Abstract
AMPs are evolutional weapons, widely used by animals and plants in their innate immune system to fend off invading microbes. The present study reports characterization of a new ALF isoform (Sc-ALF; HQ638024) and the first crustin (Sc-crustin; HQ638025) from the mud crab, Scylla serrata. The full-length cDNA of Sc-ALF consisted of 477 bp with an ORF of 123 amino acids and a putative signal peptide of 26 amino acids. Sc-ALF had a predicted molecular weight (MW) of 11.17 kDa and theoretical isoelectric point (pI) of 9.95. Two highly conserved cysteine residues and putative LPS binding domain were observed in Sc-ALF. Comparison of amino acid sequences with neighbor-joining tree indicated that Sc-ALF shared maximum similarity with ALF of S. paramamosain. Peptide model of Sc-ALF created using SWISS-MODEL server was found to consist of two α-helices crowded against a four-strand β-sheet. The full-length cDNA of Sc-crustin consisted of 433 base pairs with an ORF of 111 amino acids and a putative signal peptide of 21 amino acids. Comparison of amino acid sequences with a neighbor-joining tree revealed that Sc-crustin shared high identity with other known crustins characterized from S. paramamosain, P. trituberculatus, H. araneus, C. maenas and F. chinensis. A whey-acidic-protein domain could be detected at the C-terminus with the characteristic four disulfide core. Sc-crustin had a predicted MW of 10.24 kDa and a pI of 8.76. Peptide model of Sc-crustin created using SWISS-MODEL server indicated a random coiled structure that is with two possible β-sheets but no helices.
Keywords: Antimicrobial peptide, Scylla serrata, Crustin, Anti-lipopolysaccharide factor, Innate immunity
Highlights
► Study reports an ALF isoform and the first crustin sequence from Scylla serrata. ► Sc-ALF has a predicted MW of 11.17 kDa and theoretical pI of 9.95. ► Highly conserved cysteine residues and LPS binding domain were observed in Sc-ALF. ► Sc-crustin shared 81% identity with the crustin from Scylla paramamosain. ► Sc-crustin has a predicted MW of 10.24 kDa and a theoretical pI of 8.
1. Introduction
The economic role of marine and freshwater crustaceans as a food source in the export market demands the need to augment fishery resources through the adoption of intensive culture practices. This has led to physiological stress on cultured organisms, often increasing the incidence of diseases [1]. Despite the development of safe and potent antibiotics, bacterial diseases remain a worldwide health crisis due to the emergence of multiple drug resistant pathogens [2]. The use of antimicrobial peptides (AMPs) as a therapeutic tool has been among the most promising avenues investigated to date for addressing antibiotic resistance [3]. AMPs are found in a wide range of prokaryotic and eukaryotic organisms from plants to human beings [4–7]. In crabs, several AMPs have been isolated and characterized, viz. the 6.5 kDa AMP and a cysteine-rich 11.5 kDa AMP (carcinin) from the shore crab, Carcinus maenas [8,9], callinectin from the blue crab, Callinectes sapidus [10], scygonadin, an anionic antimicrobial peptide from the mud crab, Scylla serrata [11], anti-lipopolysaccharide factor (ALF) and crustin from the mud crab, Scylla paramamosain [12,13] and arasin and hyasin from the spider crab, Hyas araneus [14]. Hemocytes have been proved to be the site of production and storage of AMPs in invertebrates such as horseshoe crabs, mussels and decapod crustaceans [9,15–18].
The brown mud crab, Scylla serrata, is a decapod crustacean of the brachyuran family, Portunidae. They are tolerant to wide range of environmental parameters and there has been a huge interest in the aquaculture of this species due to its high demand/price, high flesh content and rapid growth rates in captivity. Mud crabs are a highly delicious seafood commodity and are therefore an important candidate species for aquaculture. S. serrata is a well known commercial species in India, Philippines and Vietnam. Like many other decapod crustaceans [8,19–21], the mud crab also possesses broad-spectrum antibacterial activity in its hemolymph that constitutes part of its nonspecific defenses. However, there are hardly any published works on major AMP families (ALF and crustins) present in this species. The present study aims at the molecular characterization and phylogenetic analysis of two major families of AMPs, viz. anti-lipopolysaccharide factor and crustin from S. serrata.
2. Materials and methods
Healthy adults of S. serrata (∼300 g body weight) were collected from the backwater stream along Vypeen, Kochi, India. Hemolymph was collected from the base of abdominal appendages using specially designed capillary tubes (RNase-free) rinsed using pre-cooled anticoagulant solution (RNase-free 10% sodium citrate, pH 7.0). Total RNA was extracted from the hemocytes using TRI reagent (Sigma) following manufacture's protocol. RNA was quantified by spectrophotometry at 260 and 280 nm. Only RNAs with absorbance ratios (A260:A280) greater than 1.8 were used for the present work. First strand cDNA was generated in a 20 μl reaction volume containing 5 μg total RNA, 1x RT buffer, 2 mM dNTP, 2 μM oligo d(T20), 20 U of RNase inhibitor and 100 U of M-MLV reverse transcriptase (Ferementas, Inc.). The reaction was conducted at 42 °C for 1 h followed by an inactivation step at 85 °C for 15 min. PCR amplification of 1 μl of cDNA was performed in a 25 μl reaction volume containing 1x standard Taq buffer (10 mM Tris–HCl, 50 mM KCl, pH 8.3), 3.5 mM MgCl2, 200 μM dNTPs, 0.4 μM each primer and 1 U Taq DNA polymerase (Fermentas Inc.). PCR primers were designed using GeneTool software based on consensus sequence. Amplifications were performed using the primers (1) Sc-ALF-F (5′-ggacagaagaaacattgaggacgacgca-3′), Sc-ALF-R (5′-ggaaatcaaaaacatccattacaggtca-3′) and (2) Sc-Crus-F (5′-gagagcagaattagacactgt-3′), Sc-Crus-R (5′-atatagtataacataaccatacttc-3′). The thermal profile used was 94 °C for 2 min followed by 35 cycles of 94 °C for 15 s, 60 °C for 30 s and 68 °C for 30 s and a final extension at 68 °C for 10 min. PCR products were analyzed by electrophoresis in 1.5% agarose gels in TBE buffer, stained with ethidium bromide and visualized under UV light. Purified PCR products were sequenced at Scigenom, India.
The sequence homology and the deduced amino acid sequence comparisons were carried out using BLAST algorithm (tblastn) at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/blast). Gene translation and prediction of deduced protein were performed with ExPASy (http://www.au.expasy.org/). The signal peptide was predicted by SignalP program (http://www.au.expasy.org/). The multiple sequence alignments were performed with amino acid sequences of known crustins and ALFs from decapod crustaceans using CLUSTALW and GENDOC. Amino acid sequences of all known crustins and ALFs were retrieved from the NCBI GenBank and phylogenetic tree was constructed by the Neighbor-Joining (NJ) method and the Maximum Likelihood (ML) method using MEGA version 4.0 [22]. The structural models of the AMPs were created using SWISS-MODEL server. The nucleotide sequences and deduced amino acid sequences of the antimicrobial peptides were submitted to GenBank.
3. Results and discussion
In the present study two AMPs belonging to ALF and crustin families were characterized from the hemocytes of S. serrata, herein after referred to as Sc-ALF and Sc-crustin, respectively. The ORF of Sc-ALF consisted of 123 amino acids (Fig. 1A). BLAST analysis of the nucleotide sequences revealed the relation of Sc-ALF to other ALFs present in other decapod crustaceans. Sc-ALF was found to be 93% similar to an ALF isoform characterized from S. serrata. However, a 100% similar ALF molecule was found to be present in S. paramamosain [12]. Sc-ALF also shared similarity to ALFs of Portunus trituberculatus (76%), Pacifastacus leniusculus (52%) and Fenneropenaeus indicus (41%) (Table 1). Analysis with the SignalP software revealed the presence of a signal peptide with 26 amino acids at the N-terminal region of the Sc-ALF (Fig. 1A). The mature peptide consisted of 97 amino acid residues with a predicted molecular weight (MW) of 11.17 kDa. The Sc-ALF was highly cationic and the isoelectric point (pI) was estimated to be 9.95 as predicted by the PROTPARAM software. The sequence was deposited in the NCBI GenBank under accession number HQ638024. Sequence comparison of Sc-ALF amino acid revealed conserved amino acid residues in the region of LPS binding domain (Fig. 1A). The deduced amino acid sequence of Sc-ALF showed a 24 amino acid domain from residue 54 through 77, which was necessary for LPS binding and neutralization [12]. The Sc-ALF molecule also showed the conservation of two cysteine residues at positions Cys55 and Cys76, important for one disulfide bond (loop) formation in the peptide (Fig. 1A). The deduced amino acid sequence of Sc-ALF was found to be rich in positively charged amino acid residues, arginine (10.3%) and lysine (7.2%) forming a cluster within the disulfide loop regarded as the functional domain of ALF as described by Imjongjirak et al. [12]. Peptide model of Sc-ALF created using SWISS-MODEL server consisted of two α-helices crowded against a four-strand β-sheet. Two of the β-strands are in turn linked by a disulfide bond to form an amphipathic loop rich in cationic amino acid side chains (Fig. 1B). Multiple alignment performed for Sc-ALF with other ALFs revealed the presence of conserved regions within the sequence (Fig. 1C). The phylogenetic relationship between Sc-ALF and other ALFs of decapod crustaceans was analyzed using the Neighbor-Joining (NJ) method (Fig. 1D). Molecular phylogenetic tree based on amino acid sequences suggests that all the ALF members possess the same ancestral origin, which has subsequently diverged at different phases of evolution. The tree could be broadly divided into two major groups, Group I included ALFs from shrimps and lobsters and Group II consisted of ALFs from crabs and crayfishes. The bootstrap distance tree calculated for the Sc-ALF clearly indicated that the Sc-ALF possessed great similarity to ALFs of other crabs (Fig. 1D).
Fig. 1.
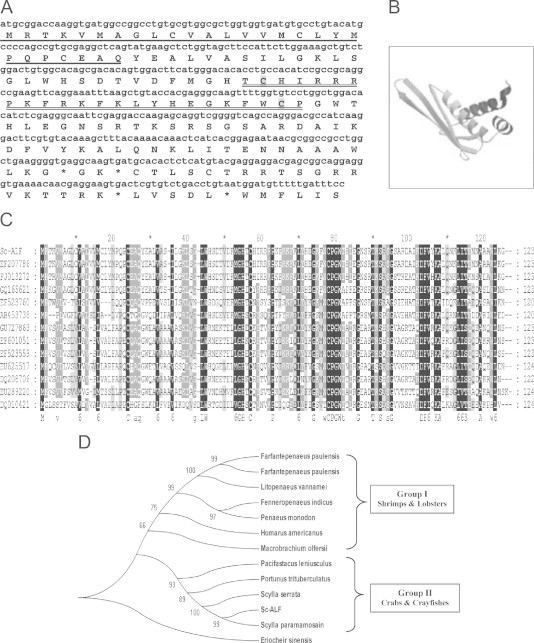
(A) Nucleotide and amino acid sequences of Sc-ALF from the haemocyte of the mud crab, Scylla serrata (HQ638024). The underlined amino acid residues indicate a putative signal sequence. LPS binding domain characteristic of the ALF family is double underlined and the two conserved cysteine residues important for one disulfide bond (loop) formation are highlighted in gray. An asterisk is the stop codon. (B) Structural model of Sc-ALF (HQ638024) of Scylla serrata created using SWISS-MODEL server. (C) Multiple alignment of nucleotide sequence of the Sc-ALF (HQ638024) with other ALFs (Scylla paramamosainEF207786, Scylla serrataFJ013272, Portunus trituberculatusGQ165621, Pacifastacus leniusculusEF523760, Marsupenaeus japonicasAB453738, Fenneropenaeus indicusGU727863, Farfantepenaeus paulensisEF601051, Penaeus monodonEF523559, Homarus americanusEU625517, Litopenaeus vannameiDQ208706, Macrobrachium olfersiiEU289220, Litopenaeus stylirostrisDQ010421) obtained using GeneDoc programme Version 2.7.0. Black and gray indicate conserved sequences. (D) A bootstrapped neighbor-joining tree obtained using MEGA version 4.0 illustrating relationships between the deduced amino acid sequence of the Sc- ALF (HQ638024) with other ALFs of decapod crustaceans (Scylla paramamosainEF207786, Scylla serrataFJ013272, Portunus trituberculatusGQ165621, Pacifastacus leniusculusEF523760, Fenneropenaeus indicusGU727863, Farfantepenaeus paulensisEF601051, Penaeus monodonEF523559, Homarus americanusEU625517, Litopenaeus vannameiDQ208706, Macrobrachium olfersiiEU289220, Farfantepenaeus paulensisEF601052. Eriocheir sinensis (GU014699) was used as an outgroup. Values at the node indicate the percentage of times that the particular node occurred in 1000 trees generated by bootstrapping the original deduced protein sequences.
Table 1.
Result of BLAST analysis of Sc-ALF (HQ638024).
| GenBank accession no. | Description | Query coverage (%) | E-value | Max identity (%) |
|---|---|---|---|---|
| EF207786 | Scylla paramamosain anti-lipopolysaccharide factor mRNA, complete cds | 100 | 2e−65 | 100 |
| FJ013272 | Scylla serrata anti-lipopolysaccharide factor precursor, mRNA, complete cds | 100 | 4e−61 | 93 |
| GQ165621 | Portunus trituberculatus anti-lipopolysaccharide factor isoform 3 (ALF) mRNA, complete cds, alternatively spliced | 98 | 6e−50 | 76 |
| EF523760 | Pacifastacus leniusculus anti-lipopolysaccharide factor mRNA, complete cds | 85 | 1e−24 | 52 |
| GU727863 | Fenneropenaeus indicus anti-lipopolysaccharide factor (ALF) mRNA, complete cds | 98 | 6e−23 | 41 |
The full-length cDNA of Sc-crustin was 433 bp in length, encoding 144 amino acids (Fig. 2A). The Sc-crustin cDNA encoded a polypeptide of 111 amino acids in the ORF with a putative signal peptide of 21 amino acid residues and a mature protein of 90 amino acids (Fig. 2A). The calculated molecular mass of the mature protein was 10.24 kDa. The isoelectric point (pI) was estimated to be 8.76 as predicted by the PROTPARAM software. The full-length sequence was deposited in the NCBI GenBank under accession number HQ638025. Sequence comparison using BLAST algorithm showed that the deduced amino acid sequence of Sc-crustin possessed an overall similarity of 81%, 62%, 73%, 56% and 39% to the crustins of S. paramamosain, P. trituberculatus, H. araneus, C. maenas and F. chinensis, respectively (Table 2). The deduced amino acid sequence of Sc-crustin was found to be rich in amino acid residues cysteine (13.3%) and proline (11.1%). A WAP domain could be detected in the C-terminus of Sc-crustin. As described by Imjongjirak et al. [13] a conserved eight-cysteine residue region responsible for the formation of 4 disulfide core (4-DSC) could also be detected in the C5–C12 position (Fig. 2A). The 12 cysteines in Sc-crustin (Fig. 2A) are considered to be important for maintaining the tertiary structure of the peptide just as that reported in the case of shrimp crustins [23,24]. Peptide model of Sc-crustin created using SWISS-MODEL server indicated a random coiled structure, that is, with two possible β-sheets but no helices (Fig. 2B). Multiple alignment performed for Sc-crustin with other crustins of decapods revealed the presence of conserved regions within the sequence (Fig. 2C).
Fig. 2.
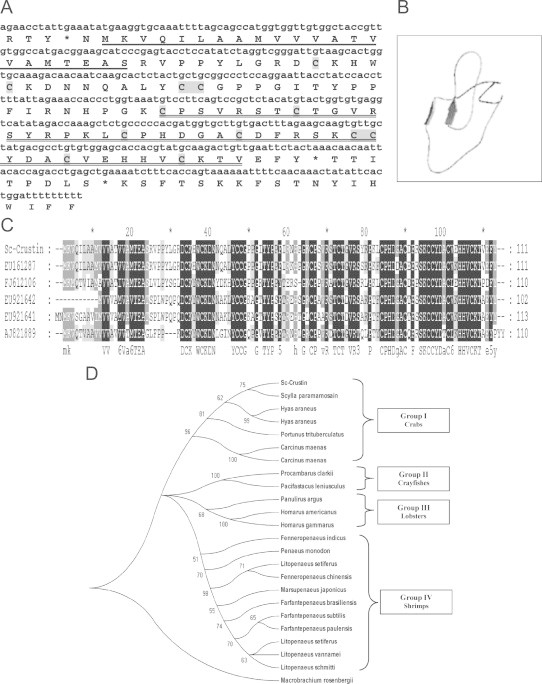
(A) Nucleotide and amino acid sequences of Sc-crustin from the haemocyte of the mud crab, Scylla serrata (HQ638025). The underlined amino acid residues indicate a putative signal sequence. Cysteine residues that participate in the formation of intramolecular disulphide bonds are highlighted in gray and the WAP domain is double underlined. An asterisk is the stop codon. (B) Structural model of Sc-crustin (HQ638025) of Scylla serrata created using SWISS-MODEL server. (C) Multiple alignment of nucleotide sequence of the Sc-crustin (HQ638025) with other crustins (Scylla paramamosainEU161287, Portunus trituberculatusFJ612106, Hyas araneusEU921642, Hyas araneusEU921641, Carcinus maenasAJ821889) obtained using GeneDoc programme Version 2.7.0. Black and gray indicate conserved sequences. (D) A bootstrapped neighbor-joining tree obtained using MEGA version 4.0 illustrating relationships between the deduced amino acid sequence of the Sc-crustin (HQ638025) with other crustins of decapod crustaceans (Scylla paramamosainEU161287, Portunus trituberculatusFJ612106, Hyas araneusEU921642, Hyas araneusEU921641, Carcinus maenasAJ821889, Carcinus maenasAJ237947, Procambarus clarkiaGQ301202, Pacifastacus leniusculusEF523613, Marsupenaeus japonicusAB121744, Litopenaeus setiferusAF430079, Penaeus monodonFJ380049, Farfantepenaeus brasiliensisEF601055, Farfantepenaeus subtilisEF450744, Fenneropenaeus chinensisFJ853147, Litopenaeus vannameiAY488497, Farfantepenaeus paulensisEF182747, Litopenaeus setiferusAF430078, Fenneropenaeus indicusGQ469987, Litopenaeus schmittiEF182748, Homarus americanusEF193003, Homarus gammarusAJ786653, Panulirus argusAY340636). Macrobrachium rosenbergii (EF364561) was used as an outgroup. Values at the node indicate the percentage of times that the particular node occurred in 1000 trees generated by bootstrapping the original deduced protein sequences.
Table 2.
Result of BLAST analysis of Sc-crustin (HQ638025).
| GenBank accession no. | Description | Query coverage (%) | E-value | Max identity (%) |
|---|---|---|---|---|
| EU161287 | Scylla paramamosain crustin antimicrobial peptide mRNA, complete cds | 100 | 3e−44 | 81 |
| FJ612106 | Portunus trituberculatus crustin antimicrobial peptide mRNA, complete cds | 100 | 1e−30 | 62 |
| EU921642 | Hyas araneus crustin Ha2 mRNA, partial cds | 73 | 3e−29 | 73 |
| AJ821889 | Carcinus maenas mRNA for carcinin-like protein, isolate IV | 99 | 2e−27 | 56 |
| FJ853148 | Fenneropenaeus chinensis crustin 3 mRNA, complete cds | 67 | 2e−08 | 39 |
In the present study, the BLAST homology search for the AMPs (Sc-ALF and Sc-crustin) from the brown mud crab, S. serrata showed maximum similarity to those from the green mud crab, S. paramamosain, a commercially important species in Queensland, Australia. The brown mud crab is a well known commercial species in India, Philippines and Vietnam. The brown mud crab differs from the green mud crab in having one less spine on the wrist and behind the fingers of the male. It also lacks the overall pale green mottling on the legs/rear paddles and is comparatively small in size. The study emphasizes the similarity in the defense peptides of these taxonomically related species.
This report presents the characterization and phylogenetic analysis of a new ALF isoform (Sc-ALF) and the first crustin sequence (Sc-crustin) from S. serrata. Sc-ALF gave 93% similarity to an ALF isoform from S. serrata. Sc-crustin is the first report of a crustin sequence from S. serrata. Discovery of novel AMPs and its antimicrobial spectrum might pave way to unravel the obscurity of crustacean immunity. Further research on the expression profile of these molecules in response to various environmental conditions and microbial infection would reveal their role in the protection of the animals from the onslaught of diseases.
Acknowledgment
The authors are grateful to the Ministry of Earth Sciences (MoES), Government of India for the research grant (MoES/10-MLR/2/2007) with which the work was carried out.
References
- 1.Lorenzon S., de Guarrini S., Smith V.J., Ferrero E.A. Effects of LPS injection on circulating haemocytes in crustaceans in vivo. Fish Shellfish Immunol. 1999;9:31–50. [Google Scholar]
- 2.Smith V., Brown J., Hauton C. Immunostimulation in crustaceans: does it really protect against infection? Fish Shellfish Immunol. 2003;15:71–90. doi: 10.1016/s1050-4648(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 3.Sheynis T., Sykora J., Benda A., Kolusheva S., Hof M., Jelinek R. Bilayer localization of membrane-active peptides studied in biomimetic vesicles by visible and fluorescence spectroscopies. Eur J Biochem. 2003;270:4478–4487. doi: 10.1046/j.1432-1033.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- 4.Bulet P., Hetru C., Dimarcq J.L., Hoffmann D. Antimicrobial peptides in insects; structure and function. Dev Comp Immunol. 1999;23:329–344. doi: 10.1016/s0145-305x(99)00015-4. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Boman H.G. Antibacterial peptides: basic facts and emerging concepts. J Int Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 7.Tincu J.A., Taylor S.W. Antimicrobial peptides from marine invertebrates. Antimicrob Agents Chemother. 2004;48:3645–3654. doi: 10.1128/AAC.48.10.3645-3654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnapp D., Kemp G.D., Smith V.J. Purification and characterization of a proline-rich antibacterial peptide, with sequence similarity to bactenecin-7, from the haemocytes of the shore crab, Carcinus maenas. Eur J Biochem. 1996;240:532–539. doi: 10.1111/j.1432-1033.1996.0532h.x. [DOI] [PubMed] [Google Scholar]
- 9.Relf J.M., Chisholm J.R., Kemp G.D., Smith V.J. Purification and characterization of a cysteine-rich 11.5-kDa antibacterial protein from the granular haemocytes of the shore crab, Carcinus maenas. Eur J Biochem. 1999;2642:350–357. doi: 10.1046/j.1432-1327.1999.00607.x. [DOI] [PubMed] [Google Scholar]
- 10.Khoo L., Robinette D.W., Callinectin Noga EJ. an antibacterial peptide from blue crab, Callinectes sapidus, haemocytes. Mar Biotechnol. 1999;1:44–51. doi: 10.1007/pl00011750. [DOI] [PubMed] [Google Scholar]
- 11.Wang K.J., Huang W.S., Yang M., Chen H.Y., Bo J., Li S.J. A male-specific expression gene, encodes a novel anionic antimicrobial peptide, scygonadin, in S. serrata. Mol Immunol. 2007;44:1961–1968. doi: 10.1016/j.molimm.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 12.Imjongjirak C., Amparyup P., Tassanakajon A., Sittipraneed S. Antilipopolysaccharide factor ALF of mud crab Scylla paramamosain: molecular cloning, genomic organization and the antimicrobial activity of its synthetic LPS binding domain. Mol Immunol. 2007;44:3195–3203. doi: 10.1016/j.molimm.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 13.Imjongjirak C., Amparyup P., Tassanakajon A., Sittipraneed S. Molecular cloning and characterization of crustin from mud crab Scylla paramamosain. Mol Biol Rep. 2009;36:841–850. doi: 10.1007/s11033-008-9253-0. [DOI] [PubMed] [Google Scholar]
- 14.Stensvag K., Haug T., Sperstad S.V., Rekdal O., Indrevoll B., Styrvold O.B. Arasin 1, a proline-arginine-rich antimicrobial peptide isolated from the spider crab, Hyas araneus. Dev Comp Immunol. 2008;32:275–285. doi: 10.1016/j.dci.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Shigenaga T., Muta T., Toh Y., Tokunaga F., Iwanaga S. Antimicrob. tachyplesin peptide precursor. cDNA cloning and cellular localization in the horseshoe crab Tachypleus tridentatus. J Biol Chem. 1990;265:21350–21354. [PubMed] [Google Scholar]
- 16.Destoumieux D., Munoz M., Bulet P., Penaeidins Bachere E. a family of antimicrobial peptides from penaeid shrimp Crustacea, Decapoda. Cell Mol Life Sci. 2000;57:1260–1271. doi: 10.1007/PL00000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitta G., Hubert F., Dyrynda E.A., Boudry P., Roch P., Mytilin B. and MGD2, two antimicrobial peptides of marine mussels: Gene structure and expression analysis. Dev Comp Immunol. 2000;24:381–393. doi: 10.1016/s0145-305x(99)00084-1. [DOI] [PubMed] [Google Scholar]
- 18.Hauton C., Brokton V., Smith V.J. Cloning of a crustin-like single whey-acidic-domain, antibacterial peptide from the haemocytes of the European lobster, Homarus gammarus, and its response to infection with bacteria. Mol Immunol. 2006;43:1490–1496. doi: 10.1016/j.molimm.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 19.Stewart J.E., Zwicker B.M. Induction and natural bactericidal activities of the lobster Homarus americanus: products of haemocyte–plasma interactions. Can J Microbiol. 1972;18:1499–1511. doi: 10.1139/m72-229. [DOI] [PubMed] [Google Scholar]
- 20.Chisholm J.R.S., Smith V.J. Antibacterial activity in the haemocytes of the shore crab, Carcinus maenas (L.) J Mar Biol Assoc UK. 1992;72:529–542. [Google Scholar]
- 21.Noga E.J., Arroll T.A., Fan Z. Specificity and some physicochemical characteristics of the antibacterial activity from blue crab Callinectes sapidus. Fish Shellfish Immunol. 1996;6:403–412. [Google Scholar]
- 22.Tamura K., Dudley J., Nei M., Kumar S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 23.Gross P.S., Barlett T.C., Browdy C.L., Chapman R.W., Warr G.W. Immune gene discovery by expressed sequence tag analysis of haemocytes and hepatopancreas in the Pacific white shrimp, Litopenaeus vannamei, and Atlantic white shrimp, Litopenaeus setiferus. Dev Comp Immunol. 2001;25:565–577. doi: 10.1016/s0145-305x(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Li F., Wang Z., Xiang J. Cloning and recombinant expression of a crustin-like gene from Chinese shrimp, Fenneropenaeus chinensis. J Biotechnol. 2007;127:605–614. doi: 10.1016/j.jbiotec.2006.08.013. [DOI] [PubMed] [Google Scholar]


