Abstract
We have constructed IgG1-Fc scaffolds with increased thermal stability by directed evolution and yeast surface display. As a basis a new selection strategy that allowed the application of yeast surface display for screening of stabilizing mutations in proteins of already high intrinsic thermal stability and Tm-values up to 85 °C was developed. Besides library construction by error prone PCR, strong heat stress at 79 °C for 10 min and screening for well-folded proteins by FACS, sorting rounds had to include an efficient plasmid DNA isolation step for amplification and further transfection. We describe the successful application of this experimental setup for selection of 17 single, double and triple IgG1-Fc variants of increased thermal stability after four selection rounds. The recombinantly produced homodimeric proteins showed a wild-type-like elution profile in size exclusion chromatography as well as content of secondary structures. Moreover, the kinetics of binding of FcRn, CD16a and Protein A to the engineered Fc-molecules was very similar to the wild-type protein. These data clearly demonstrate the importance and efficacy of the presented strategy for selection of stabilizing mutations in proteins of high intrinsic stability within reasonable time.
Abbreviations: IgG1, immunoglobulin G class 1; Fcab, antigen binding Fc protein; Fc-wt, recombinant wild-type Fc protein; scTCR, single-chain T-cell receptor; MHC, major histocompatibility complex; CDC, complement dependent cytotoxicity; ADCC, antibody dependent cell-mediated cytotoxicity; FcRn, neonatal Fc receptor; CD64, cellular leucocyte Fc receptor FcγRI; DSC, differential scanning calorimetry; ECD, electronic circular dichroism; SEC, size exclusion chromatography; SPR, surface plasmon resonance; RU, response unit; FACS, fluorescent activated cell sorting; PBS, phosphate-buffered saline
Keywords: Stability engineering, Antibody engineering, Crystallizable domain, Yeast display, Directed evolution
Highlights
► Construction of IgG1-Fc scaffolds with increased thermal. ► Selection from a yeast displayed protein-library after heat incubation. ► New protocol allowing stabilization of proteins of already high intrinsic stability.
1. Introduction
Yeast surface display has been used for a variety of applications, including the selection of proteins with improved affinity toward soluble antigens, identification of epitopes and screening of protein–protein interactions [1]. Furthermore, this technology has been utilized for the analysis of the thermal stability of proteins displayed on the yeast cells. In a previous study various single-chain Fv (scFv)- and single-chain T-cell receptor (scTCR)-mutants have been expressed on the surface of Saccharomyces cerevisiae and a strong correlation between the temperatures of irreversible denaturation of the surface-expressed proteins and of the corresponding soluble forms has been observed [2]. Moreover, Shusta and colleagues were able to stabilize an scTCR by directed evolution using yeast surface display [3]. However, the limitation of this strategy was that only unstable proteins with low denaturation temperatures, such as scTCRs, could be stabilized so far by using this method.
Immunoglobulin G class 1 (IgG1) is used as a therapeutic molecule against a variety of diseases, such as autoimmune disorders and cancer. The most important properties of IgG1 are its ability to bind virtually any desired antigen with high affinity and specificity, its long in vivo half life mediated by the interaction with the neonatal Fc receptor (FcRn) and the capability to trigger immunological clearance mechanisms. Those mechanisms include antibody dependent cell-mediated cytotoxicity (ADCC) triggered by the interaction with Fcγ receptor IIIa (FcγRIIIa — CD16a) and complement dependent cytotoxicity (CDC) initiated by binding to C1q [4]. As all those ligands (Fcγ receptors, FcRn and C1q) bind to the crystallizable fragment (Fc) of IgG1, the Fc part is responsible for all functions of IgG1 except for antigen binding. This shortcoming was recently redressed by Wozniak-Knopp and colleagues [5], who selected Her2/neu binding Fc proteins from a library containing randomized C-terminal structural loops in the CH3-domains. An affinity matured variant was shown to trigger ADCC in vitro and the in vivo half life in mice was comparable to wild-type Fc (Fc-wt), demonstrating the possibility to generate antigen binding Fc proteins (Fcab, antigen binding Fc) with all antibody properties at only one third of the size (~ 50 kDa) of a whole IgG1 molecule, which could be an advantage regarding tissue penetration. In another work the integrin-binding motive “GCRGDCL” was incorporated into the C-terminal structural loops of the CH3-domains of IgG1-Fc [6]. Integrin binding Fcabs could be expressed and purified and all variants showed wild-type like secondary and tertiary structures and still bound to CD16, FcRn and Protein A. However, both works demonstrated that engineering of C-terminal loops at the CH3-domains might decrease the thermal stability of the Fc scaffold.
The aim of the present work was the selection of stabilizing mutations within the framework of IgG1-Fc in order to compensate for the reduced stability of Fcabs comprising mutated structural loops. An IgG1-Fc-library was constructed by error prone PCR, displayed on yeast and exposed to a heat shock. Finally, the Fc-library was probed for binding to structurally specific ligands, followed by flow sorting and after 4 consecutive sorting rounds enriched Fc variants were sub-cloned and expressed solubly in Pichia pastoris. 13 out of 14 mutations stabilized the CH3-domain when measured by differential scanning calorimetry (DSC). Moreover, they showed wild-type like size exclusion chromatography (SEC) profiles and electronic circular dichroism (ECD) spectra and the kinetics of binding to CD16a, FcRn and Protein A were also similar to the wild-type protein (Fc-wt).
2. Materials and methods
2.1. Library construction and characterization
The human IgG1-Fc-gene (comprising the Hinge-region, CH2- and CH3-domains) was codon-optimized for expression in yeast and cloned into pYD1 (Invitrogen, Carlsbad, CA, USA) for Saccharomyces cerevisiae surface expression. The Fc-gene was cloned C-terminally of Aga2 by using the BamHI and NotI restriction sites, resulting in the following construct: Aga2-GlySerLinker-Xpress-Fc, as described previously [5]. Directly after the CH3-domain a stop codon was introduced in order to avoid expression of any C-terminal tags that are present on the vector. This pYD1-Fc plasmid was used as the template for error prone PCR (GeneMorph II Random Mutagenesis Kit, Stratagene, La Jolla, CA). By using different amounts of template DNA, 2 libraries differing in their mutation rates were constructed. S. cerevisiae EBY100 (Invitrogen, Carlsbad, CA, USA) was transformed with the gel-purified library inserts together with BamHI/NotI-digested pYD1 using the lithium acetate method [7]. The library inserts comprised homologous regions to the BamHI/NotI-digested pYD1 backbone on both sides in order to facilitate gap repair driven homologous recombination in S. cerevisiae. After growing the S. cerevisiae-libraries in SD-CAA-medium [20 g/l glucose, 0.1 M KH2PO4/K2HPO4, pH 6, 10 g/l (NH4)2SO4, 0.1 g/l l-leucine (all from Sigma, St. Louis, MO), 3.4 g/l yeast nitrogen base, 10 g/l bacto casamino acids (both from Difco, BD, Franklin Lakes, NJ)] at 28 °C for 48 h, pYD1 plasmid DNA was isolated by using the Zymoprep Yeast Plasmid Miniprep Kit II (Zymo Research, Orange, CA) and transformed into Escherichia coli Top 10 for sequencing. Furthermore, cell surface expression was induced and analyzed using flow cytometry as described below.
2.2. Induction of surface expression, heat shock, staining, FACS and DNA isolation
S. cerevisiae-libraries were cultured overnight in SD-CAA-medium. Subsequently the cultures were passaged to an OD600 of 1 and after shaking them at 28 °C for 4 h the cells were centrifuged and resuspended in SGR-CAA (same as SD-CAA, but 20 g/l galactose and 10 g/l raffinose instead of glucose, both from Sigma, St. Louis, MO) to an OD600 of 1. Induction was done at 20 °C for 18–20 h. The cells were centrifuged and resuspended in PBS/BSA [0.2 g/l KCl, 0.2 g/l KH2PO4, 8 g/l NaCl, 1.15 g/l Na2HPO4 anhydrous + 20 g/l bovine serum albumin (Sigma, St. Louis, MO)] to an OD600 of 3. The suspension was aliquoted into microfuge tubes and after storage on ice for 10 min they were incubated at 79 °C for 10 min in a thermomixer (Eppendorf, Hamburg, Germany) while shaking at 300 rpm. The tubes were put on ice again for at least 5 min. After centrifugation the cells were stained with 5 μg/ml anti-Xpress antibody (Invitrogen, Carlsbad, CA, USA) that had been conjugated to allophycocyanin (APC) using the LYNX Rapid APC Antibody Conjugation Kit (AbD Serotec, Kidlington, UK) and with either 2 μg/ml fluorescein isothiocyanate isomer 1-labeled anti-human IgG CH2-domain antibody (anti-CH2-FITC, clone MK 1 A6, AbD Serotec, Kidlington, UK) or 1 μg/ml His-tagged CD64 (R&D Systems, Abingdon, UK). In the case of CD64-staining, the cells were washed and incubated with 1 μg/ml Alexa Fluor 488-labeled anti-His antibody (QIAGEN, Venlo, Netherlands). All stainings were done in PBS/BSA at 4 °C for 30 min, shaking. After a further washing step the cells were resuspended in PBS/BSA and sorted on a FACS Aria cell sorter. If the cells were not sorted but only analyzed a FACSCanto II or a FACS Calibur was utilized (all FACS machines from BD, Franklin Lakes, NJ). The sorted cells were subsequently centrifuged. If the number of sorted cells was below 5 × 105, non-transformed EBY100 cells (not containing any pYD1 plasmid) were added before centrifugation in order to facilitate the formation of a cell pellet.
After washing the cells with PBS/BSA, the pYD1-Fc-library DNA was isolated from S. cerevisiae by using the Zymoprep Yeast Plasmid Miniprep Kit II with some modifications of the manufacturer's protocol. In detail, the cell suspension was incubated at 37 °C for 60 min with twice the recommended amount of zymolyase. Centrifugation after addition of the neutralization buffer was done for 10 min, followed by an additional centrifugation step of the supernatant for 5 min and finally, two elution steps with 10 μl H2O each were performed. 10% of the isolated DNA was subsequently mixed with 1 pg pUC19 plasmids and transformed into chemically competent E. coli Top 10 (both from Invitrogen, Carlsbad, CA, USA), which were plated onto LB Agar containing 100 μg/ml ampicillin and X-gal (Sigma, St. Louis, MO; 1 mg X-gal per 90 mm Petri dish). As, in contrast to pYD1, pUC19 is lac+, the ratio of pYD1/pUC19 plasmids, and thereby the number of pYD1 plasmids in the DNA preparation, could be determined by blue/white screening.
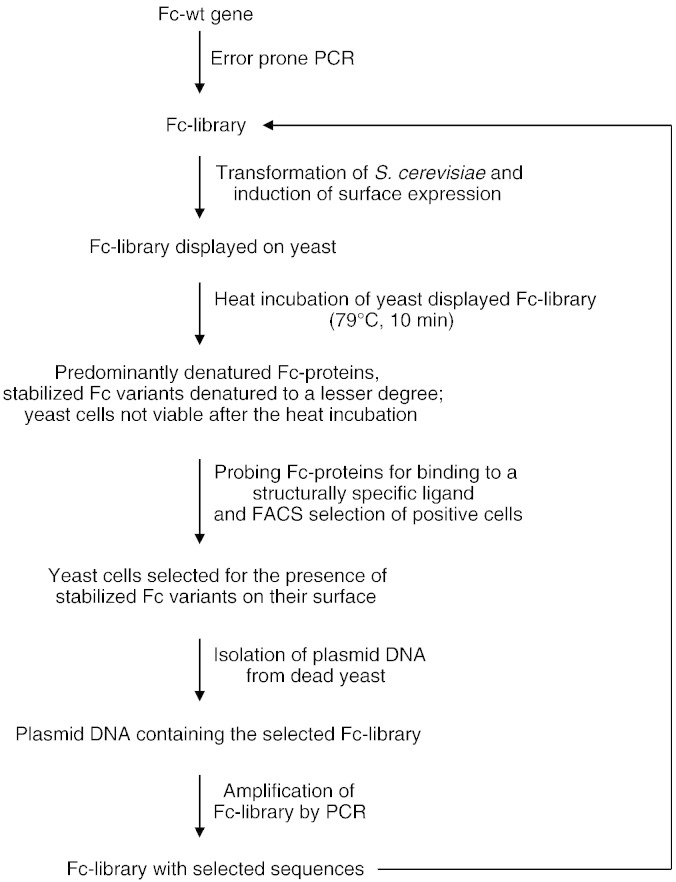
The isolated pYD1-Fc-library was used as the template for a conventional PCR with the same primers that had been used for error prone PCR before. Subsequently, S. cerevisiae was transformed with the PCR product and BamHI/NotI-digested pYD1 as described above, yielding a new yeast library. This cycle of yeast transformation, induction of surface expression, heat shock, FACS and DNA isolation was done 4 times (Fig. 2). After each cycle pYD1-Fc-library-DNA was isolated from yeast as described above, transformed into E. coli and 48 clones per library were sequenced, yielding 44–48 evaluable sequences.
Fig. 2.
Schematic overview of one round of sorting. The Fc-wt gene was used as the template for an error prone PCR, resulting in an Fc-library with point mutations randomly distributed over the entire gene. This Fc-library, together with the linearized vector pYD1, was cloned into the yeast S. cerevisiae by homologous recombination. After induction of surface expression of the Fc-library the yeast suspension was incubated for 10 min at 79 °C, followed by cooling on ice and labeling with structurally specific Fc-ligands. Positive cells were selected by FACS and since the cells did not survive the heat shock applied before, plasmid DNA had to be isolated and amplified by PCR. With this Fc-library the next sorting round was performed, starting again with the yeast transformation step.
2.3. Sub-cloning of enriched mutants for soluble expression in P. pastoris, protein purification and biochemical/biophysical characterization
Enriched Fc-mutants were sub-cloned from pYD1 into pPICZαA (Invitrogen, Carlsbad, CA, USA), a vector suitable for soluble expression in P. pastoris. pYD1-Fc vectors containing the enriched mutations in the Fc-gene were used as templates for PCRs with primers flanking the Fc-gene and containing EcoRI/NotI sites. The resulting PCR-product was subsequently EcoRI/NotI cloned into pPICZαA. P. pastoris X33 was transformed with SacI-linearized pPICZαA-Fc vectors and Fc-proteins were expressed by methanol-induction as described previously [6].
Yeast culture supernatants were supplemented with 1 M phosphate buffer, pH 7, to achieve a concentration of 100 mM. After two centrifugation steps (1500 g, 4 °C, 15 min and 12,000 g, 4 °C, 25 min), filtration (0.45 μm Durapore membrane filter, Millipore, Billerica, MA) and degassing, supernatant was loaded onto a 5 ml HiTrap Protein A HP column by using an ÄKTA purifier (GE Healthcare, Waukesha, WI). The column was washed with 100 mM phosphate buffer, pH 7, followed by elution of Fc-proteins with 100 mM glycine, pH 3.5. Immediately after elution 12 μl of 2 M Tris pH 12 buffer was added to each 2 ml fraction in order to neutralize the acidic pH from the glycine solution. Highly concentrated fractions were pooled and dialyzed against PBS at 4 °C overnight (SnakeSkin Dialysis Tubing, 10,000 MWCO, Thermo Fisher Scientific, Waltham, MA). Finally, the column was regenerated with 100 mM glycine, pH 2.5, and washed with 100 mM phosphate buffer, pH 7.
Purified and dialyzed Fc variants were analyzed by using differential scanning calorimetry (DSC). Samples were diluted to a protein concentration of 5 μM, degassed using a ThermoVac instrument and subsequent measurements were performed on a VP-DSC Capillary Cell MicroCalorimeter (MicroCal, North-hampton, MA). Temperature range and heating rate were from 20 °C to 110 °C and 1 °C/min, respectively. Buffer baselines were subtracted and data were normalized for protein concentration and fitted with a non-2-state thermal unfolding model using the software supplied by MicroCal (Origin 7).
Furthermore, the structural integrity of Fc proteins was analyzed routinely by size exclusion chromatography (SEC) and electronic circular dichroism (ECD). For SEC (Shimadzu Prominence LC-20) a TSK-Gel G3000WXL column (30 cm × 7.8 mm) at a flow rate of 1 ml/min was used. Circular dichroism measurements were done using Chirascan from Applied Photophysics (Leatherhead, UK). Protein samples were adjusted to an absorbance at 280 nm of 0.80 and measured in a 2 mm cuvette placed in a thermostatic cell holder. In the far-UV region (260–195 nm) instrument parameters were set as follows. Spectral bandwidth: 5 nm; step size: 1 nm; scan time: 120 s. Each spectrum was automatically baseline corrected to remove birefringence of the cell.
FcRn−, Protein A and CD16a-binding to recombinant IgG1-Fc proteins were analyzed by surface plasmon resonance (SPR) spectroscopy using a Biacore 3000 instrument (GE Healthcare, Waukesha, WI). FcRn was expressed in High Five cells and purified by using a 10 ml IgG Sepharose 6 Fast Flow column (GE Healthcare). For binding studies the running buffer was PBS, pH 6.0, and interactions were measured at a flow of 20 μl/min. A fixed amount of FcRn coated on a chip was incubated with 10 μg/ml IgG1-Fc in PBS, pH 6.0, for 2 min. Additionally, sample protein was injected to uncoated reference cell. After measuring the interaction the dissociation rate with PBS, pH 6.0, was measured. Finally, rapid dissociation was followed with PBS, pH 7.4, for 1 min at a flow of 30 μl/min.
For CD16-binding studies Protein A (Zymed, San Francisco, CA) was immobilized on a CM5 chip. The running buffer was HBS-EP (0.01 M HEPES, pH 7.4; 0.15 M NaCl; 3 mM EDTA; 0.005% surfactant P20; GE Healthcare, Waukesha, WI) and the flow rate was set to 20 μl/min. IgG1-Fc samples (10 μg/ml) were injected to Protein A and uncoated reference cell for 3 min. Subsequently, CD16a (25 mg/ml) was injected onto the CH5 chip for 2 min, followed by injection of HBS-EP, pH 7.4, only. Regeneration was achieved by injection of 5 mM glycine, pH 3.0, containing 1.5 M MgCl2, for 1 min at a flow rate of 30 μl/min. CD16a (produced by transient transfection of HEK293 cells and purified via His-tag) was a gift kindly provided by f-star.
3. Results
3.1. Preliminary experiments
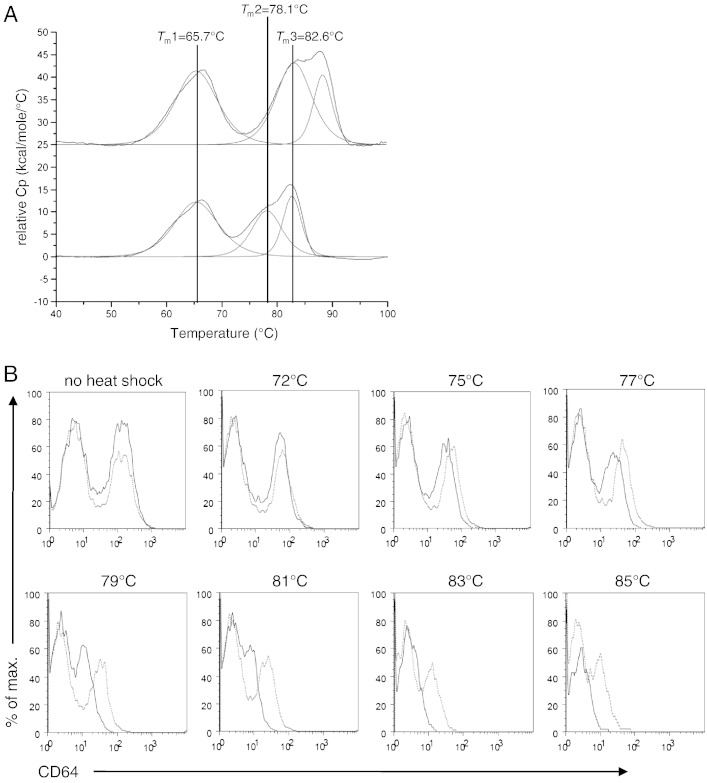
At first we investigated whether it is possible to separate a stabilized mutant from wild-type human IgG1-Fc (Fc-wt) when displayed on the yeast S. cerevisiae. For this purpose we compared Fc-wt and Fc-K370L, the latter being a rationally designed stabilized mutant of IgG1-Fc (unpublished data). Fig. 1A comparing the thermograms of Fc-wt and the variant K370L clearly demonstrates that the CH3-domain in the mutant protein has an increased thermal stability. In detail, as published recently [6], Fc-wt recombinantly produced in P. pastoris, shows a prominent heat absorption peak of the CH2-domain at 65.7 °C (Tm1, shoulder at 62 °C) and two transitions at 78.1 °C (Tm2) and 82.6 °C (Tm3), corresponding to heat absorption maxima of the CH3-domain. Upon exchange of lysine-370 by leucine both Tm-values of the CH3-domain of this Fc-variant have been elevated by 5 °C compared to Fc-wt (Fig. 1A).
Fig. 1.
(A) DSC thermograms of Fc-wt (bottom) and the K370L variant (top). For comparison, the three thermal transitions of Fc-wt are marked with a black line. For the K370L variant Tm2 and Tm3, representing the thermal denaturation of the CH3-domain, are shifted to higher temperatures, demonstrating the stabilizing effect of this mutation on the CH3-domain. (B) Comparison of yeast displayed Fc-wt and the K370L variant. Yeast cells displaying Fc-wt (black line) or the K370L variant (gray line) were incubated for 10 min at the temperatures indicated. After cooling the surface displayed Fc-proteins were probed for their binding to CD64 and analyzed by flow cytometry.
Both Fc-wt and Fc-K370L were expressed as Aga2p fusion proteins on the surface of S. cerevisiae. After induction at 20 °C the cells were washed with PBS and either incubated on ice or at temperatures ranging from 42 to 85 °C for 10, 30 or 90 min, followed by flow-cytometric analysis of folded Fc-protein with CD64 (Fcγ receptor I) or an anti-human IgG CH2-domain antibody (anti-CH2). Both ligands only bind structurally intact Fc-proteins. If no heat shock was applied or if the temperature was too low, no difference was observed between Fc-wt and Fc-K370L. However, after incubation at higher temperatures Fc-K370L showed stronger binding to CD64 (Fig. 1B) and anti-CH2 (data not shown). This first experiment demonstrated that it is possible to sort out stabilized Fc-mutants from a randomized Fc-library displayed on yeast by screening for maintained binding properties to structurally specific ligands after a heat shock at temperatures never probed so far for selection. Although the yeast cells do not survive temperatures > 50 °C even at short incubation time (Supplemental Fig. 1), this preliminary experiment unequivocally demonstrated that the displayed proteins are still structurally intact.
Since the yeast cells did not survive the applied temperatures necessary for separation of stabilized variants from Fc-wt, the plasmid DNA had to be isolated from dead yeast cells after flow-cytometric sorting, amplified by PCR and transformed again into S. cerevisiae. In order to avoid loss of library diversity it had to be ensured that enough plasmids were used for proceeding steps and for that reason we developed a method for the quantification of pYD1 in the plasmid preparation obtained after FACS. A known amount of pUC19 (lac+) was mixed with part of the pYD1 plasmid (lac−) preparation from the dead yeast cells. Both pUC19 (lac+) and pYD1 (lac−) were shown to have the same efficacy of transformation and the mixture was transformed into E. coli, followed by selection on LB Agar containing ampicillin and X-gal. As the applied number of pUC19 was known, the concentration of pYD1 could be calculated from the ratio of white (clones carrying pYD1) and blue (pUC19) colonies. Isolation of pYD1 from S. cerevisiae after incubation at high temperatures was very efficient, yielding approximately 0.5–1 plasmids per cell (data not shown).
The limiting factor of our experimental approach was the stability of the cells. Although the cells already died at much lower temperatures, they stayed morphologically intact up to temperatures of ~ 72 °C when incubated for 90 min, ~ 76 °C (30 min) and ~ 85 °C (10 min). Above these temperatures the cells bursted, resulting in gelatinization of the cell suspension.
3.2. Selection of thermally stabilized Fc-mutants
Based on this promising experimental groundwork we hypothesized that we can screen for stabilized IgG1-Fc variants by random mutagenesis of the Fc-gene, followed by heat incubation of the Fc-library displayed on the yeast surface, flow-cytometric sorting of yeast cells displaying folded Fc-protein and plasmid DNA isolation from the sorted cells. Fig. 2 summarizes this experimental approach schematically.
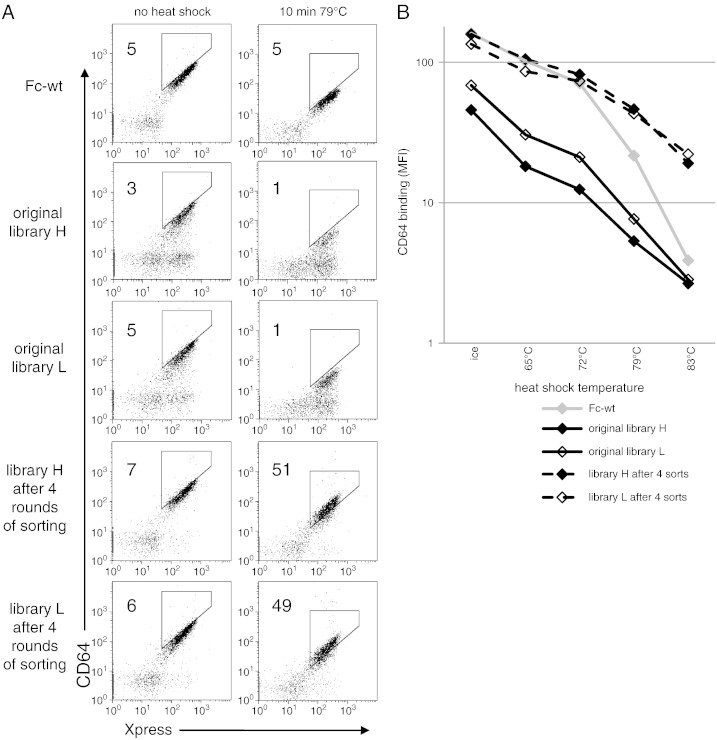
The Fc-wt-gene was randomized by error prone PCR. Two libraries differing in mutation rates were generated (termed library H and L for high and low mutation rate, respectively) and 48 clones from each library were sequenced. Analysis of the sequences showed, that the average number of DNA-point mutations per Fc-gene in the libraries H and L was 2.5 and 1.5, respectively. The mutations were distributed over the entire gene and all 12 types of mutations were present, although there was some bias toward certain mutations (C → T and G → A) (Supplemental Fig. 2). Both Fc-libraries, displayed as Aga2p fusion proteins on S. cerevisiae, were characterized by flow-cytometric analysis, demonstrating that the majority of the cells in the library bound CD64 (Fig. 3A) and anti-CH2 (Supplemental Fig. 3). However, in contrast to yeast displaying Fc-wt, a population of cells in both libraries lost these binding properties (positive for the expression marker Xpress, but CD64- and anti-CH2-negative). This is also reflected by the mean fluorescence intensity with library H being even less positive than library L due to the higher mutation rate (Fig. 3B).
Fig. 3.
Comparison of CD64 binding of surface displayed Fc-wt, original Fc-libraries and Fc-libraries after 4 rounds of sorting. (A) After induction of surface expression a heat shock was either applied (right column) or not (left column), followed by staining with CD64 and an antibody against the Xpress epitope which is used for expression normalization. For comparison, gate positions were set identically in the left and in the right column, respectively. Numbers in the dot plots indicate the percentage of cells in the gate. (B) Yeast displayed Fc-wt or Fc-libraries were incubated at the indicated temperatures for 10 min and subsequently analyzed for their binding to CD64 by flow cytometry. Mean fluorescence intensities (MFI) for CD64 binding are depicted. In A and B one representative experiment out of two is shown.
Both libraries were screened in parallel for stabilized Fc-mutants. After induction of surface expression of Aga2p-Fc fusion proteins on the yeast cells a heat shock was applied. 79 °C for 10 min was chosen as a compromise between high temperatures for good separation of Fc-wt and stabilized mutants (Fig. 1) and maintenance of the integrity of the yeast cells in order to avoid gelatinization of the cell suspension. After cooling, yeast cells were probed for binding to either CD64 or anti-CH2 and positive cells were selected by FACS. We alternated between usage of CD64 (rounds 1 and 3) and anti-CH2 (rounds 2 and 4) in order to really select for improved thermal stability instead of increased affinity toward a certain ligand. After flow sorting plasmid DNA was isolated from the dead yeast cells, followed by PCR amplification (Fig. 2). Finally, yeast was transformed again with the Fc-library. After 4 rounds of sorting the binding intensity to CD64 (Fig. 3A) and anti-CH2 (Supplemental Fig. 3) was enhanced compared to Fc-wt. However, this observation was only made after a heat shock. Furthermore, it was shown, that the enhancement in CD64-binding intensity of the Fc-libraries compared to Fc-wt increased with the temperature of the heat shock (Fig. 3B), indicating, that the effect was caused by an increase in the thermal stability of the Fc-library rather than of the affinity to CD64 or anti-CH2.
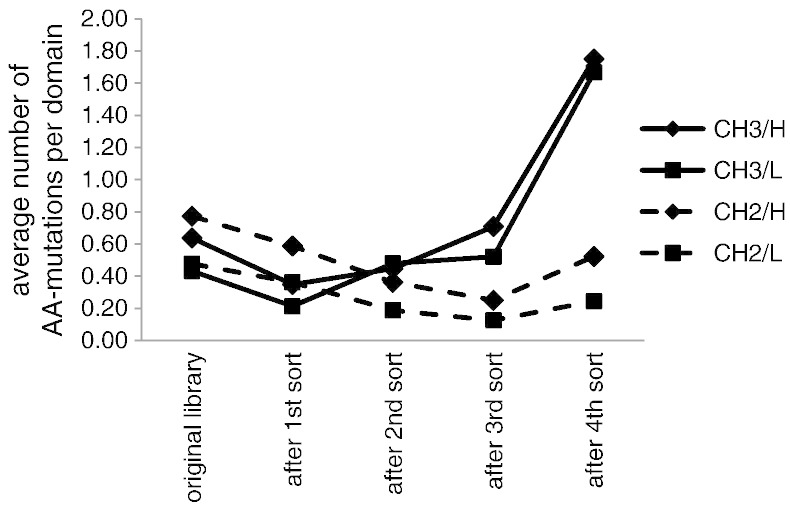
In order to investigate the changes in the mutation rates with advancing sorting rounds, 48 clones from each library (H and L) were sequenced after each cycle and the average numbers of amino acid (AA) mutations in the CH2- and CH3-domains were calculated (Fig. 4). After one round of sorting the mutation rate in both, the CH2- and CH3-domains, decreased, followed by an increase during later rounds in the case of the CH3- domain, whereas the elimination of mutations in the CH2-domain continued in the 2nd and 3rd cycle. Only in the last round the mutation rate in the CH2-domain was slightly elevated. This clearly demonstrates that mainly mutations in the CH3-, but not in the CH2-domain, were selected for.
Fig. 4.
Changes in the mutation rates in the CH2- and CH3-domains during consecutive sorting rounds. After each round of sorting plasmid DNA was isolated from the yeast libraries and 48 clones of each library were sequenced. The average numbers of amino acid (AA) mutations per domain (CH2- and CH3-domains of library H and L) are shown.
3.3. Sub-cloning of enriched Fc-variants into P. pastoris for soluble expression and biochemical/biophysical characterization
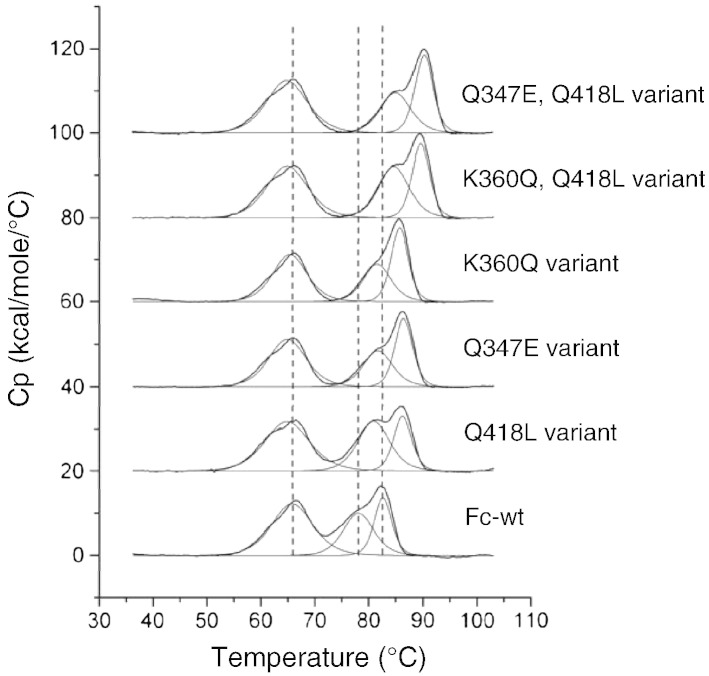
After 4 consecutive sorting rounds, 48 clones from each library were sequenced. The most frequent mutations (Table 1) and some combinations of them were sub-cloned into P. pastoris for soluble expression and probed by DSC. Representative thermograms of the three most stable single mutants and combinations of them are shown in Fig. 5, the corresponding Tm values of all variants are listed in Table 1. All mutants were stabilized in their CH3-domains compared to Fc-wt, demonstrated by higher Tm2 and Tm3 values. The only exception was the variant V263A, which is also the only enriched mutation located in the CH2-domain. In variants containing 2 or 3 stabilizing mutations the effects observed in the single mutants were additive.
Table 1.
Differential scanning calorimetric data of Fc-wt and all Fc variants that were expressed solubly in P. pastoris. Depicted are the temperatures of unfolding (Tm) and the differences in the Tm values compared to Fc-wt (ΔTm). The first transition represents the unfolding process of the CH2 and the second and third the ones of the CH3-domain. Averages and standard deviations from three experiments are shown.
| Variant | Tm1 (°C) | Tm2 (°C) | Tm3 (°C) | ΔTm1 | ΔTm2 | ΔTm3 |
|---|---|---|---|---|---|---|
| Fc-wt | 65.7 ± 0.1 | 78.1 ± 0.1 | 82.6 ± 0.0 | – | – | – |
| V263A | 63.1 ± 0.1 | 77.5 ± 0.0 | 82.2 ± 0.0 | − 2.6 | − 0.6 | − 0.4 |
| Q347E | 64.9 ± 0.1 | 81.6 ± 0.1 | 86.4 ± 0.0 | − 0.8 | 3.5 | 3.8 |
| S354P | 65.1 ± 0.2 | 80.0 ± 0.1 | 85.8 ± 0.1 | − 0.6 | 1.9 | 3.2 |
| K360E | 64.9 ± 0.2 | 80.4 ± 0.1 | 85.1 ± 0.1 | − 0.8 | 2.3 | 2.5 |
| K360Q | 65.4 ± 0.0 | 81.6 ± 0.1 | 85.7 ± 0.0 | − 0.3 | 3.4 | 3.1 |
| K392R | 64.7 ± 0.1 | 79.1 ± 0.1 | 84.4 ± 0.1 | − 1.0 | 0.9 | 1.8 |
| S400F | 65.3 ± 0.1 | 80.1 ± 0.2 | 85.4 ± 0.1 | − 0.4 | 2.0 | 2.8 |
| Q418L | 65.0 ± 0.1 | 81.0 ± 0.1 | 86.3 ± 0.1 | − 0.7 | 2.9 | 3.7 |
| N421D | 65.4 ± 0.4 | 79.5 ± 0.1 | 84.2 ± 0.0 | − 0.3 | 1.4 | 1.6 |
| S424T | 65.2 ± 0.4 | 81.0 ± 0.1 | 85.9 ± 0.1 | − 0.5 | 2.9 | 3.3 |
| T437I | 64.4 ± 0.1 | 80.2 ± 0.1 | 84.3 ± 0.0 | − 1.3 | 2.0 | 1.7 |
| Q438K | 64.0 ± 0.1 | 80.4 ± 0.1 | 85.2 ± 0.1 | − 1.7 | 2.3 | 2.6 |
| Q438R | 64.1 ± 0.1 | 79.5 ± 0.1 | 83.9 ± 0.0 | − 1.6 | 1.4 | 1.3 |
| Q347E, Q418L | 64.9 ± 0.1 | 85.0 ± 0.0 | 90.2 ± 0.0 | − 0.8 | 6.9 | 7.6 |
| K360Q, Q418L | 64.9 ± 0.1 | 84.6 ± 0.1 | 89.6 ± 0.1 | − 0.8 | 6.5 | 7.0 |
| K360Q Q438R | 63.7 ± 0.1 | 82.8 ± 0.1 | 87.2 ± 0.0 | − 2.0 | 4.7 | 4.6 |
| K360Q, Q418L, Q438R | 63.8 ± 0.0 | 86.5 ± 0.1 | 91.0 ± 0.1 | − 1.9 | 8.4 | 8.4 |
| K360Q, G446C | 64.7 ± 0.3 | 83.2 ± 0.1 | 90.4 ± 0.1 | − 1.0 | 5.0 | 7.8 |
Fig. 5.
DSC analysis of Fc-wt and selected Fc variants. Fc-wt and Fc variants containing mutations that were enriched in the library after 4 sorting rounds were expressed solubly in P. pastoris and analyzed by DSC. Protein samples were set to a concentration of 5 μM in PBS and analyzed in a temperature range from 20 to 110 °C. For comparison, the three thermal transitions of Fc-wt are marked with dashed lines. One experiment out of three is shown.
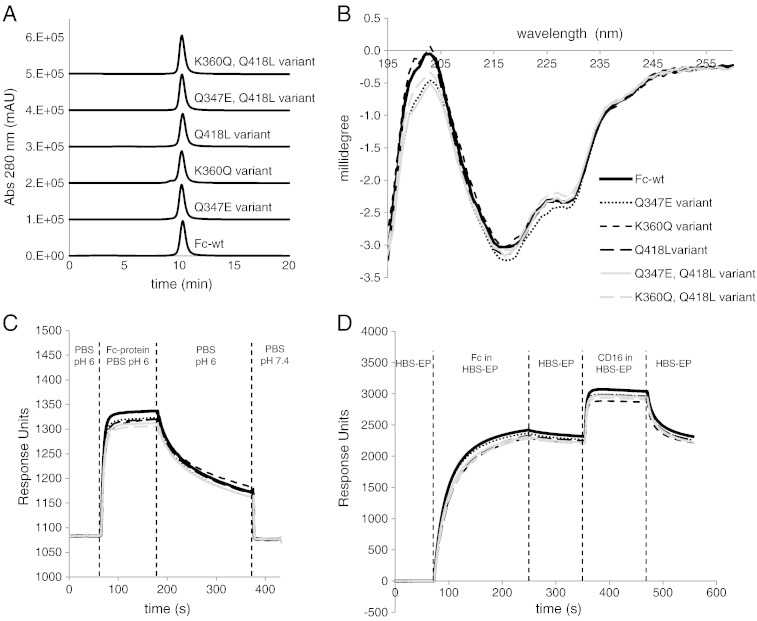
Furthermore, the Fc-proteins were analyzed by SDS-PAGE and SEC. With all selected variants SDS-PAGE under non-reducing conditions revealed a prominent band with an approximate molar mass of 55 kDa, whereas under reducing conditions a single band at about half the molar mass was detected (not shown). Size exclusion chromatography also gave only one peak at retention times almost identical to Fc-wt (Fig. 6A and Supplemental Fig. 4 for all other variants). Furthermore, the overall secondary structure was probed by electronic circular dichroism spectroscopy (ECD). Fig. 6B and Supplemental Fig. 5 demonstrate that the single, double and triple mutants showed very similar and wild-type-like far-UV ECD spectra with two prominent minima at 218 nm and 229 nm [6]. These findings underlined that the selected and stabilized IgG1-Fc variants were homodimeric and non-aggregating proteins of wild-type-like secondary structure.
Fig. 6.
Characterization of stabilized Fc variants compared to Fc-wt. (A) SEC-HPLC analysis of Fc-wt and selected Fc variants. One representative experiment out of two is shown. (B) Comparison of electronic circular dichroism (ECD) far-UV spectra of Fc-wt and stabilized Fc variants in the range of 260–195 nm. Average spectra from three experiments are shown. (C) Qualitative SPR analysis of FcRn binding of Fc-wt and stabilized Fc variants. An FcRn coated SPR-chip was incubated with PBS pH 6.0 (baseline), followed by injection of Fc-samples in PBS pH 6.0 (association), PBS pH 6.0 only and PBS pH 7.4 only for analysis of the pH-dependent dissociation. (D) Qualitative evaluation of Protein A and CD16a binding properties of Fc samples. Protein A was immobilized onto an SPR-chip. After measuring the baseline with HBS-EP buffer (pH 7.4), association with Protein A was analyzed by injection of Fc samples (HBS-EP buffer), followed by analysis of the dissociation of the Fc proteins from Protein A by injection of HBS-EP buffer only. Subsequently, association with CD16a was analyzed by incubation with the respective ligand, followed by dissociation analysis by injecting HBS-EP buffer only. In C and D average values from two experiments are shown.
In order to get further insights into the effect of mutations on the overall structure and on the binding of ligands, the stabilized Fc variants were analyzed for their ability to interact with FcRn, Protein A and CD16a by using surface plasmon resonance spectroscopy. An FcRn-coated chip was incubated with Fc variants, followed by washing with PBS, pH 6.0, and PBS pH 7.4 for evaluation of off rates at different pH values. All variants showed the typical wild-type-like pH-dependent binding to FcRn (Fig. 6C and Supplemental Fig. 6A for all other variants). For analysis of Protein A and CD16a binding a Protein A-coated chip was loaded with Fc samples for probing the interaction between Protein A and the engineered Fc-proteins. After a washing step with buffer only, the chip with bound Fc-protein was incubated with CD16a and buffer only. Fig. 6D (Supplemental Fig. 6B for all other variants) shows no striking differences in the kinetics of binding of both Protein A- and CD16a to the Fc variants compared to Fc-wt.
4. Discussion
The aim of this work was to advance yeast surface display for screening of stabilizing point mutations in proteins that have already a high intrinsic conformational and thermal stability. The chosen target protein was the crystallizable fragment of human IgG1 because of its stable scaffold and because of increasing interest in the development of alternative therapeutic antibody formats based on IgG1-Fc. Most promising in this respect is the engineering of structural loops of the CH3-domains of IgG1-Fc in the design of antigen-binding Fc-proteins, Fcabs [5,6,8].
Yeast display has already been used for stabilization of various proteins including scTCRs [3,9,10] and class I and class II major histocompatibility complex (MHC) molecules [11–13]. Moreover, stabilized mutants of monomeric enhanced green fluorescent protein (GFPM) have been evolved. These GFPM variants, in contrast to wild type GFPM, retained their fluorescence properties after insertion of random sequences into solvent-exposed loops, thereby making these stabilized scaffolds ideal starting molecules for selections of fluorescent proteins binding to a specific target [14]. In these studies the selection strategy was based on an observed correlation of protein stability and expression level mediated by the quality control system in the endoplasmic reticulum (ER) [15–17]. However, the authors argued that for proteins with higher melting temperatures the differences in expression levels are not sufficient for separation of stabilized variants [18]. This is consistent with an experiment in this study that demonstrated similar expression levels of Fc-wt and the stabilized variant K370L on the yeast surface, making the expression density-based selection strategy not suitable for stabilization of a protein of very high thermal stability (e.g. IgG1-Fc). Another strategy applied in some studies was the incubation of the yeast displayed protein-library at higher temperatures [3,12,19]. Only stabilized scTCR and MHC variants resisted thermal unfolding and still bound to a structurally specific ligand after the heat shock. However, due to the low thermal stability of those scTCR and MHC proteins only low temperatures, ranging from 40 to 46 °C, were necessary.
In contrast to scTCRs and class I and II MHC molecules, IgG1-Fc is significantly more stable with the CH2- and the CH3-domains denaturing at ~ 66 and ~ 80 °C, respectively. Since the CH2-domain unfolds reversibly, thermal unfolding of the CH3-domain has to be achieved in order to be able to discriminate between Fc-wt and stabilized variants. This hypothesis was confirmed by the fact that only at temperatures above ~ 75 °C (10 min incubation) a detectable difference in the CD64 binding intensity was observed between Fc-wt and the K370L variant displayed on yeast. It clearly demonstrated the necessity to incubate the protein library at high temperatures in order to allow for screening and selection of stabilized variants. Although this need of a high heat shock temperature resulted in complete loss of any viable yeast cells that could be cultivated again after the sort process, we were able to demonstrate that sufficient plasmid DNA can still be isolated and amplified by PCR for subsequent transformation of yeast. As a consequence, the presented experimental approach (Fig. 2) significantly expands the method introduced by Shusta and colleagues for stability engineering of proteins displayable on yeast with high intrinsic thermal stability and Tm-values similar to those of IgG1-Fcs (80–85 °C). It was interesting to see that dead yeast cells still displayed well folded Fc-proteins for screening of ligand binding by FACS. The irrevocable upper limit of this new experimental approach was gelatinization of the yeast cells at T > 85 °C (10 min incubation).
By applying this new experimental approach to an Fc-library constructed by error prone PCR we succeeded in selection of stabilizing point mutations within the Fc fragment of human IgG1. Analysis of the development of the mutation rates during 4 consecutive rounds of sorting showed that they decreased in both, the CH2- and the CH3-domain, during the first round of sorting, most probably because of elimination of mutations with strong negative impact on the stability and foldability of the protein. During later sorting rounds only in the CH3-, but not in the CH2-domain mutations accumulated. As mentioned above this reflects the unfolding pathway of IgG1-Fc [6]. The CH2-domain denatures reversibly in the presence of intact CH3-domains, whereas unfolding of CH3-domains is irreversible. In case of Tm1 < T < Tm2 (Tm3) only the CH2 domain unfolds but completely refolds by restoring room temperature. As a consequence in the present study only stabilizing mutations in the CH3-domain could be detected. On the other hand, the thermal denaturation of CH3 had to be irreversible because otherwise the protein would have refolded after the heat shock. With keeping these prerequisites in mind, a very potent method with the potential of various applications has been developed.
Another immediate benefit of the present work is the selection and characterization of 17 stabilizing single, double and triple mutants of IgG1-Fc of wild-type-like overall structure and ligand binding behavior but increased thermal stability (Table 1). Recently, it has been demonstrated that loops of IgG1-Fc can be engineered to form antigen-bindings sites. Both the grafting of a rigid disulfide-bridged cyclic heptapeptide (conferring binding to human αvβ3 integrin) into AB, CD and/or EF loops of the CH3 domains [6] as well as successful engineering of the EF and AB loop for binding of HER2/neu [5] have been reported. However, engineering of the C-terminal structural loops might decrease the conformational and thermal stability of these antibody formats and stabilizing the scaffold by the selected mutations could improve their folding. Additionally, it will improve the design of Fcab libraries and increase the portion of well-folded binders. Since without exception all introduced mutations did not negatively influence the binding of the ligands FcRn and CD16a to their respective sites on the Fc-protein, it is reasonable to assume that Fcabs having these mutations still can fulfill all properties of a Fc-protein, namely effector functions and long half-life. Both the ADCC potency of an Fcab as well as a long half-life in a mouse model have already been demonstrated recently [5].
The following are the supplementary materials related to this article.
Survival of yeast cells after a heat shock. Surface expression of Fc-wt was induced in SGR-CAA at 20 °C for 18–20 h, followed by centrifugation of the yeast cells, resuspension in PBS/2% BSA and incubation on ice for at least 10 min. Subsequently the cells were subjected to a heat shock for 10 or 30 min at various temperatures as indicated, immediately followed by incubation on ice again. Finally, various dilutions were plated on SD-CAA-Agar and the survival rate was determined by colony counting after incubation at 30 °C for 3 days.
Characterization of original libraries H and L by sequencing. 48 clones from each library were sequenced, yielding 44 evaluable sequences for each library. (A) For each library the percentage of each possible mutation (from the original nucleotide shown below to the nucleotide indicated above) is shown. (B) Distribution of detected mutations over the Fc-gene. The total number of detected mutations in both libraries among 88 analyzed sequences at each nucleotide position is shown.
Comparison of anti-CH2 binding of surface displayed Fc-wt, original Fc-libraries and Fc-libraries after 4 rounds of sorting. After induction of surface expression a heat shock was either applied (right column) or not (left column), followed by staining with anti-CH2 and an antibody against the Xpress epitope which is used for expression normalization. For comparison, gate positions were set identically in the left and in the right column, respectively. Numbers in the dot plots indicate the percentage of cells in the gate.
Characterization of stabilized Fc variants compared to Fc-wt. SEC-HPLC analysis of Fc-wt and selected Fc variants. One representative experiment out of two is shown.
Characterization of stabilized Fc variants compared to Fc-wt. Comparison of electronic circular dichroism (ECD) far-UV spectra of Fc-wt and stabilized Fc variants in the range of 260–195 nm. Average spectra from three experiments are shown.
Characterization of stabilized Fc variants compared to Fc-wt. (A) SPR analysis of FcRn binding of Fc-wt and stabilized Fc variants. An FcRn coated SPR-chip was incubated with PBS pH 6.0 (baseline), followed by injection of Fc-samples in PBS pH 6.0 (association), PBS pH 6.0 only and PBS pH 7.4 only for analysis of the pH-dependent dissociation. (B) For evaluation of Protein A and CD16a binding properties of Fc samples Protein A was immobilized onto an SPR-chip. After measuring the baseline with HBS-EP buffer (pH 7.4), association with Protein A was analyzed by injection of Fc samples (HBS-EP buffer), followed by analysis of the dissociation of the Fc proteins from Protein A by injection of HBS-EP buffer only. Subsequently, association with CD16a was analyzed by incubation with the respective ligand, followed by dissociation analysis by injecting HPS-EP buffer only. Average values from two experiments are shown.
Acknowledgements
This work was supported by the Christian Doppler Research Association (CDG) and the doctoral program BioToP — Biomolecular Technology of Proteins of the Austrian Science Funds (FWF-W1224).
References
- 1.Gai S.A., Wittrup K.D. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orr B.A., Carr L.M., L. M., Wittrup K.D., Roy E.J., Kranz D.M. Rapid method for measuring ScFv thermal stability by yeast surface display. Biotechnol. Prog. 2003;19:631–638. doi: 10.1021/bp0200797. [DOI] [PubMed] [Google Scholar]
- 3.Shusta E.V., Holler P.D., Kieke M.C., Kranz D.M., Wittrup K.D. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat. Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 4.Kubota T., Niwa R., Satoh M., Akinaga S., Shitara K., Hanai N. Engineered therapeutic antibodies with improved effector functions. Cancer Sci. 2009;100:1566–1572. doi: 10.1111/j.1349-7006.2009.01222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wozniak-Knopp G., Bartl S., Bauer A., Mostageer M., Woisetschlager M., Antes B., Ettl K., Kainer M., Weberhofer G., Wiederkum S., Himmler G., Mudde G.C., Ruker F. Introducing antigen-binding sites in structural loops of immunoglobulin constant domains: Fc fragments with engineered HER2/neu-binding sites and antibody properties. Protein Eng. Des. Sel. 2010;23:289–297. doi: 10.1093/protein/gzq005. [DOI] [PubMed] [Google Scholar]
- 6.Traxlmayr M.W., Wozniak-Knopp G., Antes B., Stadlmayr G., Ruker F., Obinger C. Integrin binding human antibody constant domains—probing the C-terminal structural loops for grafting the RGD motif. J. Biotechnol. 2011;155:193–202. doi: 10.1016/j.jbiotec.2011.06.042. [DOI] [PubMed] [Google Scholar]
- 7.Gietz R.D., Schiestl R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 8.Rüker F., Wozniak-Knopp G. Engineering of non-CDR loops in immunoglobulin domains. In: Little M., editor. Recombinant Antibodies for Immunotherapy. Cambridge University Press; New York: 2009. pp. 231–242. [Google Scholar]
- 9.Kieke M.C., Shusta E.V., Boder E.T., Teyton L., Wittrup K.D., Kranz D.M. Selection of functional T cell receptor mutants from a yeast surface-display library. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber K.S., Donermeyer D.L., Allen P.M., Kranz D.M. Class II-restricted T cell receptor engineered in vitro for higher affinity retains peptide specificity and function. Proc. Natl. Acad. Sci. U. S. A. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteban O., Zhao H. Directed evolution of soluble single-chain human class II MHC molecules. J. Mol. Biol. 2004;340:81–95. doi: 10.1016/j.jmb.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 12.Jones L.L., Brophy S.E., Bankovich A.J., Colf L.A., Hanick N.A., Garcia K.C., Kranz D.M. Engineering and characterization of a stabilized alpha1/alpha2 module of the class I major histocompatibility complex product Ld. J. Biol. Chem. 2006;281:25734–25744. doi: 10.1074/jbc.M604343200. [DOI] [PubMed] [Google Scholar]
- 13.Starwalt S.E., Masteller E.L., Bluestone J.A., Kranz D.M. Directed evolution of a single-chain class II MHC product by yeast display. Protein Eng. 2003;16:147–156. doi: 10.1093/proeng/gzg018. [DOI] [PubMed] [Google Scholar]
- 14.Pavoor T.V., Cho Y.K., Shusta E.V. Development of GFP-based biosensors possessing the binding properties of antibodies. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11895–11900. doi: 10.1073/pnas.0902828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalski J.M., Parekh R.N., Mao J., Wittrup K.D. Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J. Biol. Chem. 1998;273:19453–19458. doi: 10.1074/jbc.273.31.19453. [DOI] [PubMed] [Google Scholar]
- 16.Kowalski J.M., Parekh R.N., Wittrup K.D. Secretion efficiency in Saccharomyces cerevisiae of bovine pancreatic trypsin inhibitor mutants lacking disulfide bonds is correlated with thermodynamic stability. Biochemistry. 1998;37:1264–1273. doi: 10.1021/bi9722397. [DOI] [PubMed] [Google Scholar]
- 17.Shusta E.V., Kieke M.C., Parke E., Kranz D.M., Wittrup K.D. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J. Mol. Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 18.Park S., Xu Y., Stowell X.F., Gai F., Saven J.G., Boder E.T. Limitations of yeast surface display in engineering proteins of high thermostability. Protein Eng. Des. Sel. 2006;19:211–217. doi: 10.1093/protein/gzl003. [DOI] [PubMed] [Google Scholar]
- 19.Aggen D.H., Chervin A.S., Insaidoo F.K., Piepenbrink K.H., Baker B.M., Kranz D.M. Identification and engineering of human variable regions that allow expression of stable single-chain T cell receptors. Protein Eng. Des. Sel. 2011;24:361–372. doi: 10.1093/protein/gzq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Survival of yeast cells after a heat shock. Surface expression of Fc-wt was induced in SGR-CAA at 20 °C for 18–20 h, followed by centrifugation of the yeast cells, resuspension in PBS/2% BSA and incubation on ice for at least 10 min. Subsequently the cells were subjected to a heat shock for 10 or 30 min at various temperatures as indicated, immediately followed by incubation on ice again. Finally, various dilutions were plated on SD-CAA-Agar and the survival rate was determined by colony counting after incubation at 30 °C for 3 days.
Characterization of original libraries H and L by sequencing. 48 clones from each library were sequenced, yielding 44 evaluable sequences for each library. (A) For each library the percentage of each possible mutation (from the original nucleotide shown below to the nucleotide indicated above) is shown. (B) Distribution of detected mutations over the Fc-gene. The total number of detected mutations in both libraries among 88 analyzed sequences at each nucleotide position is shown.
Comparison of anti-CH2 binding of surface displayed Fc-wt, original Fc-libraries and Fc-libraries after 4 rounds of sorting. After induction of surface expression a heat shock was either applied (right column) or not (left column), followed by staining with anti-CH2 and an antibody against the Xpress epitope which is used for expression normalization. For comparison, gate positions were set identically in the left and in the right column, respectively. Numbers in the dot plots indicate the percentage of cells in the gate.
Characterization of stabilized Fc variants compared to Fc-wt. SEC-HPLC analysis of Fc-wt and selected Fc variants. One representative experiment out of two is shown.
Characterization of stabilized Fc variants compared to Fc-wt. Comparison of electronic circular dichroism (ECD) far-UV spectra of Fc-wt and stabilized Fc variants in the range of 260–195 nm. Average spectra from three experiments are shown.
Characterization of stabilized Fc variants compared to Fc-wt. (A) SPR analysis of FcRn binding of Fc-wt and stabilized Fc variants. An FcRn coated SPR-chip was incubated with PBS pH 6.0 (baseline), followed by injection of Fc-samples in PBS pH 6.0 (association), PBS pH 6.0 only and PBS pH 7.4 only for analysis of the pH-dependent dissociation. (B) For evaluation of Protein A and CD16a binding properties of Fc samples Protein A was immobilized onto an SPR-chip. After measuring the baseline with HBS-EP buffer (pH 7.4), association with Protein A was analyzed by injection of Fc samples (HBS-EP buffer), followed by analysis of the dissociation of the Fc proteins from Protein A by injection of HBS-EP buffer only. Subsequently, association with CD16a was analyzed by incubation with the respective ligand, followed by dissociation analysis by injecting HPS-EP buffer only. Average values from two experiments are shown.