Abstract
Stem cells generate progeny that undergo terminal differentiation. The initiation and maintenance of the differentiated status is crucial for tissue development, function and homeostasis. Drosophila neural stem cells (neuroblasts) are a model for stem cell self-renewal and differentiation; they divide asymmetrically to self-renew and generate the neurons and glia of the CNS. Here we report the identification of midlife crisis (mdlc; CG4973) as a gene required for the maintenance of neuronal differentiation and for neuroblast proliferation in Drosophila. mdlc encodes a ubiquitously expressed zinc-finger-containing protein with conserved orthologs from yeast to humans that are reported to have a role in RNA splicing. Using clonal analysis, we demonstrate that mdlc mutant neurons initiate but fail to complete differentiation, as judged by the loss of the pro-differentiation transcription factor Prospero, followed by derepression of the neuroblast factors Deadpan, Asense and Cyclin E. RNA-seq shows that loss of Mdlc decreases pros transcript levels and results in aberrant pros splicing. Importantly, misexpression of the full-length human ortholog, RNF113A, completely rescues all CNS defects in mdlc mutants. We conclude that Mdlc plays an essential role in maintaining neuronal differentiation, raising the possibility that RNF113A regulates neuronal differentiation in the human CNS.
Keywords: Neuroblast, Neuron, Prospero, Self-renewal, Splicing, Stem cell
INTRODUCTION
A defining characteristic of stem cells is the ability to produce daughters that retain stem cell fate (self-renew) as well as daughters that begin the process of differentiation. The robust adoption of these distinct fates is crucial both for the maintenance of stem cell pool size and for the production of differentiated progeny. In the Drosophila CNS, neuroblasts divide in a manner that is asymmetric in both progeny size and fate. The majority of neuroblasts - termed ‘type I’ neuroblasts - divide to generate a self-renewed neuroblast and a smaller ganglion mother cell (GMC), which divides only once more to produce neurons or glia. Additionally, there are eight bilateral ‘type II’ neuroblasts in the brain that repeatedly divide to self-renew and generate smaller intermediate neural progenitors (INPs), which each undergo a series of molecularly asymmetric divisions (similar to type I neuroblast divisions) to self-renew and generate a series of four to six GMCs (Bayraktar et al., 2010; Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Izergina et al., 2009). Type I and II neuroblasts have emerged as an important model for studying stem cell self-renewal and differentiation.
Type I neuroblast asymmetric division results in the segregation of cell fate determinants into the GMC. These fate determinants inhibit neuroblast self-renewal, direct cell cycle exit, promote neuronal differentiation and prevent tumor formation (Doe, 2008; Knoblich, 2010). Type II neuroblast lineages contain INPs that are particularly susceptible to dedifferentiation. Loss of function of earmuff (erm), barricade (barc), brain tumor (brat) or misexpression of activated Notch all lead to failure in neuronal differentiation and an expansion of type II neuroblast or INP fates (Bowman et al., 2008; Neumüller et al., 2011; Weng et al., 2010).
One of the crucial differentiation factors is the transcription factor Prospero (Pros). In type I neuroblasts, Pros protein and mRNA are asymmetrically segregated into the GMC. In the GMC, pros mRNA is translated and Pros protein is imported into the nucleus (Broadus et al., 1998; Knoblich et al., 1995; Spana and Doe, 1995), where it represses cell cycle genes and promotes differentiation (Choksi et al., 2006; Li and Vaessin, 2000). Therefore, it is essential that GMCs inherit Pros from the neuroblast; in a Pros loss-of-function mutant, GMCs fail to exit the cell cycle, derepress neuroblast fate genes and can form tumorous overgrowths (Bello et al., 2006; Betschinger et al., 2006; Choksi et al., 2006; Lee et al., 2006). In the embryo, Pros protein can be detected in the GMC and transiently in newly born embryonic neurons (Srinivasan et al., 1998). In the larval CNS, Pros is detected in nearly all postmitotic neurons. In contrast to its tumor suppressor function in the GMC, the function of Pros in postmitotic larval neurons is unknown.
Here we identify midlife crisis (mdlc; CG4973 - FlyBase) as a gene required to maintain Pros expression and neuronal differentiation in Drosophila larvae. mdlc encodes a conserved protein containing both a RING domain and a CCCH-type zinc finger. The yeast and human orthologs of Mdlc have been reported to be components of the spliceosome (Bessonov et al., 2008; Goldfeder and Oliveira, 2008). Clonal analysis of larval neuroblast lineages demonstrates that loss of mdlc function results in the loss of neuronal Pros expression followed by loss of the neuronal marker Embryonic lethal abnormal vision (Elav) and ectopic expression of the neuroblast transcription factors Asense (Ase) and Deadpan (Dpn). This results in single neuroblast clones containing multiple Dpn+ Ase+ Elav- Pros- cells, which are more like neuroblasts than neurons in terms of molecular marker expression, indicating that Mdlc promotes the maintenance of neuron fate gene expression in larval neurons and inhibits neuronal dedifferentiation. Mdlc also functions in neuroblasts to promote their characteristically rapid (∼2 hour) cell cycle. Surprisingly, these roles for Mdlc do not require the RING domain, whereas the CCCH-type zinc finger is essential for all known Mdlc CNS functions.
MATERIALS AND METHODS
Fly stocks
Fly stocks used were UAS-Dicer2 (Dcr2), inscuteable-Gal4, tubulin-Gal4, UAS-mCD8:GFP, elav-Gal4; UAS-Dcr2, acj6-Gal4, atonal-Gal4 and w1118; Df(3R)ED6027 from the Bloomington Drosophila Stock Center (BDSC); w1118; P[GD11492]v42015 and w1118; P[KK101588]VIE-260B from the Vienna Drosophila RNAi Center (VDRC); mdlcc04701 from the Exelixis collection at Harvard Medical School; worniu-Gal4 (Albertson et al., 2004); UAS-Dcr2; worniu-Gal4 asense-Gal80; UAS-mCD8:GFP (Neumüller et al., 2011). MARCM clones were generated using hs-flp70; tub-Gal4 UAS-mCD8:GFP; FRT82B tub-Gal80; this was crossed to FRT82B to generate control clones and FRT82B mdlcc04701 to generate mutant clones.
Immunostaining and confocal microscopy
Antibodies used were rat anti-Dpn (1:50; C.Q.D. lab), guinea pig anti-Miranda (Mira) (1:1000 or 1:2000; C.Q.D. lab), chicken anti-GFP (1:2000; Aves Labs), rabbit anti-Ase (1:2000) (Brand et al., 1993), mouse anti-Pros [1:1000; Developmental Studies Hybridoma Bank (DSHB) MR1A], rat anti-Elav (1:50; DSHB 7E8A10), mouse anti-Repo (1:4; DSHB), and guinea pig anti-Mdlc (1:100; this study). Secondary antibodies (Molecular Probes or Jackson ImmunoResearch) were used at 1:500. Antibody staining was performed as described (Carney et al., 2012) with the following change: the block used was 2.5% normal goat serum plus 2.5% normal donkey serum. Microscopy images were captured using either a Zeiss 700 or 710 confocal microscope.
EdU incorporation
EdU (Life Technologies) was delivered at 100 μg/ml in larval food. Detection was performed according to the manufacturer’s instructions.
RNA interference (RNAi) and mosaic analysis with a repressible cell marker (MARCM)
RNAi was performed at 30°C. MARCM clones were induced at 24±4 hours after larval hatching (ALH) by heat shock at 37°C for 20 minutes. Larvae were then raised at 23°C until late third instar.
Molecular biology and antibody generation
UAS-mdlc and UAS-RNF113A constructs were generated by PCR-mediated mutagenesis (where applicable) and cloned into a pUAST-attB vector (Bischof et al., 2007) using XhoI (5′) and XbaI (3′) overhangs. Forward primers included a Kozak sequence (CAAC) immediately upstream of the start codon, as well as sequence coding for an N-terminal hemagglutinin (HA) epitope tag. All transgenes were site-specifically inserted via ΦC31 integrase-mediated transgenesis at the VK37 site on chromosome 2L (GenetiVision Corporation, Houston, TX, USA).
For the generation of the anti-Mdlc antibody, sequence coding for the N-terminal 165 residues of Mdlc was PCR amplified and cloned into a pET15b vector (Novagen) using NdeI (5′) and XhoI (3′) overhangs. The vector has been modified to encode an N-terminal penta-histidine epitope tag and has a Tobacco Etch Virus (TEV) cleavage site between the His tag and peptide-of-interest sequence. Protein was expressed in BL21 E. coli cells and purified by adsorption to Ni-NTA agarose (Qiagen), followed by elution via cleavage with TEV protease. Antibody was raised in guinea pig (Alpha Diagnostics, San Antonio, TX, USA) and affinity purified using the ImmunoLink Plus Immobilization Kit (Thermo Scientific) according to the manufacturer’s instructions.
To verify the presence of the transposon insertion c04701, we performed PCR on genomic DNA from a single c04701 fly using primers: 5′-TACCATCACTAGCCGGGAAG-3′, which recognizes the mdlc 5′ UTR upstream of the transposon insertion site; and 5′-CCTCGATATACA - GACCGATAAAACACATG-3′, which recognizes a site near the 3′ end of the piggyBac ‘PB’ transposon (Thibault et al., 2004).
RNA-seq
Genotypes used were elav-Gal4; UAS-Dcr2/+ and elav-Gal4; UAS-Dcr2/UAS-mdlc RNAi. We performed two biological replicates for both treatments. Larvae were raised at 25°C for 48 hours and then transferred to 30°C for the remainder of their development. Total RNA was isolated from brains of wandering third instar larvae as described (Miller et al., 2009). Poly(A) RNA was purified using the Poly(A)Purist Kit (Ambion), fragmented to generate 250-400 base fragments using the NEBNext Magnesium RNA Fragmentation Module (New England BioLabs), and prepared for Illumina sequencing using the ScriptSeq v2 RNA-seq Library Preparation Kit (Epicentre Biotechnologies, Madison, WI, USA). Sequencing resulted in 16.5-29.3 million single-end 100 bp reads from each barcoded library. The reads were aligned against the Drosophila melanogaster release 5.69 genome sequence (Ensembl) using GSNAP (Wu and Nacu, 2010) (http://research-pub.gene.com/gmap/) allowing for up to seven mismatches and set to look for novel splice junctions.
Detecting differentially expressed genes
The number of reads mapping to the exons of each gene was quantified using the HTseq-count Python script in ‘union’ mode (http://www-huber.embl.de/users/anders/HTSeq/doc/index.html). The BAM alignment files for each sample and a downloaded Ensembl GTF file were used as inputs. Differentially expressed genes were called using the DEseq package (Anders and Huber, 2010) following the developer’s recommended workflow in R (http://www.R-project.org).
Detection of differential intron retention (DIR) using MISO
The software package MISO (Katz et al., 2010) (http://genes.mit.edu/burgelab/miso/docs/) was used to detect differentially regulated introns across samples. An ‘exon-centric’ MISO analysis was performed according to the developer’s recommended workflow using single-end reads. The BAM alignment files produced by GSNAP and a custom set of pros annotations as well as the alternative events file provided by MISO were used as inputs. To identify highly reliable mdlc-associated DIR events, we required the following stringent criteria to be met: (1) the absolute value of the difference (ΔΨ) >0.2; (2) the sum of inclusion and exclusion reads is greater than 10 (≥1 inclusion read and ≥1 exclusion read); (3) the Bayes factor >1000; (4) these criteria must be met for all comparisons between wild-type (wt) and mdlc RNAi samples; and (5) the event did not appear differentially regulated between wt biological replicates. Alternative events were visualized using the included sashimi_plot software from the MISO package (http://genes.mit.edu/burgelab/miso/docs/sashimi.html).
RESULTS
mdlc RNAi results in ectopic Dpn+ neuroblast-like cells
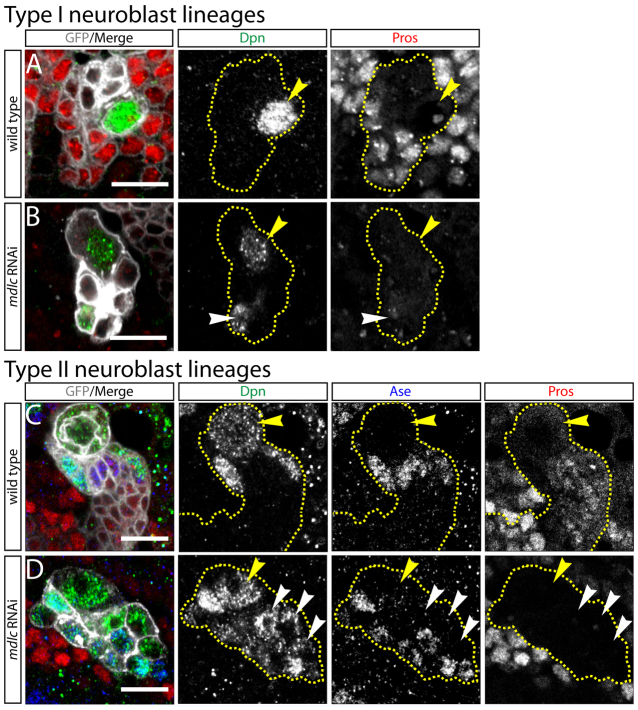
We initially observed ectopic Dpn+ cells in the mdlc RNAi central brain lobes in an RNAi screen (Carney et al., 2012). The screen used the neuroblast-specific worniu-Gal4 [wor-Gal4 (Albertson et al., 2004)] driving expression of UAS-Dcr2 to increase RNAi efficacy (Dietzl et al., 2007) and single UAS-RNAi transgenes targeting transcripts enriched in neuroblasts. Wt type I neuroblast lineages contain a single large Dpn+ neuroblast (Fig. 1A), whereas knockdown of mdlc results in small ectopic Dpn+ cells at a distance from the large parental neuroblast (Fig. 1B). This indicates that the ectopic Dpn+ cells are not the result of symmetric neuroblast divisions, which always result in adjacent Dpn+ neuroblasts (Cabernard and Doe, 2009). To determine whether the ectopic Dpn+ cells might be dedifferentiating neurons, we stained for Pros, which marks GMCs and young neurons in wt lineages (Fig. 1A). Indeed, we observed loss of Pros in mdlc knockdown brains (Fig. 1B). We conclude that mdlc RNAi leads to ectopic Dpn and loss of Pros in type I neuroblast progeny.
Fig. 1.
Knockdown of mdlc causes ectopic Dpn and loss of Pros in Drosophila larval neuroblast lineages. (A,B) Type I central brain neuroblast lineages. Yellow dashed lines indicate lineage boundaries; yellow arrowheads mark type I neuroblasts. (A) Wild-type (wt) lineages show a single large Dpn+ neuroblast and multiple smaller Pros+ neurons. (B) mdlc RNAi causes small ectopic Dpn+ cells (white arrowhead) as well as a strong loss of Pros in many neurons. (C,D) Type II central brain neuroblast lineages. Yellow dashed lines indicate lineage boundaries; yellow arrowheads mark type II neuroblasts. (C) Wt lineage showing Dpn+ type II neuroblast and Dpn+ mature INPs; Ase is absent from the type II neuroblast but present in mature INPs and GMCs. (D) mdlc RNAi lineage showing ectopic Dpn+ cells; some are Ase+ indicating an INP fate, whereas others are Ase- indicating reversion to a type II neuroblast-like fate. Pros staining is strongly reduced. Yellow arrowhead marks the presumptive type II neuroblast; white arrowheads indicate ectopic Dpn+ Ase- cells. Genotypes: (A) wor-Gal4 UAS-Dcr2; UAS-mCD8:GFP; (B) wor-Gal4 UAS-Dcr2/UAS-mdlc RNAi; UAS-mCD8:GFP; (C) UAS-Dcr2; wor-Gal4 ase-Gal80; UAS-mCD8:GFP; (D) UAS-Dcr2; wor-Gal4 ase-Gal80/UAS-mdlc RNAi; UAS-mCD8:GFP. Scale bars: 10 μm.
We next determined the mdlc knockdown phenotype in type II neuroblasts, which generate small Dpn+ INPs, each of which undergoes asymmetric cell division to generate small Dpn- Pros+ GMCs and neurons (Bayraktar et al., 2010; Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Izergina et al., 2009). We performed mdlc RNAi knockdown using UAS-Dcr2; wor-Gal4 ase-Gal80; UAS-mCD8:GFP, which results in Gal4-induced gene expression specifically in type II neuroblasts (Neumüller et al., 2011). In wt, each type II neuroblast lineage contains a single large Dpn+ Ase- neuroblast and several adjacent smaller Dpn+ Ase+ INPs (Fig. 1C). By contrast, mdlc knockdown resulted in many ectopic small Dpn+ Ase+ cells, as well as many Dpn+ Ase- cells (Fig. 1D). Thus, the ectopic Dpn+ cells could have either neuroblast or INP identity. We also observed a strong loss of Pros in the mdlc knockdown type II lineage (compare Fig. 1C,D), similar to the loss of Pros phenotype following mdlc knockdown in type I lineages (Fig. 1A,B). In addition, we observed the loss of some type II neuroblasts, fewer cells per lineage and enlarged cell size (data not shown); these phenotypes will be explored below. We conclude that mdlc knockdown causes ectopic Dpn+ neuroblast-like (or INP-like) cells and loss of Pros+ cells in both type I and type II neuroblast lineages.
mdlc RNAi results in a failure to maintain Pros in postmitotic neurons
The loss of Pros expression upon mdlc RNAi knockdown appeared more penetrant than the ectopic Dpn+ cells, suggesting that it might be the earliest detectable phenotype and might directly result in the ectopic Dpn expression. Here, we address whether mdlc knockdown leads to a failure to properly establish Pros expression in newly born GMCs or a failure to maintain Pros in mature postmitotic neurons. In subsequent sections we will address the timing of the Pros versus Dpn phenotypes and whether loss of Pros is sufficient to derepress Dpn in neurons.
To determine whether mdlc knockdown results in a failure to establish or to maintain Pros expression, we used EdU labeling to identify newly born GMCs or mature postmitotic neurons derived from type I neuroblasts. To unambiguously identify newly born GMCs, we fed larvae at 48 hours ALH with EdU for 4 hours and immediately fixed and stained brains; using this approach, only the neuroblast and newly born GMCs are EdU+. We found that both wt and mdlc knockdown brains have Pros present in all newly born GMCs (Fig. 2A,B, quantified in 2C). Similar results were observed for type II neuroblast lineages (Fig. 2G,H). Of course, this could be due to a lag in mdlc RNAi knockdown of Mdlc protein, but RNAi knockdown begins over 36 hours prior to assaying GMC gene expression and we detected no Mdlc protein over background levels (supplementary material Fig. S1). We conclude that Mdlc is not required to establish Pros expression in newly born GMCs.
Fig. 2.
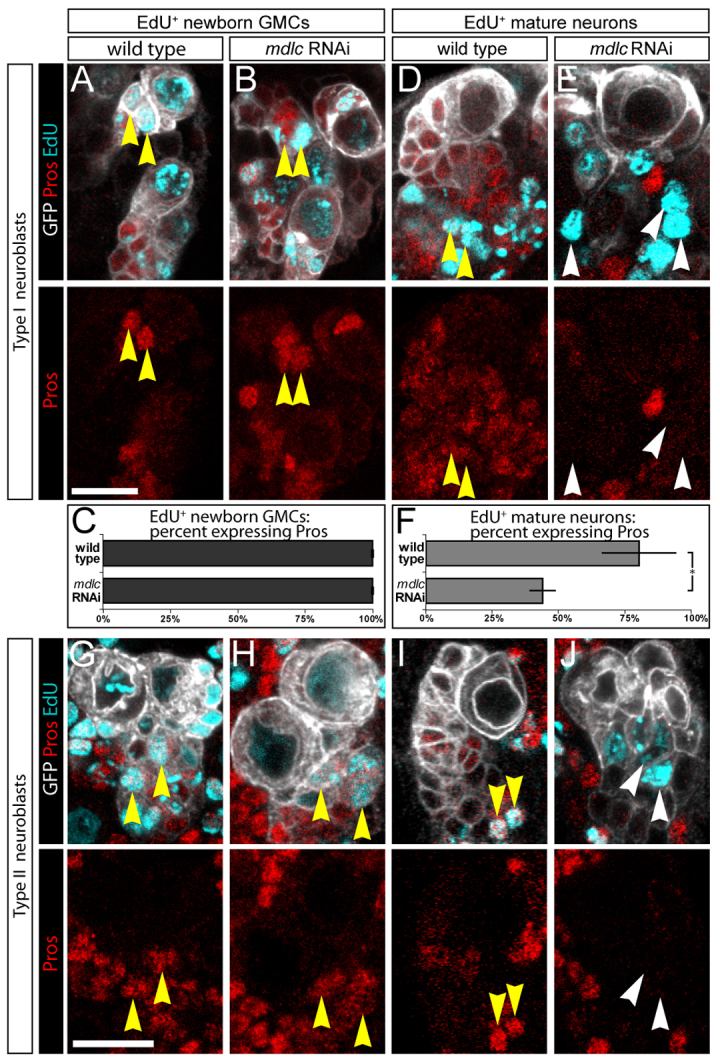
mdlc RNAi results in repression of Pros in postmitotic neurons. (A-F) Type I neuroblasts. (A-C) Newly born GMCs marked by a 4-hour pulse of EdU and immediately stained; both wt GMCs (A) and mdlc RNAi GMCs (B) are Pros+, as quantified in C. n=304 EdU+ cells in wt and n=99 EdU+ cells in mdlc RNAi, 100% of which were Pros+. (D-F) Mature neurons marked by a 4-hour pulse of EdU followed by a 36-hour EdU-free chase; wt neurons are Pros+ (D) whereas mdlc RNAi neurons (E) fail to maintain Pros, as quantified in F. n=1016 EdU+ cells in wt, 79% of which were Pros+; n=438 EdU+ cells in mdlc RNAi brains, 44% of which were Pros+. *P<0.01; error bars indicate s.d. (G-J) Type II neuroblasts. (G,H) Newly born GMCs marked by a 4-hour pulse of EdU and immediately stained; both wt GMCs (G) and mdlc RNAi GMCs (H) are Pros+. (I,J) Mature neurons marked by a 4-hour pulse of EdU followed by a 36-hour EdU-free chase; wt neurons are Pros+ (I) whereas mdlc RNAi neurons (J) fail to maintain Pros. All yellow arrowheads mark EdU+ Pros+ cells; all white arrowheads mark EdU+ Pros- cells. Scale bars: 10 μm.
To unambiguously identify mature postmitotic neurons, we fed larvae at 48 hours ALH with EdU for 4 hours and then ‘chased’ for 36 hours with EdU-negative food; this approach ensures that only postmitotic neurons are EdU+ (neuroblasts dilute out all EdU labeling, and newly born GMCs arise during the EdU-negative chase period). We found that some wt neurons lose Pros protein, as expected due to downregulation of Pros in the most mature neurons in the lineage (Fig. 2D), but significantly more mdlc knockdown neurons do not have detectable Pros protein (Fig. 2E, quantified in 2F). Similar results were observed for type II neuroblast lineages (Fig. 2I,J). We conclude that mdlc RNAi results in a failure to maintain Pros levels in ‘middle-aged’ neurons, leading to our choice of gene name (midlife crisis).
mdlc mutants fail to maintain Pros and Elav in postmitotic neurons
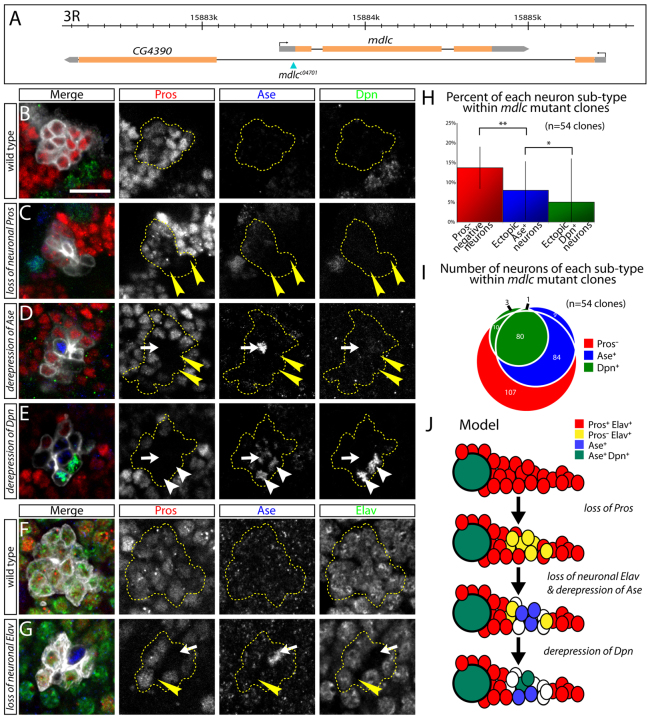
To confirm that the RNAi phenotype is due to loss of function of mdlc (and not an off-target RNAi effect) we analyzed a genetic lesion in the mdlc gene. mdlc is on chromosome 3R and resides within an intron of CG4390 on the opposite strand (Fig. 3A). We acquired a mutant allele at the mdlc locus: c04701, a piggyBac transposon from the Exelixis collection (Thibault et al., 2004). Its annotated insertion site is in the 5′ UTR, 10 bp upstream of the mdlc translation start site (Fig. 3A). We used PCR analysis to confirm the presence of c04701, which is homozygous lethal (see Materials and methods), renamed the insertion c04701 as mdlcc04701, and used it for subsequent genetic analysis of mdlc function.
Fig. 3.
mdlc mutants fail to maintain Pros and Elav in postmitotic neurons. (A) Genomic context of mdlc (CG4973), which resides in the intron of CG4390 on the opposite strand. The piggyBac transposon c04701 is inserted in the 5′ UTR of mdlc. (B) Wt MARCM clone showing Pros+ neurons lacking expression of Dpn and Ase. (C-E) mdlc mutant MARCM clones chosen to indicate the progression of neuronal phenotypes shown in J. (C) Middle-aged neurons (lying midway between the neuroblast and the oldest, most distal neurons) lack Pros expression (yellow arrowheads) but have not upregulated Dpn or Ase. (D) Middle-aged neurons lack Pros expression (yellow arrowheads) and Ase is derepressed in one of them (white arrow). (E) Middle-aged neurons lack Pros expression (arrow and arrowheads); Ase is derepressed in several of these cells (e.g. arrow), and two Ase+ cells have also derepressed Dpn (arrowheads). (F,G) mdlc mutant neurons lose Elav. (F) Wt neurons in which Pros and Elav are present but Ase is absent. (G) mdlc mutant MARCM clone showing the loss of both Pros and Elav from a small number of neurons (arrow and arrowhead); Ase is derepressed in one of these cells (white arrow). (H) Quantification of neuronal phenotypes in mdlc- clones (n=54 clones). Numbers are given as the percentage of the total number of cells in the clone. *P<0.05, **P<0.01; error bars indicate s.d. (I) Comprehensive quantification of absolute numbers of non-wt neuronal phenotypes in mdlc mutant clones, showing the overlap of Pros-, Ase+ and Dpn+ neurons (n=54 clones). This analysis demonstrates that the majority of Dpn+ cells comprise a subset of Ase+ neurons [81 of 94 Dpn+ neurons (86%) are also Ase+], and that the Ase+ cells are a subset of Pros- neurons [164 of 174 Ase+ neurons (94%) are Pros-]. (J) Model illustrating the progression of mdlc mutant clone phenotypes. Scale bar: 10 μm.
We analyzed mdlcc04701 phenotypes using the MARCM method (Lee and Luo, 2001). Wt type I neuroblast clones possess a single neuroblast that is Dpn+, Ase+ and Cyclin E (CycE)+, and all neurons in the clone are Pros+ with the exception of the oldest neurons at the most distal tip of the clone (Fig. 3B; supplementary material Movie 1; data not shown). By contrast, mdlcc04701 clones frequently have cells in the middle of the clone that have lost Pros expression (Fig. 3C), which is very rarely observed in wt neuroblast clones. This phenotype is less penetrant but remarkably similar to the mdlc RNAi phenotype (Figs 1, 2). We conclude that both mdlc RNAi and mdlc mutants lead to a failure to maintain Pros levels in middle-aged neurons.
We next aimed to determine the timing of Pros loss and Dpn derepression, as well as examining a second neuroblast marker, Ase, and a second neuronal differentiation marker, Elav, within mdlc mutant clones. First, we co-stained for Pros, Dpn and Ase. GMCs lying adjacent to the parental neuroblast typically co-express Pros and Ase and sometimes have weak Dpn due to perdurance from the neuroblast; therefore, they were not considered to be ectopically expressing cells and were excluded from our counts. We observed some clones in which the Pros- cells do not show detectable Dpn or Ase (Fig. 3C), whereas some have derepressed Ase but are Dpn- (Fig. 3D) and others are both Dpn+ and Ase+ (Fig. 3E, quantified in 3H,I; supplementary material Movie 2). Similar results were observed for the well-characterized neuronal differentiation marker Elav: it is present in all wt neurons, but absent from most (but not all) of the Pros- neurons in mdlc mutants (Fig. 3F,G). We conclude that that loss of the neuronal differentiation factor Pros precedes the loss of the neuronal marker Elav and the ectopic expression of the neuroblast markers Dpn and Ase in mdlc mutant neurons (Fig. 3J).
mdlc mutant neuroblasts have a longer cell cycle and reduced clone size
In addition to the neuronal dedifferentiation phenotype shown by mdlc RNAi and mutant clones, we noted that mdlc mutant neuroblast lineages have fewer cells than comparable wt lineages. We quantified this phenotype by counting total cells in both mutant and wt clones generated in 96 hours (slightly longer or shorter intervals were normalized to 96 hours). Wt clones averaged 89±35 cells, which is significantly more than mdlc mutant clones at 43±19 cells (P<10-5; Fig. 4A). Decrease in clone size was not due to apoptosis, as there was no increase in the apoptotic marker Caspase 3 in mdlc mutant clones (Fig. 4B). Instead, we found that the cell cycle is extended: wt neuroblast clones generate 14±4 EdU+ cells over 8 hours of EdU labeling, whereas mdlc mutants only generate 4±6 EdU+ cells during the same span (P<10-5; Fig. 4C). The variability stems from the fact that some mutant neuroblasts generate near wt numbers of progeny whereas others fail entirely to undergo S phase during the EdU incorporation period (not shown). We conclude that Mdlc promotes larval neuroblast cell cycle progression.
Fig. 4.
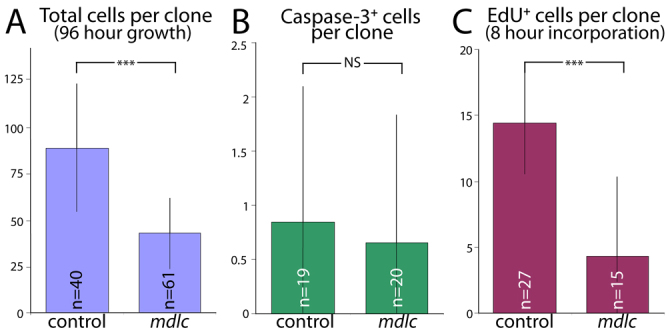
mdlc mutant neuroblasts have an extended cell cycle. (A) Comparison of wt and mdlc- clone sizes. The time between clone induction and dissection was 96 hours. (B) Number of Caspase 3+ cells per clone in wt and mdlc- clones. (C) Comparison of wt and mdlc- EdU incorporation rates. The EdU incorporation period was 8 hours prior to dissection. ***P<10-5; NS, not significantly different; error bars indicate s.d.
Mdlc is a conserved zinc-finger-containing protein with broad expression
mdlc encodes a well-conserved protein that is 70% similar (58% identical) to the human ortholog RNF113A. Both proteins have two conserved zinc-finger domains: a CCCH zinc finger, which is commonly found in RNA-binding proteins involved in splicing; and a RING domain, which is frequently found in E3 ubiquitin ligases (Fig. 5A). In order to analyze the expression patterns and subcellular localization of Mdlc, we generated an antibody against the N-terminal 165 amino acids. Immunofluorescent staining with this antibody revealed that Mdlc is a nuclear protein that is present throughout the larval CNS (Fig. 5B).
Fig. 5.
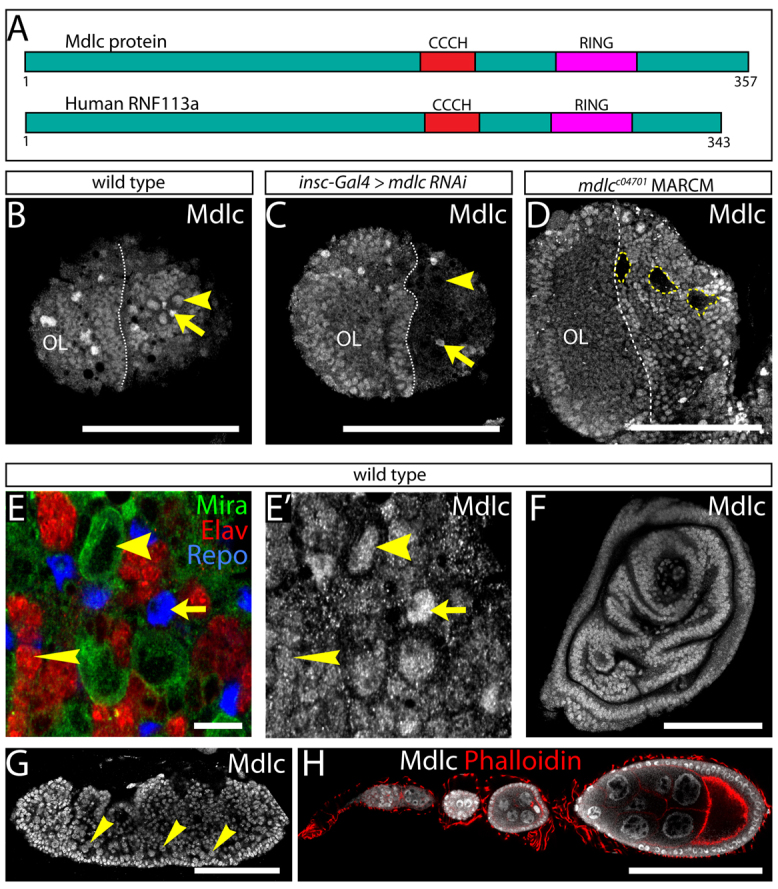
Mdlc is a zinc-finger protein with broad expression in the CNS and other tissues. (A) Mdlc protein and the human ortholog RNF113A have a conserved CCCH zinc finger and a C-terminal RING domain. Numbers indicate amino acids. (B-D) Anti-Mdlc shows ubiquitous nuclear localization of Mdlc, which is lost upon mdlc loss of function. (B) Wt Mdlc is expressed in larval brain neuroblasts (arrowhead) and glia (arrow). (C) mdlc RNAi in central brain neuroblasts and their immediate progeny reduces Mdlc protein staining in the central brain, particularly in neuroblasts (arrowhead), as identified by the presence of Dpn (not shown). Mdlc expression is still visible in glia (arrow) and in the optic lobe (OL), where the RNAi transgene was not expressed. (D) Homozygous mutant mdlcc04701 clones (yellow dashed lines) show a strong reduction in Mdlc protein. (E,E′) In the wt larval brain, Mdlc protein is detected in neuroblasts (wide arrowheads), neurons (narrow arrowheads) and glia (arrows), which can be identified based on staining with antibodies against Mira, Elav and Repo, respectively. (F-H) Mdlc is ubiquitously expressed in all tissues examined. (F) Representative imaginal disc. (G) Mdlc expression in the embryo, including in neuroblasts (arrowheads) identified by Mira expression (not shown). (H) Adult ovariole. Scale bars: 100 μm in B-D,F-H; 10 μm in E.
mdlc knockdown using inscuteable-Gal4 (insc-Gal4) to drive UAS-mdlc RNAi caused a marked decrease in Mdlc immunostaining in the larval central brain (but not in glia or optic lobe; Fig. 5C), validating the efficacy of mdlc RNAi and the specificity of the Mdlc antibody. Furthermore, mosaic clones of homozygous mdlcc04701 cells showed strongly reduced Mdlc levels compared with the surrounding wt cells, indicating that mdlcc04701 is a strong loss-of-function allele (Fig. 5D). Upon close examination of larval brains, we found that Mdlc is expressed ubiquitously, including NBs, neurons and glia, as judged by staining with antibodies against Mira, Elav and Reversed polarity (Repo), respectively (Fig. 5E,E′). Mdlc is also broadly expressed in larval imaginal discs (Fig. 5F), in the embryo (Fig. 5G) and in the adult ovary (Fig. 5H).
Mdlc zinc-finger deletions reveal different requirements for the CCCH and RING domains
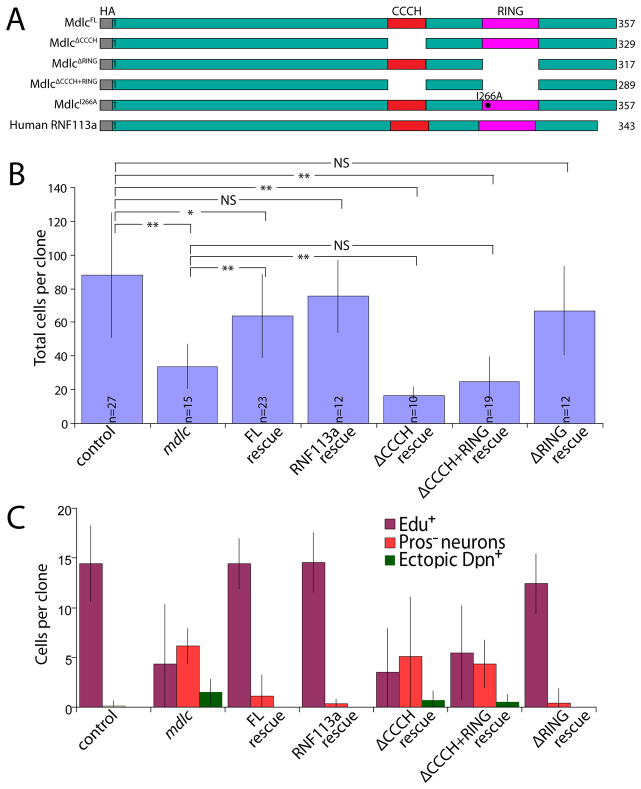
We have shown that Mdlc has context-dependent functions: it is required for neuroblast cell cycle progression as well as in middle-aged neurons to prevent dedifferentiation. Here we asked whether the CCCH and RING zinc-finger domains (which are proposed to regulate RNA splicing and ubiquitylation, respectively) have specific roles in either of these two phenotypes. We generated fly lines expressing different Mdlc domains under UAS control: full-length Mdlc (MdlcFL); Mdlc lacking the CCCH zinc finger (MdlcΔCCCH); Mdlc lacking the RING domain (MdlcΔRING); Mdlc with RING domain isoleucine 266 mutated to alanine (MdlcI266A), which is reported to interfere with E2/E3 interactions in RING domain-containing E3 ubiquitin ligases (Mulder et al., 2007); and Mdlc lacking both zinc fingers (MdlcΔCCCH+RING). In addition, we generated a transgene expressing the full-length human ortholog of Mdlc (RNF113A). Each construct included a hemagglutinin (HA) epitope tag at the N-terminus of the protein (Fig. 6A).
Fig. 6.
The CCCH zinc finger of Mdlc is crucial for CNS function whereas the RING finger is dispensable. (A) The protein truncations used in misexpression and rescue experiments. (B) Quantification of clone size in rescue experiments. The time between clone induction and brain dissection was 96 hours. Control and mdlc- clone sizes are given as references. *P<0.01, **P<10-3; NS, not statistically different. (C) Quantification of EdU cells per clone (8-hour EdU incorporation), Pros- neurons per clone and ectopic Dpn+ cells per clone in rescue experiments. Expression of MdlcFL, RNF113A or MdlcΔRING rescues all of these phenotypes whereas expression of MdlcΔCCCH or MdlcΔCCCH+RING fails to rescue. Error bars indicate s.d.
We assayed the ability of each construct to rescue mutant phenotypes by monitoring both the most penetrant neuronal phenotype (loss of Pros) and the most severe phenotype (ectopic Dpn expression), as well as neuroblast lineage size and extent of EdU incorporation. Misexpression of MdlcFL, or the full-length human ortholog RNF113A, in mdlc mutant clones significantly rescued clone size (Fig. 6B) and completely rescued the loss of Pros/ectopic Dpn+ phenotypes (Fig. 6C). In addition, ubiquitous expression of either protein using tubulin-Gal4 rescued mdlcc04701/Df(3R)ED6027 hemizygotes to viability (Table 1). These results indicate that the epitope-tagged full-length version of Mdlc is functional and that Mdlc and its human ortholog have conserved functions.
Table 1.
Summary of rescue and misexpression phenotypes
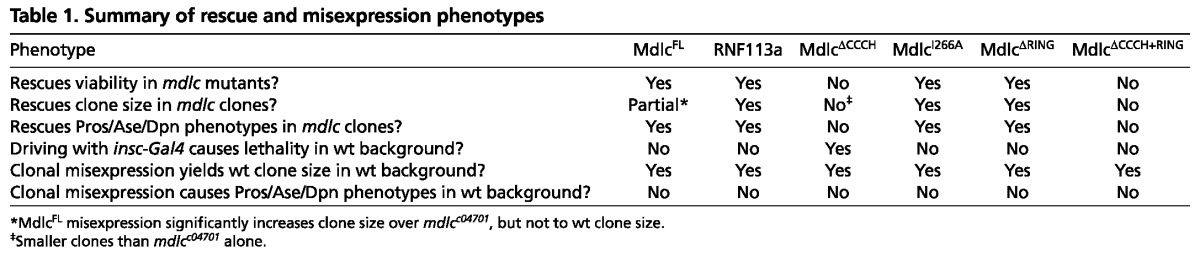
Next we tested the ability of Mdlc proteins lacking RING domain function (MdlcΔRING or MdlcI266A) to rescue mdlc mutant phenotypes. Expression of MdlcΔRING or MdlcI266A fully rescued mdlcc04701/Df(3R)ED6027 hemizygotes to viability (Table 1), as well as fully rescuing the mdlc mutant neuroblast lineage size and cell cycle phenotypes and preventing the dedifferentiation phenotypes of neurons (Fig. 6B,C; data not shown). We conclude that the RING domain, and its presumptive function in ubiquitylation, is not required for any known aspect of Mdlc neural function; the role of the conserved Mdlc RING domain therefore remains to be elucidated.
Lastly, we tested the requirement of the CCCH domain to rescue mdlc mutant phenotypes; this domain has been implicated in regulating splicing in yeast and human cells (Bessonov et al., 2008; Coltri and Oliveira, 2012; Goldfeder and Oliveira, 2008). Both MdlcΔCCCH and MdlcΔCCCH+RING failed to rescue mdlcc04701/Df(3R)ED6027 hemizygotes to viability (Table 1). In addition, both proteins failed to rescue mdlc mutant lineage size and neuronal dedifferentiation (Fig. 6B,C); in fact, expression of MdlcΔCCCH (but not MdlcΔCCCH+RING) in mdlc mutant clones resulted in even smaller clones than the mdlc mutant alone (Fig. 6B). Furthermore, misexpression of MdlcΔCCCH, but none of the other proteins, using the neuroblast driver insc-Gal4 caused pupal lethality (Table 1).
As the misexpression of MdlcΔCCCH in wt brains results in lethality, and misexpression in mdlc mutant clones causes a more severe clone size phenotype than the mdlc mutant alone, we suspected that this truncated protein might have a dominant-negative function. To investigate this, we expressed each of the Mdlc proteins in wt MARCM clones and monitored both clone size and Pros/Dpn levels in neurons. None of the constructs assayed caused cell cycle or neuronal dedifferentiation phenotypes (Table 1), suggesting that the presence of wt Mdlc is sufficient to suppress any dominant-negative effects in neuroblast lineages; it remains unclear why pan-neuroblast expression of MdlcΔCCCH causes lethality. We conclude that the Mdlc CCCH zinc-finger domain is crucial for all known neural functions of Mdlc.
RNA-seq of mdlc RNAi brains reveals pros splicing defects
We hypothesized that the most penetrant phenotype upon mdlc loss of function, which is the loss of neuronal Pros, might be due to splicing defects. We performed transcriptional and differential splicing analyses to compare wt with mdlc loss-of-function brains. We used elav-Gal4 to drive an mdlc RNAi construct in all neuroblasts and neurons of the larval brain, which strongly decreased mdlc transcript and protein levels (data not shown). We extracted mRNA from wt and mdlc RNAi brains and performed whole-transcriptome RNA-seq (see Materials and methods). Over 800 transcripts were differentially expressed in mdlc RNAi brains (supplementary material Table S1). We chose to focus here on the pros transcript. We observed a striking reduction in pros transcript levels in mdlc RNAi brains (71% reduction, P<10-26), consistent with the loss of Pros protein observed by immunostaining (Figs 1, 2, 3).
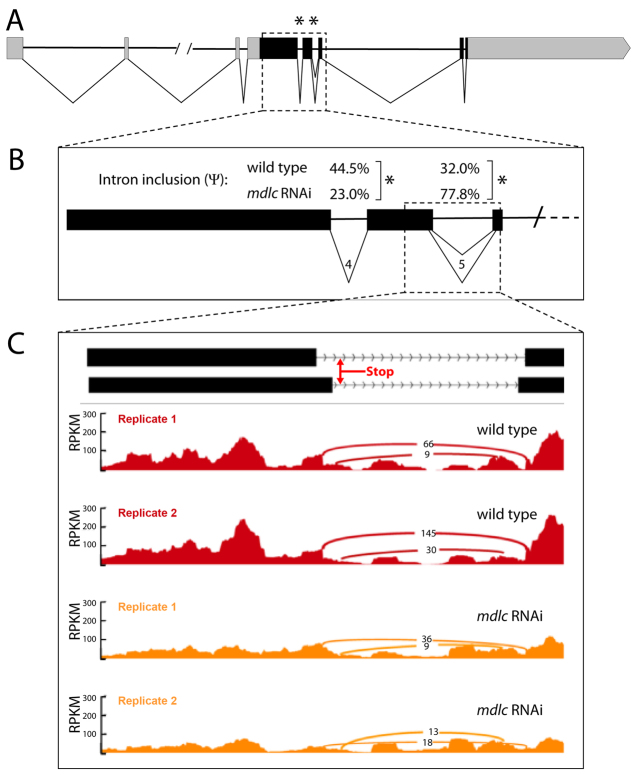
Next we assayed differential intron retention (DIR) events within the pros transcript. pros has multiple alternatively spliced isoforms with a maximum of seven introns, three of which are within the 5′ UTR in the longest isoforms (Fig. 7A). Two introns within the pros coding domain were found to be differentially spliced (asterisks in Fig. 7A,B). Interestingly, intron 4 is retained more frequently in wt than in mdlc RNAi (44.5% in wt versus 23.0% in mdlc RNAi), whereas intron 5 is retained more often in the mdlc RNAi (32.0% in wt versus 77.8% in mdlc RNAi) (Fig. 7B). The observation that the loss of Mdlc causes differential effects on introns 4 and 5 indicates that Mdlc is involved in splice-specific regulation, rather than having a general function in promoting splicing efficiency.
Fig. 7.
mdlc RNAi results in reduced levels and aberrant splicing of pros mRNA. (A) The pros locus. Black boxes indicate exons, gray boxes indicate UTRs, and horizontal lines indicate introns. Percentage spliced in (Ψ) estimates were generated by the MISO software package. Asterisks indicate introns that met our stringent criteria (see Materials and methods) for calling introns that are retained at differential levels between the wt and mdlc RNAi samples. (B) View of the region boxed in A showing pros introns 4 and 5, which exhibit differential intron retention between wt and mdlc RNAi. (C) Read density in RPKM (reads per kilobase per million mapped reads) across the pros twintron and its flanking exons. Arches represent the number of reads mapping to each junction. Retention of the twintron results in the introduction of a premature stop codon.
The intron that is retained in mdlc RNAi brains is one of a small group of Drosophila twintrons (Scamborova et al., 2004). The pros twintron is composed of two nested pairs of splice sites, the use of which is mutually exclusive. Failure to splice the twintron results in a premature stop codon (Fig. 7C), which is likely to result in the nonsense-mediated decay of the pros transcript. We utilized the sashimi_plot utility within MISO (Katz et al., 2010) to visualize the read density across the twintron in both wt and both mdlc RNAi samples (Fig. 7C). We found that reads per kilobase per million mapped reads (RPKM) across the twintron decreased more in wt than in mdlc RNAi, illustrating the more frequent retention of the twintron in the absence of Mdlc than in wt (Fig. 7C). We conclude that Mdlc is a regulator of mRNA splicing in Drosophila. Furthermore, the pros splicing defects upon mdlc loss of function reduce the number of pros transcripts, which could contribute to the loss of Pros observed in middle-aged neurons.
DISCUSSION
Function of Mdlc and Pros in larval postmitotic neurons
We have identified Mdlc as a ubiquitous nuclear protein that is required to maintain neuronal differentiation. Mdlc maintains the expression of Pros in larval postmitotic neurons and inhibits the expression of neuroblast genes, thus maintaining neuronal differentiation. Mdlc is not required for Pros expression in the oldest neurons located most distal from the neuroblast in mdlc mutant clones (not shown); there might be enough Mdlc protein or RNA present at the time of mutant clone induction to allow the first neurons born after clone induction to successfully pass through middle-age without dedifferentiation. This suggests that after a certain age, neurons do not require Mdlc to maintain neuronal differentiation. Alternatively, neurons born at early larval stages might not have the same requirement for Mdlc as neurons born at later larval stages.
Interestingly, mdlc mutant clones never show EdU incorporation in ectopic Dpn+ cells, raising the question of whether they are true neuroblasts or have a mixed neuroblast/neuronal fate. We note that loss of Mdlc results in cell cycle delay in parental neuroblasts, so it is perhaps unsurprising that the ectopic Dpn+ cells do not proliferate. Nevertheless, our data show that Mdlc is not required to suppress cell cycle entry in postmitotic neurons. Similarly, our data show that Pros is not required to suppress cell cycle entry in postmitotic neurons, as pros RNAi specifically within postmitotic neurons removes all detectable Pros protein (supplementary material Fig. S2) but does not trigger entry into the cell cycle (data not shown). This is comparable to the situation in wt embryonic neurons, which rapidly lose Pros but never re-enter the cell cycle, and contrasts with the role of Pros in GMCs, where it is required to repress neuroblast genes and promote cell cycle exit (Choksi et al., 2006). The maturation step that is taken by neurons to make them incapable of re-entering the cell cycle in the absence of Pros is not well understood.
Pros is known to bind the dpn, ase and CycE loci (Choksi et al., 2006; Southall and Brand, 2009), and is known to keep the expression of these genes low in embryos (Li and Vaessin, 2000). Does Pros directly or indirectly maintain repression of the dpn, ase and CycE neuroblast genes in larval neurons? To attempt to address this question, we used RNAi to ablate either Pros or Mdlc in postmitotic neurons, assaying for derepression of the neuroblast factor Dpn. Driving UAS-pros RNAi with either atonal-Gal4 or acj6-Gal4 specifically eliminated Pros protein in multiple clusters of postmitotic neurons, yet did not lead to the derepression of Dpn (supplementary material Fig. S2). However, RNAi knockdown of mdlc using the same Gal4 lines also did not cause Dpn derepression (data not shown). Several factors may account for this unexpected result. First, the lineages expressing atonal-Gal4 and acj6-Gal4 might not require Mdlc, consistent with our finding that some mdlcc04701 mutant clones had no Pros loss, Dpn/Ase derepression or EdU incorporation phenotypes. Alternatively, or in addition, the postmitotic Gal4 lines might have eliminated Mdlc and Pros in neurons after they passed through the susceptible middle-aged stage. However, we note that acj6-Gal4 appears to drive expression in one lineage in all ages of neurons (only excluding the neuroblast and GMCs; supplementary material Fig. S2C,D). This indicates either that this lineage does not require Pros or Mdlc to maintain Dpn repression or that Mdlc is required at the GMC level in this lineage. We lack appropriate drivers to distinguish between these possibilities. Given these caveats, we are unable to determine whether neuronal loss of Pros alone is sufficient for derepression of neuroblast genes. We think it likely that Pros does have some role in the neuronal repression of neuroblast genes, however, because we nearly always observe derepression of Dpn/Ase in neurons that have already lost Pros (Fig. 3I).
The role of Mdlc in splicing regulation
We found that the CCCH zinc-finger domain, which is implicated in RNA-binding in other proteins, is essential for Mdlc function in the nervous system and for organismal viability. The S. cerevisiae and human orthologs have roles in splicing (Bessonov et al., 2008; Chan et al., 2003; Coltri and Oliveira, 2012; Goldfeder and Oliveira, 2008; Ohi et al., 2002). In yeast, Cwc24p is reported to be a splicing efficiency factor primarily affecting primary transcripts with atypical branchpoints. For example, splicing of the transcripts snR17A and B, which encode the U3 snoRNAs, was strongly affected, resulting in defects in the processing of pre-rRNA (Coltri and Oliveira, 2012; Goldfeder and Oliveira, 2008). Our observation that loss of Mdlc causes specific splicing defects (both increased and decreased intron retention) in the pros transcript, together with our finding that RNF113A can rescue the mdlc loss-of-function phenotypes, suggest that the fly and human proteins might have a more complex role in regulating splicing than that of the yeast general splicing factor Cwc24p.
What might be the CNS splicing targets of Mdlc, in addition to pros? In mammals, alternatively spliced transcripts resulting in protein isoforms that influence stemness or differentiation are known, including within the nervous system (Lipscombe, 2005; Nelles and Yeo, 2010). Work from our laboratory has recently identified splice isoforms that are differentially regulated by the neuroblast transcription factor Wor (Lai et al., 2012). The analysis of genome-wide changes in splicing in mdlc RNAi brains is in progress but beyond the scope of this paper.
Relevance of alternative splicing of the pros twintron
The pros twintron undergoes developmentally regulated alternative splicing (Scamborova et al., 2004) to generate protein isoforms that differ by 29 amino acids at the homeodomain N-terminus (Chu-Lagraff et al., 1991). The two nested pairs of splice sites in the pros twintron are utilized mutually exclusively by two separate spliceosomes: U2 and U12 (Scamborova et al., 2004). Loss of Mdlc specifically reduces pros U12 splicing (Fig. 7), so we examined most of the other introns in Drosophila that are known to utilize the U12 spliceosome. The pros intron was the sole example of differential retention (data not shown). This indicates that Mdlc does not preferentially affect the U12 spliceosome.
A role for Mdlc as a ubiquitin ligase?
The RING-type zinc-finger proteins constitute one of the largest protein families, with over 600 members (Deshaies and Joazeiro, 2009). Many of these proteins have been shown to function as E3 ubiquitin ligases, and the presence of a RING domain is often sufficient for such an annotation. The Mdlc human ortholog RNF113A is no exception and is thought to function as an E3 ligase; consistent with this assumption, RNF113A was found to physically interact with one of the human E2 proteins, UBE2U (Li et al., 2008; van Wijk et al., 2009). Moreover, the Mdlc RING domain is very well conserved from yeast to humans, suggesting its functional importance. We were therefore surprised to find that the RING domain was completely dispensable not only for CNS function but also for organismal viability, since the ubiquitous misexpression of a version of Mdlc lacking the RING domain was able to substitute for the full-length protein.
Supplementary Material
Acknowledgments
We thank Bruce Bowerman, Judith Eisen and Tasha Joy for comments on the manuscript and Leslie Gay for assistance generating the Mdlc antibody. We thank the BDSC and VDRC for fly stocks and DSHB for antibodies.
Footnotes
Funding
This work was supported by the Howard Hughes Medical Institute, where C.Q.D. is an Investigator. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
T.D.C. and C.Q.D. conceived all experiments. T.D.C. and A.J.S. performed the RNA-seq experiments; T.D.C. performed all other experiments. C.Q.D. and T.D.C. wrote the manuscript. C.Q.D. was the principal investigator for this work.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.093781/-/DC1
References
- Albertson R., Chabu C., Sheehan A., Doe C. Q. (2004). Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J. Cell Sci. 117, 6061–6070 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar O. A., Boone J. Q., Drummond M. L., Doe C. Q. (2010). Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 5, 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B., Reichert H., Hirth F. (2006). The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133, 2639–2648 [DOI] [PubMed] [Google Scholar]
- Bello B. C., Izergina N., Caussinus E., Reichert H. (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Will C. L., Urlaub H., Lührmann R. (2008). Isolation of an active step I spliceosome and composition of its RNP core. Nature 452, 846–850 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. Q., Doe C. Q. (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68, 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G., Knoblich J. A. (2008). The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M., Jarman A. P., Jan L. Y., Jan Y. N. (1993). asense is a Drosophila neural precursor gene and is capable of initiating sense organ formation. Development 119, 1–17 [DOI] [PubMed] [Google Scholar]
- Broadus J., Fuerstenberg S., Doe C. Q. (1998). Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature 391, 792–795 [DOI] [PubMed] [Google Scholar]
- Cabernard C., Doe C. Q. (2009). Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell 17, 134–141 [DOI] [PubMed] [Google Scholar]
- Carney T. D., Miller M. R., Robinson K. J., Bayraktar O. A., Osterhout J. A., Doe C. Q. (2012). Functional genomics identifies neural stem cell sub-type expression profiles and genes regulating neuroblast homeostasis. Dev. Biol. 361, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C. (2003). The Prp19p-associated complex in spliceosome activation. Science 302, 279–282 [DOI] [PubMed] [Google Scholar]
- Choksi S. P., Southall T. D., Bossing T., Edoff K., de Wit E., Fischer B. E., van Steensel B., Micklem G., Brand A. H. (2006). Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell 11, 775–789 [DOI] [PubMed] [Google Scholar]
- Chu-Lagraff Q., Wright D. M., McNeil L. K., Doe C. Q. (1991). The prospero gene encodes a divergent homeodomain protein that controls neuronal identity in Drosophila. Development 2 Suppl. 2, 79–85 [PubMed] [Google Scholar]
- Coltri P. P., Oliveira C. C. (2012). Cwc24p is a general Saccharomyces cerevisiae splicing factor required for the stable U2 snRNP binding to primary transcripts. PLoS ONE 7, e45678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Joazeiro C. A. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 [DOI] [PubMed] [Google Scholar]
- Doe C. Q. (2008). Neural stem cells: balancing self-renewal with differentiation. Development 135, 1575–1587 [DOI] [PubMed] [Google Scholar]
- Goldfeder M. B., Oliveira C. C. (2008). Cwc24p, a novel Saccharomyces cerevisiae nuclear ring finger protein, affects pre-snoRNA U3 splicing. J. Biol. Chem. 283, 2644–2653 [DOI] [PubMed] [Google Scholar]
- Izergina N., Balmer J., Bello B., Reichert H. (2009). Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 4, 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E. T., Airoldi E. M., Burge C. B. (2010). Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A. (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A., Jan L. Y., Jan Y. N. (1995). Asymmetric segregation of Numb and Prospero during cell division. Nature 377, 624–627 [DOI] [PubMed] [Google Scholar]
- Lai S. L., Miller M. R., Robinson K. J., Doe C. Q. (2012). The Snail family member Worniu is continuously required in neuroblasts to prevent Elav-induced premature differentiation. Dev. Cell 23, 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254 [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Wilkinson B. D., Siegrist S. E., Wharton R. P., Doe C. Q. (2006). Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441–449 [DOI] [PubMed] [Google Scholar]
- Li L., Vaessin H. (2000). Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 14, 147–151 [PMC free article] [PubMed] [Google Scholar]
- Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008). Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE 3, e1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D. (2005). Neuronal proteins custom designed by alternative splicing. Curr. Opin. Neurobiol. 15, 358–363 [DOI] [PubMed] [Google Scholar]
- Miller M. R., Robinson K. J., Cleary M. D., Doe C. Q. (2009). TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat. Methods 6, 439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder K. W., Inagaki A., Cameroni E., Mousson F., Winkler G. S., De Virgilio C., Collart M. A., Timmers H. T. (2007). Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics 176, 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles D. A., Yeo G. W. (2010). Alternative splicing in stem cell self-renewal and differentiation. Adv. Exp. Med. Biol. 695, 92–104 [DOI] [PubMed] [Google Scholar]
- Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G., Knoblich J. A. (2011). Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8, 580–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M. D., Link A. J., Ren L., Jennings J. L., McDonald W. H., Gould K. L. (2002). Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22, 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scamborova P., Wong A., Steitz J. A. (2004). An intronic enhancer regulates splicing of the twintron of Drosophila melanogaster prospero pre-mRNA by two different spliceosomes. Mol. Cell. Biol. 24, 1855–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall T. D., Brand A. H. (2009). Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J. 28, 3799–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana E. P., Doe C. Q. (1995). The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development 121, 3187–3195 [DOI] [PubMed] [Google Scholar]
- Srinivasan S., Peng C. Y., Nair S., Skeath J. B., Spana E. P., Doe C. Q. (1998). Biochemical analysis of ++Prospero protein during asymmetric cell division: cortical Prospero is highly phosphorylated relative to nuclear Prospero. Dev. Biol. 204, 478–487 [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., Singh C. M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H. L., et al. (2004). A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283–287 [DOI] [PubMed] [Google Scholar]
- van Wijk S. J., de Vries S. J., Kemmeren P., Huang A., Boelens R., Bonvin A. M., Timmers H. T. (2009). A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol. Syst. Biol. 5, 295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M., Golden K. L., Lee C. Y. (2010). dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Dev. Cell 18, 126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. D., Nacu S. (2010). Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.