Abstract
The fungal cell possesses an essential carbohydrate cell wall. The outer layer, mannan, is formed by mannoproteins carrying highly mannosylated O- and N-linked glycans. Yeast mannan biosynthesis is initiated by a Golgi-located complex (M-Pol I) of two GT-62 mannosyltransferases, Mnn9p and Van1p, that are conserved in fungal pathogens. Saccharomyces cerevisiae and Candida albicans mnn9 knockouts show an aberrant cell wall and increased antibiotic sensitivity, suggesting the enzyme is a potential drug target. Here, we present the structure of ScMnn9 in complex with GDP and Mn2+, defining the fold and catalytic machinery of the GT-62 family. Compared with distantly related GT-78/GT-15 enzymes, ScMnn9 carries an unusual extension. Using a novel enzyme assay and site-directed mutagenesis, we identify conserved amino acids essential for ScMnn9 ‘priming’ α-1,6-mannosyltransferase activity. Strikingly, both the presence of the ScMnn9 protein and its product, but not ScMnn9 catalytic activity, are required to activate subsequent ScVan1 processive α-1,6-mannosyltransferase activity in the M-Pol I complex. These results reveal the molecular basis of mannan synthesis and will aid development of inhibitors targeting this process.
Keywords: cell wall, glycobiology, glycosyltransferase, mannan, M-Pol I, protein crystallography
2. Introduction
N-linked glycosylation is an abundant post-translational protein modification on secreted proteins in both bacteria and eukaryotes [1]. The synthesis of eukaryotic N-linked glycans is initiated at the luminal side of the endoplasmic reticulum (ER). Here, the core glycan GlcNAc2Man9Glc3 is transferred onto the asparagine in the sequon N–X–S/T. This core glycan is then trimmed by mannosidases and glucosidases, a process conserved across the eukaryotic kingdom and used as protein-folding quality control [2,3]. The glycosylated proteins are then transported to the Golgi apparatus, where numerous glycosyltransferases (GTs) extend the core glycan to form a rich, cell-type-specific diversity of N-linked glycosylated proteins. While the ER-localized part of the pathway is well conserved across most eukaryotes, the type and activity of the Golgi GTs is cell-type-specific [4–7]. In addition, differential regulation and protein acceptor specificity of these GTs also lead to different types of glycosylation of different proteins in the same cell. The precise activities, substrate specificities and molecular mechanisms for most of these GTs remain largely unknown.
The synthesis of N-linked glycans and their decoration in the Golgi apparatus has been extensively studied in the yeast Saccharomyces cerevisiae. Like many other yeasts and fungi, S. cerevisiae possesses a core cell wall structure of chitin and glucan, further decorated by elaborate glycans in the outer cell wall [8,9]. Mannose and phosphomannose derivatives are the only sugars added to glycans in the yeast Golgi, a process performed by mannosyltransferases that use GDP-mannose (GDP-Man) as the donor substrate [10,11]. Upon entry into the cis-Golgi, core N-linked glycans receive an α-1,6-linked mannose by the transferase Och1p [12,13] (figure 1). This residue is extended with an α-1,2 and α-1,3-linked mannose in proteins targeted for retention in cellular organelles [14]. While the α-1,2-mannosyltransferase remains unknown, Mnn1p has been identified as the GT-71 mannosyltransferase that attaches the terminal α-1,3-linked mannose [15]. The second class of glycosylated proteins in S. cerevisiae is composed of mannoproteins. These proteins form mannan, the outermost layer of the fungal cell wall that can make up to 40% of the cell dry weight [13,16,17]. Mannoproteins have been shown to carry up to 200 mannose residues on each of their N-linked glycans [18]. The synthesis of mannoproteins starts by the extension of the α-1,6-linked mannose to form a backbone of at least 10 α-1,6-linked mannose residues [16] (figure 1). This reaction is carried out by the M-Pol I heterodimeric complex, composed of the mannosyltransferases ScMnn9p and ScVan1p [19]. This backbone is further extended with α-1,6-linked mannose residues by the heteropentameric mannosyltransferase complex M-Pol II [19–21] (figure 1). Additional mannosyltransferases add branches of α-1,2- or α-1,3-linked mannose and phosphomannose to this backbone [11,22]. Finally, mannoproteins leave the trans-Golgi in vesicles that fuse with the membrane and release their content to the extracellular space where the mannoproteins either associate with or are transglycosylated into β-glucan to form the outer layer of the fungal cell wall [8].
Figure 1.
Diagram of mannosylation pathways in the yeast Golgi. N-linked glycosylated proteins arrive from the ER and are extended with a single α-1,6-linked mannose by Och1p. This residue either can be further elongated to a primary α-1,6-linked mannose backbone by Mnn9p and Van1p (M-Pol I, top) to form the mannan backbone on mannoproteins or is further extended by an unknown mannosyltransferase with an α-1,2-linked mannose that eventually leads to a core glycosylation structure for proteins that are retained in internal compartments (bottom).
It remains unclear how the S. cerevisiae Golgi apparatus targets only certain N-linked glycans towards the synthesis of mannoproteins. Stolz & Munro [23] showed that an immunoprecipitated heterodimeric M-Pol I complex of active ScMnn9p and inactive ScVan1p extends the pseudo-acceptor α-1,6-mannobiose with a single α-1,2-linked mannose in vitro, whereas an immunoprecipitated heterodimeric M-Pol I complex of inactive ScMnn9p and active ScVan1p extends the same acceptor with several α-1,6-linked mannose residues. However, when a core glycosylated protein was used as the acceptor, a mannose backbone was only formed with an M-Pol I complex possessing active ScMnn9p and active ScVan1p. The authors suggested that an N-glycosylated substrate protein arrives at the complex and interacts with ScMnn9p, which in turn changes its mannosyltransferase activity according to the type of acceptor protein. If the protein is to remain in the internal compartments, then ScMnn9p would attach an α-1,2-linked mannose to the α-1,6-linked mannose to prevent it from further elongation to a mannose backbone, whereas mannoproteins targeted for secretion would receive an α-1,6-linked mannose from ScMnn9p that would be extended by ScVan1p to form the primary mannan backbone (figure 1). However, when S. cerevisiae Mnn9p and Van1p were coexpressed in Pichia pastoris, purified and used for an in vitro assay, the only observed reaction product was an α-1,6-linked mannose backbone attached to mannobiose [24]. It is possible that the immunoprecipitated samples used in the study by Stolz & Munro [23] were contaminated with an α-1,2-mannosyltransferase and that M-Pol I is solely active on mannoproteins. In vitro studies with the individually expressed components will help resolve this controversy, and aid in the dissection of the glycosyltransferase and activating functions of this pair of enzymes in the M-Pol I complex.
ScMnn9p and ScVan1p are type II membrane proteins, possessing a short cytosolic N-terminal domain followed by a transmembrane domain that is required for anchoring to the Golgi membrane, but not for interaction of the two enzymes [20]. The transmembrane domain is followed by a linker and the C-terminal globular catalytic domain. ScMnn9p and ScVan1p belong to the CAZy glycosyltransferase family GT-62. This family contains metal-dependent, retaining mannosyltransferases that are predicted to possess the GT-A fold [25] and use GDP-Man as their donor substrate. There is currently no structural information available for any GT-62 family member, limiting our understanding of the molecular details of the interaction between the M-Pol I enzymes and their acceptor proteins, as well as the catalytic mechanism.
Here, we report the structure of the mannosyltransferase domain of ScMnn9 in complex with manganese and GDP. The structure reveals that ScMnn9 possesses the GT-A fold and shows structural similarity to GT families 15 and 78. However, ScMnn9 possesses a unique extrusion that may act as a molecular ruler or multimerization domain. Using a novel fluorimetric coupled enzyme assay, we determine steady-state kinetics of wild-type ScMnn9 and identify conserved residues that are necessary for activity. Furthermore, we show that ScMnn9 and its product are necessary for processive M-Pol I mannosyltransferase activity in vitro. Using complementation studies in a yeast ΔMNN9 strain, we demonstrate that the presence of inactive full-length ScMnn9 partially rescues the ΔMNN9 phenotype. The results presented here show that both the presence and the priming activity of ScMnn9 are required for the formation of the α-1,6-mannose backbone of mannan proteins.
3. Results and discussion
3.1. ScMnn9 is structurally similar to GT-15 and GT-78 mannosyltransferases
Mnn9 proteins from yeasts and filamentous fungi possess high levels of sequence conservation (figure 2a). To study the molecular basis of α-1,6-mannosyl transfer activity of Mnn9, we determined the crystal structure of the enzyme from S. cerevisiae. An Escherichia coli expression construct (ScMnn9) was created covering the glycosyltransferase core (93–395) only (figure 2a). Expression of this construct, followed by Ni–IMAC, anion exchange and size-exclusion chromatography, yielded 5 mg of pure protein sample per litre of bacterial culture. The recombinant protein was used to grow crystals from ammonium sulfate solutions. The ScMnn9 structure was solved by a 2.2 Å single-wavelength anomalous dispersion (SAD) phasing experiment with a mercury derivative and refined against 2.0 Å synchrotron diffraction data of a mutant (D236N) in the presumed GT-A DxD catalytic motif, in complex with GDP and Mn2+, yielding a final R/Rfree of 0.19/0.24 (table 1). The structure reveals that ScMnn9, and by extension the entire GT-62 glycosyltransferase family, adopts the GT-A fold as previously proposed by sequence analysis [25] (figure 2b). Seven β-strands form a sheet that is covered on both sides by α-helices. Using the DALI server (http://ekhidna.biocenter.helsinki.fi/dali_server), the Rhodothermus marinus mannosylglycerate synthase from GT-76 (RmMGS, PDB: 2Y4M; figure 2b) [26] as well as the S. cerevisiae α-1,2-mannosyltransferase ScKre2p/Mnt1p from GT-15 (PDB: 1S4O; figure 2b) [27] were identified as structural homologues (r.m.s.d. = 2.4 Å on 158 equivalent Cα atoms or r.m.s.d. = 3.4 Å on 167 equivalent Cα atoms, respectively). RmMGS uses GDP-Man as the donor substrate to transfer mannose onto d-glycerate, d-lactate or glycolate [26]. ScKre2p/Mnt1p synthesizes O-linked oligomannose and the terminal oligomannose decorations on mannoproteins [22]. Superposition of ScMnn9, RmMGS and ScKre2p/Mnt1p reveals the structural similarity derived from the GT-A fold around the catalytic site (figure 2b). However, ScMnn9 has a unique hairpin loop formed by two additional β-strands (I262-N283; figure 2a,b). This loop is positioned in line with the active site and could serve a number of purposes. In order to address this question, we replaced this loop with a tetraglycine stretch. However, this construct did not produce soluble protein in E. coli, suggesting that the protein lacking the hairpin loop is misfolded and/or degraded. The loop could potentially act as a molecular ruler for the formation of a mannose backbone of defined length or act as a guide to recognize and correctly position protein N-linked glycans for mannosyl transfer. By contrast, RmMGS has a more extended C-terminus formed by six helices and a short β-strand, whereas ScKre2p/Mnt1p does not contain any protruding features. Interestingly, the interactions between GDP and the enzymes are very similar (figure 3). The guanine forms hydrogen bonds between the N1 amine and the amide oxygen of a glutamine (ScMnn9 and RmMGS) or an aspartic acid (ScKre2p/Mnt1p). In all three enzymes, the guanine is buried by residues with relatively long side chains (i.e. Q124 in ScMnn9, K9 in RmMGS and R130 in ScKre2p/Mnt1p; figure 3). The ribofuranose forms extensive hydrogen bonds with side-chain residues forming the active site. The metal is coordinated by a histidine side chain, a carboxylate and the pyrophosphate moiety of GDP. The histidine, common to many retaining GT-A GTs, occupies a similar position within the active site of all three transferases (figure 3) and is one of the very few (five) conserved side chains in the sequences of the GT-15, GT-62 and GT-76 GTs compared here (figure 3). The carboxylate metal ligand is part of the canonical GT-A fold DxD motif (figures 2a and 3). Both the α- and β-phosphates of the GDP interact with the metal, similar to the RmMGS enzyme (figure 3).
Figure 2.
Structure and conservation of Mnn9. (a) Protein sequence alignment of Mnn9 proteins of fungal species. Red barrels indicate α-helices, blue arrows indicate β-strands. Black shading indicates completely conserved residues, grey shading indicates residues that are conserved between two of the three species. Purple shading indicates residues that form the active site of ScMnn9. The cyan box indicates the stretch that forms the extension in ScMnn9. Residue numbers are shown for ScMnn9. (b) (i) Stereoscopic cartoon/surface representation of the structures of ScMnn9 in complex with GDP and Mn2+, RmMGS in complex with GDP-Man and Mg2+, and ScKre2p/Mnt1p in complex with GDP and Mn2+. Structural similarities between all three transferases are shown in red. Unique features are shown in cyan (hairpin loop of ScMnn9) and purple (dimerization domain of RmMGS). GDP and GDP-Man are shown as sticks and metals are shown as brown spheres. (ii) Topology of ScMnn9, RmMGS and ScKre2p/Mnt1p with the same colour codes as in the cartoon representations. Disordered regions of the proteins are shown as dashed lines.
Table 1.
Details of data collection and structure refinement. Values between parentheses correspond to the highest resolution shell.
| ScMnn9 + Hg-SAD | ScMnn9-D236N + Mn2+ + GDP | |
|---|---|---|
| space group | P6522 | P6522 |
| cell dimensions | ||
| a, b, c (Å) | 57.05, 57.05, 330.17 | 57.09, 57.09, 330.91 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| resolution range (Å) | 25.0–2.2 | 55.0–2.0 |
| number of observed reflections | 1 342 417 | 366 234 |
| number of unique reflections | 19 947 | 23 775 |
| redundancy | 37.8 (33.4) | 9.7 (10.1) |
| I/σ(I) | 37.0 (13.6) | 11.8 (5.0) |
| completeness (%) | 99.9 (100.0) | 100.0 (100.0) |
| Rmerge | 0.148 (0.579) | 0.149 (0.482) |
| number of protein residues | 288 | 288 |
| number of water molecules | — | 148 |
| Rwork, Rfree | — | 0.19, 0.24 |
| r.m.s.d. from ideal geometry | ||
| bond lengths (Å) | — | 0.012 |
| bond angles (°) | — | 1.43 |
| B-factors (Å2) | ||
| protein | — | 17.9 |
| ligand | — | GDP: 34.7 Mn2+: 45.2 |
| water | — | 19.7 |
Figure 3.
Active site of ScMnn9, RmMGS and ScKre2p/Mnt1p. A close-up of the active sites of ScMnn9 and structurally related enzymes shown in figure 2b. ScMnn9 residues within 6 Å of GDP are shown as sticks with purple carbon atoms. The unbiased |FO|-|FC| map (1.75σ) is shown as cyan mesh around GDP and Mn2+ in ScMnn9. Mn2+ and Mg2+ are shown as brown spheres. Hydrogen bonds are shown as dashed black lines.
Despite the fact that ScMnn9-D236N was soaked or co-crystallized with GDP-Man, electron density for mannose was not observed. The mannose of GDP-Man in the RmMGS complex forms hydrogen bonds with K76 and D192, and several backbone amides (figure 3). A mannose may be similarly coordinated in the ScMnn9 through the structurally equivalent R209 and E364/T365, respectively. Despite the striking similarities between ScMnn9 and ScKre2p/Mnt1p, the reactions catalysed of both GTs result in a different glycosidic bond. Owing to the lack of a binary GDP-Man complex structure of both GTs, we can speculate that only the positions of the side chains in the pocket around residues R209, R210, D280, E305 and T365 result in the formation of an α-1,6-glycosidic bond.
3.2. Recombinant ScMnn9 possesses mannosyltransferase activity in vitro
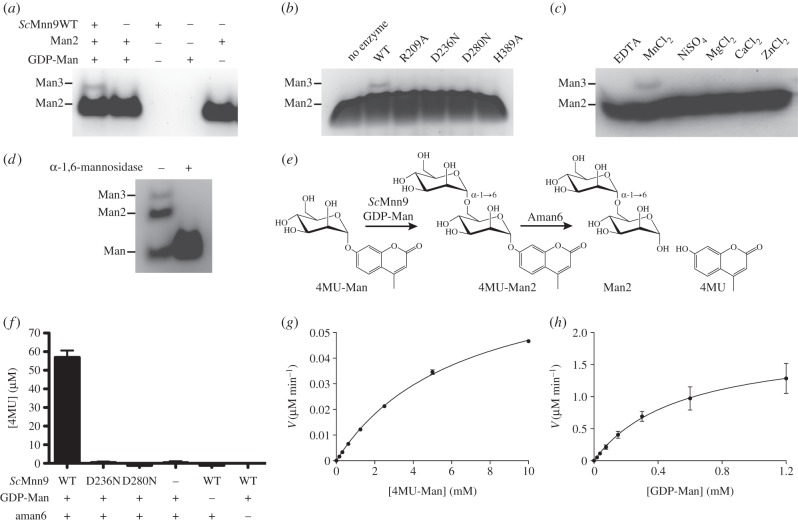
To date, enzyme activity of ScMnn9p has been demonstrated only in the presence of ScVan1p [23,24]. We tested the activity of bacterially expressed ScMnn9 in the presence of manganese, GDP-Man, and the model acceptor substrates mannose and α-1,6-linked mannobiose. Reaction products were separated and observed by fluorophore-assisted carbohydrate gel electrophoresis (FACE; figure 4a,d), showing that ScMnn9 alone is able to transfer mannose from the sugar donor onto the acceptor substrates, forming mannotriose. We then investigated the role of conserved amino acid side chains in the active site (figures 2 and 4b). R209 lines the putative mannose binding site, and mutation to alanine results in the loss of ScMnn9 activity (figure 4b). Similar effects were reported for the equivalent K76A mutation in RmMGS [26]. ScMnn9 D236 is the first aspartic acid in the canonical GT-A DxD catalytic motif, and mutation to the isosteric asparagine results in the loss of activity (figure 4b), similar to the previously reported less conservative D236A mutation [23]. The equivalent RmMGS Asp100 forms a hydrogen bond with the mannose O3 hydroxyl (figure 2b). Although we were unable to obtain a complex of ScMnn9 in complex with an intact donor, inspection of the superimposed RmMGS complex suggests that ScMnn9 Asp280 would also line the putative mannose binding site, positioned close to the O2 hydroxyl group. Mutation of this aspartic acid to asparagine (D280N) results in an inactive enzyme (figure 4b). ScMnn9 His389 coordinates the manganese and is indispensable for catalytic activity (figure 4b). Manganese is required for activity, and cannot be substituted by other divalent cations, such as magnesium or calcium (figure 4c). Interestingly, ScMnn9 was able to extend mannose to α-1,6-mannobiose and -mannotriose, as shown by treatment with a commercially available α-1,6-specific mannosidase (figure 4d).
Figure 4.
Activity assay of ScMnn9 wild-type and mutants. (a) FACE gel of the reaction products of ScMnn9 wild-type incubated with GDP-Man, α-1,6-mannobiose (Man2) and MnCl2 or controls. (b) Same as (a), but incubated with wild-type or mutants of ScMnn9. (c) Same as (a), but incubated with 10 mM of MnCl2, other divalent cations or EDTA. (d) FACE gel of a reaction containing ScMnn9, mannose, GDP-Man and MnCl2. The reaction products were either treated or not treated with an α-1,6-mannosidase. (e) Diagram of the chemical reaction that is the foundation for the coupled enzyme assay to determine steady-state kinetics for ScMnn9. 4MU-Man is extended by ScMnn9 to 4MU-Man2, which in turn acts as a minimal substrate for B. circulans Aman6, an α-1,6-mannosidase. The release of fluorescent 4MU was measured at an excitation wavelength of 360 nm and an emission wavelength of 460 nm. (f) Bar chart of the measured 4MU released after the reaction of ScMnn9 wild-type, D236N and D280N in presence and absence of GDP-Man and Aman6 mannosidase. Error bars indicate the standard error of the mean (s.e.m.), n = 3. (g,h) Steady-state kinetics of ScMnn9 (g) in the presence of GDP-Man (1.2 mM) and variable 4MU-Man concentrations or (h) in the presence of 4MU-Man (10 mM) and variable GDP-Man concentrations. Error bars indicate s.e.m., n = 3.
To study ScMnn9 steady-state kinetics, we developed a novel coupled enzyme assay that involves only one additional enzyme, in contrast to the established glycosyltransferase assays where the release of GDP is measured by NADH oxidation through pyruvate kinase and lactate dehydrogenase [28]. Here, we used the gene product of Bacillus subtilis TN-31 aman6 (Aman6), an α-1,6-mannosidase [29,30], as a coupling enzyme (figure 4e). This mannosidase has been reported to act on mannotriose as a minimal substrate [29]. We discovered that this enzyme also liberates 4-methylumbelliferone (4-MU) from 4MU-α-d-Man-(1 → 6)-d-Man (4MU-Man2), but crucially not from 4MU-Man. Thus, only in the presence of active ScMnn9, which transfers a mannose onto 4MU-Man, does the Aman6 mannosidase liberate fluorescent 4MU from the resultant 4MU-Man2 product (figure 4f).
This assay was used to establish steady-state kinetics for wild-type ScMnn9 (figure 4g,h). The KM,app and Vmax determined for the 4MU-Man acceptor are 6.5 (±0.3) mM and 77.7 (±2) nM min−1, respectively, resulting in a kcat of 0.2 min−1. The KM,app and Vmax for GDP-Man are 0.5 (± 0.2) mM and 1.9 (±0.3) µM min−1, respectively, resulting in a kcat of 3.8 min−1. Compared with the Km and Vmax values measured for RmMGS using glycerate as an acceptor, ScMnn9 seems to have low affinity for both of its substrates [26,31]. Interestingly, the kcat for ScMnn9 and GDP-Man is comparable to kcat of RmMGS and GDP-Man (1.9 min−1 [26]). By contrast, ScKre2p/Mnt1p is considerably faster than ScMnn9, with kcat of 12.8 s−1 for GDP-Man and 10.8 s−1 for methyl-α-mannoside, whereas with a Km of 26 mM, the acceptor analogue methyl-α-mannoside appears to be a poor substrate for ScKre2p/Mnt1p [27]. The low substrate affinity of ScMnn9 and ScKre2p/Mnt1p in vitro might be a result of the artificial acceptor substrates used. ScKrep2/Mnt1p is not only involved in O-linked mannosylation of serines and threonines but also attaches α-1,2-linked mannose to the α-1,6-mannose backbone formed by M-Pol I and II [32–34]. Hence, it recognizes rather complex substrates. The substrate of ScMnn9 is an N-linked core glycan extended with a mannose attached by Och1p [19]—structurally rather dissimilar from the 4MU-Man pseudo-acceptor used here. Furthermore, ScMnn9p is found in the multimeric glycosyltransferase complexes M-Pol I and II [19,21], and intermolecular interactions in these complexes may well increase the affinity of ScMnn9p for its substrates. It is also possible that ScMnn9 has a comparatively low affinity for its substrates in order to limit its consumption of cellular GDP-Man, which may be particularly important as M-Pol I activity is the starting point of extensive additional mannosylation [21], requiring large amounts of additional GDP-Man.
3.3. ScMnn9 is required as a priming enzyme and an allosteric activator for ScVan1 polymerase activity
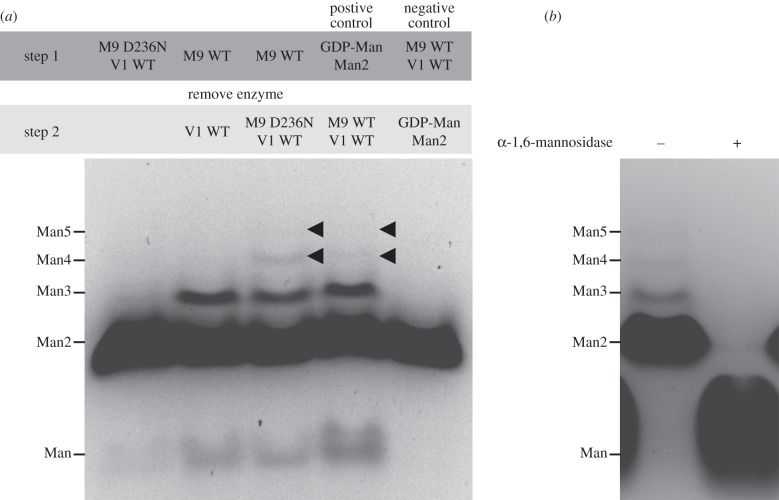
The formation of oligomannose by M-Pol I has been demonstrated in vitro [19,23,24]. However, it remains unclear how Mnn9p and Van1p act synergistically. In addition to recombinant ScMnn9, we were also able to separately express and purify the ScVan1 glycosyltransferase domain (residues 87–535) using similar procedures as described for ScMnn9 earlier. In order to ensure that the truncations of ScMnn9 and ScVan1 are still able to form a complex, we measured the interaction between the two proteins using biolayer interferometry, yielding a Kd of 480 nM (kon 6.2 × 102 M−1 s−1; koff 3.2 × 10−4 s−1). We then tested whether ScMnn9 is required as a ‘priming’ α-1,6-mannosyltransferase and/or essential for extension of the α-1,6 mannan backbone with a polymerase-like activity. Recombinant ScVan1 and GDP-Man were mixed either with the reaction product of ScMnn9 (α-1,6-mannotriose) alone or with the reaction product together with the catalytically inactive ScMnn9-D236N mutant. The reaction products were labelled with 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) and observed on a FACE gel (figure 5a). The resultant oligomers were of α-1,6-linked mannose, as demonstrated by susceptibility to α-1,6-mannosidase treatment (figure 5b). Strikingly, ScMnn9-priming mannosyltransferase activity is required to allow ScVan1 polymerase activity. However, it is the ScMnn9 protein, but not its catalytic activity, that is necessary and sufficient for this ScVan1 polymerase activity, suggesting the involvement of an allosteric mechanism in the M-Pol I complex (figure 5a). This mechanism has been proposed previously using complexes immunoprecipitated from S. cerevisiae where either ScMnn9p or ScVan1p were inactive [23]. However, the same report showed that ScVan1p is able to form oligomannose without the product of ScMnn9p, a result we (and others [24]) were not able to reproduce with recombinant ScMnn9. Our data suggest that in the context of the M-Pol I complex, ScMnn9 acts as both a priming glycosyltransferase and an allosteric activator, whereas ScVan1 is the polymerase synthesizing the α-1,6 mannose backbone on mannoproteins (figure 1). The formation of free mannose in lanes 2 and 3 of figure 5a is likely to be the result of GDP-Man hydrolysis (i.e. glycosyltransfer onto a water molecule acceptor).
Figure 5.
Synergy of ScMnn9 and ScVan1. (a) FACE gel of ANTS-labelled reaction products of the reaction of ScMnn9 (M9), ScVan1 (V1), GDP-Man, α-1,6-mannobiose (Man2) and MnCl2. First lane: ScMnn9-D236N and ScVan1 wild-type were incubated together, the reaction was stopped and the products were labelled before the separation on a FACE gel. The faint band above Man2 is assumed to be leakage from lane 2 (noted on other gels, not shown). Second and third lanes: ScMnn9 WT was used in absence of ScVan1; ScMnn9 WT was then removed using a 10 000 MWCO filter, and ScVan1 WT alone (second lane) or ScMnn9-D236N and ScVan1 WT (third lane) were added to the reaction. The second step of the reaction was stopped and the products were labelled before being separated on a FACE gel. Fourth lane: control in which the substrates were incubated in the absence of enzymes, spun through the filter and subsequently the reaction was carried out by addition of the enzymes. The products formed due to the presence of substrates and GTs. Fifth lane: control in which the enzymes were incubated in the absence of the substrates, removed by the filter and the filtrate was enriched with the substrates. Products did not form due to the absence of GTs. (b) ScMnn9, ScVan1, GDP-Man, α-1,6-mannobiose and MnCl2 were incubated, and the reaction products were either treated (+) or not treated (–) with the B. circulans Aman6 α-1,6-mannosidase. The products after the reaction were labelled with ANTS and run on a FACE gel.
3.4. ScMnn9p catalytic activity is indispensable for mannoprotein synthesis in yeast
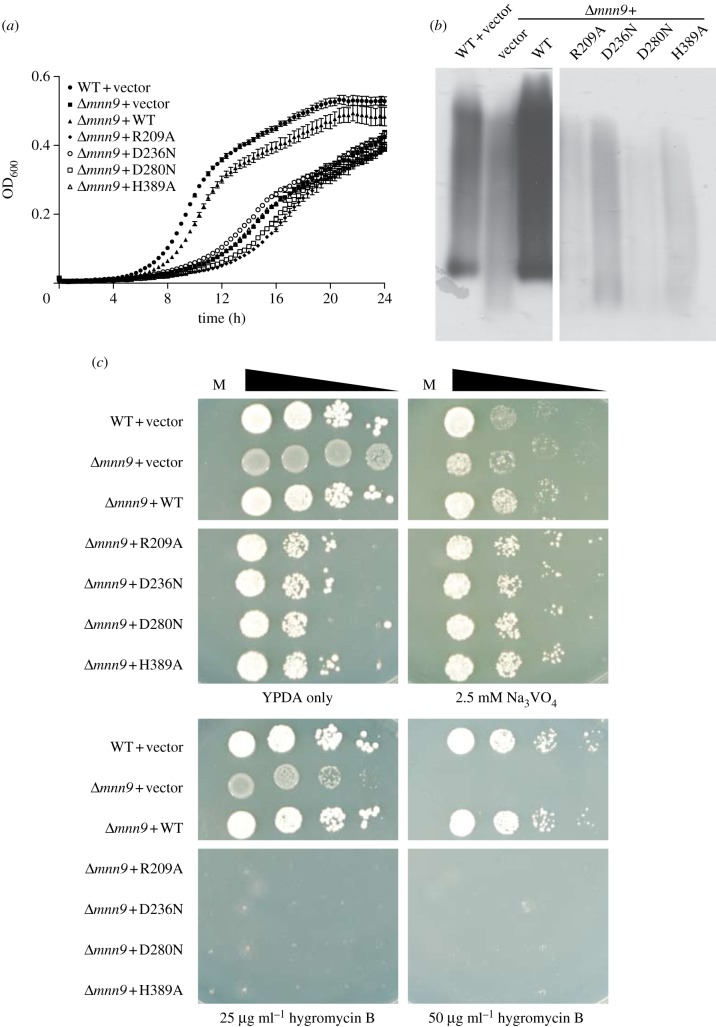
Strains of Saccharomyces cerevisiae and Candida albicans with defects in mannan synthesis show hypersensitivity to hygromycin B and reduced sensitivity to sodium orthovanadate [35,36]. Guided by the crystal structure, we have identified catalytically inactive mutants of ScMnn9 (figure 4b) that can now be used to dissect the function of the protein with the help of a ΔMNN9 strain of S. cerevisiae. Wild-type and point mutants R209A, D236N, D280N and H389A of the MNN9 gene, including 5′- and 3′-UTR, were cloned into the yeast expression plasmid pRS315 [37]. Saccharomyces cerevisiae BY4741 wild-type and ΔMNN9 cells were transformed with these plasmids. Successfully transformed cells were selected on DO-Leu(–) plates. ΔMNN9 cells transformed with wild-type MNN9 grew after a longer lag phase at a similar rate, but to slightly lower density, in YPD compared with wild-type cells carrying the empty pRS315 vector control (figure 6a). Similar observations were made in complementation experiments of the C. albicans ΔMNN9 mutant [38]. Cells lacking MNN9 or carrying MNN9 with a point mutation grew notably slower and did not reach stationary phase after 24 h, in contrast to wild-type and reconstituted MNN9 (figure 6a). Thus, cells expressing catalytically inactive ScMnn9p show growth kinetics comparable with those measured for cells lacking MNN9. Furthermore, wild-type cells and ΔMNN9 cells carrying the wild-type MNN9 showed similar glycosylation patterns of the extracellular invertase, whereas the ΔMNN9 and cells containing the MNN9 mutants showed very low glycosylation (figure 6b; electronic supplementary material, figure S1). This indicates that the point mutations in MNN9 lead to the same effect as the deletion of MNN9 in YPD. On YPD plates, the ΔMNN9 phenotype is presented as an increased sensitivity to hygromycin B and reduced susceptibility to Na3VO4 (figure 6b). The susceptibility to Na3VO4 was not decreased in cells carrying the point mutations in MNN9 compared with WT cells. (figure 6b). However, cells lacking MNN9 or carrying the point mutants were susceptible to lower concentrations of hygromycin B than wild-type or reconstituted cells (figure 6b). Notably, cells expressing ScMnn9p with a point mutation did not grow at the lowest concentration of hygromycin B, whereas MNN9 knockout cells were still able to grow, suggesting that the complete loss of MNN9 may activate rescue pathways for cell survival, whereas these pathways are not being activated in the presence of inactive ScMnn9p. In contrast to the results obtained in the invertase assay (figure 6b), this result indicates that the presence of an inactive form of ScMnn9p has a more severe impact on cell wall architecture than the complete absence of the transferase. The difference between the two results could be explained by the additional pressure through the presence of hygromycin B that may activate additional rescue pathways. The results gathered from the in vivo experiments are similar to previous reports, although these only covered either complete knockouts or a single point mutant [23,38].
Figure 6.
Phenotypic analysis of native and complemented Saccharomyces cerevisiae ΔMNN9 strains. (a) Growth curve of S. cerevisiae BY4741 wild-type or ΔMNN9 transformed with either pRS315 vector control (vector) or different mutations of the MNN9 gene. Growth of S. cerevisiae was followed by measurement of absorption at 600 nm. Error bars indicate s.e.m., n = 3. (b) 2,3,5-triphenyltetrazolium-labelled PAGE gel to determine the state of glycosylation of extracellular invertase of the S. cerevisiae transformants indicated. A Coomassie brilliant blue-stained PAGE gel as loading control can be found in the electronic supplementary material, figure S1. (c) Saccharomyces cerevisiae BY4741 wild-type or ΔMNN9 transformed with the same constructs as used in panel (a) grown on YPD plates either without any supplement or with 2.5 mM Na3VO4, or 25 and 50 µg ml−1 hygromycin B. Cells were spotted in a serial dilution on plates, M indicates the medium-only control.
4. Concluding remarks
The ScMnn9 glycosyltransferase is essential for the formation of mannosylated proteins that form mannan in the yeast cell wall. The results presented here define the structure of this key enzyme, the first example of a structure of the GT-62 family of GTs. ScMnn9 adopts a GT-A fold with an unusual hairpin loop extension. Residues that form the active site and are conserved across fungal species are important for activity. Furthermore, we show that ScVan1 is shifted into an active conformation only upon binding of ScMnn9 and requires the priming product of ScMnn9 for GT polymerase activity. However, it has to be noted that this report uses a non-physiological substrate and that the activities of ScMnn9 and ScVan1 may well be different with native acceptors (i.e. N-glycosylated proteins). Nevertheless, these results indicate the importance of ScMnn9p for the formation of the mannan backbone. Further work is required to capture ScMnn9 in complex with ScVan1 or in complex with a physiologically relevant substrate such as an N-linked glycosylated peptide or protein. Previous reports have suggested that knockouts of MNN9 in C. albicans show phenotypes that are inconsistent with virulence, making ScMnn9p a potential drug target [38]. Future studies will exploit the data presented here to identify potent inhibitors that may serve as lead compounds for new therapeutics.
5. Material and methods
5.1. Cloning and protein expression
The genes encoding ScMnn9p and ScVan1p lacking the first 92 amino acid residues (ScMnn9) or the first 86 amino acid residues (ScVan1), excluding the N-terminal cytoplasmic domain, the transmembrane domain and the predicted disordered region, were cloned from genomic DNA of S. cerevisiae S288c into pNIFTY/maltose-binding protein (MBP), introducing a MBP, a hexahistidine stretch and a tobacco etch virus (TEV) protease recognition sequence at the N-terminus using the oligonucleotides 5′-ATGGCGCCGAAGGTCATATTGCACATTATGATTTGAACAAATTGC-3′ (forward) and 5′-ATGGATCCATCAATGGTTCTCTTCCTCTATGTGATAAACC-3′ (reverse) for ScMnn9, and 5′-GCGGCGCCATGGGCATTGGTGTATCCACGC-3′(forward) and 5′-ATGGATCCATTACTCTGATTGTCTTCTCTTCTCTCTTTCCC (reverse) for ScVan1 (restriction sites KasI and BamHI, respectively, shown in bold). A plasmid containing ScMnn9 wild-type was used to introduce different point mutations by site-directed mutagenesis. To do so, the following oligonucleotides were used: R209, 5′-GCTTTAGATGTTCAAAAGGAAGCTCGTGCAGC AATGGCTTTGGCG-3′ and 5′-CGCCAAAGCCATTGCTGCACGAGCTTCCTTTTGAACATCTAAAGC-3′; D236N, 5′-GGTGCTGTGGCTAAATGCCGATATTATAGAGACACC-3′ and 5′-GGTGTCTCTATAATATCGGCATTTAGCCACAGCACC-3′; D280N, 5′-ATCAGACCATACAATTTCAACAACTGG-3′ and 5′-CCAGTTGTTGAAATTGTATGGTCTGAT-3′; and H389A, 5′-GGCTTACCAAACTATTTGGTTTATGCTATAGAGGAAGAGAACCATTGATGGATCC-3′ and 5′-GGATCCATCAATGGTTCTCTTCCTCTATAGCATAAACCAAATAGTTTGGTAAGCC-3′.
Escherichia coli BL21(DE3) pLysS cells were transformed with pNIFTY/MBP containing the gene of interest. A fresh overnight culture of E. coli BL21 transformants was diluted 1 : 50 in autoinduction medium [39] containing 100 µg ml−1 carbenicillin and grown at 18°C for 24 h. Cells were harvested by centrifugation at 3300g for 30 min at 4°C, and the pellet was resuspended in lysis buffer (25 mM Tris–HCl, pH 7.5, 250 mM NaCl, 30 mM imidazole) and kept frozen at −80°C until lysis.
5.2. Cell lysis and protein purification
Resuspended cells were supplemented with lysozyme and DNase I (Sigma-Aldrich), and lysed on a constant cell disruptor system at 30 kpsi (Avestin). The lysate was spun down for 30 min at 31 000g and 4°C. The soluble fraction was passed through a 0.2 μm filter and bound to Ni2+-charged IMAC resin (GE Healthcare) by gravity flow. Unspecific proteins were washed off by applying 10 column volumes (CVs) of lysis buffer. The protein of interest was eluted with 3 CV lysis buffer supplemented with 200 mM imidazole. The eluate was dialysed against buffer A (25 mM Tris–HCl, pH 7.5) for 2 h at room temperature (RT). The tag was cleaved off by adding 500 μg TEV protease and incubation for 16 h at 4°C. Cleaved protein was injected onto a 5 ml HiTRAP Q FF (GE Healthcare) equilibrated in buffer A. The MBP tag was removed by washing with 2 CV buffer A containing 150 mM NaCl. ScMnn9 or ScVan1 was eluted using 3 CV buffer A containing 400 mM NaCl. Fractions containing protein of interest were pooled and dialysed against buffer B (25 mM Tris–HCl, pH 7.5, 150 mM NaCl and 2 mM MnCl2) for 2 h at RT. The sample was concentrated to 1 ml and injected onto a Superdex 75 size-exclusion column equilibrated in buffer B. Fractions containing ScMnn9 or ScVan1 were pooled, concentrated to 5 mg ml−1, flash frozen in liquid nitrogen and stored at −80°C.
5.3. Protein crystallization, data collection and refinement
Octahedral ScMnn9 crystals were grown by vapour diffusion in 1 µl sitting drops containing 0.5 µl protein and 0.5 µl mother liquor (0.1 M HEPES, pH 7.5 and 2 M ammonium sulfate). Crystals were transferred to 50% Na–malonate, pH 7.5, containing mersalyl acid and soaked for 16 h at 20°C. Soaked crystals were directly frozen in liquid nitrogen, because the Na–malonate acted as cryoprotectant [40]. A 38-fold redundant 2.2 Å dataset collected at beamline ID14-4 at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) was used for SAD. Initial phases were calculated from a single Hg site using the SHELX program suite [41]. Solvent flattening was also performed with SHELX, which gave a good-quality map showing protein/solvent boundaries and some secondary structure elements. The map was used as input for warpNtrace [42], which built 200 of 305 residues with one molecule in the asymmetric unit. Model building and refinement was continued in COOT [43] and REFMAC [44], yielding the final model with statistics shown in table 1.
Crystals of ScMnn9-D236N were transferred to 50% Na–malonate, pH 7.5 and soaked with 3 mM GDP and 10 mM MnCl2 for 10 min at 20°C and then flash frozen in liquid nitrogen. Diffraction data were collected at beamline ID14-4 at the ESRF to 2.0 Å. Refinement was initiated from the native structure. This revealed well-defined |FO| − |FC| electron density for GDP and Mn2+. Model building and refinement was continued as described above and resulted in the statistics of the final model as shown in table 1.
5.4. Coupled fluorimetric ScMnn9 enzyme assay
To measure enzyme kinetics 500 nM ScMnn9 was incubated in 20 mM HEPES, pH 7.5, with 10 mM MnCl2, 0.2 mg ml−1 BSA and 10 µM of the α-1,6-specific mannosidase Aman6 [29]. Initial rates of mannotriose formation were measured with substrate concentrations ranging from 0 to 1.2 mM GDP-Man and 10 mM 4-methylumbelliferyl-α-d-mannopyranoside. Initial rates of mannosyltransfer were measured with concentrations ranging from 0 to 10 mM 4MU-Man and 1.2 mM GDP-Man. Product formation was determined fluorimetrically at λex = 360 nm and λem = 460 nm.
5.5. Binding kinetics of ScMnn9 and ScVan1
The Kd of ScMnn9 and ScVan1 was determined on an Octet RED384 (Fortebio). ScMnn9 was biotinylated with EZ-link NHS-PEG4-Biotin (Thermo Scientific) according to the manufacturer's instructions, and the activity of ScMnn9 was confirmed by FACE. ScMnn9 was bound at 25 µg ml−1 to streptavidin sensors in buffer C (100 mM HEPES, pH 7.5, 5 mM MnCl2, 1 mg ml−1 BSA, 0.015% BRIJ 35). The loaded sensors were incubated with ScVan1 at concentrations ranging from 0 to 15 µM in buffer C. Association was measured over 10 min and dissociation in buffer C was measured over 20 min. The measurements were performed in triplicate and full kinetic fit was used to determine kinetics.
5.6. In vitro mannosyltransferase assay
Mannosyltransferase assays contained 500 nM ScMnn9 and/or 500 nM ScVan1, 20 mM HEPES, pH 7.5, 10 mM MnCl2, 10 mM α-1,6-d-mannobiose and 1.2 mM GDP-Man. The reaction was incubated for 16 h at 30°C and stopped by adding three volumes of ice-cold ethanol. The reaction products were labelled with 750 nmol ANTS and used in a FACE, as described elsewhere [45]. For the reaction containing mannose as acceptor substrate, reactions were as described above, except for 1 mM mannose and 20 mM GDP-Man as acceptor and donor, respectively. The products were digested with α-1,6-mannosidase (NEB) according to the manufacturer's instructions.
5.7. Transformation of Saccharomyces cerevisiae with MNN9 and in vivo assays
Saccharomyces cerevisiae BY4741 wild-type (MAT a; his3Δ 1; leu2Δ 0; met15Δ 0; ura3Δ 0) and ΔMNN9 (Mat a; his3D1; leu2D0; met15D0; ura3D0; YPL050c::kanMX4) were obtained from EUROSCARF. The yeast expression plasmid pRS315 was a kind gift of Michael Stark (University of Dundee). The gene of MNN9 including the 5′- and 3′-untranslated regions, according to the yeast promoter atlas, was cloned into pRS315 using the oligonucleotides 5′-AAAGGATCCATCACAGAACCGGAAAATAGTAGCCAC-3′ and 5′-TTTGAGCTCCTCAAGCTCAGAAATTAGTTGTTGTAGC-3′ (restriction sites for BamHI and SacI shown in bold, respectively). A single colony of S. cerevisiae was used to inoculate 20 ml YPD (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose) medium and the cells were grown for 16 h at 30°C and 220 r.p.m. The culture was spun down for 5 min at 1000g, and the pellet was washed twice in 5 ml sterile ddH2O. For transformation, 100 µl of resuspended cells were mixed with 2 µg pRS315 [37] containing the gene of interest and 300 µl of transformation buffer (40% PEG 3350, 120 mM lithium acetate, 0.8 mg ml−1 single-stranded salmon sperm DNA). The transformation mixture was incubated at 42°C for 45 min. Cells were spun down for 5 min at 6000g, the supernatant was aspirated and the pellet was resuspended in 100 µl sterile ddH2O. The transformed cells were spread on DO-agar plates (0.7% (w/v) yeast nitrogen base, 2% (w/v) glucose, 0.07% (w/v) adenine, complete supplement mixture DCS0099 (Formedium), 2% (w/v) agar) lacking leucine and incubated for 2 days at 30°C. Single colonies of transformed cells were used to inoculate 1 ml YPD medium, and the culture was left to grow for 8 h at 30°C. Growth curves were determined measuring the absorption at 600 nm, starting by diluting cells to OD600 = 0.05 in YPD followed by growth for 12 h at 30°C under agitation. Mannosylation defects were determined by diluting S. cerevisiae to OD600 = 0.2, making 1 : 10 serial dilutions and spotting 2 µl of cells onto YPD plates containing either no supplement, sodium orthovanadate (2.5 mM, Sigma-Aldrich) or hygromycin B (25 and 50 µg ml−1, Sigma-Aldrich). Cells were grown for 2 days at 30°C.
5.8. Invertase assay
Detection of mannosylation defects in S. cerevisiae transformants was carried out as described by Ballou [46]. Briefly, S. cerevisiae cells were grown to mid-log phase (OD600 ∼ 0.35) in YPD at 30°C and 220 r.p.m. The medium was changed to YPD containing only 0.05% glucose, and cells were grown for 3 h at RT. Cell pellets were washed with 20 mM sodium azide and resuspended in TBS + 1 mM PMSF. Cells were lysed with 0.45 µm glass beads four times by vortexing vigorously for 30 s, keeping the cells on ice for 2 min between each lysis step. The samples were run over an 8% polyacrylamide gel for 2.5 h at 20 mA at RT. The gel was incubated for 10 min in an ice-cold 0.1 M sucrose solution in 0.1 M sodium acetate, followed by incubation in the same solution at 37°C. The gel was rinsed twice with water, and a solution of 2,3,5-triphenyltetrazolium in 0.5 M NaOH was added and heated until staining was sufficient. The same samples were run again, and the gel was stained in Coomassie brilliant blue for loading control.
Supplementary Material
Acknowledgements
We thank beamline staff for technical assistance and support at ID14-4 beamline, European Synchrotron Radiation Facility. Furthermore, we also thank Mike Stark for providing the pRS315 plasmid and for helpful advice. A.S. performed and analysed experiments, prepared figures, and wrote the manuscript. A.W.S. and D.M.F.v.A. conceived the project, analysed experiments, prepared figures and wrote the manuscript. The authors declare that they have no conflict of interest.
Data accessibility
The coordinates and structure factors for ScMnn9-D236N + Mn2+ + GDP have been deposited with the PDBe and can be accessed under entry code 3ZF8.
Funding statement
This work was supported by an MRC Programme grant no. G0900138. D.M.F.v.A. is supported by a Wellcome Trust Senior Research Fellowship (WT087590MA). A.S. is supported by a SULSA/BBSRC studentship.
References
- 1.Helenius A, Aebi M. 2001. Intracellular functions of N-linked glycans. Science 5512, 2364–2369 (doi:10.1126/science.291.5512.2364) [DOI] [PubMed] [Google Scholar]
- 2.Lodish HF, Kong N. 1984. Glucose removal from N-linked oligosaccharides is required for efficient maturation of certain secretory glycoproteins from the rough endoplasmic reticulum to the Golgi complex. J. Cell Biol. 5, 1720–1729 (doi:10.1083/jcb.98.5.1720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore SE, Spiro RG. 1993. Inhibition of glucose trimming by castanospermine results in rapid degradation of unassembled major histocompatibility complex class I molecules. J. Biol. Chem. 6, 3809–3812 [PubMed] [Google Scholar]
- 4.Shibata N, et al. 1995. Existence of branched side chains in the cell wall mannan of pathogenic yeast, Candida albicans. Structure–antigenicity relationship between the cell wall mannans of Candida albicans and Candida parapsilosis. J. Biol. Chem. 3, 1113–1122 [DOI] [PubMed] [Google Scholar]
- 5.Ballou L, Ballou C. 1995. Schizosaccharomyces pombe mutants that are defective in glycoprotein galactosylation. Proc. Natl Acad. Sci. USA 7, 2790–2794 (doi:10.1073/pnas.92.7.2790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gemmill TR, Trimble RB. 1996. Schizosaccharomyces pombe produces novel pyruvate-containing N-linked oligosaccharides. J. Biol. Chem. 42, 25 945–25 949 [DOI] [PubMed] [Google Scholar]
- 7.Gemmill TR, Trimble RB. 1999. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 2, 227–237 (doi:10.1016/S0304-4165(98)00126-3) [DOI] [PubMed] [Google Scholar]
- 8.Kollár R, Reinhold BB, Petráková E, Yeh HJC, Ashwell G, Drgonová J, Kapteyn JC, Klis FM, Cabib E. 1997. Architecture of the yeast cell wall: β-(1→6)-glucan interconnects mannoprotein, β-(1→3)-glucan, and chitin. J. Biol. Chem. 28, 17 762–17 775 (doi:10.1074/jbc.272.28.17762) [DOI] [PubMed] [Google Scholar]
- 9.Latgé J-P 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 2, 279–290 (doi:10.1111/j.1365-2958.2007.05872.x) [DOI] [PubMed] [Google Scholar]
- 10.Ballou CE, Kern KA, Raschke WC. 1973. Genetic control of yeast mannan structure. J. Biol. Chem. 13, 4667–4671 [PubMed] [Google Scholar]
- 11.Rayner JC, Munro S. 1998. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J. Biol. Chem. 41, 26836–26843 (doi:10.1074/jbc.273.41.26836) [DOI] [PubMed] [Google Scholar]
- 12.Nakayama K, Nagasu T, Shimma Y, Kuromitsu J, Jigami Y. 1992. OCH1 encodes a novel membrane bound mannosyltransferase: outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 7, 2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakanishi-Shindo Y, Nakayama K, Tanaka A, Toda Y, Jigami Y. 1993. Structure of the N-linked oligosaccharides that show the complete loss of alpha-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 35, 26 338–26 345 [PubMed] [Google Scholar]
- 14.Hernández LM, Ballou L, Alvarado E, Gillece-Castro BL, Burlingame AL, Ballou CE. 1989. A new Saccharomyces cerevisiae mnn mutant N-linked oligosaccharide structure. J. Biol. Chem. 20, 11 849–11 856 [PubMed] [Google Scholar]
- 15.Ballou L, Hernández LM, Alvarado E, Ballou CE. 1990. Revision of the oligosaccharide structures of yeast carboxypeptidase Y. Proc. Natl Acad. Sci. USA 9, 3368–3372 (doi:10.1073/pnas.87.9.3368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ballou L, Cohen RE, Ballou CE. 1980. Saccharomyces cerevisiae mutants that make mannoproteins with a truncated carbohydrate outer chain. J. Biol. Chem. 12, 5986–5991 [PubMed] [Google Scholar]
- 17.Klis FM, de Jong M, Brul S, de Groot PWJ. 2007. Extraction of cell surface-associated proteins from living yeast cells. Yeast 4, 253–258 (doi:10.1002/yea.1476) [DOI] [PubMed] [Google Scholar]
- 18.Herscovics A, Orlean P. 1993. Glycoprotein biosynthesis in yeast. FASEB J. 6, 540–550 [DOI] [PubMed] [Google Scholar]
- 19.Jungmann J, Munro S. 1998. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 2, 423–434 (doi:10.1093/emboj/17.2.423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojima H, Hashimoto H, Yoda K. 1999. Interaction among the subunits of Golgi membrane mannosyltransferase complexes of the yeast Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 11, 1970–1976 (doi:10.1271/bbb.63.1970) [DOI] [PubMed] [Google Scholar]
- 21.Jungmann J, Rayner JC, Munro S. 1999. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 10, 6579–6585 (doi:10.1074/jbc.274.10.6579) [DOI] [PubMed] [Google Scholar]
- 22.Lussier M, Sdicu AM, Bussey H. 1999. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 2, 323–334 (doi:10.1016/s0304-4165(98)00133-0) [DOI] [PubMed] [Google Scholar]
- 23.Stolz J, Munro S. 2002. The components of the Saccharomyces cerevisiae mannosyltransferase complex M-Pol I have distinct functions in mannan synthesis. J. Biol. Chem. 47, 44 801–44 808 (doi:10.1074/jbc.M208023200) [DOI] [PubMed] [Google Scholar]
- 24.Rodionov D, Romero PA, Berghuis AM, Herscovics A. 2009. Expression and purification of recombinant M-Pol I from Saccharomyces cerevisiae with α-1, 6 mannosylpolymerase activity. Protein Expr. Purif. 1, 1–6 (doi:10.1016/j.pep.2009.02.013) [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Mushegian A. 2003. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 7, 1418–1431 (doi:10.1110/ps.0302103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen MM, et al. 2011. Substrate and metal ion promiscuity in mannosylglycerate synthase. J. Biol. Chem. 17, 15 155–15 164 (doi:10.1074/jbc.M110.199844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobsanov YD, Romero PA, Sleno B, Yu B, Yip P, Herscovics A, Howell PL. 2004. Structure of Kre2p/Mnt1p: a yeast α-1,2-mannosyltransferase involved in mannoprotein biosynthesis. J. Biol. Chem. 17, 17 921–17 931 (doi:10.1074/jbc.M312720200) [DOI] [PubMed] [Google Scholar]
- 28.Gosselin S, Alhussaini M, Streiff MB, Takabayashi K, Palcic MM. 1994. A continuous spectrophotometric assay for glycosyltransferases. Anal. Biochem. 1, 92–97 (doi:10.1006/abio.1994.1303) [DOI] [PubMed] [Google Scholar]
- 29.Nakajima T, Maitra SK, Ballou CE. 1976. An endo-alpha-1-6-D-mannanase from a soil bacterium: purification, properties, and mode of action. J. Biol. Chem. 1, 174–181 [PubMed] [Google Scholar]
- 30.Maruyama Y, Nakajima T. 2000. The aman6 gene encoding a yeast mannan backbone degrading 1,6-α-D-mannanase in Bacillus circulans: cloning, sequence analysis, and expression. Biosci. Biotechnol. Biochem. 9, 2018–2020 (doi:10.1271/bbb.64.2018) [DOI] [PubMed] [Google Scholar]
- 31.Flint J, et al. 2005. Structural dissection and high-throughput screening of mannosylglycerate synthase. Nat. Struct. Mol. Biol. 7, 608–614 (doi:10.1038/nsmb950) [DOI] [PubMed] [Google Scholar]
- 32.Häusler A, Ballou L, Ballou CE, Robbins PW. 1992. Yeast glycoprotein biosynthesis: MNT1 encodes an alpha-1,2-mannosyltransferase involved in O-glycosylation. Proc. Natl Acad. Sci. USA 15, 6846–6850 (doi:10.1073/pnas.89.15.6846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Häusler A, Robbins PW. 1992. Glycosylation in Saccharomyces cerevisiae: cloning and characterization of an α-1,2-mannosyltransferase structural gene. Glycobiology 1, 77–84 (doi:10.1093/glycob/2.1.77) [DOI] [PubMed] [Google Scholar]
- 34.Lussier M, Sdicu AM, Bussereau F, Jacquet M, Bussey H. 1997. The Ktr1p, Ktr3p, and Kre2p/Mnt1p mannosyltransferases participate in the elaboration of yeast O- and N-linked carbohydrate chains. J. Biol. Chem. 24, 15 527–15 531 (doi:10.1074/jbc.272.24.15527) [DOI] [PubMed] [Google Scholar]
- 35.Dean N. 1995. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl Acad. Sci. USA 5, 1287–1291 (doi:10.1073/pnas.92.5.1287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballou L, Hitzeman RA, Lewis MS, Ballou CE. 1991. Vanadate-resistant yeast mutants are defective in protein glycosylation. Proc. Natl Acad. Sci. USA 8, 3209–3212 (doi:10.1073/pnas.88.8.3209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Southard SB, Specht CA, Mishra C, Chen-Weiner J, Robbins PW. 1999. Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins. J. Bacteriol. 24, 7439–7448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 1, 207–234 (doi:10.1016/j.pep.2005.01.016) [DOI] [PubMed] [Google Scholar]
- 40.Holyoak T, Fenn TD, Wilson MA, Moulin AG, Ringe D, Petsko GA. 2003. Malonate: a versatile cryoprotectant and stabilizing solution for salt-grown macromolecular crystals. Acta Crystallogr. D, Biol. Crystallogr. 59, 2356–2358 (doi:10.1107/S0907444903021784) [DOI] [PubMed] [Google Scholar]
- 41.Sheldrick GM. 2010. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D, Biol. Crystallogr. D66, 479–485 (doi:10.1107/S0907444909038360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perrakis A, Morris R, Lamzin VS. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 5, 458–463 (doi:10.1038/8263) [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D, Biol. Crystallogr. 60, 2126–2132 (doi:10.1107/S0907444904019158) [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, Biol. Crystallogr. 53, 240–255 (doi:10.1107/S0907444996012255) [DOI] [PubMed] [Google Scholar]
- 45.Jackson P. 1990. The use of polyacrylamide-gel electrophoresis for the high-resolution separation of reducing saccharides labelled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid. Detection of picomolar quantities by an imaging system based on a cooled charge-coupled device. Biochem. J. 3, 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballou CE. 1990. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 185, 440–470 (doi:10.1016/0076-6879(90)85038-P) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for ScMnn9-D236N + Mn2+ + GDP have been deposited with the PDBe and can be accessed under entry code 3ZF8.