective log fold change ranging from lowest (most downregulated) to highest (mostupregulated). Gene counts for each of the 10 × 10 bins were displayed as a heatmap. Similar heatmaps were generated for log fold changes after Cnot1, 2, and 3 knockdown, respectively, against log fold changes after 4 days in N2B27 (N4) and 4 days after Gata6 overexpression (G4) from Aiba et al.
For Figure 4B and Supporting Information Fig. S9, published ESC reference datasets were downloaded from GEO using respective accession numbers (Supporting Information Table 2). Where available, raw data files were reprocessed using RMA [36] and Entrez gene-based custom chip definition files, version 13 [37], resulting in a single-expression intensity measure per Entrez gene ID. Otherwise, the respective already preprocessed expression values and corresponding probe annotations were down loaded. All data were transformed to log 2 scale. Each probe identifier was mapped to an Entrez gene ID by matching annotations in the following order until an Entrez gene ID was found: Entrez gene ID, RefSeq ID, gene symbol or alias, gene name, unigene ID, and Ensembl transcript ID. Probes mapping to the same Entrez gene ID were averaged (after log transformation). Fold changes and adjusted p values (knockdown/knockout vs. control) were computed for each dataset independently again using moderated t statistic described above. Datasets were then combined using Entrez gene IDs to map probes/probe sets.
Figure 4.
The Cnot genes maintain self-renewal by repressing early trophectoderm (TE) transcription factors. (A): Cnot1, Cnot2, and Cnot3 knockdown did not immediately affect known self-renewal factors and pathways. Oct4GiP cells were transfected with control-siRNA (Control), Cnot1-siRNA1 (Cnot1-KD), Cnot2-siRNA2 (Cnot2-KD), or Cnot3-siRNA2 (Cnot3-KD) in M15 medium. Cells were collected 48 hours after transfection, and total Stat3, Smad1, b-Catenin as well as phospho-Stat3, phospho-Smad1, phosphor-b-Catenin, Oct4, and Nanog levels were determined by Western blot. Starved: control-transfected ESCs cultured in serum-free and LIF-free medium for additional 4 hours. (B): Comparing gene expression changes caused by perturbations of known self-renewal factors: Cnot1, 2, and 3 silencing induced similar changes to those of Oct4 or Sox2 silencing. Pearson's correlation coefficients were calculated between microarray datasets and depicted in a heatmap. The self-renewal factors were clustered by unsupervised hierarchical clustering based on the correlation coefficients. Microarray datasets used for this plot are listed in Supporting Information Table 2. (C): Cnot2 or Cnot3 overexpression cannot rescue Oct4 or Sox2 silencing-induced differentiation. Oct4GiP cells and Oct4GiP cells overexpressing Cnot2 (Cnot2-Rescue, same as in Fig. 1C) or Cnot3 (Cnot3-Rescue, same as in Fig. 1C) were transfected with control, Oct4 (Oct4-KD), or Sox2 (Sox2-KD) siRNAs, and the % differentiation was determined by the Oct4GiP reporter assay. (D): Cnot1, Cnot2, and Cnot3 knockdown induced TE differentiation in the presence of sustained Oct4 expression. ZHBTc4 cells that constitu-tively express Oct4 at the normal level from a Tet-Off promoter were transfected with control or Cnot1-siRNA1 (Cnot1-KD), Cnot2-siRNA2 (Cnot2-KD), Cnot3-siRNA2 (Cnot3-KD), and the expression of TE markers Cdx2 and Gata3 was determined by qRT-PCR after 4 days. (E): Cdx2 deletion partially rescued Cnot1, Cnot2, and Cnot3 silencing-induced differentiation. Oct4GiP (WT) or dKO23-5 (Cdx2-/- ) cells were transfected with Control-siRNA (Control), Cnot1-siRNA1 (Cnot1-KD), Cnot2-siRNA2 (Cnot2-KD), or Cnot3-siRNA2 (Cnot3-KD), and the expression of lineage markers was determined by qRT-PCR 96-hour after transfection. Abbreviations: ESC, embryonic stem cell; KD, Knockdown; WT, wild type.
Human ESC Culture and siRNA Transfection
Human ESC line H1 (WA01) and H9 (WA09) were received from WiCell Research Institute. Cells were maintained undifferentiated on Matrigel-coated tissue culture plates in mTeSR1 media or E8 media [38] and passaged every 4 days using EDTA. All the experiments were performed with cells between passages 29 and 40.
Human ESCs were reverse-transfected with 100 nM siRNAs using Dharmafect 1 transfection reagent according to the manufacturer's instructions. To verify knockdown efficiency, RNA was extracted 2 days after transfection. For lineage marker analysis, the RNA was extracted 6 days after transfection.
Immunostaining
Human ESCs were washed in phosphate buffered saline (PBS), fixed in 4% formaldehyde (EMD Chemicals) at 4°C for 30 minutes, washed in PBS, blocked and permeabilized in PBS with 5% goat serum and 0.3% Triton X-100 at room temperature for 1 hour. Cells were then incubated in primary antibody overnight in the blocking buffer. The following antibodies at 1:100 dilutions were used: anti-Oct4 (Abcam), anti-Cdx2 (Cell Signaling), and anti-Gata3 (Abcam). The antibody-antigen complexes were visualized using Alexa Fluor 488 goat anti-rabbit IgG at 1:1,000 dilutions.
Results
Cnot1, Cnot2, and Cnot3 Play Critical Roles in Mouse ESC Maintenance As a Protein Complex
We previously identified Cnot3 as a regulator of mouse ESC self-renewal in a genome-wide RNAi screen [16]. Because Cnot3 is part of the CCR4-Not complex, we tested if other components are also important for ESC maintenance using the Oct4GiP reporter assay we previously developed. The Oct4GiP cells express the green fluorescent protein (GFP) from an ESC-specific gene promoter, the Oct4 promoter. As a result, the GFP expression faithfully correlates with the ESC identity and can be used to determine the self-renewal and differentiation status [39]. We silenced the CCR4-Not com plex components with siRNAs individually or in combination in the Oct4GiP cells and measured the percentage of differentiated cells by fluorescence-activated cell sorting (FACS) analysis. We found that only silencing Cnot1, Cnot2, or Cnot3, but not the other components of the Ccr4-Not complex, resulted in a significant increase in the percentage of differentiated cells (Fig. 1A, Supporting Information Fig. S1A, S1B). To rule out siRNA off-target effects, we used two different siRNAs to knockdown each gene and confirmed that the siRNAs efficiently silenced their targets by qRT-PCR (Supporting Information Fig. S2A) and Western blot (Supporting Information Fig. S2B). Furthermore, we generated ESC lines that expressed siRNA-resistant Cnot2 or Cnot3, and showed that these lines were able to fully rescue the differentiation induced by Cnot2 or Cnot3 siRNA transfections, respectively (Fig. 1B, Supporting Information Fig. S3). Expression of siRNA-resistant Cnot2 or Cnot3 was not able to rescue the differentiation caused by silencing of the other two Cnot genes (Fig. 1B), indicating that Cnot1, Cnot2, and Cnot3 are all required to prevent differentiation.
Figure 1.
Silencing Cnot1, Cnot2, or Cnot3 led to mouse embryonic stem cell (ESC) differentiation. (A): Silencing Cnot1, Cnot2, or Cnot3 resulted in ESC differentiation based on the Oct4GiP reporter assay. Oct4GiP ESCs were transfected with indicated siRNAs (two different siR NAs for each CCr4-Not complex gene) in M15 medium and cultured for 4 days. The percentage of differentiated cells (% differentiation) was determined by measuring the percentage of green fluorescent protein-negative cells by fluorescence-activated cell sorting (FACS) at the end of the culture. (B): Expression of siRNA-resistant Cnot2 or Cnot3 rescued the differentiation caused by Cnot2 or Cnot3 knockdown, respectively. Oct4GiP cells or Oct4GiP cells expressing siRNA-resistant Cnot2 (Cnot2-Rescue) or Cnot3 (Cnot3-Rescue) were transfected with Control, Cnot1-siRNA1, Cnot2-siRNA2, or Cnot3-siRNA2, and the percentage of differentiated cells was determined by the Oct4GiP reporter assays. Note that Cnot2-Rescue cells were not able to rescue the differentiation caused by Cnot1 or Cnot3 silencing, and Cnot3-Rescue cells were not able to rescue Cnot1 or Cnot2 silencing. ***, p < .001. (C): Silencing Cnot1, Cnot2, or Cnot3 resulted in morphological changes and loss of alkaline phosphatase (AP) staining in ESCs. Oct4GiP cells were transfected with the indicated siRNAs and cultured in the M15 medium. Cells were stained with the AP staining kit and imaged 4 days after transfection. (D): Cnot1, Cnot2, or Cnot3 silencing led to downregulation of ESC marker and upregulation of differentiation markers. Oct4GiP cells were transfected with the indicated siRNAs and cultured in the M15 medium. Cells were harvested for quantitative real-time PCR (qRT-PCR) analysis 4 days after transfection. ESC marker: Oct4; differentiation markers: Cdx2, Eomes, Gata3, Hand1, and Krt8. (E): Cnot1, Cnot2, or Cnot3 silencing reduced cell proliferation or viability in 2i medium. Oct4GiP cells were transfected with control-siRNA (Control), Cnot1-siRNA1 (Cnot1-KD), Cnot2-siRNA2 (Cnot2-KD), or Cnot3-siRNA2 (Cnot3-KD) and cul tured in 2i medium. Cell numbers were counted by FACS 4 days after transfection and normalized to control-transfected cells. (F): Cnot1, Cnot2, or Cnot3 silencing led to differentiation in 2i medium. Oct4GiP cells were transfected with indicated siRNAs and cultured in 2i medium. Cells were harvested for qRT-PCR analysis 4 days after transfection. (G): Expression of C-terminally HA-tagged Cnot2 (Cnot2-HA) in E14Tg2a cells. Expression of the exogenous Cnot2-HA was detected in Western blot with the HA-antibody, and Ran was used as a loading control. Expression of total (endogenous and exogenous) Cnot2 was determined by qPCR in wild-type E14Tg2a (E14) and Cnot2-HA expressing cells. The expression of the Cnot2-HA was estimated to be ∼2-fold of the endogenous Cnot2 on the mRNA level. (H): Identification of Cnot1 and Cnot3 in Cnot2-HA immunoprecipitation. HA-pull-down was carried out in E14Tg2a cells expressing Cnot2-HA. The presence of Cnot1, Cnot2-HA, and Cnot3 in the total lysate and pull-down sample (HA-beads) were detected by Western blot. Note that Oct4 was not detected in the pull down sample. As a negative control, protein-A beads were used in an independent pull-down. Abbreviations: HA, hemagglutinin; IP, immunoprecipitation; KD, knockdown.
To confirm the result of the reporter assays, we performed AP staining in Oct4GiP and two additional ESC lines E14tg2a and J1. Cells transfected with a nontargeting control-siRNA formed typical compact ESC colonies with high AP staining, but Cnot1, Cnot2, or Cnot3 siRNA transfected cells showed significant changes in cell morphology and loss of AP staining (Fig. 1C, Supporting Information Fig. S4). Interestingly, Cnot1 silencing led to different morphological changes from those caused by Cnot2 or Cnot3 silencing as well as slight difference in lineage marker expressions (Fig. 1D). This is possibly because Cnot1 is the scaffold protein in the Ccr4-Not complex and its silencing may have a more profound effect. Alternatively, Cnot1 may have nonoverlapping functions with Cnot2 and Cnot3. In addition to the AP staining, we performed qRT-PCRs to deter mine the expression of lineage marker genes. Cnot1, Cnot2, or Cnot3 silencing resulted in a modest decrease in the expression of ESC markers Oct4, Lin28, and Tcl1 (Fig. 1D, Supporting Information Fig. S5), consistent with the Oct4GiP reporter assay. Furthermore, the silencing led to markedly increased expression of differentiation markers, such as Cdx2, Gata3, Eomes, Krt8, and Hand1 (Fig. 1D, Supporting Information Figs. S5, S6). These results clearly demonstrated that Cnot1, Cnot2, and Cnot3 play important roles in ESC maintenance.
Double inhibition of Erk and GSK-3β signaling by the “2i” medium has been shown to maintain mouse ESCs in a naïve pluripotent state or the ground state [40]. We found that silencing Cnot1, Cnot2, or Cnot3 reduced ESC proliferation or viability in the ground state (Fig. 1E), consistent with recent findings that they are involved in cell proliferation and apoptosis [41, 42]. More importantly, the silencing led to downregulation of Oct4 and upregulation of Cdx2 and Gata3 (Fig. 1F). Therefore, Cnot1, Cnot2, and Cnot3 are required for the main tenance of ESC identity in the ground state and are likely an essential component of the core self-renewal circuitry.
To test whether Cnot1, Cnot2, and Cnot3 exist in a complex in ESCs, we generated a mouse ESC line that expresses HA-tagged Cnot2 (Cnot2-HA) (Fig. 1G). We found that the exoge nously expressed Cnot2-HA coimmunoprecipitated with endogenous Cnot1 and Cnot3 but not Oct4 (Fig. 1H). Because Cnot1, Cnot2, and Cnot3 have all been suggested to be transcription factors [43–45], it is possible that they may function as a transcription factor complex to regulate ESC self-renewal.
Cnot1, Cnot2, and Cnot3 Expression Closely Correlates with the Pluripotency State
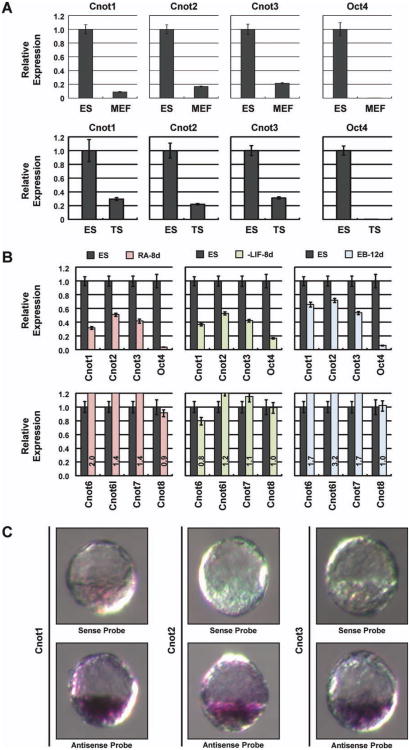
To further support the above findings, we investigated whether the expression of Cnot1, Cnot2, and Cnot3 positively correlates with the pluripotency state. We first determined their relative expression by qRT-PCR between ESCs, MEFs, and TS cells. We found that they are expressed at much higher levels in ESCs compared to the other two cell types (Fig. 2A). We also determined their expression during ESC differentiation induced by retinoid acid treatment, EB formation, and LIF withdrawal. During differentiation, the expression of Cnot1, 2, and 3 decreased (Fig. 2B). In contrast, the expression of other CCR4-Not complex components that did not score in the Oct4GiP reporter assay, such as Cnot6, 6L, 7, and 8, did not decrease significantly or even increased (Fig. 2B, Supporting Information Fig. S7A). This is consistent with our finding that only Cnot1, Cnot2, and Cnot3 play important roles in ESC maintenance. Finally, because ESCs are derived from the ICM of the blastocyst embryos, we examined the expression of Cnot1, Cnot2, and Cnot3 in E3.5 mouse embryos by in situ hybridization. We found that they are enriched in the ICM of the blastocysts, suggesting that they are likely important for the maintenance of the ICM cells in vivo (Fig. 2C). In agreement with these findings, Cnot1, Cnot2, Cnot3 are highly expressed in oocyte, fertilized eggs, and ESCs in comparison to other somatic cell types and tissues based on the Bio-GPS database [46](Supporting Information Fig. S7B). Thus, the expression of Cnot1, Cnot2, and Cnot3 highly correlates with the state of pluripotency, supporting the notion that they are important for the maintenance of pluripotency.
Figure 2.
Cnot1, Cnot2, and Cnot3 are expressed in mouse ESCs and the inner cell mass (ICM) of mouse blastocysts. (A): Cnot gene expression in mouse ES, MEF, and TS cells. The expression of the Cnot genes was determined by quantitative real-time PCR (qRT-PCR), and the ESC-specific marker Oct4 was used as a reference. (B): Cnot gene expression during ESC differentiation induced by embryoid body formation, retinoid acid treatment, and LIF withdrawal. The expression of the Cnot genes was determined by qRT-PCR at the indicated time points. Oct4 was used as a reference. (C): Detection of Cnot1, Cnot2, and Cnot3 transcripts in mouse E3.5 embryo via whole-mount in situ hybridization. Sense probes were used as negative controls, and the antisense probes showed the predominant expression of Cnot1, Cnot2, and Cnot3 in the ICM. Abbreviations: ES, embryonic stem; MEF, mouse embryonic fibroblast; TS, trophoblast stem.
Cnot1, Cnot2, and Cnot3 Silencing in Mouse ESCs Led to Trophectoderm Differentiation
To identify genes affected by Cnot1, Cnot2, or Cnot3 silencing in ESCs, we determined the resulting global gene expression changes by microarrays. We found that Cnot1, Cnot2, or Cnot3 silencing each affected a large number of genes, among which there is a significant overlap (Fig. 3A, < 2 × 10−308). In the commonly affected genes, the upregulated ones were highly enriched for genes involved in cytoskeleton and cellular morphogenesis, and the downregulated ones were highly enriched for genes involved in histone assembly, chromatin modification, and transcription (Fig. 3B, Supporting Information Table 3).
To better understand the consequence of Cnot1, Cnot2, and Cnot3 silencing, we compared our microarray results with previously published datasets of ESC differentiation into three specific lineages: the primitive endoderm (PE), trophectoderm (TE), and neuroectoderm (NE) [34, 35]. PCA indicated that gene expression signatures induced by Cnot1, Cnot2, or Cnot3 knockdown are similar to those found during TE differentiation, especially to ESCs overexpressing Cdx2 (Fig. 3C). In fact, Cnot gene silencing and Cdx2 overexpression led to very similar changes in the global transcriptional profile (Fig. 3D). In contrast, Cnot gene silencing produced different expression changes from cells undergoing NE or PE differentiation (Supporting Information Fig. S8). qRT-PCRs con firmed that Cnot1, Cnot2, or Cnot3 knockdown induced high expression of TE lineage markers, including Cdx2, Eomes, Gata3, Hand1, Krt8, and Aqp3 (Fig. 1D, Supporting Information Fig. S5). On the contrary, the knockdowns only induced the expression of very few ectoderm, mesoderm, and endo-derm genes at much lower levels (Supporting Information Fig. S5). Importantly, the knockdowns induced TE markers in the 2i medium that maintains ESCs in the ground state (Fig. 1F). Thus, Cnot1, Cnot2, or Cnot3 silencing led to a differentiation pattern that is consistent with the TE lineage.
The Cnot Genes Maintain Mouse ESC Self-Renewal by Repressing TE Transcription Factors
In mouse ESCs, self-renewal is supported by signaling pathways such as the LIF-Stat3, BMP-Smad, and Wnt-β-catenin/ Tcf3. We found that silencing Cnot1, Cnot2, and Cnot3 did not significantly inhibit the expression or phosphorylation of Stat3, Smad1, and β-catenin (Fig. 4A), indicating that the silencing did not impair the LIF, BMP, or Wnt signaling.
Besides the signal transduction pathways, ESC self-renewal is also maintained by a list of pluripotency transcription factors. We compared gene expression changes that are caused by the perturbation of Cnot1, Cnot2, Cnot3 and other self-renewal genes. We calculated correlation coefficients for every two self-renewal genes based on the similarity in the gene expression changes caused by their perturbation, and we clustered all the genes by unsupervised hierarchical clustering of the correlation coefficients. We identified different clusters among these self-renewal genes, which may represent different functional modules for maintaining self-renewal (Fig. 4B, Supporting Information Fig. S9). For example, Oct4 and Sox2 clustered together, consistent with their known biochemical and genetic interactions [12, 47]. Stat3 and Brg1 clustered together, consistent with the recent report that Brg1 potentiates Stat3 signaling [48]. Importantly, Cnot1, Cnot2, and Cnot3 clustered as a group, again supporting our finding that they function as a complex to maintain self-renewal. However, Cnot1, Cnot2, and Cnot3 did not cluster closely with Tcf3, Sall4, Ring1b, Tip60, Tcl1, Esrrb, Tbx2, Paf1, Klf2/4/5, Stat3, Brg1, and Nanog (Fig. 4B, Supporting Information Fig. S9), suggesting that the Cnot1-2-3 complex does not have strong functional overlap with these self-renewal genes.
Interestingly, Cnot1, Cnot2, and Cnot3 clustered closely with Oct4 and Sox2 (Fig. 4B, Supporting Information Fig. S9). To understand the relationship between the Cnot1-2-3 and Oct4-Sox2 complexes, we first tested whether they are physically associated with each other. By immunoprecipitation, we were not able to detect interactions between the Cnotproteins and Oct4 (Fig. 1H). In addition, previous proteomic studies also did not identify Cnot1, Cnot2, or Cnot3 as Oct4 interacting proteins [47, 49, 50]. We next tested whether Cnot1, Cnot2, and Cnot3 have genetic interactions with Oct4 and Sox2. In the Oct4GiP cells, Cnot2 and Cnot3 overexpression was able to rescue Cnot2 and Cnot3 siRNA-induced differentiation, respectively (Fig. 1B). However, the overexpression was not able to suppress Oct4 or Sox2 siRNA-induced differentiation (Fig. 4C), indicating that the Cnot genes most likely do not operate downstream of Oct4 or Sox2. In addition, sustained expression of Oct4 in the ZHBTc4 ESCs [51] was not able to suppress Cnot1, Cnot2, or Cnot3 siRNA-induced differentiation (Fig. 4D), indicating that Cnot1, Cnot2, or Cnot3 silencing do not induce differentiation by downregulation of Oct4. Consistent with these results, Oct4 or Sox2 knockdowns did not lead to immediate changes in Cnot1, Cnot2, or Cnot3 expression on the mRNA levels (Supporting Information Fig. S10), and Cnot1, Cnot2, or Cnot3 knockdowns did not lead to immediate changes in Oct4 or Sox2 expression either (Fig. 4A, Supporting Information Fig. S10). Thus, it is unlikely that Cnot1-2-3 and Oct4-Sox2 directly regulate each other on the transcription level. Together, we were not able to find obvious biochemical or genetic interactions between Cnot1-2-3 and Oct4-Sox2. Because silencing any of these genes leads to TE differentiation, we believe that the similarities in gene expression changes are most likely a consequence of the differentiation.
Based on the above results, we concluded that Cnot1, Cnot2, and Cnot3 do not impinge on the known self-renewal pathways and factors, and they may therefore regulate ESC self-renewal through a different mechanism. We noticed that silencing of Cnot1, Cnot2, or Cnot3 induced the expression of several early TE transcription factors, including Cdx2, Eomes and Gata3 (Fig. 1D). All these TE transcription factors can induce TE differentiation when overexpressed in ESCs [35, 52, 53]. In addition, gene expression changes caused by Cnot1, Cnot2, or Cnot3 silencing highly resembled those caused by Cdx2 overexpression (Fig. 3D). Therefore, we hypothesized that the Cnot genes may maintain self-renewal by repressing early TE transcription factors and restrict TE differentiation. To test our hypothesis, we silenced the Cnot genes in the Cdx2-/- ESCs [35, 52, 53]. We found that Cdx2 deletion rescued the loss of pluripotency marker Oct4 due to Cnot1, Cnot2, and Cnot3 silencing. Furthermore, Cdx2 deletion partially rescued the induction of TE markers caused by Cnot gene silencing (Fig. 4E). This result strongly supports our hypothesis that the Cnot genes maintain the ESC state by inhibiting early TE factors and TE differentiation.
Cnot1, Cnot2, and Cnot3 Are Important for Human ESC Self-Renewal
Human ESCs, like mouse ESCs, are derived from the ICM of the preimplantation embryo. However, they are significantly different from mouse ESCs with respect to the requirement for culture conditions and extrinsic signaling pathways. Thus far, only the core self-renewal transcription factors, Nanog, Oct4, and Sox2, have been shown to be shared between human and mouse ESCs for maintenance. We found that Cnot1, Cnot2, and Cnot3 are highly expressed in the H1 human ESCs and markedly downregulated during BMP4-induced differentiation (Fig. 5A). Silencing the Cnot genes in the H1 cells (Supporting Information Fig. S11) led to changes in cellular morphology that is reminiscent of extraembryonic differentiation (Fig. 5B). Interestingly, unlike in the mouse ESCs, Cnot1 silencing in human ESCs resulted in more subtle phenotypes compared to Cnot2 and Cnot3 silencing. By immunofluorescence staining and qRT-PCR analysis, Cnot gene silencing largely induced the expression of TE and PE markers such as Cdx2, Eomes, Gata3, Hand1, Cga, Gata4, and Gata6, with much less induction of the mesoderm and endoderm markers (Fig. 5C, 5D, Supporting Information Fig. S12). As a comparison, Oct4 knockdown resulted in similar levels of Cdx2 expression by immunofluorescence (Fig. 5C). We silenced Cnot2 and Cnot3 in another human ESC line H9 and observed similar lineage marker expression (Supporting Information Fig. S13). Therefore, our data indicate that Cnot1, Cnot2, and Cnot3 are important for human ESC maintenance and they mainly, but not exclusively, inhibit differentiation into the extraembryonic lineages. Based on these findings, we propose that the functionality of the Cnot1-2-3 complex is likely conserved between mouse and human ESCs, and it may represent a critical component in the selfrenewal regulatory network in pluripotent stem cells in general (Fig. 6).
Figure 5.
Cnot1, Cnot2, and Cnot3 are required for human embryonic stem cell (ESC) self-renewal. (A): Cnot1, Cnot2, and Cnot3 were down-regulated during human ESC differentiation. H1 human ESCs were differentiated for 7 days using 100 ng/ml human recombinant BMP4. The expression levels of Cnot1, Cnot2, and Cnot3 as well as Oct4 and differentiation markers Cdx2 and Hand1 were determined by quantitative realtime PCR (qRT-PCR). (B): Silencing of Cnot1, Cnot2, or Cnot3 led to morphological changes of human ESCs. H1 cells were imaged 6 days after transfection. Phase-contrast images highlight the undifferentiated morphology of human ESCs in the lipids-only transfected cells (mock) versus the differentiated phenotype in the Cnot1, Cnot2, or Cnot3 siRNA transfected cells. (C): Silencing of the Cnot genes led to upregulation of the Cdx2 and Gata3 proteins. H1 cells were transfected with lipids-only (mock), Oct4, Cnot2, or Cnot3 siRNAs. Cells were fixed and stained for Cdx2 or Gata3 expression by immunofluorescence staining 6 days after transfection. (D): Silencing of the Cnot genes led to downregulation of the ESC marker and upregulation of the extraembryonic markers. H1 cells were harvested 6 days after transfection and marker expression was determined by qRT-PCR. Abbreviations: BMP, bone morphogenetic protein; DAPI, 4′-6-diamidino-2-phenylindole.
Figure 6.
Proposed model of Cnot1, Cnot2, and Cnot3 in ESC maintenance. Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and self-renewal by inhibiting differentiation into the extraembryonic lineages. Abbreviations: BMP, bone morphogenetic protein; ESC, embryonic stem cell; FGF, fibroblast growth factor; LIF, leukemia inhibitory factor.
Discussion
In this study, we presented detailed evidence that Cnot1, Cnot2, and Cnot3 of the Ccr4-Not complex are key players in the maintenance of self-renewal in both mouse and human ESCs, possibly through the inhibition of extraembryonic lineage differentiation. The Ccr4-Not complex is important for the regulation of mRNA synthesis and decay in eukaryotic cells [26–28, 54, 55]. Interestingly, different members of the complex have been implicated in different cellular functions [26–28, 54, 55], and distinct Ccr4-Not complexes with different subunit compositions have been discovered [56, 57]. Cnot1, Cnot2, and Cnot3 were initially identified as negative regulators of transcription in yeast [58]. In human cells, they can act as transcription repressors or activators to regulate nuclear receptor-mediated transcription as well [43–45]. Cnot6, Cnot6L, Cnot7, and Cnot8 are deadenylases and play major roles in the deadenylation of mRNAs [26, 28]. In this study, we found that only silencing Cnot1, Cnot2, or Cnot3, but not other known Ccr4-Not complex members, led to significant differentiation of ESCs. Silencing the deadenylase subunits, either individually or in combination, did not result in differentiation (Fig. 1B, Supporting Information Fig. S2), suggesting that the deadenylation activity of the Ccr4-Not complex may not be rate-limiting in self-renewal. Our finding is consistent with the phenotypes of the Cnot gene knockout mice. Specifically, Cnot3 deletion in mice resulted in embryonic lethality at the implantation stage, and the Cnot3 null blastocysts had defective ICM outgrowth [59, 60]. On the contrary, Cnot7 deletion resulted in defects in spermatogenesis but not early embryonic development [61]. Because Cnot1, Cnot2, and Cnot3 have been associated with transcription regulation and because they function as a complex in ESCs, we propose that they may maintain self-renewal by regulating gene expression as a transcription factor complex. Consistent with this hypothesis, we previously showed that Cnot3 occupies the promoter regions of many genes in ESCs and forms a novel transcription module with several other self-renewal factors [16].
Suppressing Erk activation can maintain mouse ESCs in the ground state of self-renewal and pluripotency, presumably by insulating the cells from extrinsic signalling-induced differentiation. Inhibition of Gsk3 further reinforces this effect by relieving the repression of pluripotency genes by the Tcf3 transcription factor [40, 62–64]. In the “2i” condition, some factors that are otherwise necessary for self-renewal in serum containing medium, such as BMP4 [40]and Myc/Max [65], are no longer required. In contrast, we found that Cnot1, Cnot2, and Cnot3 are indispensible for ESC maintenance even in the 2i medium, as their knockdowns resulted in reduced cell proliferation or viability and loss of ESC state. Therefore, Cnot1, Cnot2, and Cnot3 are critical components of the self-renewal circuitry.
Mouse ESCs are derived from ICM that is established after the segregation of the TE. They do not differentiate into TE cells under normal culture conditions, and they do not contribute to the TE lineage after blastocyst injection. However, TE differentiation can be initiated by a number of genetic manipulations including the repression of self-renewal genes such as Oct4 or Sox2 and the activation of TE transcription factors such as Cdx2, Eomes, or Gata3. Our results showed that Cnot1, Cnot2, or Cnot3 silencing also induced differentiation primarily into the TE lineage. TE marker expression occurred in the normal ESC culture condition as well as the 2i medium that maintains ground state pluripotency. However, protein interaction, gene expression, and epistasis analysis indicated that Cnot1-2-3 and Oct4-Sox2 most likely act independently, and our previous ChIP-chip data also suggested that Cnot3 and Oct4 belong to different modules in the self-renewal transcription network [16]. On the other hand, silencing of Cnot1, Cnot2, or Cnot3 induced the expression of several early TE transcription factors, including Cdx2, Eomes, and Gata3. Cdx2 deletion can partially rescue the phenotype of Cnot2 or Cnot3 knockdowns. Therefore, we propose that Cnot1, Cnot2, and Cnot3 maintain self-renewal by inhibiting the TE markers and preventing TE differentiation in mouse ESCs. Interestingly, Cnot3 does not appear to bind to the promoter regions of Cdx2, Eomes, or Gata3 based on our previous result (Supporting Information Fig. S12) [16]. There may be other unknown factors mediating the function of Cnot1, Cnot2, and Cnot3 in this process.
Finally, it has been proposed that there are two phases or states of pluripotency: the naïve pluripotency and the primed pluripotency. Both states share the following characteristics: differentiation into three germ layers, teratomaformation, and expression of and requirement for Nanog, Oct4, and Sox2. However, the naïve pluripotency is further characterized by the competency to contribute to blastocyst chimeras. Mouse ESCs are thought to represent the naïve pluripotency state, while human ESCs represent the primed pluripotency [66], and they require different factors for their maintenance. Our data suggest that like Nanog, Oct4, and Sox2, Cnot1, Cnot2, and Cnot3 are the very few factors identified so far that have conserved functions in both mouse and human ESCs, and therefore in the maintenance of the two different states of pluripotency (Fig. 6). They provide a new opportunity for dissecting the similarities and differences between mouse and human ESCs and the differences between naïve and primed pluripotency on the molecular level.
Summary
We showed that Cnot1, Cnot2, and Cnot3 are highly expressed in ESCs and the ICM of the blastocyst-stage embryos in comparison to other cell types. They form a complex and play critical roles in the maintenance of both mouse and human ESCs. In mouse ESCs, Cnot1, Cnot2, and Cnot3 act independently of known self-renewal genes and pathways, and they regulate self-renewal by inhibiting TE differentiation through the repression of early TE transcription factors. Our data suggest that Cnot1, Cnot2, and Cnot3 represent a novel component in the core self-renewal circuitry.
Supplementary Material
Acknowledgments
We thank Drs. Thomas Kunkel, Paul Wade, Carmen Williams (NIEHS), Rene Maehr (UMass Med), and Guokai Chen (NHLBI) for providing insightful comments and suggestions on the manuscript. We thank Dr. Austin Smith for providing the Oct4GiP cells, Dr. Hitoshi Niwa for providing the Cdx2-/- ESCs, Dr. Janet Rossant for providing the mouse TS cells, Dr. G. Sebastiaan Winkler (University of Nottingham) and Dr. H. Th. Marc Timmers (University Medical Centre, Utrecht) for providing the Cnot1 antibody and technical guidance on detecting the Cnot proteins. We thank the FACS, microarray, and animal facilities at NIEHS for assistance with the experiments. This research was supported by the National Institute of Environmental Health Sciences, National Institutes of Health Intramural Research Program Z01ES102745 (to G.H.), Z01ES071006-11 (to T.K.A.), and 1ZIAES102625-02 (to R.J.).
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no conflicts of interest related to this work.
See www.StemCells.com for supporting information available online.
References
- 1.Keller G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 2.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Loh YH, Yang L, Yang JC, et al. Genomic approaches to deconstruct pluripotency. Annu Rev Genomics Hum Genet. 2011;12:165–185. doi: 10.1146/annurev-genom-082410-101506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loh KM, Lim B. A precarious balance: Pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 6.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 7.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niwa H, Burdon T, Chambers I, et al. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying QL, Nichols J, Chambers I, et al. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 10.Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 11.Kunath T, Saba-El-Leil MK, Almousailleakh M, et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 12.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 13.Mathur D, Danford TW, Boyer LA, et al. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Chu J, Shen X, et al. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu G, Kim J, Xu Q, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levenstein ME, Ludwig TE, Xu RH, et al. Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells. 2006;24:568–574. doi: 10.1634/stemcells.2005-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 19.Adachi K, Suemori H, Yasuda SY, et al. Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes Cells. 2010;15:455–470. doi: 10.1111/j.1365-2443.2010.01400.x. [DOI] [PubMed] [Google Scholar]
- 20.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaehres H, Lensch MW, Daheron L, et al. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005;23:299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
- 22.Chia NY, Chan YS, Feng B, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 23.Ding L, Paszkowski-Rogacz M, Nitzsche A, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanova N, Dobrin R, Lu R, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 26.Bartlam M, Yamamoto T. The structural basis for deadenylation by the CCR4-NOT complex. Protein Cell. 2010;1:443–452. doi: 10.1007/s13238-010-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- 28.Wiederhold K, Passmore LA. Cytoplasmic deadenylation: Regulation of mRNA fate. Biochem Soc Trans. 2010;38:1531–1536. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka S, Kunath T, Hadjantonakis AK, et al. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 30.Piette D, Hendrickx M, Willems E, et al. An optimized procedure for whole-mount in situ hybridization on mouse embryos and embryoid bodies. Nat Protoc. 2008;3:1194–1201. doi: 10.1038/nprot.2008.103. [DOI] [PubMed] [Google Scholar]
- 31.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: Open software development for computational biology and bioinformatics. GenomeBiol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet MolBiol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 33.Freudenberg JM, Joshi VK, Hu Z, et al. CLEAN: Clustering enrichment analysis. BMC Bioinformatics. 2009;10:234. doi: 10.1186/1471-2105-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aiba K, Nedorezov T, Piao Y, et al. Defining developmental potency and cell lineage trajectories by expression profiling of differentiating mouse embryonic stem cells. DNA Res. 2009;16:73–80. doi: 10.1093/dnares/dsn035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishiyama A, Xin L, Sharov AA, et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell. 2009;5:420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 37.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen G, Gulbranson DR, Hou Z, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- 40.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito K, Inoue T, Yokoyama K, et al. CNOT2 depletion disrupts and inhibits the CCR4-NOT deadenylase complex and induces apoptotic cell death. Genes Cells. 2011;16:368–379. doi: 10.1111/j.1365-2443.2011.01492.x. [DOI] [PubMed] [Google Scholar]
- 42.Ito K, Takahashi A, Morita M, et al. The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell. 2011;2:755–763. doi: 10.1007/s13238-011-1092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garapaty S, Mahajan MA, Samuels HH. Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting Factor NIF-1. J Biol Chem. 2008;283:6806–6816. doi: 10.1074/jbc.M706986200. [DOI] [PubMed] [Google Scholar]
- 44.Winkler GS, Mulder KW, Bardwell VJ, et al. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006;25:3089–3099. doi: 10.1038/sj.emboj.7601194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zwartjes CG, Jayne S, van den Berg DL, et al. Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4-not complex. J Biol Chem. 2004;279:10848–10854. doi: 10.1074/jbc.M311747200. [DOI] [PubMed] [Google Scholar]
- 46.Wu C, Orozco C, Boyer J, et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Rao S, Chu J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 48.Ho L, Miller EL, Ronan JL, et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardo M, Lang B, Yu L, et al. An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van den Berg DL, Snoek T, Mullin NP, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 52.Niwa H, Toyooka Y, Shimosato D, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 53.Ralston A, Cox BJ, Nishioka N, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- 54.Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: A regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 55.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 56.Chen J, Rappsilber J, Chiang YC, et al. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 57.Lau NC, Kolkman A, van Schaik FM, et al. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J. 2009;422:443–453. doi: 10.1042/BJ20090500. [DOI] [PubMed] [Google Scholar]
- 58.Collart MA, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 59.Morita M, Oike Y, Nagashima T, et al. Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3(+/™) mice. EMBO J. 2011;30:4678–4691. doi: 10.1038/emboj.2011.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neely GG, Kuba K, Cammarato A, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berthet C, Morera AM, Asensio MJ, et al. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol Cell Biol. 2004;24:5808–5820. doi: 10.1128/MCB.24.13.5808-5820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silva J, Barrandon O, Nichols J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 64.Wray J, Kalkan T, Gomez-Lopez S, et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hishida T, Nozaki Y, Nakachi Y, et al. Indefinite self-renewal of ESCs through Myc/Max transcriptional complex-independent mechanisms. Cell Stem Cell. 2011;9:37–49. doi: 10.1016/j.stem.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 66.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.