Abstract
S-adenosyl-L-methionine (AdoMet)-dependent methylation is central to the regulation of many biological processes: more than 50 AdoMet-dependent methyltransferases methylate a broad spectrum of cellular compounds including nucleic acids, proteins and lipids. Common to all AdoMet-dependent methyltransferase reactions is the release of the strong product inhibitor S-adenosyl-L-homocysteine (AdoHcy), as a by-product of the reaction. S-adenosyl-L-homocysteine hydrolase is the only eukaryotic enzyme capable of reversible AdoHcy hydrolysis to adenosine and homocysteine and, thus, relief from AdoHcy inhibition. Impaired S-adenosyl-L-homocysteine hydrolase activity in humans results in AdoHcy accumulation and severe pathological consequences. Hyperhomocysteinemia, which is characterized by elevated levels of homocysteine in blood, also exhibits a similar phenotype of AdoHcy accumulation due to the reversal of the direction of the S-adenosyl-L-homocysteine hydrolase reaction. Inhibition of S-adenosyl-L-homocysteine hydrolase is also linked to antiviral effects. In this review the advantages of yeast as an experimental system to understand pathologies associated with AdoHcy accumulation will be discussed.
Keywords: AdoMet, AdoHcy, Homocysteine, S-adenosyl-L-homocysteine hydrolase
Highlights
► AdoHcy is a potent product inhibitor of AdoMet-dependent methyltransferases. ► AdoHcy accumulates in hyperhomocysteinemia. ► Yeast is an advantageous system to understand AdoHcy toxicity. ► Lipid metabolism is deregulated in response to AdoHcy accumulation.
1. AdoMet — a principal methyl group donor and more
Beyond its role in protein synthesis and structure, methionine, after its activation to AdoMet by methionine adenosyltransferase, plays a crucial role in many aspects of cellular metabolism. AdoMet is synthesized in virtually all living organisms [1]. It is the second most widely used enzyme substrate after ATP [2] and, thus, one of the most versatile biomolecules. Being a high-energy sulfonium compound AdoMet can serve as a source of all three ligands linked to the sulfur atom. AdoMet is the methyl group donor used by all organisms in most methyl transfer reactions [3]. It was also shown to be used in a number of other reactions, serving as a source of propylamine for the synthesis of spermidine and spermine, amino groups for the synthesis of biotin, or ribosyl groups in the synthesis of queuosine, a hypermodified tRNA, as well as 5′ deoxyadenosyl radicals required for a number of radical reactions [3], [4].
The fact that under normal conditions more than 90% of total AdoMet in mammalian cells is used for methylation reactions catalyzed by AdoMet-dependent methyltransferases in which AdoMet donates its methyl group to a large variety of acceptors including nucleic acids, proteins and lipids underscores the importance of methylation in downstream reactions of AdoMet [3], [5]. AdoHcy, which is released as a by-product of AdoMet-dependent methyl transfer reactions, is a strong product inhibitor of AdoMet-dependent methyltransferases and is hydrolyzed by S-adenosyl-L-homocysteine hydrolase to homocysteine and adenosine (Fig. 1a). In mammals, homocysteine produced by this reaction can be remethylated to methionine by methionine synthase or betaine–homocysteine methyltransferase and, thus, retained in the methylation cycle. Alternatively, it can be withdrawn from the methylation cycle by its conversion to cysteine, the precursor of glutathione, via the two-step transsulfuration pathway. Cystathionine β-synthase is the first and rate-limiting enzyme of the latter. Both cystathionine β-synthase and the enzyme catalyzing the second step of the transsulfuration pathway, cystathionine γ-lyase, are vitamin B6 requiring enzymes.
Fig. 1.
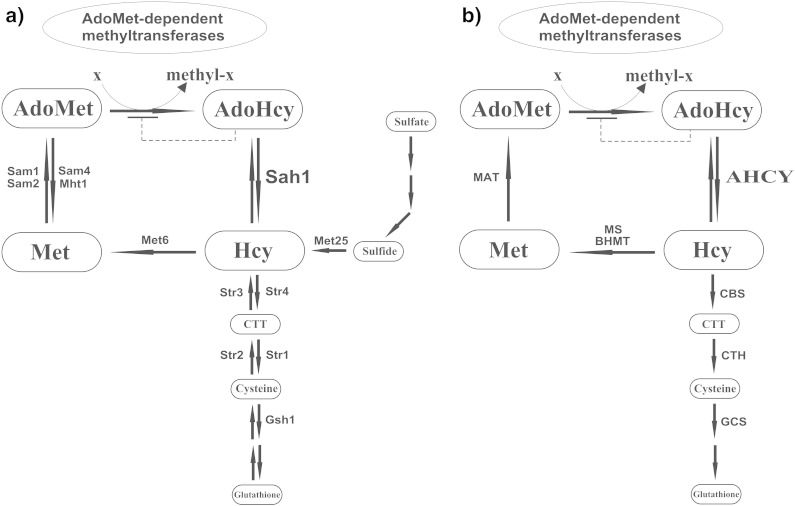
AdoMet-dependent methylation: the role of AdoHcy and S-adenosyl-L-homocysteine hydrolase a) in yeast and b) in mammals. Reverse transsulfuration pathway in yeast converts cysteine into homocysteine via cystathionine γ-synthase, Str2, and cystathionine β-lyase, Str3. AdoMet, S-adenosyl-L-methionine; AdoHcy, S-adenosyl-L-homocysteine; Hcy, homocysteine; Met, methionine; Sah1, S-adenosyl-L-homocysteine hydrolase in yeast; AHCY, S-adenosyl-L-homocysteine hydrolase in mammals; CTT, cystathionine; Sam1, AdoMet synthetase 1; Sam2, AdoMet synthetase 2; Sam4, AdoMet-homocysteine methyltransferase; Mht1, S-methylmethionine–homocysteine methyltransferase; Met6, methionine synthase; Met25, O-acetylhomoserine sulfhydrylase; Str1, cystathionine γ-lyase; Str2, cystathionine γ-synthase; Str3, cystathionine β-lyase; Str4, cystathionine β-synthase; Gsh1, γ-glutamylcysteine synthetase; MAT, methionine adenosyltransferase; MS, methionine synthase; BHMT, betaine-homocysteine methyltransferase; CBS, cystathionine β-synthase; CTH, cystathionine γ-lyase; GCS, γ-glutamylcysteine synthetase.
AdoMet is synthesized by methionine adenosyltransferase from ATP and methionine. In mammals, methionine can be either ingested in the diet or formed by methylation of homocysteine using either a methyl group of betaine or that of 5-methyltetrahydrofolate. The latter is formed by methylenetetrahydrofolate reductase from 5,10-methylenetetrahydrofolate in a cobalamin-dependent reaction [5]. Deficiency of methionine synthase that catalyzes remethylation of homocysteine to methionine using 5-methyltetrahydrofolate links homocysteine and folate metabolism leading to accumulation of methyltetrahydrofolate at the expense of the methylenetetrahydrofolate and tetrahydrofolate pools required for thymidylate and purine biosynthesis [6].
The enzymes required for AdoMet synthesis, AdoHcy catabolism, homocysteine remethylation and transsulfuration are conserved between yeast and mammals except for betaine–homocysteine methyltransferase and the reverse transsulfuration pathway (Fig. 1a, b). A further difference between sulfur metabolism in mammals and in yeast is the presence of the sulfur assimilation pathway in the latter (Fig. 1b); however, this pathway can be efficiently eliminated in yeast by the introduction of a mutation disrupting the pathway [7]. Deficiencies in the enzymes involved in the methylation cycle or homocysteine catabolism e.g. cystathionine β-synthase, methionine adenosyltransferase, methylenetetrahydrofolate reductase and S-adenosyl-L-homocysteine hydrolase show that many of them are linked to a number of pathologies including deregulation of lipid metabolism [7], [8], [9], [10], [11], [12], [13], neurological abnormalities [10], [14], [15], [16], tumorigenicity [8], [9], [11], oxidative stress [8] and myopathy [14].
2. AdoMet-dependent methyltransferases
Computational analysis of the human proteome revealed 208 known and putative human AdoMet-dependent methyltransferases that make up about 0.9% of genes in the human genome [17]. Noteworthy, 30% of these proteins have been linked to disease states e.g. cancer and mental disorders [17]. Analysis of the yeast proteome revealed 81 known and putative AdoMet-dependent methyltransferases, making up about 1.2% of genes in the yeast genome [18]. The increased number of AdoMet-dependent methyltransferases in humans as compared to yeast reflects partial redundancy as well as novel functions present in the human methyltransferasome in comparison with yeast [17]. AdoMet-dependent methyltransferases such as DNA-, glycine-, guanidinoacetate- and isoaspartyl methyltransferases are found only in humans. However, sterol-24C-methyltransferase, Erg6p, is found only in yeast. Nevertheless, the majority of AdoMet-dependent methyltransferases are conserved between yeast and humans. In Table 1, yeast and human AdoMet-dependent methyltransferases grouped by their substrate specificity are compared. The whole list of known and putative yeast and human AdoMet-dependent methyltransferases can be found in [18], [17], respectively.
Table 1.
Comparison of yeast and human AdoMet-dependent methyltransferases grouped by substrate specificity. The whole list of known and putative yeast and human AdoMet-dependent methyltransferases can be found in [18], [17], respectively.
| AdoMet-dependent methyltransferases | Biological function/process | Yeast | Mammalian orthologs |
|---|---|---|---|
| DNA | |||
| DNA (cytosine-5-)-methyltransferases | Gene silencing | – | DNMT1, DNMT2, DNMT3a, DNMT3b |
| RNA | |||
| mRNA (guanine-N7-)-methyltransferases | Methylation of the 5′ cap structure of mRNA | Abd1 | RNMT |
| mRNA (adenosine-N6)-methyltransferases | Entry into meiosis | Ime4 | METTL3 |
| Trimethylguanosine synthases | Hypermethylation of m(7)G to the m(2,2,7)G 5′ cap of snRNAs, snoRNAs and telomerase TLC1 RNA | Tgs1 | TGS1 |
| Probable rRNA (cytosine-5-)-methyltransferases | 27S pre-rRNA processing and 60S ribosome biogenesis | Nop2 | NOP2 |
| rRNA (guanine-N7)-methyltransferases | rRNA processing | Bud23 | WBSCR22 |
| rRNA (adenine-N6,N6-)-dimethyltransferases | 18S pre-ribosomal rRNA processing | Dim1 | DIMT1, TFB1M |
| rRNA (2′-O-ribose)-methyltransferases | 27S pre-rRNA and 25S rRNA processing, 60S ribosomal subunit maturation | Spb1 | FTSJ1, FTSJ2, FTSJ3 |
| Mitochondrial rRNA (2′-O-ribose)-methyltransferases | Mitochondrial 21S rRNA processing | Mrm1, Mrm2 | MRM1 |
| tRNA (guanine)-methyltransferases | tRNA modification | Trm1, Trm5, Trm10, Trm8/Trm82, Trm11/Trm112, Trm12 | TRMT1, TRMTL1, TRMT5, RG9MTD2, METTL1, TRMT11, TRMT12 |
| tRNA (uracil-5-)-methyltransferases | tRNA modification | Trm2, Trm9 | TRMT2A, ALKBH8, KIAA145 |
| tRNA (cytosine-5-)-methyltransferases | tRNA modification | Ncl1 (Trm4) | NSUN2, NSUN3, NSUN5 |
| tRNA (adenine-N1-)-methyltransferases | Maturation of initiator methionyl-tRNA | Trm6/Trm61 | TRMT6 |
| tRNA (2′-O-ribose)-methyltransferases | tRNA modification | Trm3, Trm7, Trm13, Trm44 | TARBP1, FTSJ1, CCDC76 |
| tRNA methyltransferases | Methylation of N-4 position of yW-86 in wybutosine biosynthesis | Tyw3 | TYW3 |
| Carboxyl methyltransferases | Methylation of the α-carboxy group of yW-72 in wybutosine biosynthesis | Ppm2 | LCMT2 |
| Proteins | |||
| Histone lysine N-methyltransferases | Transcriptional elongation and silencing | Set1, Set2, Dot1 | SET superfamily consisting of SUV39, SET1, SET2, RIZ, SMYD, EZ and SUV4-20 families, SET7/9, SET8, DOT1L |
| Ribosomal protein lysine N-methyltransferases | Ribosome biogenesis | Rkm1, Rkm2, Rkm3, Rkm4, Rkm5 | SETD6 |
| ω-NG-monomethylarginine and asymmetric ω-NG,NG-dimethylarginine methyltransferases | Methylation of hnRNPs affecting their activity and nuclear export; methylation of U1 snRNP protein Snp1p and ribosomal protein Rps2p | Hmt1 (Rmt1) | PRMT1, PRMT3, PRMT4 (CARM1), PRMT6, PRMT8 |
| ω-NG-monomethylarginine and symmetric ω-NG,NG-dimethylarginine methyltransferases | Recruitment and inactivation of Swe1 (Wee1 ortholog) during mitotic entry | Hsl7 | PRMT5 (JBP1), PRMT7 |
| δ-NG-monomethylarginine methyltransferase | Methylation of ribosomal protein Rpl12 | Rmt2 | – |
| Protein histidine methyltransferases | 3-Methylhistidine modification of ribosomal protein Rpl3p | Hpm1 | METTL18 |
| N-terminal methyltransferases | Methylation of ribosomal proteins Rpl12 and Rps25 | Tae1 | METTL11A |
| C-terminal leucine carboxyl methyltransferases | Methylation of C-terminal leucine of PP2A catalytic subunit, complex formation | Ppm1 | LCMT1 |
| Protein S-isoprenylcysteine O-methyltransferases | C-terminal methylation of CAAX proteins | Ste14 | ICMT |
| Protein-l-isoaspartate (d-aspartate) O-methyltransferase | Repair of damaged proteins | − | PCMT1 |
| Cytochrome c lysine N-methyltransferase | Trimethylation of cytochrome c, not required for respiratory growth | Ctm1 | − |
| eRF1 methyltransferases | Peptidyl-glutamine methylation, translation release | Mtq2 | N6AMT1 |
| Mrf1methyltransferases | Peptidyl-glutamine methylation, mitochondrial translation release | Mtq1 | HEMK1 |
| Lipids | |||
| Phospholipid N-methyltransferases | Phosphatidylcholine de novo synthesis | Cho2, Opi3 | PEMT |
| Sterol 24-C-methyltransferase | Ergosterol biosynthesis | Erg6 | – |
| Others | |||
| S-AdoMet-homocysteine S-methyltransferase | Regulation of methionine/AdoMet ratio | Sam4 | – |
| Trans-aconitate methyltransferase | Leucine biosynthesis | Tmt1 | – |
| Uroporphyrin-III C-methyltransferase | Siroheme biosynthesis | Met1 | – |
| Glycine/sarcosine N-methyltransferase | Regulation of hepatic AdoMet metabolism | – | GNMT |
| Guanidinoacetate N-methyltransferase | Creatine synthesis | – | GAMT |
| Ubiquinone methyltransferases | Ubiquinone biosynthesis, respiration | Coq3, Coq5 | COQ3, COQ5 |
| Phenylethanolamine N-methyltransferase | Adrenaline (epinephrine) biosynthesis | – | PNMT |
| Acetylserotonin O-methyltransferase | Melatonin biosynthesis | – | ASMT |
| Histamine N-methyltransferase | Histamine degradation | – | HNMT |
| Catechol O-methyltransferase | Degradation of catecholamines | – | COMT |
| Indolethylamine N-methyltransferase | Tryptamine methylation | – | INMT |
| Nicotinamide N-methyltransferases | Nicotinamide metabolism | Nnt1 | NNMT |
The largest group of AdoMet-dependent methyltransferases methylates proteins, predominantly at the ε-amine group of lysine and ω- or δ-guanidine groups of arginine as well as on the C-terminal leucine and isoprenylated cysteine residues. Targets of AdoMet-dependent protein methyltransferases include histones, ribosomal proteins, transcription and translation factors, signal transduction proteins etc. [19], [20], [21], [22]. For instance, the SET domain protein lysine methyltransferase family, which makes up 27% of human methyltransferasome and 14% of yeast methyltransferasome, methylates predominantly histones at lysine residues [17], [23]. Comparison of a number of histone lysine methyltransferases from various organisms ranging from yeast to human underlines the similarity of their roles in the maintenance of normal cellular functions in eukaryotes [24].
Protein arginine methyltransferases methylate in addition to histones [19], [20] numerous non-histone proteins [20], [22], [25] both in yeast and in mammals, including heterogeneous nuclear riboproteins (hnRNPs) required for eukaryotic transcription elongation, mRNA processing and export [26], [27], [28]. Further targets for AdoMet-dependent methylation are the protein phosphatase 2A (PP2A), which is methylated at the highly conserved C-terminal leucine of its catalytic C subunits [29], and G proteins, in particular Ras proteins, that are methylated at C-terminal isoprenylcysteine residues [30].
A big group of AdoMet-dependent methyltransferases methylate nucleic acids. However, while many AdoMet-dependent methyltransferases are involved in the methylation of mRNA, rRNA and tRNA both in yeast and in mammals, there are no orthologs to mammalian DNA (cytosine-5)-methyltransferases that methylate DNA in yeast. While methylation of rRNA is crucial for ribosomal processing and maturation [31], methylation of tRNA helps to control tRNA folding and to ensure decoding specificity and efficiency [32]. Cap methylation at the 5′-terminal guanosine by mRNA (guanine-7-)-methyltransferase is an essential process both in yeast and mammals [33], [34].
In addition to proteins and nucleic acids AdoMet-dependent methyltransferases methylate lipids and a number of small molecules, in particular, phosphatidylethanolamine, ubiquinone and nicotinamide (Table 1). It is noteworthy that de novo biosynthesis of phosphatidylcholine from phosphatidylethanolamine catalyzed by AdoMet-dependent phospholipid methyltransferases is the major AdoMet consumer in mice and humans [35], [36] as well as in yeast in the absence of choline/ethanolamine supplementation.
3. The role of S-adenosyl-L-homocysteine hydrolase in AdoMet-dependent methylation
AdoHcy that is released as a by-product of AdoMet-dependent methyltransferase reactions, is a potent product inhibitor of most AdoMet-dependent methyltransferases with Ki values in the submicromolar to low micromolar range [37]. AdoHcy was shown to inhibit a number of AdoMet-dependent methyltransferases in vitro, such as mammalian DNA (cytosine-5-)-methyltransferase, mRNA cap (guanine-N7-)-methyltransferase, tRNA (uracil-5-)-methyltransferase, protein isoprenylcysteine carboxylmethyltransferase and phospholipid methyltransferases [38], [39], [40], [41], [42], [43], [44], [45], and in vivo, such as DNA (cytosine-5-)-methyltransferase and phospholipid methyltransferases [7], [46], [47]. However, not all AdoMet-dependent methyltransferases exhibit equal sensitivity against AdoHcy. While some AdoMet-dependent methyltransferases have a Ki value for AdoHcy that is lower than the Km value for AdoMet and are especially sensitive to AdoHcy [37], others, in particular glycine N-methyltransferase, which regulates AdoMet levels in mammals [1], are only weakly inhibited by AdoHcy [48]. Being a potent product inhibitor for most AdoMet-dependent methyltransferases, AdoHcy is a sensitive marker for the cellular methylation status that is reflected by AdoMet/AdoHcy ratio, and can be used to predict methylation deficiency e.g. DNA hypomethylation [49].
The only eukaryotic enzyme capable of AdoHcy catabolism both in yeast and mammals is S-adenosyl-L-homocysteine hydrolase (Sah1 in yeast, AHCY in mammals), which catalyzes the reversible hydrolysis of AdoHcy to homocysteine and adenosine. Similarly to AdoHcy accumulation, interference with S-adenosyl-L-homocysteine hydrolase was shown to affect methylation of different targets of AdoMet-dependent methyltransferases. In particular, inhibition of S-adenosyl-L-homocysteine hydrolase and the resulting increase of AdoHcy levels were shown to be associated with DNA hypomethylation in cultured endothelial cells [46] and to interfere with RNA methylation in cultured lymphocytic leukemia cells [50]. Furthermore, inhibition of S-adenosyl-L-homocysteine hydrolase was also shown to result in decreased methylation of poly(A)(+) RNA in Xenopus laevis oocytes [51] as well as in inhibition of phospholipid methylation in yeast [7]. In accordance with the crucial role of AdoMet-dependent methylation in many biological processes, accumulation of AdoHcy and/or S-adenosyl-L-homocysteine hydrolase dysfunction is linked to numerous pathologies including neurological and vascular disorders, myopathy, fatty liver, cancer, renal insufficiency and diabetic nephropathy [11], [14], [52], [53], [54], [55], [56], [57], [58]. The pathological consequences of AdoHcy accumulation as well as the molecular mechanisms triggered by elevated AdoHcy levels are largely not understood. Considering the role of AdoMet-dependent methylation in biological processes, inhibition of a number of AdoMet-dependent methyltransferase reactions appears to play a central role in AdoHcy-associated pathology. However, evidence of in vivo sensitivity of individual AdoMet-dependent methyltransferase reactions as well as the responsiveness of methylation-dependent biological processes to AdoHcy accumulation is limited. Since S-adenosyl-L-homocysteine hydrolase is a crucial physiological regulator of AdoHcy levels we will discuss dysfunction of S-adenosyl-L-homocysteine hydrolase, its evolutional conservation, structural organization and regulation in the next sections.
4. S-adenosyl-L-homocysteine hydrolase dysfunction
Deletion of a locus overlapping the AHCY gene in mouse results in embryonic death [59]. Homozygous insertion mutations in the S-adenosyl-L-homocysteine hydrolase gene in Arabidopsis thaliana result in zygotic lethality [60]. In contrast, yeast mutants lacking Sah1 are viable, due to the presence of an alternative pathway for homocysteine synthesis via the sulfur assimilation. Homocysteine that is synthesized by this pathway is further used both for the synthesis of cysteine and glutathione as well as for the synthesis of methionine and AdoMet (Fig. 1) [7]. Introduction of an additional mutation in the sulfur assimilation pathway indeed renders the resulting yeast sah1 mutants inviable [7], consistent with an essential function of S-adenosyl-L-homocysteine hydrolase.
S-adenosyl-L-homocysteine hydrolase deficiency in humans, a rare genetic disorder, is characterized by up to 150-fold elevated plasma AdoHcy levels with a significant decrease in the AdoMet/AdoHcy ratio and severe pathological consequences that can lead to death during childhood [14], [61], [62], [63], [64]. In these patients, three tissues are predominantly affected: muscles, brain and liver, manifested by severe myopathy, slow myelination, developmental delay [61], [62], [63] as well as mild hepatitis [61] and lipid droplet accumulation in the liver [63]. Biochemical analyses revealed that AHCY deficient patients display 3–20% of mean control S-adenosyl-L-homocysteine hydrolase activity in the liver, red blood cells and fibroblasts [61], [62], [63]. They exhibit a decrease of phosphatidylcholine, choline and albumin levels as well as an elevation of creatine kinase, alanine transferase, aspartate transaminase [61], [62], [63] and lactate dehydrogenase [63] levels in the plasma. At the cellular level these patients show a proliferation of smooth and a decrease of rough endoplasmic reticulum [61], altered mitochondria [61], [63] as well as hypermethylation of leukocyte DNA [61], [62], [63].
5. S-adenosyl-L-homocysteine hydrolase: conservation, structure and catalytic mechanism
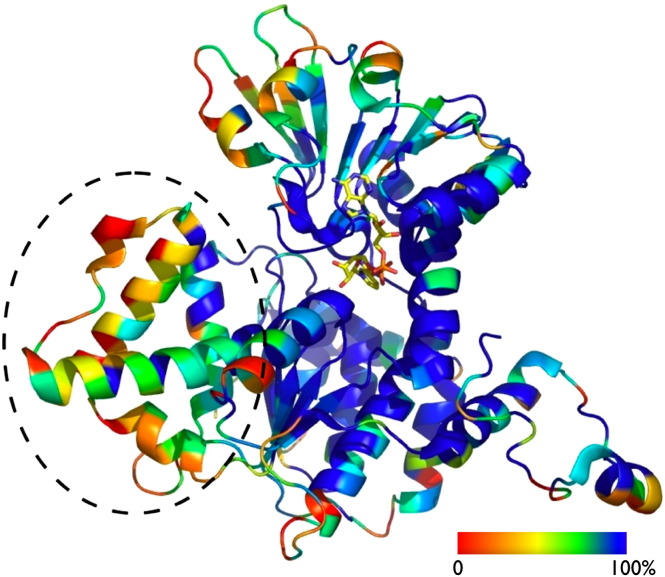
S-adenosyl-L-homocysteine hydrolase is an exceptionally well-conserved enzyme exhibiting over 70% identity at the protein level between human and yeast orthologs, and is among the 90 most highly conserved yeast proteins, including actin (90%), ubiquitin (90%), histones (70–90%), ribosomal (70%) and heat shock proteins (70%) [65]. Fig. 2 displays a comparison of S-adenosyl-L-homocysteine hydrolase sequences from Archaea, Bacteria and Eucarya. Emphasizing the similarity of yeast and human orthologs, many bacterial and some eukaryotic sequences, but not the mammalian and yeast proteins, exhibit an insertion segment of 40 amino acids of as yet unknown function in the catalytic domain of the enzyme [66].
Fig. 2.
S-adenosyl-L-homocysteine hydrolase: sequence and structural conservation. The sequence conservation based on ConSurf calculations [160] using 166 unique S-adenosyl-L-homocysteine hydrolase sequences is color coded (from blue – identity to red – least conservation) and mapped onto the structure of the Pf-SAHH (PDB 1v8b [77]). Shown is one monomer of the functional tetrameric S-adenosyl-L-homocysteine hydrolase protein in cartoon drawing, the NAD+ cofactor (yellow) is shown in a stick representation and the substrate (Ado) was omitted for clarity. Note that the 40 amino acid insertion consisting of 3 helices and a loop (black circle), which is not present in mammalian and yeast S-adenosyl-L-homocysteine hydrolases, shows the least degree of conservation.
S-adenosyl-L-homocysteine hydrolase belongs to the large family of NAD(P)H/NAD(P)+-binding proteins that share a Rossmann-fold [67]. The NAD(P)H/NAD(P)+ binding domain is found in numerous dehydrogenases as well as in many other redox enzymes, but is rather unusual for a hydrolase. The atomic structures of S-adenosyl-L-homocysteine hydrolase from different sources have been resolved: Homo sapiens (PDB 1a7a [68], PDB 1li4 [69] and PDB 3nj4 [70]), Rattus norvegicus (PDB 1b3r [71], PDB 1ky4 and PDB 1ky5 [72], PDB 1k0u [73], PDB 2h5l [74], PDB 1xwf [75] and PDB 1d4f [76]), Plasmodium falciparum (PDB 1v8b [77]), Mycobacterium tuberculosis (PDB 3ce6, PDB 3dhy, PDB 2zj1, PDB 2zj0 and PDB 2ziz [78]), Burkholderia pseudomallei (PDB 3d64), Trypanosoma brucei (PDB 3h9u) and Leishmania major (PDB 3g1u) and the plant Lupinus luteus (PDB 3ond, PDB 3one and PDB 3onf [79]).
All structurally characterized Sah1/AHCY proteins except plant S-adenosyl-L-homocysteine hydrolase are tetramers with NADH/NAD+ cofactor bound in the active site of each subunit [80], [81]. Plant S-adenosyl-L-homocysteine hydrolase from L. luteus has been shown to function as a homodimer [79]. The monomeric subunits of the protein contain three domains: N-terminal substrate-binding domain, cofactor-binding domain and C-terminal tail. The C-terminal tails of two subunits reciprocally protrude into opposite subunits and form a part of their cofactor-binding sites [68]. The two dimers then form a tetramer with the four cofactor-binding domains molding the central core of the tetramer structure, and the substrate-binding domains being exposed on the surface.
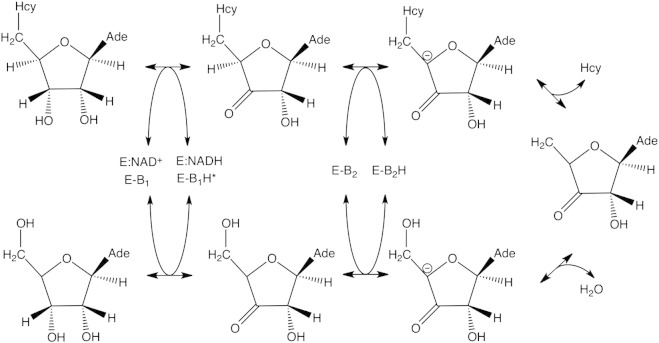
The thermodynamic equilibrium of the reaction catalyzed by S-adenosyl-L-homocysteine hydrolase favors the synthesis of AdoHcy from adenosine and homocysteine in vitro [82]. In vivo rapid enzymatic removal of homocysteine (Hcy) and adenosine (Ado) enables the net hydrolysis of AdoHcy [82], [83]. The reaction cycle requires reciprocal oxidation–reduction of the substrate and NAD+ [84]. In the first step, the Ado 3′ hydroxyl group of adenosine (synthetic reaction: Ado + Hcy → AdoHcy) or AdoHcy (hydrolytic reaction: AdoHcy → Ado + Hcy) is oxidized to ketone by NAD+ resulting in the formation of NADH (Fig. 3). In the next steps the proton is removed from C4′ forming the carbanion intermediate, followed by its cleavage and the release of water or Hcy, respectively. The catalytic cycle is finished by the addition of Hcy or water to the C4′ C5′ double bond and reduction of the 3′ keto group under regeneration of NAD+, forming AdoHcy or Hcy, respectively. Depending on the presence of the substrate the protein undergoes large conformational rearrangements [71], [85], [86], [87]. Substrate (AdoHcy/Ado) binding induces a structural transition from the open to the closed form of the enzyme, and product (Ado/AdoHcy) release induces the transition back to the open conformation [85].
Fig. 3.
Catalytic activity of S-adenosyl-L-homocysteine hydrolase.
6. S-adenosyl-L-homocysteine hydrolase: regulation and localization
S-adenosyl-L-homocysteine hydrolase requires NAD+ to maintain its quaternary structure and catalytic activity, making the enzyme sensitive to the redox status of the cell that is reflected by the NAD+/NADH ratio. As described above NAD+ bound to S-adenosyl-L-homocysteine hydrolase is subject to reduction–oxidation cycles during the reaction [84] (Fig. 3). Moreover, the form of NAD bound to the enzyme determines binding of adenosine, the substrate for the synthetic reaction and the inhibitor of the hydrolytic reaction. It was shown that S-adenosyl-L-homocysteine hydrolase from bovine kidney possesses two binding sites for adenosine: a low affinity binding site located in the catalytic domain and a high affinity binding site located in the NAD+/NADH binding domain of the enzyme [88]. The NAD+ form of S-adenosyl-L-homocysteine hydrolase binds adenosine with low affinity leading to the formation of 3′-keto-adenosine, reduction of the tightly bound NAD+ to NADH and adoption of the closed conformation by the enzyme [88]. However, the enzymatically inactive NADH form of the enzyme binds adenosine with high affinity, suggesting a role for adenosine in NADH-induced inhibition of S-adenosyl-L-homocysteine hydrolase and potential function of the enzyme as intracellular Ado-binding protein [88].
S-adenosyl-L-homocysteine hydrolase was also shown to bind copper with high affinity and is indeed the major copper binding protein in mouse liver [89], [90]. It has a similar dissociation constant for copper as albumin [89] and was suggested to serve as an intracellular copper transporter with a putative function in delivering copper to superoxide dismutase [91]. Copper inhibits S-adenosyl-L-homocysteine hydrolase activity in a non-competitive manner due to the release of NAD+ [92] that results from interruption of subunit interactions, which leads to a large decrease in the affinity for the NAD+ cofactor [92]. However, the significance of regulation of S-adenosyl-L-homocysteine hydrolase by NAD+, adenosine and copper is not clear.
In yeast S-adenosyl-L-homocysteine hydrolase is primarily found in the cytosol [93]. However, AdoMet-dependent methylation takes place in different cellular compartments e.g. nucleus and ER, suggesting that subcellular translocation of the enzyme may play a regulatory role. Indeed, S-adenosyl-L-homocysteine hydrolase was found in the nucleus in association with mRNA (guanine-7-)-methyltransferase and RNA polymerase II in transcriptionally active X. laevis oocytes [51], [94]. S-adenosyl-L-homocysteine hydrolase was also shown to translocate into the nucleus in adult mammalian cells under hypoxia conditions known to induce transcriptional activity [95]. Localization analyses in A. thaliana also demonstrated that the enzyme is capable of localizing to the cytoplasm and the nucleus [96], and was proposed to be targeted to the nucleus in a complex with adenosine kinase, another enzyme required for AdoHcy catabolism [96]. The S-adenosyl-L-homocysteine hydrolase/adenosine kinase complex is, presumably, targeted to the nucleus by the nuclear localization signal of mRNA (guanine-7-)-methyltransferase, with which it interacts [96].
7. AdoHcy in hyperhomocysteinemia
The pathological accumulation of homocysteine above the normal plasma homocysteine level of 10 μmol/L, hyperhomocysteinemia, can be caused by genetic or nutritional factors. Deficiency in cystathionine β-synthase (Fig. 1) or genetic defects of folate and cobalamin metabolisms lead to severe hyperhomocysteinemia characterized by the plasma homocysteine levels of more than 100 μmol/L [97] and are rare in comparison to mild hyperhomocysteinemia. Mild hyperhomocysteinemia, in turn, may be due to moderate deficiencies of key enzymes involved in homocysteine metabolism, such as the thermolabile variant of 5,10-methylenetetrahydrofolate reductase gene that has been reported in 5–15% of the general population [98]. Additionally, mild hyperhomocysteinemia may be a result of dietary deficiencies of vitamin cofactors required for homocysteine catabolism — folic acid, vitamins B6 and B12, and is characterized by the plasma total homocysteine levels of 15–25 μmol/L [99]. Hyperhomocysteinemia is linked to a number of disorders. It is recognized as a strong, independent and causal risk factor for cardiovascular and vascular diseases [100], [101], [102], [103]. In particular, it has been estimated that a 2.5 μM rise in homocysteine levels corresponds to a 10% increase in cardiovascular disease risk [104] and it was shown that about 40% of patients diagnosed with premature coronary artery disease, peripheral vascular disease or venous thrombosis exhibit hyperhomocysteinemia [105]. In addition to vascular diseases hyperhomocysteinemia is also associated with a number of other pathologies including neurological disorders, cancer, renal insufficiency, diabetes, birth defects and aging [55], [56], [104], [106], [107], [108], [109]. A number of potential mechanisms that are aimed at the explanation of the pathological consequences resulting from homocysteine accumulation were proposed [100], [105], [110]; however, there is still a controversy on the underlying metabolic connections [111], [112], [113].
Since certain vitamin deficiencies are characterized by mild hyperhomocysteinemia the possibility that vitamin supplementation could reduce/prevent cardiovascular events was studied in several large trials. However, it was observed that vitamins, while capable of lowering elevated plasma homocysteine levels, do not reduce the rates of vascular events [114]. This result may be due to the promotion of cell proliferation by folic acid through its role in the synthesis of thymidine, the increase of the methylation potential leading to changes in gene expression, or an increase in the levels of asymmetric dimethylarginine that inhibits the activity of nitric oxide synthase, offsetting any positive effect of homocysteine lowering [114]. An additional possibility is that a related metabolite rather than homocysteine could be a trigger of some pathological changes associated with elevated homocysteine levels. Since the thermodynamic equilibrium of the S-adenosyl-L-homocysteine hydrolase reaction favors the synthesis of AdoHcy [82], the accumulation of hydrolytic products of the reaction, in particular, homocysteine leads to the reversal of the physiological direction of the Sah1 catalyzed reaction, resulting in AdoHcy synthesis and its accumulation in vivo, as shown both in mammals as well as in yeast [7], [83], [115]. Indeed, patients with even mild hyperhomocysteinemia (95% CI values for total homocysteine 11.0–14.7 μmol/L) that overlap those of controls (95% CI values 9.8–12.2 μmol/L) exhibit significant elevation of plasma AdoHcy (95% CI values 32.3–47.7 nmol/L) compared to controls (95% CI values 24.9–30.7 nmol/L) [116]. Therefore, even mild hyperhomocysteinemia is characterized by an accumulation of AdoHcy, which may lead to a decreased methylation potential and the inhibition of a number of AdoMet-dependent methyltransferases and, thus, contribute to the pathology associated with homocysteine accumulation.
Notably, the mechanisms that have been proposed to explain the pathological changes associated with elevated homocysteine levels do not address the accumulation of AdoHcy and the inhibition of AdoMet-dependent methylation in homocysteine-associated pathology, with the exception of an inhibition of DNA methylation [117], [118], [119]. However, AdoHcy rather than homocysteine was shown to be a more sensitive indicator of cardiovascular disease [116], [120] as well as of renal insufficiency [56] suggesting that AdoHcy is indeed a crucial pathological factor in homocysteine-associated disorders. It was also reported that the supplementation with B-vitamins including folate does not efficiently lower plasma AdoHcy levels in contrast to homocysteine levels in elderly people with hyperhomocysteinemia [121], possibly explaining the failure of homocysteine lowering vitamins to reduce vascular events mentioned earlier. Furthermore, supporting the pathogenic role of AdoHcy, studies in yeast showed that indeed AdoHcy but not homocysteine is more toxic to cells that are deficient in homocysteine catabolism [122].
8. Induced AdoHcy accumulation as a strategy towards antiviral therapy
During the last three decades S-adenosyl-L-homocysteine hydrolase was intensively studied as a target for antiviral drug design [123]. Inhibitors that block the enzyme are efficient against many types of viruses, including Ebola, that are dependent, in particular, on mRNA methylation [123], [124], [125]. In addition to antiviral activity inhibitors targeted at S-adenosyl-L-homocysteine hydrolase show also other effects of pharmacological importance, for instance, antimalarial and immunosuppressive activity [126], [127]. Although AdoHcy accumulation may be responsible for a number of the effects of inhibitors targeted at S-adenosyl-L-homocysteine hydrolase, the ability of some of these inhibitors to undergo metabolic phosphorylation to nucleotides may account for a part of their biological activities, making it difficult to validate the mode of their action [128]. Furthermore, the interference with central metabolic pathways associated with high cytotoxicity is an additional hindrance in the utilization of S-adenosyl-L-homocysteine hydrolase inhibitors in the potential future clinical applications [125]. Nevertheless, suppression of viral replication in plants by controlled downregulation of S-adenosyl-L-homocysteine hydrolase confirms the antiviral activity as a result of AdoHcy accumulation [129].
9. Yeast as a model system to study methylation deficiency
Numerous disorders associated with AdoHcy accumulation suggest that this metabolite is capable of triggering multiple pathological mechanisms, presumably via inhibition of different AdoMet-dependent methyltransferases. However, the role of AdoHcy in these diseases as well as the AdoHcy-dependent molecular mechanisms contributing to the consequences inherent to these pathologies is poorly understood. Of course, understanding the responsiveness of methylation-dependent processes requiring AdoMet-dependent methyltransferases that are present only in mammals, e. g. protein L-isoaspartate methyltransferase, requires the use of a mammalian model system. However, due to the complexity of the methylation metabolism its dissection in a multicellular organism as well as in higher cells is rather difficult.
The yeast Saccharomyces cerevisiae is a unicellular eukaryote with an about 4 times lower number of genes compared to humans, but shares the complexity of the cellular architecture of higher cells. Yeast is especially amenable to experimentation, in particular for genetic manipulation and whole genome studies. As a result, this model system is prized with the highest genome annotation level and was successfully used to characterize a number of fundamental biological processes, including secretion, organelle biogenesis and cell cycle [130], [131], [132], [133]. Yeast exhibits a highly conserved methylation metabolism [17], [65] and as such is an advantageous system to understand fundamental toxicity of AdoHcy at the cellular level. For instance, yeast mutants capable of reproducible down-regulation of S-adenosyl-L-homocysteine hydrolase and, thus, in vivo modulation of AdoHcy levels can be used as a valuable tool to understand downstream mechanisms triggered by AdoHcy accumulation. Usage of a yeast mutant that is deficient in homocysteine remethylation to methionine, met6, provides another independent system to understand the role of AdoHcy. In this mutant, grown under homocysteine supplementation, elevated homocysteine leads to AdoHcy synthesis via the reversal of S-adenosyl-L-homocysteine hydrolase reaction resulting in an accumulation of the latter. However, in this mutant elevated homocysteine cannot be remethylated to methionine, impeding an additional increase in AdoMet levels and an elevation of the AdoMet/AdoHcy ratio. Further advantage of this system is that it uncouples the S-adenosyl-L-homocysteine hydrolase function in the regulation of AdoMet-dependent methylation from its potential effect on glutathione levels. Both systems were already shown to help in the identification and characterization of critical cellular processes that respond to AdoHcy accumulation [7]. In the next section the deregulation of lipid metabolism, the major consumer of AdoMet both in yeast and in mammals, will be discussed, as a special aspect of AdoHcy toxicity.
10. Role of AdoHcy in lipid metabolism
Changes at the epigenetic level are the most extensively studied consequences of methylation deficiency [117], [118], [119]. However, quantitatively the most important AdoMet-dependent methyltransferases take part in the primary metabolism. Phospholipid methylation requires three sequential AdoMet-dependent methylation steps to synthesize one molecule of phosphatidylcholine (PC) from phosphatidylethanolamine (PE) de novo. Whereas in yeast phospholipid methylation is the predominant way to synthesize PC (in particular, in the absence of choline/ethanolamine in the culture medium), phospholipid methylation in mammals that occurs primarily in the liver covers only 30% of hepatic PC synthesis and accounts for estimated 10 μmol and 1.65 mmol PC secreted into bile per day in mice and humans, respectively [36]. Phospholipid methylation is the major AdoMet consumer in yeast in the absence of choline/ethanolamine supplementation. Despite of the relatively low contribution of phospholipid de novo synthesis to PC production in mammals this reaction is also the major consumer of AdoMet in mice [35]. Reexamination of the methylation metabolism in humans also revealed that phospholipid methylation, but not creatine synthesis as was assumed previously, accounts for the major part of AdoMet being utilized in the human body [36].
10.1. AdoHcy in yeast lipid metabolism
The involvement of S-adenosyl-L-homocysteine hydrolase in the regulation of phospholipid biosynthesis in yeast was found by showing that yeast SAH1 is regulated at the transcriptional level in coordination with phospholipid biosynthesis [134]. S-adenosyl-L-homocysteine hydrolase controls phospholipid de novo methylation in yeast by regulating the hydrolysis of AdoHcy, which is a competitive inhibitor of Cho2 and Opi3 phospholipid methyltransferases that catalyze methylation of PE to PC [7], [45] (Fig. 4). However, not only phospholipid methylation, but also a methylation-independent branch of lipid metabolism is affected by AdoHcy accumulation in yeast: yeast cells deficient in AdoHcy catabolism massively accumulate triacylglycerols (TAG) [7]. Supporting the causal role of impaired phospholipid methylation in the deregulation of TAG metabolism in response to AdoHcy accumulation, it was found that yeast mutants that are deficient in the enzymatic activities required for methylation of PE to PC, cho2 and opi3, also accumulate TAG [7]. Additionally, homocysteine supplementation in yeast that leads to a 10-fold accumulation of AdoHcy results in the inhibition of phospholipid methylation and accumulation of TAG, confirming a crosstalk between homocysteine and lipid metabolism [7].
Fig. 4.
Role of AdoHcy and S-adenosyl-L-homocysteine hydrolase in lipid metabolism in yeast. AdoMet, S-adenosyl-L-methionine; AdoHcy, S-adenosyl-L-homocysteine; Hcy, homocysteine; Met, methionine; Sah1, S-adenosyl-L-homocysteine hydrolase; CTT, cystathionine; Sam1, AdoMet synthetase 1; Sam2, AdoMet synthetase 2; Sam4, AdoMet-homocysteine methyltransferase; Mht1, S-methylmethionine–homocysteine methyltransferase; Met6, methionine synthase; Met25, O-acetylhomoserine sulfhydrylase; Str1, cystathionine γ-lyase; Str2, cystathionine γ-synthase; Str3, cystathionine β-lyase; Str4, cystathionine β-synthase; Gsh1, γ-glutamylcysteine synthetase; Gro, glycerol; DHAP, dihydroacetone phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; CDP-DAG, CDP-diacylglycerol; Cho, choline; Etn, ethanolamine; CL, cardiolipin; PG, phosphatidylglycerol; PGP, phosphatidylglycerol phosphate; Glc, glucose; Ins, inositol; PI, phosphatidylinositol; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; Cho2, phosphatidylethanolamine methyltransferase catalyzing first methylation from PE to PC; Opi3, phospholipid methyltransferase catalyzing two last methylation steps from PE to PC.
In addition to TAG metabolism, transcriptional regulation of phospholipid biosynthesis is also affected in mutants impaired in AdoHcy catabolism. Deficient phospholipid methylation in Sah1 depleted mutant cells unable to hydrolyze AdoHcy, or in cho2 and opi3 mutants, leads to induced expression of phospholipid biosynthetic (UASINO-responsive) genes, indicating accumulation of the phospholipid precursor, phosphatidic acid, in the endoplasmic reticulum (ER) [7]. ACC1 encoding acetyl-CoA carboxylase, the first and rate-limiting enzyme of fatty acid biosynthesis, is also subject to UASINO-mediated regulation. Therefore, the inhibition of phospholipid methylation in mutants deficient in AdoHcy catabolism as well as in cells supplemented with Hcy appear to result in an accumulation of fatty acids and their channeling into TAG, which play a crucial role in buffering excess fatty acids [135]. Moreover, in addition to a deregulated phospholipid and triacylglycerol metabolism, down-regulation of SAH1 expression in yeast also impairs sterol synthesis leading to 4-fold elevated squalene levels. This suggests an accumulation of early precursors of ergosterol biosynthesis under these conditions, presumably as a result of inhibition of sterol 24-C-methyltransferase (Tehlivets, Kohlwein, unpublished).
10.2. AdoHcy and homocysteine in lipid metabolism in mammals
AdoHcy also inhibits mammalian phosphatidylethanolamine N-methyltransferase (PEMT) [136], [137]. Similarly as in yeast, deficiency of phospholipid methylation in PEMT−/− knockout mice leads to a rapid decrease of the hepatic PC/PE ratio and accumulation of TAG in the liver, in the absence of choline supplementation [138]. However, TAG accumulation in the livers of these animals is also due to a decreased TAG secretion from hepatocytes [139].
Elevated levels of homocysteine are also linked to the deregulation of lipid metabolism in mammals. Cystathionine β-synthase−/− knockout mice exhibit severe hyperhomocysteinemia and accumulate AdoHcy in all tissues tested [53], [140]. These mutant animals show elevated TAG and nonesterified fatty acid levels in the liver and serum, and develop hepatic steatosis [12]. Another genetic disorder that results in moderately elevated homocysteine levels, methylenetetrahydrofolate reductase deficiency, leads to fatty liver development as well as to neuropathology and aortic lipid deposition in mouse model systems [10], [141]. Additionally, dietary-induced hyperhomocysteinemia in mice also causes fatty liver [142] suggesting that AdoHcy accumulation and impaired phospholipid methylation are common denominators in homocysteine-associated pathologies.
10.3. Activation of UPR and deregulation of unsaturated fatty acid metabolism in AdoHcy-related deficiencies
Supporting the role of elevated homocysteine levels in the deregulation of lipid metabolism in mammals, homocysteine supplementation was shown to lead to the activation of the sterol regulatory element-binding proteins (SREBPs), which function to activate genes encoding enzymes in cholesterol, fatty acid and triacylglycerol biosynthesis and uptake pathways, both in cultured mammalian cell lines as well as in the livers of CBS−/− mice that lack cystathionine β-synthase [142], [143]. The finding that overexpression of the ER chaperone GRP78/BiP inhibits homocysteine-induced gene expression [142], [144] suggests that unfolded protein response (UPR) induction plays a direct role in the activation of triacylglycerol and cholesterol biosynthesis in response to homocysteine accumulation in mammals. One of the possible mechanisms responsible for the induction of ER stress and UPR activation in response to homocysteine accumulation is the accumulation of saturated fatty acids in membrane phospholipids [145], [146], [147]. In accordance, overexpression of stearoyl-CoA desaturase was shown to attenuate palmitate-induced ER stress and protect from lipoapoptosis in mammalian cells [148], [149], [150].
Phospholipid methyltransferases preferentially catalyze synthesis of PC containing di-C16:1 species in yeast both in vitro and in vivo [151]. In line, the PEMT-derived PC pool is also enriched in unsaturated fatty acids [152], [153]. Supporting the role of PEMT in the metabolism of unsaturated fatty acids, PEMT−/− knockout mice were reported to accumulate more saturated PC molecular species in the liver compared with the control littermates [154], and to exhibit dramatically reduced concentrations of polyunsaturated fatty acids in the plasma and in hepatic PC, independently of the choline status [155]. Moreover, both AdoHcy and homocysteine accumulation are linked to decreased levels of unsaturated fatty acids. It was shown in particular, that elevated plasma AdoHcy levels are negatively correlated with both PC content and the level of polyunsaturated fatty acids in PC in red blood cells in Alzheimer patients [156]. Furthermore, elevated plasma homocysteine levels were shown to be associated with a decrease in polyunsaturated (docosahexaenoic) fatty acids in the plasma of healthy humans [157] and in the plasma and erythrocytes of cystic fibrosis patients [158]. It was proposed that deficiency of phospholipid methylation due to AdoHcy accumulation leads to an increase in saturated PC molecular species in ER membranes followed by ER stress, protein misfolding, induction of UPR and activation of lipid metabolism [159].
11. Concluding remarks and future perspectives
With 70% identity between yeast and human orthologs S-adenosyl-L-homocysteine hydrolase, which catalyzes the reversible hydrolysis of AdoHcy to homocysteine and adenosine, is among the most conserved S. cerevisiae proteins, pointing to its central role in the cellular metabolism. Moreover, accumulation of homocysteine, characteristic to the common pathological condition, hyperhomocysteinemia, “mimics” S-adenosyl-L-homocysteine hydrolase dysfunction, due to the reversal of the S-adenosyl-L-homocysteine hydrolase reaction resulting in the synthesis and accumulation of AdoHcy.
Homocysteine is linked to numerous diseases, however, the mechanisms responsible for the pathological consequences associated with these disorders are still elusive. Most importantly, AdoHcy rather than homocysteine is recognized since recently as a more sensitive marker of several homocysteine-associated pathologies.
As a product inhibitor of a number of AdoMet-dependent methyltransferases AdoHcy can trigger multiple pathological mechanisms. Due to the complexity of methylation metabolism comprehension of these mechanisms requires a systematic approach. Yeast as a validated unicellular model system particularly amenable to genomic approaches offers new possibilities to understand cellular responses to AdoHcy accumulation. Our recent work confirms that this system can be successfully used for identification and characterization of AdoHcy responsive cellular processes that are relevant for humans.
Acknowledgements
We thank Dr. Sepp D. Kohlwein, Institute of Molecular Biosciences, University of Graz for critically reading this manuscript and for helpful comments. T.P-K. is supported by the Federal Ministry of Economy, Family and Youth (BMWFJ), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol and the ZIT Technology Agency of the City of Vienna through the COMET Funding Program managed by the Austrian Research Promotion Agency FFG.
Work in the author's laboratory is supported by the Austrian Science Fund FWF, projects P24216-B21 and P18094-B14 to O.T.
References
- 1.Luka Z., Mudd S.H., Wagner C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J. Biol. Chem. 2009;284:22507–22511. doi: 10.1074/jbc.R109.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantoni G.L. Biological methylation: selected aspects. Annu. Rev. Biochem. 1975;44:435–451. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- 3.Lu S.C. S-adenosylmethionine. Int. J. Biochem. Cell Biol. 2000;32:391–395. doi: 10.1016/s1357-2725(99)00139-9. [DOI] [PubMed] [Google Scholar]
- 4.Fontecave M., Atta M., Mulliez E. S-adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Finkelstein J.D. The metabolism of homocysteine: pathways and regulation. Eur. J. Pediatr. 1998;157:S40–S44. doi: 10.1007/pl00014300. [DOI] [PubMed] [Google Scholar]
- 6.Elmore C.L., Wu X., Leclerc D., Watson E.D., Bottiglieri T., Krupenko N.I., Krupenko S.A., Cross J.C., Rozen R., Gravel R.A., Matthews R.G. Metabolic derangement of methionine and folate metabolism in mice deficient in methionine synthase reductase. Mol. Genet. Metab. 2007;91:85–97. doi: 10.1016/j.ymgme.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malanovic N., Streith I., Wolinski H., Rechberger G., Kohlwein S.D., Tehlivets O. S-adenosyl-l-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J. Biol. Chem. 2008;283:23989–23999. doi: 10.1074/jbc.M800830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Chantar M.L., Corrales F.J., Martinez-Cruz L.A., Garcia-Trevijano E.R., Huang Z.Z., Chen L., Kanel G., Avila M.A., Mato J.M., Lu S.C. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Chantar M.L., Vazquez-Chantada M., Ariz U., Martinez N., Varela M., Luka Z., Capdevila A., Rodriguez J., Aransay A.M., Matthiesen R., Yang H., Calvisi D.F., Esteller M., Fraga M., Lu S.C., Wagner C., Mato J.M. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z., Karaplis A.C., Ackerman S.L., Pogribny I.P., Melnyk S., Lussier-Cacan S., Chen M.F., Pai A., John S.W., Smith R.S., Bottiglieri T., Bagley P., Selhub J., Rudnicki M.A., James S.J., Rozen R. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum. Mol. Genet. 2001;10:433–443. doi: 10.1093/hmg/10.5.433. [DOI] [PubMed] [Google Scholar]
- 11.Teng Y.W., Mehedint M.G., Garrow T.A., Zeisel S.H. Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and 1-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J. Biol. Chem. 2011;286:36258–36267. doi: 10.1074/jbc.M111.265348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Namekata K., Enokido Y., Ishii I., Nagai Y., Harada T., Kimura H. Abnormal lipid metabolism in cystathionine beta-synthase-deficient mice, an animal model for hyperhomocysteinemia. J. Biol. Chem. 2004;279:52961–52969. doi: 10.1074/jbc.M406820200. [DOI] [PubMed] [Google Scholar]
- 13.Cano A., Buque X., Martinez-Una M., Aurrekoetxea I., Menor A., Garcia-Rodriguez J.L., Lu S.C., Martinez-Chantar M.L., Mato J.M., Ochoa B., Aspichueta P. Methionine adenosyltransferase 1A gene deletion disrupts hepatic very low-density lipoprotein assembly in mice. Hepatology. 2011;54:1975–1986. doi: 10.1002/hep.24607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baric I. Inherited disorders in the conversion of methionine to homocysteine. J. Inherit. Metab. Dis. 2009;32:459–471. doi: 10.1007/s10545-009-1146-4. [DOI] [PubMed] [Google Scholar]
- 15.Chamberlin M.E., Ubagai T., Mudd S.H., Wilson W.G., Leonard J.V., Chou J.Y. Demyelination of the brain is associated with methionine adenosyltransferase I/III deficiency. J. Clin. Invest. 1996;98:1021–1027. doi: 10.1172/JCI118862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mita T., Kawazu I., Hirano H., Ohmori O., Janjua N., Shibata K. E1 mice epilepsy shows genetic polymorphism for S-adenosyl-l-homocysteine hydrolase. Neurochem. Int. 2001;38:349–357. doi: 10.1016/s0197-0186(00)00089-9. [DOI] [PubMed] [Google Scholar]
- 17.Petrossian T.C., Clarke S.G. Uncovering the human methyltransferasome. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.000976. (M110 000976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrossian T., Clarke S. Bioinformatic identification of novel methyltransferases. Epigenomics. 2009;1:163–175. doi: 10.2217/epi.09.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grillo M.A., Colombatto S. S-adenosylmethionine and protein methylation. Amino acids. 2005;28:357–362. doi: 10.1007/s00726-005-0197-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee D.Y., Teyssier C., Strahl B.D., Stallcup M.R. Role of protein methylation in regulation of transcription. Endocr. Rev. 2005;26:147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- 21.Polevoda B., Sherman F. Methylation of proteins involved in translation. Mol. Microbiol. 2007;65:590–606. doi: 10.1111/j.1365-2958.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y.H., Stallcup M.R. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dillon S.C., Zhang X., Trievel R.C., Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik S., Bhaumik S.R. Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J. 2011;277:1805–1821. doi: 10.1111/j.1742-4658.2010.07607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chern M.K., Chang K.N., Liu L.F., Tam T.C., Liu Y.C., Liang Y.L., Tam M.F. Yeast ribosomal protein L12 is a substrate of protein–arginine methyltransferase 2. J. Biol. Chem. 2002;277:15345–15353. doi: 10.1074/jbc.M111379200. [DOI] [PubMed] [Google Scholar]
- 26.Wong C.M., Tang H.M., Kong K.Y., Wong G.W., Qiu H., Jin D.Y., Hinnebusch A.G. Yeast arginine methyltransferase Hmt1p regulates transcription elongation and termination by methylating Npl3p. Nucleic Acids Res. 2010;38:2217–2228. doi: 10.1093/nar/gkp1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu M.C., Bachand F., McBride A.E., Komili S., Casolari J.M., Silver P.A. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q., Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolstykh T., Lee J., Vafai S., Stock J.B. Carboxyl methylation regulates phosphoprotein phosphatase 2A by controlling the association of regulatory B subunits. EMBO J. 2000;19:5682–5691. doi: 10.1093/emboj/19.21.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrycyna C.A., Clarke S. Modification of eukaryotic signaling proteins by C-terminal methylation reactions. Pharmacol. Ther. 1993;59:281–300. doi: 10.1016/0163-7258(93)90071-k. [DOI] [PubMed] [Google Scholar]
- 31.Chow C.S., Lamichhane T.N., Mahto S.K. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou Y.M., Perona J.J. Stereochemical mechanisms of tRNA methyltransferases. FEBS Lett. 2011;584:278–286. doi: 10.1016/j.febslet.2009.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao X., Schwer B., Shuman S. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: cap methyltransferase activity is essential for cell growth. Mol. Cell. Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukamoto T., Shibagaki Y., Niikura Y., Mizumoto K. Cloning and characterization of three human cDNAs encoding mRNA (guanine-7-)-methyltransferase, an mRNA cap methylase. Biochem. Biophys. Res. Commun. 1998;251:27–34. doi: 10.1006/bbrc.1998.9402. [DOI] [PubMed] [Google Scholar]
- 35.Noga A.A., Stead L.M., Zhao Y., Brosnan M.E., Brosnan J.T., Vance D.E. Plasma homocysteine is regulated by phospholipid methylation. J. Biol. Chem. 2003;278:5952–5955. doi: 10.1074/jbc.M212194200. [DOI] [PubMed] [Google Scholar]
- 36.Stead L.M., Brosnan J.T., Brosnan M.E., Vance D.E., Jacobs R.L. Is it time to reevaluate methyl balance in humans? Am. J. Clin. Nutr. 2006;83:5–10. doi: 10.1093/ajcn/83.1.5. [DOI] [PubMed] [Google Scholar]
- 37.Clarke S., Banfield K. In: Homocysteine in Health and Disease. Carmel R., Jackobsen D.W., editors. Cambridge University Press; 2001. S-adenosylmethionine-dependent methyltransferases; pp. 63–78. [Google Scholar]
- 38.Bacolla A., Pradhan S., Roberts R.J., Wells R.D. Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated DNA. J. Biol. Chem. 1999;274:33011–33019. doi: 10.1074/jbc.274.46.33011. [DOI] [PubMed] [Google Scholar]
- 39.Mao X., Schwer B., Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hausmann S., Zheng S., Fabrega C., Schneller S.W., Lima C.D., Shuman S. Encephalitozoon cuniculi mRNA cap (guanine N-7) methyltransferase: methyl acceptor specificity, inhibition BY S-adenosylmethionine analogs, and structure-guided mutational analysis. J. Biol. Chem. 2005;280:20404–20412. doi: 10.1074/jbc.M501073200. [DOI] [PubMed] [Google Scholar]
- 41.Shugart L., Chastain B. Escherichia coli tRNA (uracil-5-)-methyltransferase: inhibition by analogues of adenosylhomocysteine. Enzyme. 1979;24:353–357. doi: 10.1159/000458689. [DOI] [PubMed] [Google Scholar]
- 42.Baron R.A., Casey P.J. Analysis of the kinetic mechanism of recombinant human isoprenylcysteine carboxylmethyltransferase (Icmt) BMC Biochem. 2004;5:19. doi: 10.1186/1471-2091-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson B.A., Aswad D.W. Kinetic properties of bovine brain protein l-isoaspartyl methyltransferase determined using a synthetic isoaspartyl peptide substrate. Neurochem. Res. 1993;18:87–94. doi: 10.1007/BF00966926. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman D.R., Haning J.A., Cornatzer W.E. Microsomal phosphatidylethanolamine methyltransferase: inhibition by S-adenosylhomocysteine. Lipids. 1981;16:561–567. doi: 10.1007/BF02534900. [DOI] [PubMed] [Google Scholar]
- 45.Gaynor P.M., Carman G.M. Phosphatidylethanolamine methyltransferase and phospholipid methyltransferase activities from Saccharomyces cerevisiae. Enzymological and kinetic properties. Biochim. Biophys. Acta. 1990;1045:156–163. doi: 10.1016/0005-2760(90)90145-n. [DOI] [PubMed] [Google Scholar]
- 46.Castro R., Rivera I., Martins C., Struys E.A., Jansen E.E., Clode N., Graca L.M., Blom H.J., Jakobs C., de Almeida I.T. Intracellular S-adenosylhomocysteine increased levels are associated with DNA hypomethylation in HUVEC. J. Mol. Med. 2005;83:831–836. doi: 10.1007/s00109-005-0679-8. [DOI] [PubMed] [Google Scholar]
- 47.Schanche J.S., Schanche T., Ueland P.M. Inhibition of phospholipid methylation in isolated rat hepatocytes by analogues of adenosine and S-adenosylhomocysteine. Biochim. Biophys. Acta. 1982;721:399–407. doi: 10.1016/0167-4889(82)90095-7. [DOI] [PubMed] [Google Scholar]
- 48.Huang Y., Komoto J., Konishi K., Takata Y., Ogawa H., Gomi T., Fujioka M., Takusagawa F. Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes. J. Mol. Biol. 2000;298:149–162. doi: 10.1006/jmbi.2000.3637. [DOI] [PubMed] [Google Scholar]
- 49.Caudill M.A., Wang J.C., Melnyk S., Pogribny I.P., Jernigan S., Collins M.D., Santos-Guzman J., Swendseid M.E., Cogger E.A., James S.J. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J. Nutr. 2001;131:2811–2818. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- 50.Kramer D.L., Porter C.W., Borchardt R.T., Sufrin J.R. Combined modulation of S-adenosylmethionine biosynthesis and S-adenosylhomocysteine metabolism enhances inhibition of nucleic acid methylation and L1210 cell growth. Cancer Res. 1990;50:3838–3842. [PubMed] [Google Scholar]
- 51.Radomski N., Kaufmann C., Dreyer C. Nuclear accumulation of S-adenosylhomocysteine hydrolase in transcriptionally active cells during development of Xenopus laevis. Mol. Biol. Cell. 1999;10:4283–4298. doi: 10.1091/mbc.10.12.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjursell M.K., Blom H.J., Cayuela J.A., Engvall M.L., Lesko N., Balasubramaniam S., Brandberg G., Halldin M., Falkenberg M., Jakobs C., Smith D., Struys E., von Dobeln U., Gustafsson C.M., Lundeberg J., Wedell A. Adenosine kinase deficiency disrupts the methionine cycle and causes hypermethioninemia, encephalopathy, and abnormal liver function. Am. J. Hum. Genet. 2011;89:507–515. doi: 10.1016/j.ajhg.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dayal S., Bottiglieri T., Arning E., Maeda N., Malinow M.R., Sigmund C.D., Heistad D.D., Faraci F.M., Lentz S.R. Endothelial dysfunction and elevation of S-adenosylhomocysteine in cystathionine beta-synthase-deficient mice. Circ. Res. 2001;88:1203–1209. doi: 10.1161/hh1101.092180. [DOI] [PubMed] [Google Scholar]
- 54.Loehrer F.M., Tschopl M., Angst C.P., Litynski P., Jager K., Fowler B., Haefeli W.E. Disturbed ratio of erythrocyte and plasma S-adenosylmethionine/S-adenosylhomocysteine in peripheral arterial occlusive disease. Atherosclerosis. 2001;154:147–154. doi: 10.1016/s0021-9150(00)00449-4. [DOI] [PubMed] [Google Scholar]
- 55.Herrmann W., Schorr H., Obeid R., Makowski J., Fowler B., Kuhlmann M.K. Disturbed homocysteine and methionine cycle intermediates S-adenosylhomocysteine and S-adenosylmethionine are related to degree of renal insufficiency in type 2 diabetes. Clin. Chem. 2005;51:891–897. doi: 10.1373/clinchem.2004.044453. [DOI] [PubMed] [Google Scholar]
- 56.Jabs K., Koury M.J., Dupont W.D., Wagner C. Relationship between plasma S-adenosylhomocysteine concentration and glomerular filtration rate in children. Metabolism. 2006;55:252–257. doi: 10.1016/j.metabol.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 57.Poirier L.A., Brown A.T., Fink L.M., Wise C.K., Randolph C.J., Delongchamp R.R., Fonseca V.A. Blood S-adenosylmethionine concentrations and lymphocyte methylenetetrahydrofolate reductase activity in diabetes mellitus and diabetic nephropathy. Metabolism. 2001;50:1014–1018. doi: 10.1053/meta.2001.25655. [DOI] [PubMed] [Google Scholar]
- 58.Leal J., Ferrer I., Blanco-Aparicio C., Hernandez-Losa J., Ramon Y.C., Carnero A., Lleonart M. S-adenosylhomocysteine hydrolase downregulation contributes to tumorigenesis. Carcinogenesis. 2008;29:2089–2095. doi: 10.1093/carcin/bgn198. [DOI] [PubMed] [Google Scholar]
- 59.Miller M.W., Duhl D.M., Winkes B.M., Arredondo-Vega F., Saxon P.J., Wolff G.L., Epstein C.J., Hershfield M.S., Barsh G.S. The mouse lethal nonagouti (a(x)) mutation deletes the S-adenosylhomocysteine hydrolase (Ahcy) gene. EMBO J. 1994;13:1806–1816. doi: 10.1002/j.1460-2075.1994.tb06449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rocha P.S., Sheikh M., Melchiorre R., Fagard M., Boutet S., Loach R., Moffatt B., Wagner C., Vaucheret H., Furner I. The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 gene codes for an S-adenosyl-l-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell. 2005;17:404–417. doi: 10.1105/tpc.104.028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baric I., Fumic K., Glenn B., Cuk M., Schulze A., Finkelstein J.D., James S.J., Mejaski-Bosnjak V., Pazanin L., Pogribny I.P., Rados M., Sarnavka V., Scukanec-Spoljar M., Allen R.H., Stabler S., Uzelac L., Vugrek O., Wagner C., Zeisel S., Mudd S.H. S-adenosylhomocysteine hydrolase deficiency in a human: a genetic disorder of methionine metabolism. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4234–4239. doi: 10.1073/pnas.0400658101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baric I., Cuk M., Fumic K., Vugrek O., Allen R.H., Glenn B., Maradin M., Pazanin L., Pogribny I., Rados M., Sarnavka V., Schulze A., Stabler S., Wagner C., Zeisel S.H., Mudd S.H. S-adenosylhomocysteine hydrolase deficiency: a second patient, the younger brother of the index patient, and outcomes during therapy. J. Inherit. Metab. Dis. 2005;28:885–902. doi: 10.1007/s10545-005-0192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buist N.R., Glenn B., Vugrek O., Wagner C., Stabler S., Allen R.H., Pogribny I., Schulze A., Zeisel S.H., Baric I., Mudd S.H. S-adenosylhomocysteine hydrolase deficiency in a 26-year-old man. J. Inherit. Metab. Dis. 2006;29:538–545. doi: 10.1007/s10545-006-0240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grubbs R., Vugrek O., Deisch J., Wagner C., Stabler S., Allen R., Baric I., Rados M., Mudd S.H. S-adenosylhomocysteine hydrolase deficiency: two siblings with fetal hydrops and fatal outcomes. J. Inherit. Metab. Dis. 2010;33:705–713. doi: 10.1007/s10545-010-9171-x. [DOI] [PubMed] [Google Scholar]
- 65.Mushegian A.R., Garey J.R., Martin J., Liu L.X. Large-scale taxonomic profiling of eukaryotic model organisms: a comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res. 1998;8:590–598. doi: 10.1101/gr.8.6.590. [DOI] [PubMed] [Google Scholar]
- 66.Stepkowski T., Brzezinski K., Legocki A.B., Jaskolski M., Bena G. Bayesian phylogenetic analysis reveals two-domain topology of S-adenosylhomocysteine hydrolase protein sequences. Mol. Phylogenet. Evol. 2005;34:15–28. doi: 10.1016/j.ympev.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 67.Rao S.T., Rossmann M.G. Comparison of super-secondary structures in proteins. J. Mol. Biol. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 68.Turner M.A., Yuan C.S., Borchardt R.T., Hershfield M.S., Smith G.D., Howell P.L. Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength. Nat. Struct. Biol. 1998;5:369–376. doi: 10.1038/nsb0598-369. [DOI] [PubMed] [Google Scholar]
- 69.Yang X., Hu Y., Yin D.H., Turner M.A., Wang M., Borchardt R.T., Howell P.L., Kuczera K., Schowen R.L. Catalytic strategy of S-adenosyl-l-homocysteine hydrolase: transition-state stabilization and the avoidance of abortive reactions. Biochemistry. 2003;42:1900–1909. doi: 10.1021/bi0262350. [DOI] [PubMed] [Google Scholar]
- 70.Lee K.M., Choi W.J., Lee Y., Lee H.J., Zhao L.X., Lee H.W., Park J.G., Kim H.O., Hwang K.Y., Heo Y.S., Choi S., Jeong L.S. X-ray crystal structure and binding mode analysis of human S-adenosylhomocysteine hydrolase complexed with novel mechanism-based inhibitors, haloneplanocin A analogues. J. Med. Chem. 2011;54:930–938. doi: 10.1021/jm1010836. [DOI] [PubMed] [Google Scholar]
- 71.Hu Y., Komoto J., Huang Y., Gomi T., Ogawa H., Takata Y., Fujioka M., Takusagawa F. Crystal structure of S-adenosylhomocysteine hydrolase from rat liver. Biochemistry. 1999;38:8323–8333. doi: 10.1021/bi990332k. [DOI] [PubMed] [Google Scholar]
- 72.Takata Y., Yamada T., Huang Y., Komoto J., Gomi T., Ogawa H., Fujioka M., Takusagawa F. Catalytic mechanism of S-adenosylhomocysteine hydrolase. Site-directed mutagenesis of Asp-130, Lys-185, Asp-189, and Asn-190. J. Biol. Chem. 2002;277:22670–22676. doi: 10.1074/jbc.M201116200. [DOI] [PubMed] [Google Scholar]
- 73.Huang Y., Komoto J., Takata Y., Powell D.R., Gomi T., Ogawa H., Fujioka M., Takusagawa F. Inhibition of S-adenosylhomocysteine hydrolase by acyclic sugar adenosine analogue d-eritadenine. Crystal structure of S-adenosylhomocysteine hydrolase complexed with d-eritadenine. J. Biol. Chem. 2002;277:7477–7482. doi: 10.1074/jbc.M109187200. [DOI] [PubMed] [Google Scholar]
- 74.Yamada T., Komoto J., Lou K., Ueki A., Hua D.H., Sugiyama K., Takata Y., Ogawa H., Takusagawa F. Structure and function of eritadenine and its 3-deaza analogues: potent inhibitors of S-adenosylhomocysteine hydrolase and hypocholesterolemic agents. Biochem. Pharmacol. 2007;73:981–989. doi: 10.1016/j.bcp.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 75.Yamada T., Takata Y., Komoto J., Gomi T., Ogawa H., Fujioka M., Takusagawa F. Catalytic mechanism of S-adenosylhomocysteine hydrolase: roles of His 54, Asp130, Glu155, Lys185, and Aspl89. Int. J. Biochem. Cell Biol. 2005;37:2417–2435. doi: 10.1016/j.biocel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 76.Komoto J., Huang Y., Gomi T., Ogawa H., Takata Y., Fujioka M., Takusagawa F. Effects of site-directed mutagenesis on structure and function of recombinant rat liver S-adenosylhomocysteine hydrolase. Crystal structure of D244E mutant enzyme. J. Biol. Chem. 2000;275:32147–32156. doi: 10.1074/jbc.M003725200. [DOI] [PubMed] [Google Scholar]
- 77.Tanaka N., Nakanishi M., Kusakabe Y., Shiraiwa K., Yabe S., Ito Y., Kitade Y., Nakamura K.T. Crystal structure of S-adenosyl-l-homocysteine hydrolase from the human malaria parasite Plasmodium falciparum. J. Mol. Biol. 2004;343:1007–1017. doi: 10.1016/j.jmb.2004.08.104. [DOI] [PubMed] [Google Scholar]
- 78.Reddy M.C., Kuppan G., Shetty N.D., Owen J.L., Ioerger T.R., Sacchettini J.C. Crystal structures of Mycobacterium tuberculosis S-adenosyl-l-homocysteine hydrolase in ternary complex with substrate and inhibitors. Protein Sci. 2008;17:2134–2144. doi: 10.1110/ps.038125.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brzezinski K., Dauter Z., Jaskolski M. High-resolution structures of complexes of plant S-adenosyl-l-homocysteine hydrolase (Lupinus luteus) Acta Crystallogr. D. 2012;68:218–231. doi: 10.1107/S0907444911055090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brzezinski K., Bujacz G., Jaskolski M. Purification, crystallization and preliminary crystallographic studies of plant S-adenosyl-l-homocysteine hydrolase (Lupinus luteus) Acta Crystallogr. F. 2008;64:671–673. doi: 10.1107/S1744309108017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guranowski A., Pawelkiewicz J. Adenosylhomocysteinase from yellow lupin seeds. Purification and properties. Eur. J. Biochem. 1977;80:517–523. doi: 10.1111/j.1432-1033.1977.tb11907.x. [DOI] [PubMed] [Google Scholar]
- 82.De La Haba G., Cantoni G.L. The enzymatic synthesis of S-adenosyl-l-homocysteine from adenosine and homocysteine. J. Biol. Chem. 1959;234:603–608. [PubMed] [Google Scholar]
- 83.Hoffman D.R., Marion D.W., Cornatzer W.E., Duerre J.A. S-adenosylmethionine and S-adenosylhomocystein metabolism in isolated rat liver. Effects of l-methionine, l-homocystein, and adenosine. J. Biol. Chem. 1980;255:10822–10827. [PubMed] [Google Scholar]
- 84.Palmer J.L., Abeles R.H. The mechanism of action of S-adenosylhomocysteinase. J. Biol. Chem. 1979;254:1217–1226. [PubMed] [Google Scholar]
- 85.Wang M., Borchardt R.T., Schowen R.L., Kuczera K. Domain motions and the open-to-closed conformational transition of an enzyme: a normal mode analysis of S-adenosyl-l-homocysteine hydrolase. Biochemistry. 2005;44:7228–7239. doi: 10.1021/bi047524m. [DOI] [PubMed] [Google Scholar]
- 86.Wang M., Unruh J.R., Johnson C.K., Kuczera K., Schowen R.L., Borchardt R.T. Effects of ligand binding and oxidation on hinge-bending motions in S-adenosyl-l-homocysteine hydrolase. Biochemistry. 2006;45:7778–7786. doi: 10.1021/bi0523106. [DOI] [PubMed] [Google Scholar]
- 87.Hu C., Fang J., Borchardt R.T., Schowen R.L., Kuczera K. Molecular dynamics simulations of domain motions of substrate-free S-adenosyl-l-homocysteine hydrolase in solution. Proteins. 2008;71:131–143. doi: 10.1002/prot.21664. [DOI] [PubMed] [Google Scholar]
- 88.Kloor D., Ludtke A., Stoeva S., Osswald H. Adenosine binding sites at S-adenosylhomocysteine hydrolase are controlled by the NAD +/NADH ratio of the enzyme. Biochem. Pharmacol. 2003;66:2117–2123. doi: 10.1016/s0006-2952(03)00581-1. [DOI] [PubMed] [Google Scholar]
- 89.Bethin K.E., Cimato T.R., Ettinger M.J. Copper binding to mouse liver S-adenosylhomocysteine hydrolase and the effects of copper on its levels. J. Biol. Chem. 1995;270:20703–20711. doi: 10.1074/jbc.270.35.20703. [DOI] [PubMed] [Google Scholar]
- 90.Bethin K.E., Petrovic N., Ettinger M.J. Identification of a major hepatic copper binding protein as S-adenosylhomocysteine hydrolase. J. Biol. Chem. 1995;270:20698–20702. doi: 10.1074/jbc.270.35.20698. [DOI] [PubMed] [Google Scholar]
- 91.Petrovic N., Comi A., Ettinger M.J. Copper incorporation into superoxide dismutase in Menkes lymphoblasts. J. Biol. Chem. 1996;271:28335–28340. doi: 10.1074/jbc.271.45.28335. [DOI] [PubMed] [Google Scholar]
- 92.Li M., Li Y., Chen J., Wei W., Pan X., Liu J., Liu Q., Leu W., Zhang L., Yang X., Lu J., Wang K. Copper ions inhibit S-adenosylhomocysteine hydrolase by causing dissociation of NAD + cofactor. Biochemistry. 2007;46:11451–11458. doi: 10.1021/bi700395d. [DOI] [PubMed] [Google Scholar]
- 93.Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 94.Radomski N., Barreto G., Kaufmann C., Yokoska J., Mizumoto K., Dreyer C. Interaction of S-adenosylhomocysteine hydrolase of Xenopus laevis with mRNA(guanine-7-)methyltransferase: implication on its nuclear compartmentalisation and on cap methylation of hnRNA. Biochim. Biophys. Acta. 2002;1590:93–102. doi: 10.1016/s0167-4889(02)00205-7. [DOI] [PubMed] [Google Scholar]
- 95.Kloor D., Hermes M., Fink K., Schmid H., Klingel K., Mack A., Grenz A., Osswald H. Expression and localization of S-adenosylhomocysteine-hydrolase in the rat kidney following carbon monoxide induced hypoxia. Cell. Physiol. Biochem. 2007;19:57–66. doi: 10.1159/000099192. [DOI] [PubMed] [Google Scholar]
- 96.Lee S., Doxey A.C., McConkey B.J., Moffatt B.A. Nuclear targeting of methyl-recycling enzymes in Arabidopsis thaliana is mediated by specific protein interactions. Mol. Plant. 2012;5:231–248. doi: 10.1093/mp/ssr083. [DOI] [PubMed] [Google Scholar]
- 97.Finkelstein J.D., Martin J.J. Homocysteine. Int. J. Biochem. Cell Biol. 2000;32:385–389. doi: 10.1016/s1357-2725(99)00138-7. [DOI] [PubMed] [Google Scholar]
- 98.Golbahar J., Fathi Z., Tamadon M. Distribution of 5,10-methylenetetrahydrofolate reductase (C667T) polymorphism and its association with red blood cell 5-methyltetrahydrofolate in the healthy Iranians. Clin. Nutr. 2005;24:83–87. doi: 10.1016/j.clnu.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 99.Jacobsen D.W. Homocysteine and vitamins in cardiovascular disease. Clin. Chem. 1998;44:1833–1843. [PubMed] [Google Scholar]
- 100.McCully K.S. Homocysteine, vitamins, and vascular disease prevention. Am. J. Clin. Nutr. 2007;86:1563S–1568S. doi: 10.1093/ajcn/86.5.1563S. [DOI] [PubMed] [Google Scholar]
- 101.Refsum H., Ueland P.M., Nygard O., Vollset S.E. Homocysteine and cardiovascular disease. Annu. Rev. Med. 1998;49:31–62. doi: 10.1146/annurev.med.49.1.31. [DOI] [PubMed] [Google Scholar]
- 102.Bostom A.G., Rosenberg I.H., Silbershatz H., Jacques P.F., Selhub J., D'Agostino R.B., Wilson P.W., Wolf P.A. Nonfasting plasma total homocysteine levels and stroke incidence in elderly persons: the Framingham Study. Ann. Intern. Med. 1999;131:352–355. doi: 10.7326/0003-4819-131-5-199909070-00006. [DOI] [PubMed] [Google Scholar]
- 103.Wald D.S., Law M., Morris J.K. Homocysteine and cardiovascular disease: evidence on causality from a meta-analysis. BMJ. 2002;325:1202. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams K.T., Schalinske K.L. Homocysteine metabolism and its relation to health and disease. Biofactors. 2010;36:19–24. doi: 10.1002/biof.71. [DOI] [PubMed] [Google Scholar]
- 105.Lawrence de Koning A.B., Werstuck G.H., Zhou J., Austin R.C. Hyperhomocysteinemia and its role in the development of atherosclerosis. Clin. Biochem. 2003;36:431–441. doi: 10.1016/s0009-9120(03)00062-6. [DOI] [PubMed] [Google Scholar]
- 106.Seshadri S., Beiser A., Selhub J., Jacques P.F., Rosenberg I.H., D'Agostino R.B., Wilson P.W., Wolf P.A. Plasma homocysteine as a risk factor for dementia and Alzheimer's disease. N. Engl. J. Med. 2002;346:476–483. doi: 10.1056/NEJMoa011613. [DOI] [PubMed] [Google Scholar]
- 107.Mattson M.P., Shea T.B. Folate and homocysteine metabolism in neural plasticity and neurodegenerative disorders. Trends Neurosci. 2003;26:137–146. doi: 10.1016/S0166-2236(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 108.Kim Y.I. Folate and cancer prevention: a new medical application of folate beyond hyperhomocysteinemia and neural tube defects. Nutr. Rev. 1999;57:314–321. doi: 10.1111/j.1753-4887.1999.tb06905.x. [DOI] [PubMed] [Google Scholar]
- 109.Herrmann W., Quast S., Ullrich M., Schultze H., Bodis M., Geisel J. Hyperhomocysteinemia in high-aged subjects: relation of B-vitamins, folic acid, renal function and the methylenetetrahydrofolate reductase mutation. Atherosclerosis. 1999;144:91–101. doi: 10.1016/s0021-9150(99)00036-2. [DOI] [PubMed] [Google Scholar]
- 110.Glushchenko A.V., Jacobsen D.W. Molecular targeting of proteins by l-homocysteine: mechanistic implications for vascular disease. Antioxid. Redox Signal. 2007;9:1883–1898. doi: 10.1089/ars.2007.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ueland P.M., Refsum H., Beresford S.A., Vollset S.E. The controversy over homocysteine and cardiovascular risk. Am. J. Clin. Nutr. 2000;72:324–332. doi: 10.1093/ajcn/72.2.324. [DOI] [PubMed] [Google Scholar]
- 112.Jacobsen D.W. Hyperhomocysteinemia and oxidative stress: time for a reality check? Arterioscler. Thromb. Vasc. Biol. 2000;20:1182–1184. doi: 10.1161/01.atv.20.5.1182. [DOI] [PubMed] [Google Scholar]
- 113.Outinen P.A., Sood S.K., Liaw P.C., Sarge K.D., Maeda N., Hirsh J., Ribau J., Podor T.J., Weitz J.I., Austin R.C. Characterization of the stress-inducing effects of homocysteine. Biochem. J. 1998;332:213–221. doi: 10.1042/bj3320213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Loscalzo J. Homocysteine trials—clear outcomes for complex reasons. N. Engl. J. Med. 2006;354:1629–1632. doi: 10.1056/NEJMe068060. [DOI] [PubMed] [Google Scholar]
- 115.Isa Y., Mishima T., Tsuge H., Hayakawa T. Increase in S-adenosylhomocysteine content and its effect on the S-adenosylhomocysteine hydrolase activity under transient high plasma homocysteine levels in rats. J. Nutr. Sci. Vitaminol. 2006;52:479–482. doi: 10.3177/jnsv.52.479. [DOI] [PubMed] [Google Scholar]
- 116.Kerins D.M., Koury M.J., Capdevila A., Rana S., Wagner C. Plasma S-adenosylhomocysteine is a more sensitive indicator of cardiovascular disease than plasma homocysteine. Am. J. Clin. Nutr. 2001;74:723–729. doi: 10.1093/ajcn/74.6.723. [DOI] [PubMed] [Google Scholar]
- 117.Jamaluddin M.S., Yang X., Wang H. Hyperhomocysteinemia, DNA methylation and vascular disease. Clin. Chem. Lab. Med. 2007;45:1660–1666. doi: 10.1515/CCLM.2007.350. [DOI] [PubMed] [Google Scholar]
- 118.James S.J., Melnyk S., Pogribna M., Pogribny I.P., Caudill M.A. Elevation in S-adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. J. Nutr. 2002;132:2361S–2366S. doi: 10.1093/jn/132.8.2361S. [DOI] [PubMed] [Google Scholar]
- 119.Ingrosso D., Perna A.F. Epigenetics in hyperhomocysteinemic states. A special focus on uremia. Biochim. Biophys. Acta. 2009;1790:892–899. doi: 10.1016/j.bbagen.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 120.Liu C., Wang Q., Guo H., Xia M., Yuan Q., Hu Y., Zhu H., Hou M., Ma J., Tang Z., Ling W. Plasma S-adenosylhomocysteine is a better biomarker of atherosclerosis than homocysteine in apolipoprotein E-deficient mice fed high dietary methionine. J. Nutr. 2008;138:311–315. doi: 10.1093/jn/138.2.311. [DOI] [PubMed] [Google Scholar]
- 121.Green T.J., Skeaff C.M., McMahon J.A., Venn B.J., Williams S.M., Devlin A.M., Innis S.M. Homocysteine-lowering vitamins do not lower plasma S-adenosylhomocysteine in older people with elevated homocysteine concentrations. Br. J. Nutr. 2010;103:1629–1634. doi: 10.1017/S0007114509993552. [DOI] [PubMed] [Google Scholar]
- 122.Christopher S.A., Melnyk S., James S.J., Kruger W.D. S-adenosylhomocysteine, but not homocysteine, is toxic to yeast lacking cystathionine beta-synthase. Mol. Genet. Metab. 2002;75:335–343. doi: 10.1016/S1096-7192(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 123.De Clercq E. John Montgomery's legacy: carbocyclic adenosine analogues as SAH hydrolase inhibitors with broad-spectrum antiviral activity. Nucleos. Nucleot. Nucl. 2005;24:1395–1415. doi: 10.1080/15257770500265638. [DOI] [PubMed] [Google Scholar]
- 124.Huggins J., Zhang Z.X., Bray M. Antiviral drug therapy of filovirus infections: S-adenosylhomocysteine hydrolase inhibitors inhibit Ebola virus in vitro and in a lethal mouse model. J. Infect. Dis. 1999;179:S240–S247. doi: 10.1086/514316. [DOI] [PubMed] [Google Scholar]
- 125.Wolfe M.S., Borchardt R.T. S-adenosyl-l-homocysteine hydrolase as a target for antiviral chemotherapy. J. Med. Chem. 1991;34:1521–1530. doi: 10.1021/jm00109a001. [DOI] [PubMed] [Google Scholar]
- 126.Shuto S., Minakawa N., Niizuma S., Kim H.S., Wataya Y., Matsuda A. New neplanocin analogues. 12. Alternative synthesis and antimalarial effect of (6′R)-6′-C-methylneplanocin A, a potent AdoHcy hydrolase inhibitor. J. Med. Chem. 2002;45:748–751. doi: 10.1021/jm010374i. [DOI] [PubMed] [Google Scholar]
- 127.Wolos J.A., Frondorf K.A., Davis G.F., Jarvi E.T., McCarthy J.R., Bowlin T.L. Selective inhibition of T cell activation by an inhibitor of S-adenosyl-l-homocysteine hydrolase. J. Immunol. 1993;150:3264–3273. [PubMed] [Google Scholar]
- 128.Chiang P.K. Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacol. Ther. 1998;77:115–134. doi: 10.1016/s0163-7258(97)00089-2. [DOI] [PubMed] [Google Scholar]
- 129.Masuta C., Tanaka H., Uehara K., Kuwata S., Koiwai A., Noma M. Broad resistance to plant viruses in transgenic plants conferred by antisense inhibition of a host gene essential in S-adenosylmethionine-dependent transmethylation reactions. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6117–6121. doi: 10.1073/pnas.92.13.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zanetti G., Pahuja K.B., Studer S., Shim S., Schekman R. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 2011;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- 131.Nuttall J.M., Motley A., Hettema E.H. Peroxisome biogenesis: recent advances. Curr. Opin. Cell Biol. 2011;23:421–426. doi: 10.1016/j.ceb.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 132.Barros M.H., da Cunha F.M., Oliveira G.A., Tahara E.B., Kowaltowski A.J. Yeast as a model to study mitochondrial mechanisms in ageing. Mech. Ageing Dev. 2010;131:494–502. doi: 10.1016/j.mad.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 133.Alberghina L., Mavelli G., Drovandi G., Palumbo P., Pessina S., Tripodi F., Coccetti P., Vanoni M. Cell growth and cell cycle in Saccharomyces cerevisiae: basic regulatory design and protein–protein interaction network. Biotechnol. Adv. 2012;30:52–72. doi: 10.1016/j.biotechadv.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 134.Tehlivets O., Hasslacher M., Kohlwein S.D. S-adenosyl-l-homocysteine hydrolase in yeast: key enzyme of methylation metabolism and coordinated regulation with phospholipid synthesis. FEBS Lett. 2004;577:501–506. doi: 10.1016/j.febslet.2004.10.057. [DOI] [PubMed] [Google Scholar]
- 135.Kohlwein S.D. Obese and anorexic yeasts: experimental models to understand the metabolic syndrome and lipotoxicity. Biochim. Biophys. Acta. 2010;1801:222–229. doi: 10.1016/j.bbalip.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 136.Reitz R.C., Mead D.J., Bjur R.A., Greenhouse A.H., Welch W.H., Jr. Phosphatidylethanolamine N-methyltransferase in human red blood cell membrane preparations. Kinetic mechanism. J. Biol. Chem. 1989;264:8097–8106. [PubMed] [Google Scholar]
- 137.Reitz R.C., Mead D.J., Welch W.H., Jr. Phospholipid methylation in brain membrane preparations: kinetic mechanism. Biochim. Biophys. Acta. 1993;1166:139–144. doi: 10.1016/0005-2760(93)90089-r. [DOI] [PubMed] [Google Scholar]
- 138.Li Z., Agellon L.B., Allen T.M., Umeda M., Jewell L., Mason A., Vance D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 139.Noga A.A., Zhao Y., Vance D.E. An unexpected requirement for phosphatidylethanolamine N-methyltransferase in the secretion of very low density lipoproteins. J. Biol. Chem. 2002;277:42358–42365. doi: 10.1074/jbc.M204542200. [DOI] [PubMed] [Google Scholar]
- 140.Choumenkovitch S.F., Selhub J., Bagley P.J., Maeda N., Nadeau M.R., Smith D.E., Choi S.W. In the cystathionine beta-synthase knockout mouse, elevations in total plasma homocysteine increase tissue S-adenosylhomocysteine, but responses of S-adenosylmethionine and DNA methylation are tissue specific. J. Nutr. 2002;132:2157–2160. doi: 10.1093/jn/132.8.2157. [DOI] [PubMed] [Google Scholar]
- 141.Schwahn B.C., Chen Z., Laryea M.D., Wendel U., Lussier-Cacan S., Genest J., Jr., Mar M.H., Zeisel S.H., Castro C., Garrow T., Rozen R. Homocysteine–betaine interactions in a murine model of 5,10-methylenetetrahydrofolate reductase deficiency. FASEB J. 2003;17:512–514. doi: 10.1096/fj.02-0456fje. [DOI] [PubMed] [Google Scholar]
- 142.Werstuck G.H., Lentz S.R., Dayal S., Hossain G.S., Sood S.K., Shi Y.Y., Zhou J., Maeda N., Krisans S.K., Malinow M.R., Austin R.C. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J. Clin. Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hamelet J., Demuth K., Paul J.L., Delabar J.M., Janel N. Hyperhomocysteinemia due to cystathionine beta synthase deficiency induces dysregulation of genes involved in hepatic lipid homeostasis in mice. J. Hepatol. 2007;46:151–159. doi: 10.1016/j.jhep.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 144.Kammoun H.L., Chabanon H., Hainault I., Luquet S., Magnan C., Koike T., Ferre P., Foufelle F. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest. 2009;119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Borradaile N.M., Han X., Harp J.D., Gale S.E., Ory D.S., Schaffer J.E. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J. Lipid Res. 2006;47:2726–2737. doi: 10.1194/jlr.M600299-JLR200. [DOI] [PubMed] [Google Scholar]
- 146.Pineau L., Colas J., Dupont S., Beney L., Fleurat-Lessard P., Berjeaud J.M., Berges T., Ferreira T. Lipid-induced ER stress: synergistic effects of sterols and saturated fatty acids. Traffic. 2009;10:673–690. doi: 10.1111/j.1600-0854.2009.00903.x. [DOI] [PubMed] [Google Scholar]
- 147.Ariyama H., Kono N., Matsuda S., Inoue T., Arai H. Decrease in membrane phospholipid unsaturation induces unfolded protein response. J. Biol. Chem. 2010;285:22027–22035. doi: 10.1074/jbc.M110.126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Peter A., Weigert C., Staiger H., Machicao F., Schick F., Machann J., Stefan N., Thamer C., Haring H.U., Schleicher E. Individual stearoyl-coa desaturase 1 expression modulates endoplasmic reticulum stress and inflammation in human myotubes and is associated with skeletal muscle lipid storage and insulin sensitivity in vivo. Diabetes. 2009;58:1757–1765. doi: 10.2337/db09-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Green C.D., Olson L.K. Modulation of palmitate-induced endoplasmic reticulum stress and apoptosis in pancreatic {beta}-cells by stearoyl-CoA desaturase and Elovl6. Am. J. Physiol. 2011;300:E640–E649. doi: 10.1152/ajpendo.00544.2010. [DOI] [PubMed] [Google Scholar]
- 150.Busch A.K., Gurisik E., Cordery D.V., Sudlow M., Denyer G.S., Laybutt D.R., Hughes W.E., Biden T.J. Increased fatty acid desaturation and enhanced expression of stearoyl coenzyme A desaturase protects pancreatic beta-cells from lipoapoptosis. Diabetes. 2005;54:2917–2924. doi: 10.2337/diabetes.54.10.2917. [DOI] [PubMed] [Google Scholar]
- 151.Boumann H.A., Chin P.T., Heck A.J., De Kruijff B., De Kroon A.I. The yeast phospholipid N-methyltransferases catalyzing the synthesis of phosphatidylcholine preferentially convert di-C16:1 substrates both in vivo and in vitro. J. Biol. Chem. 2004;279:40314–40319. doi: 10.1074/jbc.M406517200. [DOI] [PubMed] [Google Scholar]
- 152.Tacconi M., Wurtman R.J. Phosphatidylcholine produced in rat synaptosomes by N-methylation is enriched in polyunsaturated fatty acids. Proc. Natl. Acad. Sci. U. S. A. 1985;82:4828–4831. doi: 10.1073/pnas.82.14.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.DeLong C.J., Shen Y.J., Thomas M.J., Cui Z. Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway. J. Biol. Chem. 1999;274:29683–29688. doi: 10.1074/jbc.274.42.29683. [DOI] [PubMed] [Google Scholar]
- 154.Jacobs R.L., Zhao Y., Koonen D.P., Sletten T., Su B., Lingrell S., Cao G., Peake D.A., Kuo M.S., Proctor S.D., Kennedy B.P., Dyck J.R., Vance D.E. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J. Biol. Chem. 2010;285:22403–22413. doi: 10.1074/jbc.M110.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Watkins S.M., Zhu X., Zeisel S.H. Phosphatidylethanolamine-N-methyltransferase activity and dietary choline regulate liver-plasma lipid flux and essential fatty acid metabolism in mice. J. Nutr. 2003;133:3386–3391. doi: 10.1093/jn/133.11.3386. [DOI] [PubMed] [Google Scholar]
- 156.Selley M.L. A metabolic link between S-adenosylhomocysteine and polyunsaturated fatty acid metabolism in Alzheimer's disease. Neurobiol. Aging. 2007;28:1834–1839. doi: 10.1016/j.neurobiolaging.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 157.Li D., Mann N.J., Sinclair A.J. A significant inverse relationship between concentrations of plasma homocysteine and phospholipid docosahexaenoic acid in healthy male subjects. Lipids. 2006;41:85–89. doi: 10.1007/s11745-006-5074-x. [DOI] [PubMed] [Google Scholar]
- 158.Innis S.M., Davidson A.G., Chen A., Dyer R., Melnyk S., James S.J. Increased plasma homocysteine and S-adenosylhomocysteine and decreased methionine is associated with altered phosphatidylcholine and phosphatidylethanolamine in cystic fibrosis. J. Pediatr. 2003;143:351–356. doi: 10.1067/S0022-3476(03)00326-3. [DOI] [PubMed] [Google Scholar]
- 159.Tehlivets O. Homocysteine as a risk factor for atherosclerosis: is its conversion to S-adenosyl-l-homocysteine the key to deregulated lipid metabolism? J. Lipid. 2011;2011 doi: 10.1155/2011/702853. (702853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]