Abstract
Mutations in the CDKL5 (cyclin-dependent kinase-like 5) gene are associated with a severe epileptic encephalopathy (early infantile epileptic encephalopathy type 2, EIEE2) characterized by early-onset intractable seizures, infantile spasms, severe developmental delay, intellectual disability, and Rett syndrome (RTT)-like features. Despite the clear involvement of CDKL5 mutations in intellectual disability, the function of this protein during brain development and the molecular mechanisms involved in its regulation are still unknown. Using human neuroblastoma cells as a model system we found that an increase in CDKL5 expression caused an arrest of the cell cycle in the G0/G1 phases and induced cellular differentiation. Interestingly, CDKL5 expression was inhibited by MYCN, a transcription factor that promotes cell proliferation during brain development and plays a relevant role in neuroblastoma biology. Through a combination of different and complementary molecular and cellular approaches we could show that MYCN acts as a direct repressor of the CDKL5 promoter. Overall our findings unveil a functional axis between MYCN and CDKL5 governing both neuron proliferation rate and differentiation. The fact that CDKL5 is involved in the control of both neuron proliferation and differentiation may help understand the early appearance of neurological symptoms in patients with mutations in CDKL5.
Keywords: Rett's syndrome, CDKL5, MYCN, Neurogenesis, Differentiation
Highlights
► CDKL5 enhances neuronal differentiation. ► CDKL5 arrests cell cycle of neuronal precursor cells. ► MYCN directly represses transcription of CDKL5. ► These results may help explain the neurological symptoms of RTT patients.
1. Introduction
Epilepsy, and in particular intractable epilepsy in infancy, often results in an encephalopathic picture, known under these circumstances as an epileptic encephalopathy. A severe epileptic encephalopathy (early infantile epileptic encephalopathy type 2 (EIEE2); OMIN 300672) that is characterized by intractable seizures with infantile spasms, intellectual disability, and a Rett's syndrome (RTT)-like phenotype, is attributable to cyclin-dependent kinase-like 5 mutations [4,6,18,29,37,42,44]. This condition affects mostly girls and presents as an atypical RTT picture. It has features of RTT, such as hyperventilation, hand stereotypies, and kyphoscoliosis, hypotonia, that are superimposed on a typical epileptic encephalopathy picture, with a clinical presentation dominated by early seizures (onset before age six months) [4,14,37,41,44].
The vast majority of RTT cases (> 95%) are caused by mutations in the MECP2 gene [2]. MECP2, which maps to chromosome Xq28 and is not expressed from the inactive X chromosome, encodes for methyl-CpG-binding protein 2 (MeCP2). Although patients carrying MECP2 mutations as well as Mecp2 knockout mice display a normal brain cytoarchitecture without detectable loss of neurons [3,9], recent studies indicate that MeCP2 regulates the number and function of excitatory synapses in the brain [7], suggesting that changes in neuronal functions may represent the primary cause of the neurological phenotype in RTT.
The CDKL5 gene maps to chromosome Xp22 and encodes a protein with kinase activity that is a member of the serine–threonine (Ser/Thr) protein kinase family. It is a large protein which is composed by a conserved N-terminal (Ser/Thr) kinase domain responsible for the catalytic activity of the protein, and a large C-terminal region involved in the regulation of CDKL5 kinase activity [24] and in the nuclear localization of the protein [5,28]. At the molecular level, the only in vitro CDKL5 substrates identified so far are DNA methyltransferase 1 (DNMT1) and MeCP2 [5,19,28], suggesting that CDKL5 may regulate DNA methylation and the binding of MeCP2 to DNA. Interestingly, CDKL5 has been also shown to play a role in the dynamic regulation of nuclear speckles, which are implicated in the regulation of mRNA splicing [35].
Studies in rodents have established that CDKL5 is expressed in the developing and adult brain [8,36], suggesting a role of CDKL5 in neuronal development and function. It has been shown that CDKL5 can shuttle between the nucleus and the cytoplasm and that its subcellular localization in the brain is developmentally regulated. Along with its proposed function in the nucleus, CDKL5 appears to regulate dendritic development, through a cytoplasmic mechanism involving the BDNF-Rac1 signaling [8].
Despite the clear involvement of CDKL5 mutations in intellectual disability, the function/s of this protein and the molecular mechanisms involved in its regulation are poorly unknown. Herein, we show that CDKL5 expression affects neuronal proliferation and differentiation, two processes closely related during brain development, and that CDKL5 is negatively regulated by MYCN, a key player in neuronal development.
2. Materials and methods
2.1. Cell cultures
Human neuroblastoma cell lines SH-SY5Y and SKNBE, obtained from ATCC (Manassas, VA, USA), and human Tet21/N cells, derived from SHEP neuroblastoma cell line that stably express MYCN under the control of tetracycline (Tet-off) [26], were maintained in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated FBS, 2 mM of glutamine and antibiotics (penicillin, 100 U/ml; streptomycin, 100 μg/ml), in a humidified atmosphere of 5% of CO2 in air at 37 °C. Cell medium was replaced every 3 days and the cells were sub-cultured once they reached 90% confluence. SH-SY5Y cells were transfected with the plasmid pCMV14-3XFLAG-MYCN that contains the MYCN gene under the control of the human cytomegalovirus promoter, and neoR, that confers neomycin resistance. Clones were selected in the presence of 600 μg/ml of G418. The Tet21/N cells were treated with tetracycline at a final concentration of 2 μg/ml for the indicated time.
Primary cultures of cerebellar granular cell precursors (GCPs) were prepared from the cerebella of 7-day-old C57BL/6J mice as previously described [15]. Briefly, cerebella were removed and dissected from their meninges in Krebs buffer containing 0.3% BSA. Tissue was dissociated with trypsin at 37 °C for 15 min and triturated 15 times using a Pasteur pipette in a DNAase/soybean trypsin inhibitor solution. Dissociated cells were plated on poly-d-lysine (20 μM, Sigma)-coated dishes at a density of 2 × 103 cells/mm2 and maintained in basal modified Eagle's medium (BME) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 25 mM KCl, 2 mM glutamine and 0.05 mg/ml gentamicin (Sigma).
2.2. Plasmids
Human CDKL5 cDNA was kindly provided by Marsha Rich Rosner (University of Chicago) [24], CDKL5 cDNA was PCR amplified and cloned into pCMV14–3xFLAG plasmid (Sigma). CDKL5-3xFLAG sequence was subcloned into the bicistronic pIRES2-EGFP plasmid (Clontech) to obtain a GFP and CDKL5-FLAG co-expression vector (pGFP/CDKL5-FLAG).
2.3. Immunocytochemistry
For cell proliferation analyses SH-SY5Y cells were plated onto poly-d-lysine coated slides in 6-well plated at density of 3 × 105 cells per well and transfected with 3 μg of CDKL5-FLAG expression plasmid using lipofectamin (Roche). Twenty-four hours after transfection cells were fixed in a 4% paraformaldehyde 4% glucose solution at 37 °C for 30 min. For immunofluorescence studies the following antibodies were used. Primary antibodies: anti-Ki-67 rabbit monoclonal (1:100, Thermo Scientific), anti-BrdU rat monoclonal (1:100, AbD Serotec), anti-cleaved caspase-3 (asp175) rabbit polyclonal (1:100, Cell Signaling Technology), anti-β-tubulin III rabbit polyclonal (TubJ, 1:500, Sigma) and anti-FLAG M2 mouse monoclonal (1:1000; Sigma). Secondary antibodies: FITC-conjugated anti-rat antibody (1:200, Jackson Immuno Research Laboratories), FITC-conjugated anti-rabbit antibody (1:200, Jackson Immuno Research Laboratories) FITC-conjugated anti-mouse antibody (1:200, Jackson Immuno Research Laboratories) and Cy3-conjugated anti-rabbit antibody (1:200, Jackson Immuno Research Laboratories). Double immunofluorescence images, taken from random microscopic fields (10–12 for each coverslip), were superimposed and used to determine the labeling index (LI), defined as percentage of cells co-labeled with: anti-BrdU and anti-FLAG or anti-cleaved caspase-3 and anti-FLAG or anti-Ki-67 and anti-FLAG antibodies.
In each experimental condition we randomly analyzed a total of 600 cells. Fluorescence images were taken on an Eclipse TE 2000-S microscope (Nikon, Tokyo, Japan) equipped with a digital camera Sight DS-2MBW (Nikon).
2.4. Western blotting
The following antibodies were used: anti-CDKL5 (1:500; Sigma), anti-FLAG M2 (1:1000; Sigma) and anti-GADPH (1:1000; Sigma). For the preparation of total cell extracts, cells were lysed in RIPA lysis buffer (Tris–HCl 50 mM, NaCl 150 mM, Triton X-100 1%, sodium deoxycholate 0.5%, SDS 0.1%, protease and phosphatase inhibitors cocktails 1%; Sigma). For the preparation of nuclear extracts cells were allowed to swell and lysed in hypotonic buffer (Hepes 10 mM, NaCl 50 mM, EDTA 1 mM, NP-40 0.1%, DTT 1 mM, PMSF 1 mM, pH 8) for 10 min at 4 °C. After centrifugation, nuclei were extracted with hypertonic salt buffer (Hepes 20 mM, NaCl 420 mM, EDTA 1 mM, DTT 1 mM, glicerol 10%, PMSF 1 mM, pH 8). Cell extracts were immediately processed by Western blot or kept frozen (− 80 °C) until assayed. Sample protein concentration was estimated by the Lowry method [25]. Equivalent amounts (50 μg) of protein were subjected to electrophoresis on a 10% SDS-polyacrylamide gel. Densitometric analysis of digitized images was performed with Scion Image software (Scion Corporation, Frederick, MD, USA) and intensity for each band was normalized to the intensity of the corresponding β-actin or GAPDH band.
2.5. Analysis of neurite outgrowth
For differentiation analyses neuroblastoma cells were plated onto poly-d-lysine coated slides in a 24-well plated at density of 2 × 104 cells per well. After 24 h of cell plating, differentiation was induced by retinoic acid (RA; 10 μM) for the indicated time. This treatment was replaced each 2 days to replenish RA in culture media. Phase contrast photographs of the cultures were taken at various time intervals with an Eclipse TE 2000-S microscope (Nikon, Tokyo, Japan) equipped with an AxioCam MRm (Zeiss, Oberkochen, Germany) digital camera. Ten different areas were randomly selected and neurite outgrowth was measured using the image analysis system Image Pro Plus (Media Cybernetics, Silver Spring, MD 20910, USA). Only cells with neurites longer than one cell body diameter were considered as neurite-bearing cells. All experiments were performed at least three times. In each experiment we analyzed a total of around 450 cells. The total length of neurites was divided for the total number of cells counted in the areas.
2.6. Flow cytometric analysis
SH-SY5Y cells were harvested 24 h after transfection with pGFP or pGFP/CDKL5-FLAG plasmids, collected by trypsinization, pelleted and resuspended in phosphate buffered saline (PBS) to a final concentration of 1 × 106 cells/ml. Transfected cells were then analyzed on a flow cytometer (FACSCalibur, Becton Dickinson, San Jose, CA, USA) and sorted by GFP fluorescence (detection filter set at 525 nm). Cell aggregates were gated out, and 10,000 events were analyzed. Untransfected SH-SY5Y cells were used to establish a threshold for green fluorescence (up to 102 arbitrary fluorescence units in a typical case), and which was taken as a threshold for positivity (Fig. 4C). For differentiation analysis an aliquot of GFP sorted cells was plated in 6-well at the density of 3 × 105 cells per well in the presence or absence of RA (10 μM).
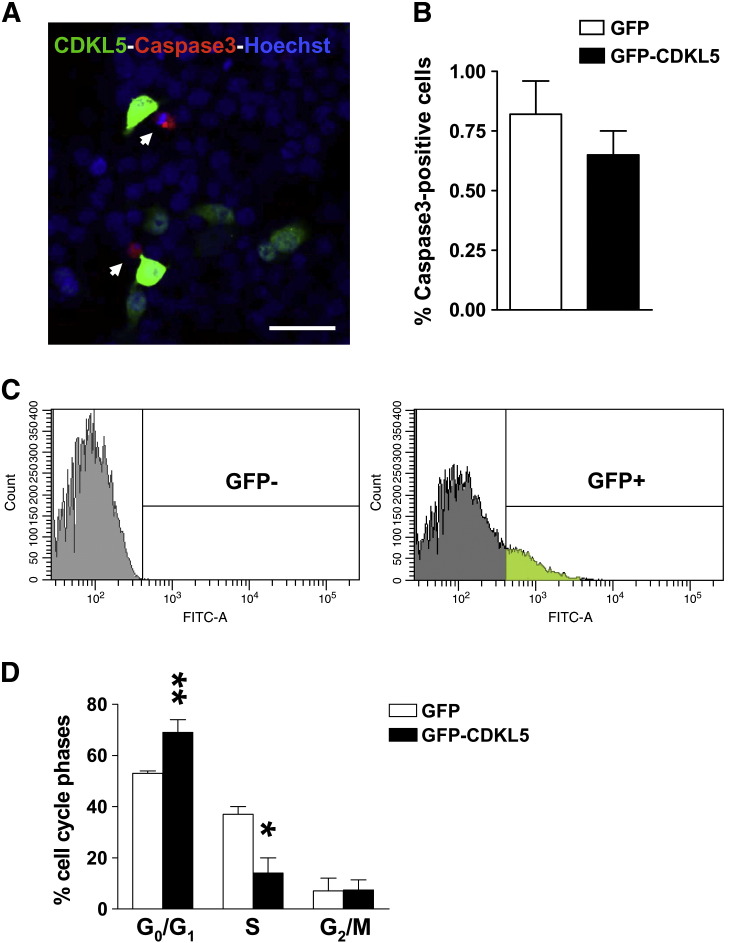
Fig. 4.
Expression of CDKL5 does not induce cell death but blocks cell cycle progression in SH-SY5Y neuroblastoma cells.
A: Example of immunofluorescence image of SH-SY5Y neuroblastoma cells transfected with GFP-CDKL5. Twenty-four hours after transfection, SH-SY5Y cells were immunostained for cleaved caspase-3 (red signal). Cell nuclei were stained with Hoechst dye (blue signal). Scale bar: 25 μm. B: Percentage of cleaved caspase-3-positive cells over total number of GFP-CDKL5-positive cells and GFP-positive cells. Data are expressed as mean ± SE of 3 independent experiments. A minimum of 450 cells were evaluated in each experiment for each condition. C: Twenty-four hours after transfection with GFP-CDKL5 or GFP alone, SH-SY5Y cells were FACS sorted for GFP expression. The threshold for positivity for GFP was established as the levels corresponding to untransfected cells (histogram on the left). The green area in the histogram on the right shows fluorescence intensity of sorted GFP-CDKL5 positive cells. D: Distribution of cell populations in the G0/1, S, or G2/M phases of the cell cycle, identified by flow cytometry analysis. Data, given as percentage of either GFP-CDKL5 or GFP cells in each phase of the cell cycle, are expressed as mean ± SE of 3 independent experiments, *P < 0.05, **P < 0.01, two-tailed t-test.
For cell cycle analysis cells were fixed in 70% ethanol in PBS at − 20 °C for at least 1 h, washed several times with cold PBS, treated with RNaseA (50 μg/ml) for 30 min at 37 °C and incubated with propidium iodide (30 μg/ml). GFP-positive and -negative populations were analyzed separately for DNA content (filter set at 675 nm) and assigned to specific cell-cycle phases by applying the Cell Quest Software (Becton Dickinson, San Jose, CA, USA).
2.7. Small interfering RNA assay
The siRNA oligonucleotides used for silencing CDKL5 were purchased from QIAGEN and were: Hs-CDKL5-5 cat. No:SI02223116 (sense; Si1) and Hs-CDKL5-10 cat. No:SI004437244 (sense; Si2). Control cells were transfected with a scramble siRNA duplex; AllStars Negative Control siRNA (siSCR QIAGEN), which does not present homology with any other human mRNAs. SH-SY5Y cells were transfected with HiPerFect Transfection Reagent (QIAGEN) with 50 nM siRNA (final concentration). For differentiation experiments retinoic acid was added to the cells 6 h post transfection and the cells harvested after further 42 h. For proliferation experiments 10 μM BrdU was added to the cells 46 h post transfection and the cells harvested after further 2 h.
2.8. Quantitative real time PCR and standard reverse transcription-PCR
RNA samples were prepared using Tri-Reagent (Sigma) and treated with DNase (DNA-freeTM; Ambion). Reverse transcription was performed using a SuperScript reverse transcription-PCR kit (Invitrogen). Real time quantitative PCR (RT-qPCR) was performed using a SYBR Premix Ex Taq kit (Takara, Shiga, Japan) and the iQ5 thermocycler (Bio-Rad). The efficiency of the used primers was evaluated by calculating the linear regression of Ct data points obtained with a series of different primer dilutions and inferring the efficiency from the slope of the line. We used the primers that gave efficiency close to 100%. Primers for RT-qPCR are listed in Supplemental Table 1A. Quantifications were always normalized using endogenous control GAPDH.
2.9. Bioinformatic analysis of the human and mouse CDKL5 promoter and chromatin immunoprecipitation (ChIP) Assay
5000 bp upstream and downstream the start site of the human and mouse CDKL5 genomic sequence was analyzed for the presence of MYCN canonical (CACGTG) and non canonical (CACGCG and CATGTG) ebox and of SP1 binding site (GGGCGG) by the DNA analysis software pDRAW32. Primer pairs were designed to amplify these regions (see Supplementary Table 1B and Figs. 6 and 8).
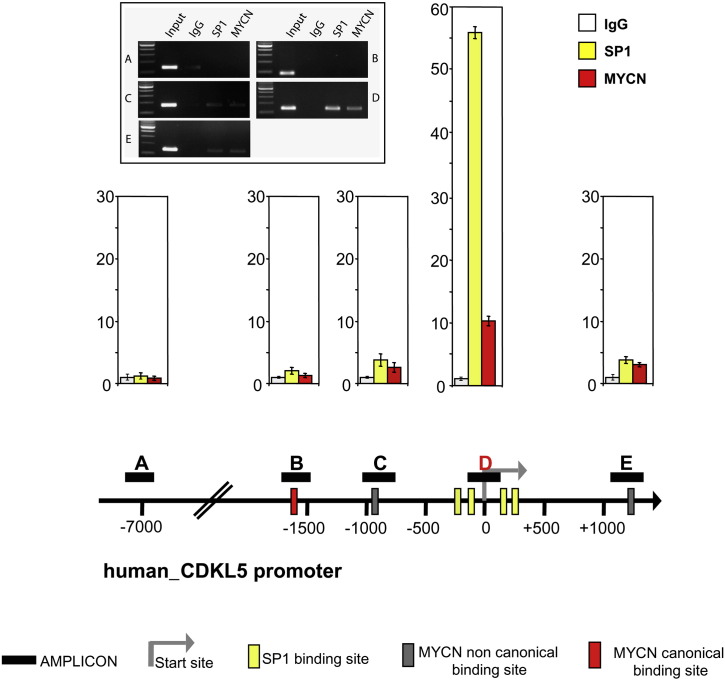
Fig. 6.
MYCN binds CDKL5 promoter.
Dual cross-linking ChIP was performed in SH-SY5Y(++MYCN) neuroblastoma cells. DNA/protein complexes were immunoprecipitated with pre-immune serum (IgG), anti-SP1 and anti-MYCN antibodies. Real-time PCR of immunoprecipitated DNA with primers targeting the negative control region (Amplicon A) or four regions overlapping MYCN-canonical or non canonical binding site (Amplicons B,C,E) or SP1-binding sites (Amplicon D) were performed. The graphs show fold enrichment of selected DNA regions of the human CDKL5 gene promoter, calculated as the logarithm of the difference between the cycle-threshold obtained with pre-immune serum (IgG control) and the cycle-threshold obtained with the specific antibody. The panel on the left shows the DNA Amplicons obtained by quantitative PCR at the 30th cycle of reaction, run on a 2% agarose gel and visualized by ethidium bromide. Values represent mean ± SE of three experiments. The lower scheme gives a schematic representation of the human CDKL5 gene promoter with indicated the positions of the SP1 and MYCN canonical and non-canonical binding sites.
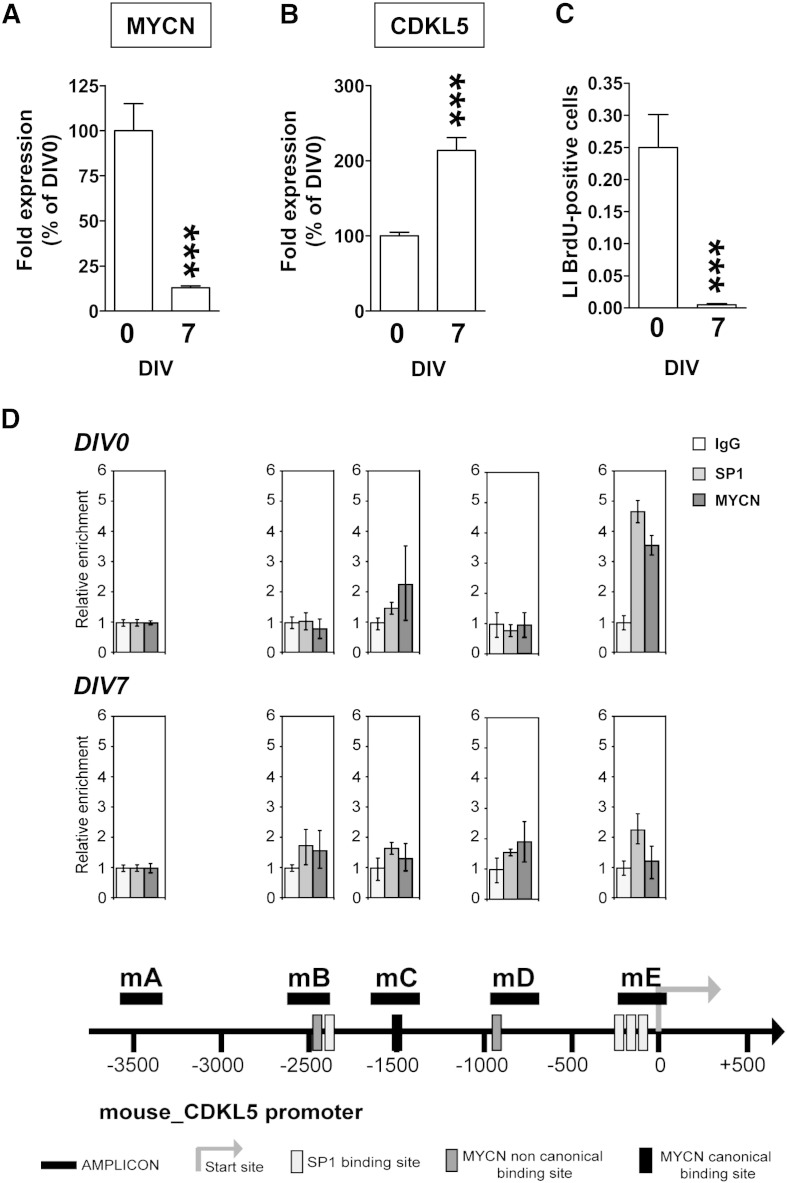
Fig. 8.
Inverse relationship between MYCN and CDKL5 expression in CGPs.
A,B: Quantification by RT-qPCR of MYCN (A) and CDKL5 (B) expression in freshly dissociated cerebellar granular cell precursors (DIV0) and after 7 days in vitro (DIV7). Data, in A and B are given as percentage of DIV0 condition and are expressed as the mean ± SE of 3 independent experiments. ***P < 0.001; two-tailed t-test. C: Labeling index (LI) was determined for cerebellar granular cells 3 h after plating (indicated as DIV0) and at DIV7. Cultures were treated with BrdU for 2 h, fixed and processed for immunofluorescence with an anti-BrdU antibody. Data are expressed as mean ± SE. ***P < 0.001, two-tailed t-test. D: Dual cross-linking ChIP and quantitative RT-PCR were applied in either freshly dissociated cerebellar granular precursors (DIV0) or after 7 days in vitro (DIV7). Real-time PCR with primers targeting the negative control region (Amplicon mA) or four regions overlapping MYCN-canonical or non canonical binding site (amplicons mB–mD) or SP1-binding sites (amplicons mB,mE) were performed. Fold enrichment of mouse Cdkl5 promoter regions immunoprecipitated by pre-immune serum (IgG), anti-SP1 and anti-MYCN antibodies was calculated as the logarithm of the difference between the cycle-threshold obtained with pre-immune serum and the cycle-threshold obtained with the specific antibody. Schematic representation of the mouse Cdkl5 gene promoter containing the SP1 binding sites.
Dual ChIP cross-linking was performed as previously described [45]. The antibodies employed in this study were: IgG (sc-2027, Santa Cruz); anti-MYCN monoclonal antibody (sc-53993, Santa Cruz), and anti-SP1 (Upstate 07‐124). Specific pairs of primers used for quantitative ChIP are listed in Supplementary Table 1B.
2.10. Luciferase assay
The pGL3-basic and Renilla-TK vectors were obtained from Promega. Promoter regions of the CDKL5 gene were obtained using PCR and cloned into the pGL3-basic vector. The activity of firefly or Renilla luciferase was measured with a dual luciferase assay kit (Promega) according to the instructions.
2.11. Statistical analysis
Results are presented as the mean ± standard error of the mean (SEM). Statistical significance was assessed by two-way analysis of variance (ANOVA), followed by Bonferroni's post hoc test or by the two-tailed Student's t-test. For the publicly available microarray dataset, clinical, pathological, and gene expression data for 102 primary neuroblastoma tumors [31] were obtained through the http://pob.abcc.ncifcrf.gov/cgi-bin/JK web site. Survival probabilities in neuroblastoma subgroups were estimated according to the methods of Kaplan and Meier [13]. Survival distributions were compared using log-rank tests [27]. A probability level of P < 0.05 was considered to be statistically significant.
3. Results
3.1. CDKL5 is up-regulated during neuroblatoma cell line differentiation
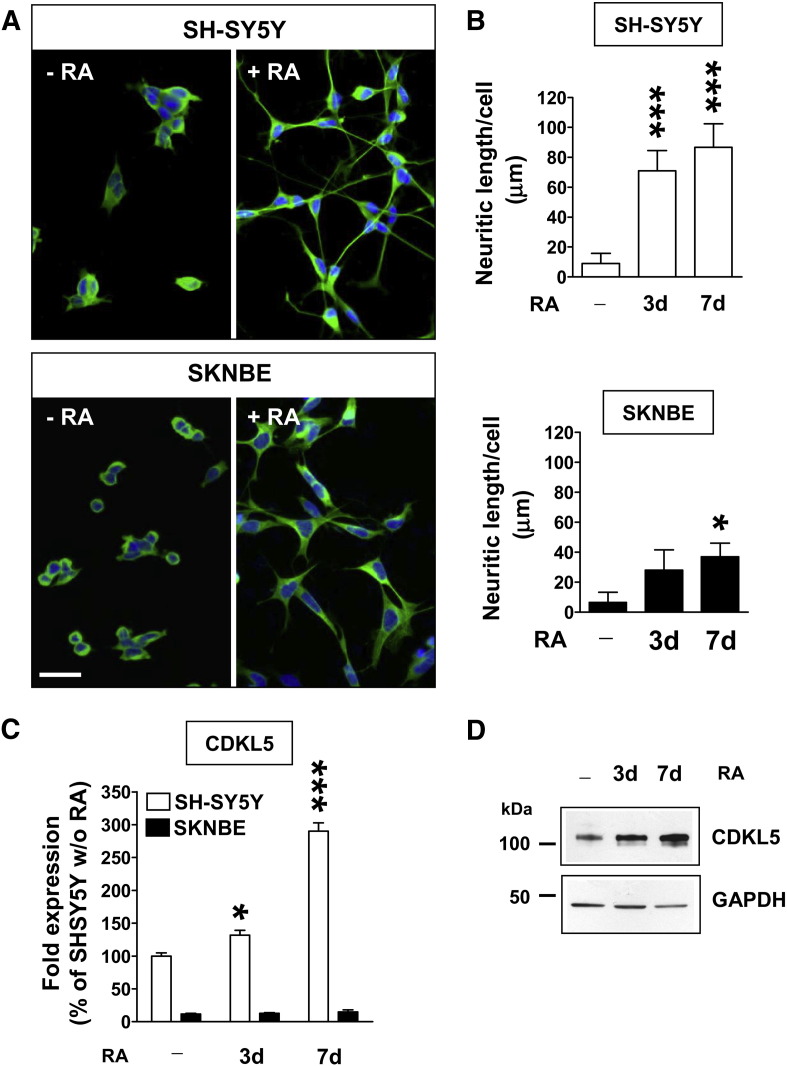
Neuroblastoma cells share several features with normal neurons and thus are considered a good in vitro model to study the biochemical and functional properties of neuronal cells, particularly when they are induced to differentiate upon treatment with agents such as retinoic acid (RA) [30,40]. For these reasons neurobastoma cells have been here employed to study the CDKL5 function in vitro. We first sought to establish whether human neuroblastoma cell lines exhibit a positive correlation between CDKL5 expression and neuronal differentiation similar to those observed in vivo during brain development [8,36]. We evaluated morphological differentiation and CDKL5 expression in two neuroblastoma cell lines, SH-SY5Y and SKNBE, following RA treatment. SH-SY5Y cells treated with RA resulted in a quick and large change in their morphology when compared to SKNBE cells kept under similar conditions. Within 3 days after addition of RA, SH-SY5Y cells extended long branched processes measuring up to 5–6 fold the length of the cell body, while SKNBE cells were still undifferentiated (Fig. 1A,B). Consistent with previous studies [10], RA-mediated differentiation of SKNBE cells starts only after 7 days from RA exposure (Fig. 1A,B). Although basal CDKL5 expression was notably higher in untreated SH-SY5Y than in untreated SKNBE cells (Fig. 1C), differentiating SH-SY5Y cells exhibited strong up-regulation of CDKL5 expression (Fig. 1C), whereas SKNBE cells did not show any change in CDKL5 expression within the same RA-treatment period (Fig. 1C). The up-regulation of CDKL5 expression in differentiating SH-SY5Y cells was confirmed also at the protein level. We found that CDKL5 expression increased after 3 (+40%) and 7 (+100%) days of RA exposure (Fig. 1D). In our experimental conditions, CDKL5 protein levels in SKNBE cells were below detection limits (data not shown), confirming the low basal expression of CDKL5 in SKNBE compared to SH-SY5Y cells.
Fig. 1.
CDKL5 expression during differentiation in neuroblastoma cell lines.
A: Immunofluorescence images showing the morphology of SH-SY5Y (upper panel) and SKNBE (lower panel) cells after 7 days of treatment with (+RA) or without (− RA) retinoic acid. Cells were stained for β-tubulin III (green) and nuclei were counterstained with Hoechst dye (blue). Scale bar: 30 μm. B: Quantification of neurite outgrowth of SH-SY5Y (upper histogram) and SKNBE (lower histogram) cells that were either untreated or treated with RA (10 μM) for 3 and 7 days. Neurite outgrowth was expressed as mean neurite length (μm) per cell. Data are expressed as mean ± SE of 4 independent experiments. A minimum of 400 cells were evaluated in each experiment for each condition. *P < 0.05; ***P < 0.001, treated vs. untreated condition (Bonferroni's test after ANOVA). C: Quantification by RT-qPCR of CDKL5 expression in SH-SY5Y and SKNBE cells that were either untreated or treated with RA (10 μM) for 3 and 7 days. Data, given as percentage of untreated SH-SY5Y cells, are expressed as mean ± SE. The asterisks indicate a significant difference between treated vs. untreated condition, *P < 0.05, ***P < 0.001 (Bonferroni's test after ANOVA). D: CDKL5 protein expression in SH-SY5Y cells either untreated or treated with RA (10 μM) for 3 and 7 days was analyzed by immunoblot with a CDKL5 specific antibody.
The correlation between CDKL5 expression and SH-SY5Y differentiation indicates that SH-SY5Y neuroblastoma cells may represent a suitable model to study the role of CDKL5 in neuronal proliferation/differentiation.
3.2. CDKL5 promotes neuronal differentiation in the SH-SY5Y neuroblastoma cell line
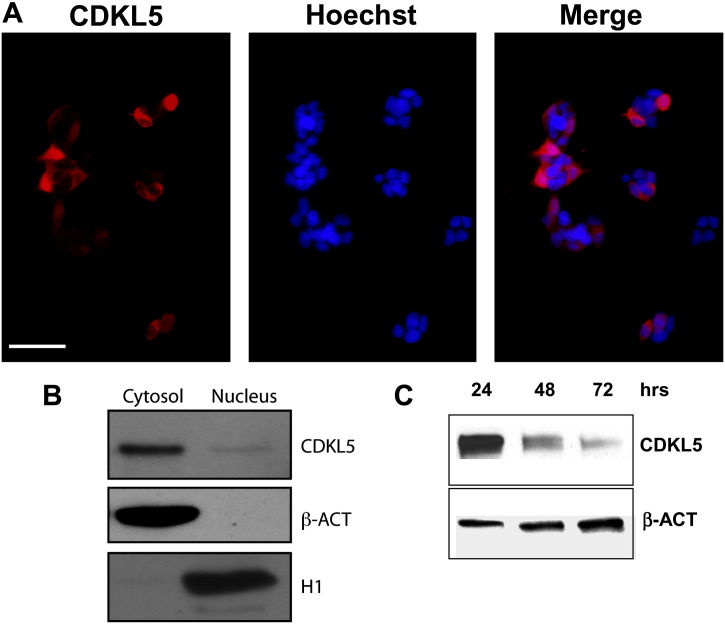
To demonstrate that CDKL5 can play some role in neuronal differentiation, we over-expressed the CDKL5 protein in SH-SY5Y cells by transient transfection of a pGFP/CDKL5-FLAG expression vector (Supplementary Fig. 1C — Analysis of the time-course of over-expression of CDKL5-FLAG showed that it was highly expressed at 24 h after transfection and that its expression was notably reduced at 72 h). As CDKL5 has been reported to localize both in the nuclear and cytoplasmic compartments [5,8,24,35,36], we first determined its cellular localization. As shown in Supplementary Fig. 1A, CDKL5 localized in both the cytoplasm and the nucleus (Supplementary Fig. 1A, panels). As previously reported [36,47], Western blot analysis of fractionated extracts showed that CDKL5 was preferentially present in the cytoplasm (Supplementary Fig. 1B).
Supplementary Fig. 1.
A: Sub-cellular localization of CDKL5. Representative fluorescence image of SH-SY5Y cells expressing CDKL5-FLAG. Twenty-four hours after transfection, SH-SY5Y cells were immunostained for FLAG (red signal) and Hoechst dye (blue signal). Cells show both CDKL5 nuclear and cytoplasmic localization. Scale bar: 40 μm. B: FLAG-tagged CDKL5 was transiently over-expressed in SH-SY5Y cells and detected in nuclear and cytoplasmatic extracts 24 h after transfection. Example of Western blot with anti-FLAG antibodies. Histon H1 (H1) and β-actina (β-ACT) were used respectively as nuclear and cytoplasmic markers. C: FLAG-tagged CDKL5 was transiently over-expressed in SH-SY5Y cells and detected 24, 48 and 72 h after transfection. Total cell extracts were analyzed by Western blotting with anti-FLAG and β-actina antibodies.
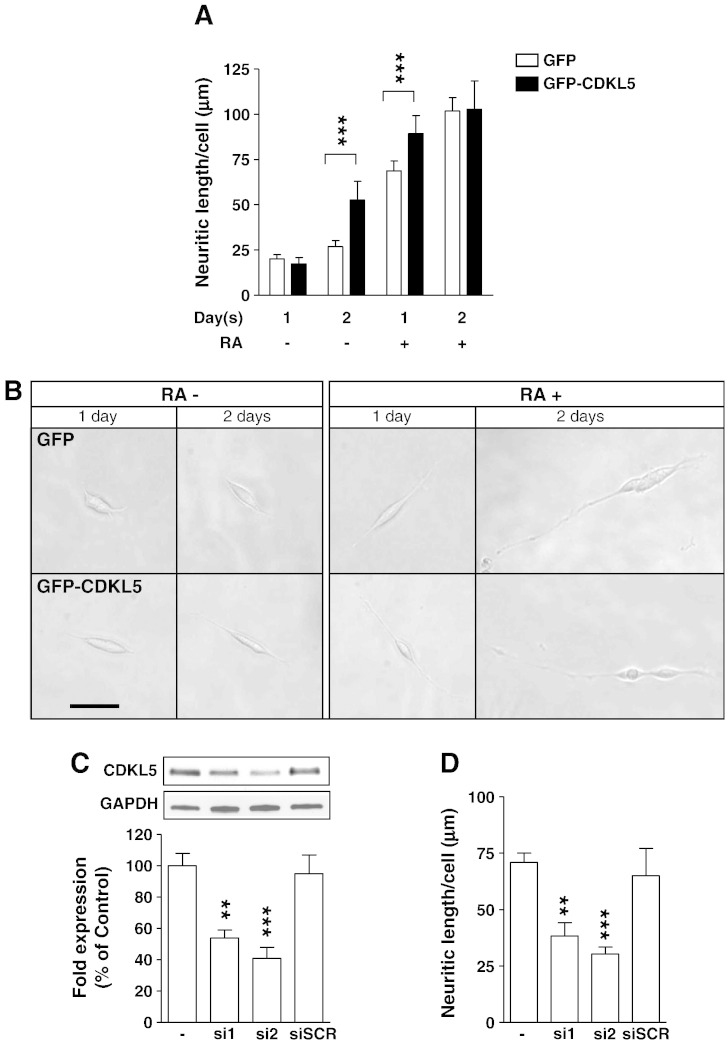
We next examined neuritic growth in GFP- and GFP-CDKL5-positive cells isolated by cell sorting 24 h after transfection and then grown for 1–2 days either in the absence or presence of RA. We found that CDKL5 was able to induce a beginning of differentiation, as assessed by evaluation of neuritic length (Fig. 2A,B). While cells expressing GFP alone (controls) occasionally emitted very short processes (Fig. 2B, left panel), cells expressing GFP-CDKL5 had longer processes with a length increase by +100% after 2 days (Fig. 2A; Fig. 2B, left panel). To analyse the effects of CDKL5 expression on RA-induced differentiation, GFP- and GFP-CDKL5-positive cells were cultured for 1–2 days in the presence of RA (Fig. 2A,B). After 1 day of RA treatment, CDKL5 expressing cells exhibited a greater neurite length as compared to control cells (+30%, Fig. 2A,B). This difference was no longer detectable after 2 days of RA treatment (Fig. 2A,B). The lack of difference between control and transfected cells after 2 days of RA treatment (i.e. 72 h post transfection) is most likely attributable to the time-dependent reduction of exogenous CDKL5 levels (Supplementary Fig. 1C). Though the effect of CDKL5 was smaller than that induced by RA (Fig. 2A), these data clearly show that CDKL5 can promote neuronal differentiation in the absence of pro-differentiative stimuli. To further corroborate this finding, we tested whether a reduction in CDKL5 expression interfered with RA-induced differentiation. To this purpose SH-SY5Y cells were transfected with two different siRNAs against CDKL5 (si1, si2) to inhibit CDKL5 expression and treated with RA. After 48 h of treatment, the reduced expression of CDKL5 induced by the siRNAs against CDKL5 (− 45% and − 60%, Fig. 2C) was paralleled by a reduction (− 46% and − 58%, Fig. 2D) in neurite elongation. Transfection with a scrambled siRNA (siScr), as negative control, had no effect on CDKL5 expression and RA-induced cell differentiation (Fig. 2C,D).
Fig. 2.
CDKL5 induces differentiation in the SH-SY5Y neuroblastoma cell line.
A: Quantification of neurite outgrowth of SH-SY5Y cells transfected with either GFP-CDKL5 or GFP. Twenty four hours from transfection, cells were FACS sorted for GFP fluorescence and cultured for 1–2 days with or without retinoic acid (RA; 10 μM). Neurite outgrowth was expressed as mean neurite length (μm) per cell. Data are expressed as mean ± SE of 3–4 independent experiments. A minimum of 400 cells were evaluated in each experiment for each condition. The asterisks indicate a significant difference between cells expressing GFP-CDKL5 vs. cells expressing GFP alone. ***P < 0.001; two-tailed t-test. B: Representative phase-contrast images of cells expressing GFP-CDKL5 or GFP and cultured for 1 or 2 days in absence of RA (RA−) or in presence of 10 μM RA (RA+). Scale bar: 15 μm C: SH-SY5Y cells were transfected with two different siRNA against CDKL5 (si1 and si2; 50 nM) or with scramble siRNA (siScr; 50 nM). Quantification by Western blots of CDKL5 expression was performed 48 h after transfection. D: Quantification of neurite outgrowth of SH-SY5Y cells transfected with si1, si2 or siSCR (50 nM). Six hours after siRNA transfection cells were treated with RA (10 μM) and analyzed 42 h later for neurite outgrowth. Data are expressed as mean ± SE of 3 independent experiments. **P < 0.01, two-tailed t-test.
These data indicate that CDKL5 alone can trigger differentiation of neuroblastoma cells and enhance the differentiation induced by RA, confirming the role of CDKL5 in neuron differentiation.
3.3. CDKL5 negatively regulates cell proliferation in the SH-SY5Y neuroblastoma cell line
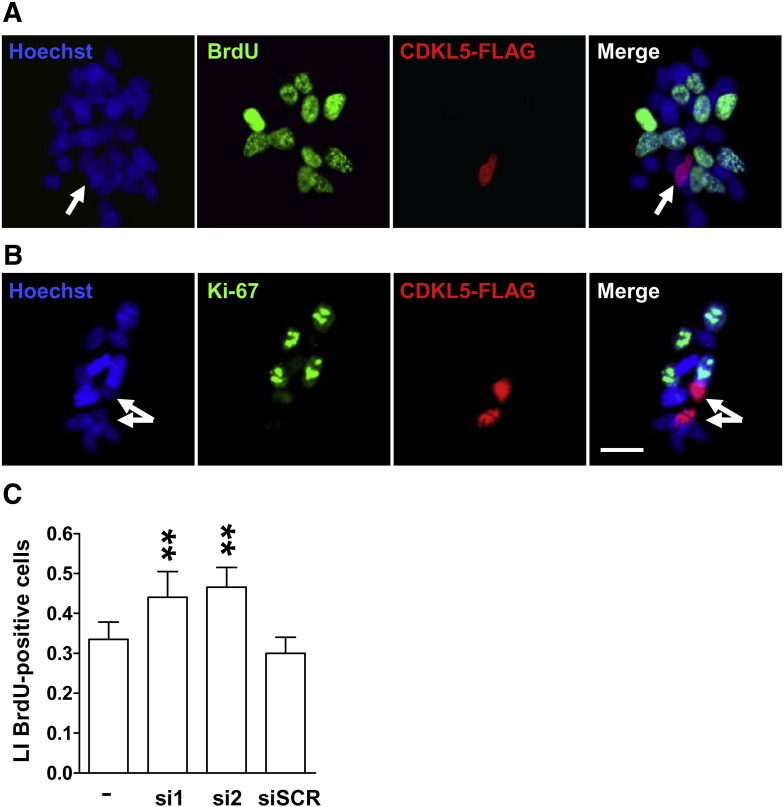
It has been previously shown that CDKL5 expression peaks during late embryonic stage and the first postnatal period [8], when most of the neuronal progenitors stop proliferating and enter the differentiated state, suggesting that it may negatively control neuron proliferation. To address this point, SH-SY5Y cells transiently expressing the CDKL5-FLAG fusion protein were evaluated for proliferation rate by a bromodeoxyuridine (BrdU) incorporation assay, in which the thymidine analogue is incorporated into DNA during the S-phase of the cell cycle. The effect of CDKL5 on cell proliferation was determined by evaluating the number of proliferating cells (BrdU positive cells) that expressed CDKL5 (CDKL5-FLAG positive cells). Interestingly, we could never observe cells that, in addition to express CDKL5, were also BrdU positive, supporting our hypothesis that CDKL5 may inhibit cell proliferation (Fig. 3A). As a further control, cells were also immunostained for Ki-67 a typical proliferation marker that is expressed in cell nuclei of dividing cells through late-G1 + S + G2 + M but not G0 and early G1 phases of cell cycle [38]. As expected no CDKL5-positive cells expressed Ki-67 (Fig. 3B). To exclude non-specific effects due to protein over‐expression, SH-SY5Y cells were transfected with a CMV-FLAG-Luciferase control vector. We have chosen Luciferase in that its expression should not affect cell cycle dynamics. Indeed, we found that about 30% of BrdU positive cells also expressed Luciferase (data not shown). Our data indicate that CDKL5 over-expression specifically blocks cell proliferation of neuroblatoma cells.
Fig. 3.
Effect of CDKL5 expression on proliferation in the SH-SY5Y neuroblastoma cell line.
A,B: Immunofluorescence images of SH-SY5Y neuroblastoma cells transfected with CDKL5-FLAG. Twenty-four hours after transfection, SH-SY5Y cells were treated with BrdU (10 μM) for 2 h and thereafter cells were processed for double immunocytochemistry. Cells were immunostained for FLAG (red signal) and BrdU (green signal in A) or FLAG and Ki-67 (green signal in B). Cell nuclei were stained with Hoechst dye (blue signal). The arrows in Hoechst and merge images indicate the nuclei of cells expressing exogenous CDKL5. Three independent experiments were performed and a minimum of 600 cells were evaluated in each experiment for each condition. Scale bar: 20 μm. C: Labeling index (LI), defined as percentage of BrdU positive cells over total cell number, was determined for SH-SY5Y transfected two different siRNA against CDKL5 (si1 and si2; 50 nM) or with scramble siRNA (siSCR; 50 nM). Forty six hours after transfection cells were exposed to BrdU (10 μM) for the last 2 h. Data, given as percentage of control condition, are expressed as mean ± SE of 3 independent experiments. **P < 0.01; two-tailed t-test.
To confirm the anti-proliferative role of CDKL5, the endogenous expression of the protein was silenced using two siRNAs directed against CDKL5 (si1, si2). We found that a reduced expression of CDKL5 after siRNAs transfection corresponded to an increase in cell proliferation (+31% si1, +39% si2, Fig. 3C), thus supporting the idea that CDKL5 can inhibit cell proliferation. Treatment with a scrambled siRNA (siScr), as negative control, had no effect on cell proliferation (data not shown).
3.4. CDKL5 does not induce cell death in the SH-SY5Y neuroblastoma cell line
We also investigated whether CDKL5 expression may trigger cell death in neuroblastoma cells. When we evaluated the percentage of apoptosis by immunostaining of intracellular cleaved caspase-3 we found no difference in the number of apoptotic cells between CDKL5 positive and negative cells (Fig. 4A,B), indicating that CDKL5 over-expression did not induce apoptotic cell death.
3.5. CDKL5 blocks cell cycle progression in the SH-SY5Y neuroblastoma cell line
The fact that CDKL5 can inhibit proliferation leads us to speculate that CDKL5 may affect the cell cycle dynamics. To clarify this point, we compared the cell cycle profile of GFP vs. GFP-CDKL5 transfected SH-SY5Y cells. FACS sorted GFP positive cells (Fig. 4C) were treated with propidium iodide to stain DNA and analyzed by flow cytometry. We found that the fraction of cells in G0/G1 was significantly increased in CDKL5 over-expressing cells as compared to control cells and, as a consequence, the fraction of S phase cells was decreased. In CDKL5 over-expressing cells, the percentage of G0/G1 became approximately 70%, suggesting that CDKL5 blocks cells in G0/G1 phase (Fig. 4D). We found no differences in the percentage of cells in the sub-G1, which is considered to indicate the proportion of apoptotic cells over total (data not shown), indicating that CDKL5 does not induce apoptosis in neuroblastoma cells. These data suggest that the inhibition of cell proliferation following expression of exogenous CDKL5 is due to arrest in the G0/G1 phase of the cell cycle.
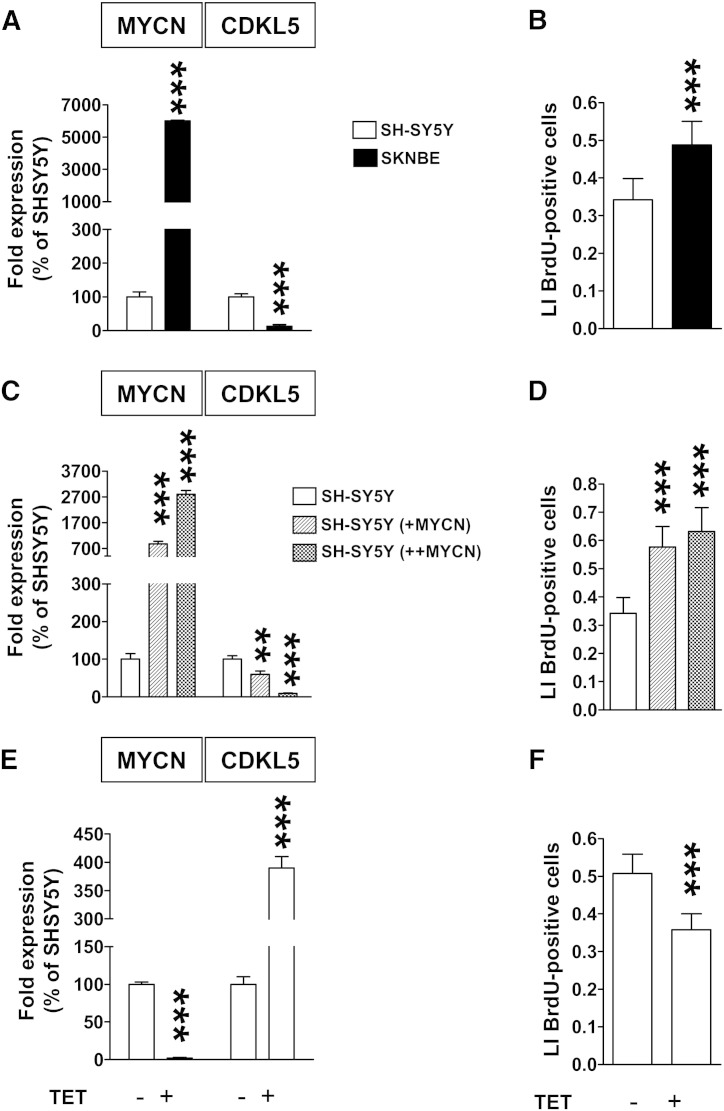
3.6. Inverse relationship between CDKL5 and MYCN expression in neuroblastoma cell lines and neuroblastomas
The oncogene MYCN is emerging as an important regulator of neuron expansion during brain development that acts by increasing cell proliferation and preventing cell differentiation [17,23]. Due to its antiproliferative/prodifferentiative role, CDKL5 can be a good candidate as one of the gene negatively regulated by MYCN during brain development. To determine a correlation between CDKL5 and MYCN we evaluated CDKL5 expression in a MYCN amplified neuroblastoma cell line (SKNBE) and in a MYCN non-amplified neuroblastoma cell line (SH-SY5Y). CDKL5 expression was found to inversely correlate with MYCN expression (Fig. 5A) and with the proliferation potency of these cells (Fig. 5B). To rule out that the inverse relationship between CDKL5 and MYCN expression observed in the SKNBE and SH-SY5Y cell lines is due to their different origins, we generated stable SH-SY5Y neuroblastoma cell clones expressing different levels of MYCN protein. CDKL5 expression in the various SH-SY5Y-derived cell lines showed a close inverse correlation with the levels of MYCN expression (Fig. 5C). This suggests that MYCN over-expression reduces CDKL5 expression. In agreement with the pro-proliferative nature of MYCN, MYCN over‐expressing cell lines, showed an increase in the proliferation rate that correlated with MYCN expression levels (Fig. 5D). To confirm the inverse relationship between CDKL5 and MYCN expression, the conditional MYCN expressing SHEP Tet21/N cell line [26] was treated with tetracycline (TET) to inhibit MYCN expression. Tetracycline treatment led to an increase in CDKL5 expression (Fig. 5E) and a concomitant decrease in cell proliferation (Fig. 5F), further supporting MYCN regulation of CDKL5 expression.
Fig. 5.
Inverse relationship between MYCN and CDKL5 expression.
A: Quantification by RT-qPCR of MYCN and CDKL5 expression in SH-SY5Y and SKNBE cells. B: Labeling index (LI) of SH-SY5Y and SKNBE cells. Cell cultures were treated with BrdU for 2 h and processed for immunofluorescence with an anti-BrdU antibody. LI is defined as the number of BrdU-positive cells over total cell number, identified by Hoechst staining. C: Quantification by RT-qPCR of MYCN and CDKL5 expression in SH-SY5Y cells and in SH-SY5Y cells engineered to express higher levels of MYCN (+MYCN, ++MYCN). D: LI of SH-SY5Y, SH-SY5Y(+MYCN) and SH-SY5Y(++MYCN). E: Quantification, by RT-qPCR, of MYCN and CDKL5 expression in Tet21/N neuroblastoma cells treated or untreated with tetracycline (TET). F: LI of Tet21/N neuroblastoma cells treated as reported in (E). Data in A–F are expressed as mean ± SE of 3 independent experiments. **P < 0.01, ***P < 0.001, two-tailed t-test.
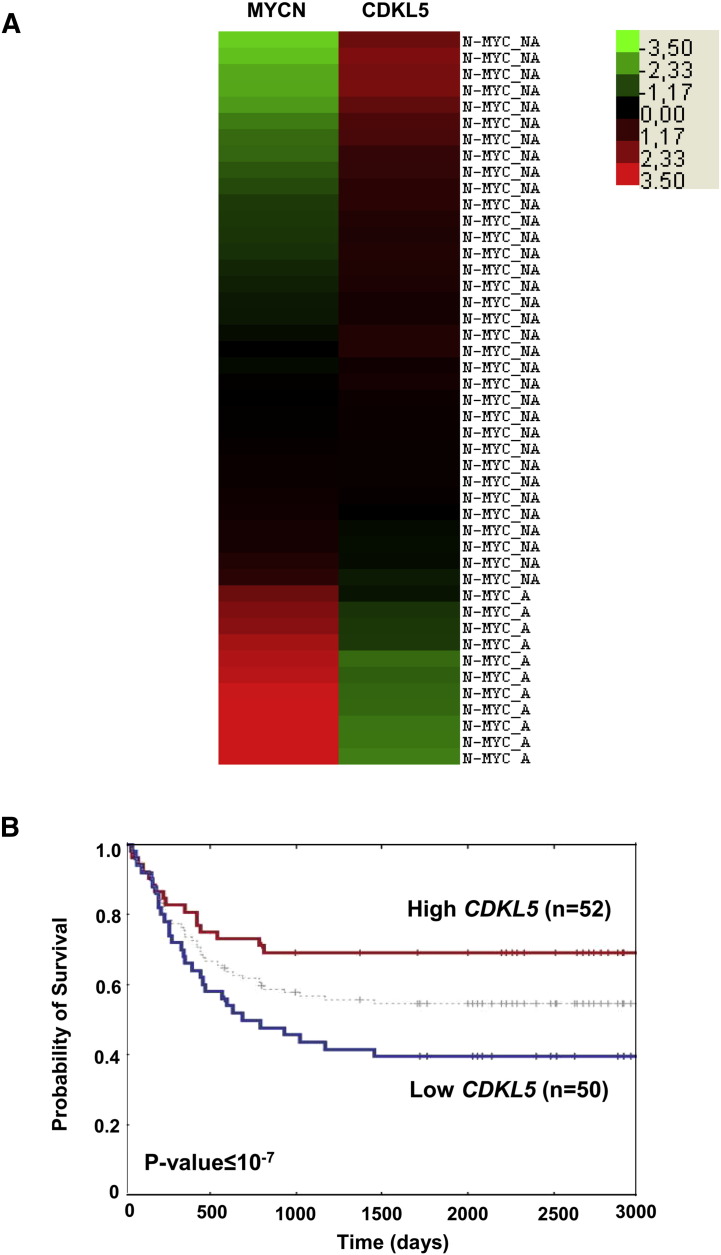
A bioinformatic meta-analysis of human neuroblastoma transcriptome databases [31] was used to further investigate CDKL5 expression in relation to MYCN expression. We found higher levels of CDKL5 expression in neuroblastomas without MYCN amplification (n = 11), compared with those with MYCN amplification (n = 34) (Supplementary Fig. 2A), indicating that in neuroblastoma patients, similarly to neuroblastoma cell lines, CDKL5 expression is inversely associated with MYCN amplification and expression. Kaplan–Meier survival analysis shows that a high CDKL5 expression is associated with a good disease outcome of neuroblastomas (Supplementary Fig. 2B), which is in line with the anti-proliferative role of CDKL5 demonstrated here in neuroblastoma cell lines.
Supplementary Fig. 2.
A: Bioinformatic analysis of CDKL5 expression in transcriptome databases of human neuroblastoma tissue samples classified on the basis of MYCN amplification. Differentially expressed genes are defined by a two-fold-change cut-off. The color-coded scale (green = down-regulation and red = up-regulation) for the normalized expression value is indicated at the top of the figure. The hierarchical clustering of the samples was done using Cluster 3 software. B: Survival probabilities of two groups of subjects with neuroblastomas with low- and high-levels of CDKL5 expression. Survival probability was estimated by the method of Kaplan–Meier. The median expression value of CDKL5 based on the entire cohort (n = 102) was used as a cut-off to stratify high- and low-expression subgroups. Five-year survival was calculated for each group, and the log-rank test was used to compare survival probabilities of the two groups.
3.7. CDKL5 expression is transcriptionally inhibited by MYCN
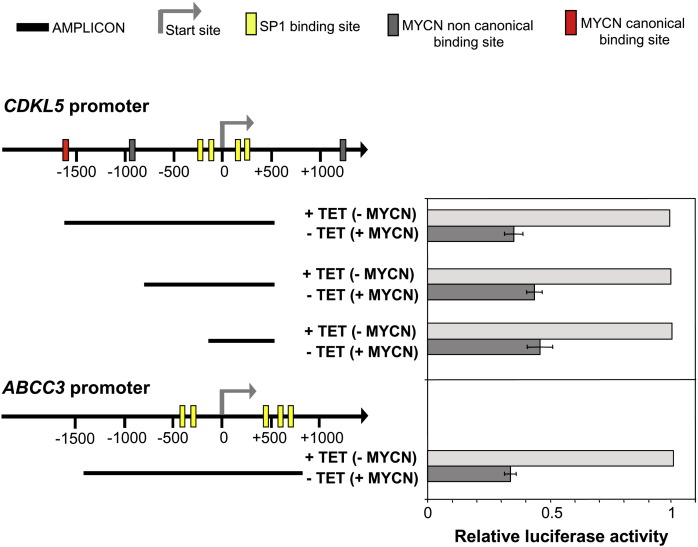
It has been recently shown that MYCN can bind and repress gene transcription by interacting with the transcription factor SP1 [34]. Bioinformatic analysis showed an SP1-rich region proximal to the start site of the CDKL5 promoter. Dual ChIP assay was performed in a SH-SY5Y cell clone engineered to constitutively express high levels of MYCN (SH-SY5Y++MYCN). We found that MYCN binds to the SP1-reach region of the CDKL5 promoter (Fig. 6). As expected, a strong binding of SP1 was also present in the same region (Fig. 6).
To establish whether MYCN can regulate CDKL5 at the promoter level, CDKL5 reporter promoter constructs were transiently transfected into Tet21/N (+MYCN) and Tet21/N (− MYCN) cells. CDKL5 promoter constructs incorporating the SP1-reach region exhibited a significant reduction in the Luciferase activity in the presence of MYCN (+MYCN cells), indicating that MYCN inhibits CDKL5 transcription (Fig. 7). A promoter construct limited to the SP1-reach region was indeed sufficient to inhibit transcription, indicating that MYCN could inhibit CDKL5 transcription through the SP1-reach region, possibly by interacting with SP1 as previously established [28]. The extent of MYCN-mediated repression was comparable to that observed for the ABCC3 promoter (Fig. 7), another gene known to be repressed by MYCN through interaction with SP1.
Fig. 7.
MYCN inhibits CDKL5 expression.
Tet21/N neuroblastoma cells were transfected with luciferase reporter constructs containing different regions of the CDKL5 promoter. The cloned DNA regions (bp) are indicated below the promoter map. The firefly luciferase activity was determined in the presence (+TET) or absence (− TET) of MYCN expression. Luciferase reporter analysis of ABCC3 promoter is shown as control. The results are the means ± SE of three independent transfections.
3.8. MYCN-dependent CDKL5 expression in primary cerebellar granule cells
We have recently shown that cerebellar granule cell precursors (GCPs) exhibit high levels of MYCN expression, that progressively decrease with granule cell differentiation both in vivo and in vitro [10,11]. To establish whether an inverse relationship between MYCN and CDKL5 expression is present in neuronal precursors, we measured their expression in primary cultures of cerebellar granule cells (CGCs) before and after in vitro differentiation. These cells are a homogeneous neuronal population (90–95% of the total cells in the culture) that replicates many of the biochemical and physiological features of native cerebellar development. GCPs undergo differentiation during the first week in culture and a fully differentiated state is reached by 7 days in vitro (DIV). We found that in freshly dissociated GCPs (DIV0) there was a higher expression of MYCN than after induction of differentiation (DIV 7) (Fig. 8A). Conversely, CDKL5 expression was lower at DIV0 than at DIV 7 (Fig. 8B). In parallel, as previously reported [10], proliferation was completely inhibited in differentiated CGCs (Fig. 8C).
To establish whether the inverse relationship between MycN and Cdkl5 protein expression during CGC differentiation underlies a MycN-dependent transcriptional inhibition of Cdkl5 expression, we performed ChIP experiments on undifferentiated CGPs vs. differentiated CGCs. Bioinformatic analysis of the mouse Cdkl5 gene promoter identified, similarly to the human CDKL5 promoter, one region proximal to the transcription start site enriched for Sp1-binding sites (Fig. 8D). Dual cross-linking ChIP assay in CGPs showed that antibodies against MycN and Sp1 can efficiently immunoprecipitate the region of Cdkl5 gene promoter carrying Sp1-binding sites (Fig. 8D). In contrast, no MycN-enrichment was found in differentiated CGCs (Fig. 8D). These data confirm a direct role of MycN on CDKL5 expression in primary neurons.
4. Discussion
In spite of the clear importance of CDKL5 for the central nervous system, the biological functions of this kinase remain largely unknown. Here we show for the first time that this gene affects both proliferation and differentiation of neural cells. Moreover, our results show that CDKL5 expression is regulated by MYCN and suggest that CDKL5 is one of the genes through which MYCN prevents proliferation and differentiation of neural precursors.
4.1. CDKL5 induces differentiation and inhibits proliferation of the SH-SY5Y neuroblastoma cell line
We provide evidence that in the SH-SY5Y neuroblastoma cell line there is a direct correlation between neurite elongation and CDKL5 expression. This is consistent with recent lines of evidence showing that CDKL5 regulates neurite growth and dendritic arborization of cortical rat neurons [8]. Our data strengthen the role of CDKL5 during development as a pro-differentiating gene. Impaired brain development, including dendritic pathology and synaptic imbalance between excitatory and inhibitory neurons, may contribute to epilepsy. There are a number of neurodevelopment disorders, such as RTT, Fragile X syndrome and Down syndrome, that directly affect dendritic structure, and which have a very high prevalence of epilepsy. Our finding that CDKL5 is important for neuronal maturation provides novel evidence for cellular mechanisms that may contribute to the EIEE2 disease phenotypes.
We found that in the SH-SY5Y cell line induction of CDKL5 expression caused a strong inhibition of cell proliferation with no increase in apoptotic cell death. Inhibition of proliferation was due to a block of cell cycle progression in the G0/1 phase, supporting the view that CDKL5 can function as an anti-proliferative gene. A recent study showed that CDKL5 is highly expressed in some clinical samples of adult T-cell leukemia (ATL) and in several ATL cell lines [20]. The role played by CDKL5 on cell proliferation in these and other tumor cells remains to be established.
Because neuronal differentiation requires the progenitor to exit the cell cycle, it is generally assumed that the processes of proliferation and differentiation are co-regulated. These processes intersect at the regulation of cell cycle regulatory proteins. Proliferation promotes positive regulation of cell cycle proteins (e.g. cyclins and cyclin-dependent kinases (Cdk)), whereas differentiation results in inhibition of these cell cycle proteins [32]. Progression of the cell cycle to the G0 phase leads to the cell cycle exit and differentiation. Our findings show that CDKL5 acts both as a cell cycle inhibitor and a pro-differentiating gene and are in line with several arguments supporting a general interdependence of proliferation and differentiation control. Very recently, it has been shown that induced Pluripotent Stem Cells (iPSCs) from CDKL5 mutated patients can be differentiated into neurons [1], suggesting that CDKL5 is not per se necessary for acquisition of a neuronal phenotype and that CDKL5 mediated differentiation may be a secondary effect of the CDKL5 block of cells in the G0–G1 phase. For this reason CDKL5-mutated iPSCs could be a suitable model to confirm the anti-proliferative and pro-differentiative role of CDKL5 in human neuronal cells, found here in a neuroblastoma cell line.
Consistently with data in other cell lines and in vivo, we found that CDKL5 localizes both at the cytoplasmic and nuclear level [8,36]. The cytoplasmic localization of CDKL5 and its catalytic activity are essential for the effects of this kinase on dendritic maturation [8]. CDKL5 appears to interact with nuclear factors, such as MeCP2 [24,28] and DNA methyltransferase I [19], and associates with nuclear speckles involved in RNA splicing [35]. It remains to be established whether CDKL5 nuclear localization plays a role in the observed regulation of cell proliferation.
4.2. CDKL5 is a target of MYCN
While the vast majority of studies on MYCN have been conducted in tumor cells, growing evidence suggests a key role for MYCN during both murine and human brain development [23,43]. During brain development, MYCN plays an important role to direct brain growth [17,23], consistently with its widespread expression pattern. In agreement with the pro-proliferative role of MYCN, conditional disruption of Mycn in neural precursor cells severely disrupts brain growth, particularly that of the cerebellum [17]. While this evidence clearly indicates a role for MYCN during brain development, the mechanisms remain somewhat unclear.
By using stable and conditional cell lines that express high and low levels of MYCN, we found that inhibition of MYCN expression correlated to marked increase of CDKL5 expression. Conversely, induction of MYCN expression led to inhibition of CDKL5 expression. Although MYCN activates transcription of pro-proliferative cellular networks, it is increasingly clear that it may exert its pro-proliferative action also through repression of specific target genes. Our data indicate that MYCN binds with the CDKL5 promoter region that is recognized by SP1 and represses CDKL5 expression. This is consistent with recent evidence showing that a direct inhibitory interaction between MYC and SP1 can promote repression of several genes involved in cell cycle control and differentiation [16]. Our study establishes a functional axis between MYCN and CDKL5 in which MYCN antagonizes the pro-differentiation activity of CDKL5 during neuron maturation.
MYCN plays a central role in the biology of neuroblastoma and several other cancers as a driving oncogene for tumorigenesis [39]. MYCN amplification remains the strongest negative prognostic indicator for neuroblastoma [12]. Interestingly, the Kaplan–Meier survival analysis shows that CDKL5 expression correlates with less aggressive tumors (Supplementary Fig. 2B), suggesting that the expression of CDKL5 may be a favorable predictor of clinical outcome. This hypothesis is fully in agreement with the anti-proliferative role of CDKL5 observed here in neuroblastoma cell lines. The present work raises the issue of whether MYCN may contribute to the progression of neuroblastoma though repression of CDKL5 expression. Future studies, exploiting the transgenic MYCN mouse model TH-MYCN, a native neuroblastoma model in which tumorigenesis is driven by targeted expression of MYCN in the neural crest [46], would help understand the MYCN-CDKL5 relationship in the neuroblastoma pathogenesis, in vivo.
4.3. The MYCN/CDKL5 system appears to be involved in cerebellar development
MYCN appears to play an important role in cerebellar development. It promotes rapid cell division of cerebellar granule neural precursors [21,22] and in Mycn-KO mice there is a strong reduction in the number of cerebellar granule precursors and mature granule neurons [23]. The search for genes that are regulated by MYC has yielded several hundred candidates. Recently, at least a dozen microarray-based screens, mostly in tumors, have added over 600 genes to the list of potential MYC targets [33]. With a few notable exceptions, these kinds of studies have not identified genes that have obvious links to the physiological processes typically associated with MYC functions (for example, cell-cycle progression and maintenance of pluripotency).
The expression of CDKL5 in the cerebellum has been shown to be only minimally detectable at embryonic stage E16.5, but to be strongly up-regulated in differentiating cerebellar granule cells, during postnatal development [36]. This expression pattern suggests that CDKL5 may be one of the genes regulated by MYCN, involved in the shift from proliferation to differentiation of cerebellar granule cell precursors. Consistently with this idea, we found a relatively low expression of CDKL5 in granule cell precursors at 0 DIV, a time at which there was a high expression of MYCN, and a strong increase after 7 DIV, concomitantly with a reduction in the expression of MYCN. At this stage, neurons have reached what is considered under several parameters a differentiate state, comparable to that of in vivo granule cells. The expression pattern of the MYCN/CDKL5 system during differentiation of cerebellar granule cells is in line with that observed in differentiating neuroblastoma cells. Taken together our data suggest that the MYCN-inhibition of CDKL5 expression may be a generalized mechanism across different neuronal precursors.
4.4. Conclusion
The connection between CDKL5 mutation genotype and disease phenotype is hindered by the lack of information regarding the role of CDKL5 in neuronal development and function. Our finding that CDKL5 is important for neuronal proliferation/differentiation provides novel evidence for cellular mechanisms that may contribute to the EIEE2 disease phenotypes.
The following are the supplementary data related to this article.
Primers used in the present study.
Acknowledgements
This work was supported by the Telethon grant (GGP11147) to EC and GP, by the Italian “CDKL5 associazione volontariato” to EC and GP and by the Fondazione del Monte di Bologna e Ravenna to EC.
Contributor Information
Giovanni Perini, Email: giovanni.perini@unibo.it.
Elisabetta Ciani, Email: elisabetta.ciani@unibo.it.
References
- 1.Amenduni M. De, Filippis R., Cheung A.Y., Disciglio V., Epistolato M.C., Ariani F., Mari F., Mencarelli M.A., Hayek Y., Renieri A., Ellis J., Meloni I. iPS cells to model CDKL5-related disorders. Eur. J. Hum. Genet. 2012;19:1246–1255. doi: 10.1038/ejhg.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amir R.E. Van, den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong D.D. Neuropathology of Rett syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 2002;8:72–76. doi: 10.1002/mrdd.10027. [DOI] [PubMed] [Google Scholar]
- 4.Bahi-Buisson N., Kaminska A., Boddaert N., Rio M., Afenjar A., Gerard M., Giuliano F., Motte J., Heron D., Morel M.A., Plouin P., Richelme C., des Portes V., Dulac O., Philippe C., Chiron C., Nabbout R., Bienvenu T. The three stages of epilepsy in patients with CDKL5 mutations. Epilepsia. 2008;49:1027–1037. doi: 10.1111/j.1528-1167.2007.01520.x. [DOI] [PubMed] [Google Scholar]
- 5.Bertani I., Rusconi L., Bolognese F., Forlani G., Conca B. De, Monte L., Badaracco G., Landsberger N., Kilstrup-Nielsen C. Functional consequences of mutations in CDKL5, an X-linked gene involved in infantile spasms and mental retardation. J. Biol. Chem. 2006;281:32048–32056. doi: 10.1074/jbc.M606325200. [DOI] [PubMed] [Google Scholar]
- 6.Castren M., Gaily E., Tengstrom C., Lahdetie J., Archer H., Ala-Mello S. Epilepsy caused by CDKL5 mutations. Eur. J. Paediatr. Neurol. 2011;15:65–69. doi: 10.1016/j.ejpn.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Chao H.T., Zoghbi H.Y., Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q., Zhu Y.C., Yu J., Miao S., Zheng J., Xu L., Zhou Y., Li D., Zhang C., Tao J., Xiong Z.Q. CDKL5, a protein associated with rett syndrome, regulates neuronal morphogenesis via Rac1 signaling. J. Neurosci. 2010;30:12777–12786. doi: 10.1523/JNEUROSCI.1102-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R.Z., Akbarian S., Tudor M., Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 10.Ciani E., Severi S., Contestabile A., Bartesaghi R. Nitric oxide negatively regulates proliferation and promotes neuronal differentiation through N-Myc downregulation. J. Cell Sci. 2004;117:4727–4737. doi: 10.1242/jcs.01348. [DOI] [PubMed] [Google Scholar]
- 11.Ciani E., Calvanese V., Crochemore C., Bartesaghi R., Contestabile A. Proliferation of cerebellar precursor cells is negatively regulated by nitric oxide in newborn rat. J. Cell Sci. 2006;119:3161–3170. doi: 10.1242/jcs.03042. [DOI] [PubMed] [Google Scholar]
- 12.Cohn S.L., Pearson A.D., London W.B., Monclair T., Ambros P.F., Brodeur G.M., Faldum A., Hero B., Iehara T., Machin D., Mosseri V., Simon T., Garaventa A., Castel V., Matthay K.K. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J. Clin. Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinse G.E., Lagakos S.W. Nonparametric estimation of lifetime and disease onset distributions from incomplete observations. Biometrics. 1982;38:921–932. [PubMed] [Google Scholar]
- 14.Evans J.C., Archer H.L., Colley J.P., Ravn K., Nielsen J.B., Kerr A., Williams E., Christodoulou J., Gecz J., Jardine P.E., Wright M.J., Pilz D.T., Lazarou L., Cooper D.N., Sampson J.R., Butler R., Whatley S.D., Clarke A.J. Early onset seizures and Rett-like features associated with mutations in CDKL5. Eur. J. Hum. Genet. 2005;13:1113–1120. doi: 10.1038/sj.ejhg.5201451. [DOI] [PubMed] [Google Scholar]
- 15.Gallo V., Ciotti M.T., Coletti A., Aloisi F., Levi G. Selective release of glutamate from cerebellar granule cells differentiating in culture. Proc. Natl. Acad. Sci. U. S. A. 1982;79:7919–7923. doi: 10.1073/pnas.79.24.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gartel A.L., Ye X., Goufman E., Shianov P., Hay N., Najmabadi F., Tyner A.L. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatton B.A., Knoepfler P.S., Kenney A.M., de Rowitch D.H., Alboran I.M., Olson J.M., Eisenman R.N. N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res. 2006;66:8655–8661. doi: 10.1158/0008-5472.CAN-06-1621. [DOI] [PubMed] [Google Scholar]
- 18.Kalscheuer V.M., Tao J., Donnelly A., Hollway G., Schwinger E., Kubart S., Menzel C., Hoeltzenbein M., Tommerup N., Eyre H., Harbord M., Haan E., Sutherland G.R., Ropers H.H., Gecz J. Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am. J. Hum. Genet. 2003;72:1401–1411. doi: 10.1086/375538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kameshita I., Sekiguchi M., Hamasaki D., Sugiyama Y., Hatano N., Suetake I., Tajima S., Sueyoshi N. Cyclin-dependent kinase-like 5 binds and phosphorylates DNA methyltransferase 1. Biochem. Biophys. Res. Commun. 2008;377:1162–1167. doi: 10.1016/j.bbrc.2008.10.113. [DOI] [PubMed] [Google Scholar]
- 20.Kawahara M., Hori T., Matsubara Y., Okawa K., Uchiyama T. Cyclin-dependent kinaselike 5 is a novel target of immunotherapy in adult T-cell leukemia. J. Immunother. 2007;30:499–505. doi: 10.1097/CJI.0b013e3180336771. [DOI] [PubMed] [Google Scholar]
- 21.Kenney A.M., Cole M.D., Rowitch D.H. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 22.Kenney A.M., Widlund H.R., Rowitch D.H. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development. 2004;131:217–228. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- 23.Knoepfler P.S., Cheng P.F., Eisenman R.N. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C., Franco B., Rosner M.R. CDKL5/Stk9 kinase inactivation is associated with neuronal developmental disorders. Hum. Mol. Genet. 2005;14:3775–3786. doi: 10.1093/hmg/ddi391. [DOI] [PubMed] [Google Scholar]
- 25.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.Lutz W., Stohr M., Schurmann J., Wenzel A., Lohr A., Schwab M. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene. 1996;13:803–812. [PubMed] [Google Scholar]
- 27.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother. Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 28.Mari F., Azimonti S., Bertani I., Bolognese F., Colombo E., Caselli R., Scala E., Longo I., Grosso S., Pescucci C., Ariani F., Hayek G., Balestri P., Bergo A., Badaracco G., Zappella M., Broccoli V., Renieri A., Kilstrup-Nielsen C., Landsberger N. CDKL5 belongs to the same molecular pathway of MeCP2 and it is responsible for the early-onset seizure variant of Rett syndrome. Hum. Mol. Genet. 2005;14:1935–1946. doi: 10.1093/hmg/ddi198. [DOI] [PubMed] [Google Scholar]
- 29.Mastrangelo M., Leuzzi V. Genes of early-onset epileptic encephalopathies: from genotype to phenotype. Pediatr. Neurol. 2011;46:24–31. doi: 10.1016/j.pediatrneurol.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Melino G., Thiele C.J., Knight R.A., Piacentini M. Retinoids and the control of growth/death decisions in human neuroblastoma cell lines. J. Neurooncol. 1997;31:65–83. doi: 10.1023/a:1005733430435. [DOI] [PubMed] [Google Scholar]
- 31.Oberthuer A., Berthold F., Warnat P., Hero B., Kahlert Y., Spitz R., Ernestus K., Konig R., Haas S., Eils R., Schwab M., Brors B., Westermann F., Fischer M. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J. Clin. Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 32.Ohnuma S., Philpott A., Harris W.A. Cell cycle and cell fate in the nervous system. Curr. Opin. Neurobiol. 2001;11:66–73. doi: 10.1016/s0959-4388(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 33.Patel J.H., Loboda A.P., Showe M.K., Showe L.C., McMahon S.B. Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 34.Porro A., Haber M., Diolaiti D., Iraci N., Henderson M., Gherardi S., Valli E., Munoz M.A., Xue C., Flemming C., Schwab M., Wong J.H., Marshall G.M. Della, Valle G., Norris M.D., Perini G. Direct and coordinate regulation of ATP-binding cassette transporter genes by Myc factors generates specific transcription signatures that significantly affect the chemoresistance phenotype of cancer cells. J. Biol. Chem. 2011;285:19532–19543. doi: 10.1074/jbc.M109.078584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricciardi S., Kilstrup-Nielsen C., Bienvenu T., Jacquette A., Landsberger N., Broccoli V. CDKL5 influences RNA splicing activity by its association to the nuclear speckle molecular machinery. Hum. Mol. Genet. 2009;18:4590–4602. doi: 10.1093/hmg/ddp426. [DOI] [PubMed] [Google Scholar]
- 36.Rusconi L., Salvatoni L., Giudici L., Bertani I., Kilstrup-Nielsen C., Broccoli V., Landsberger N. CDKL5 expression is modulated during neuronal development and its subcellular distribution is tightly regulated by the C-terminal tail. J. Biol. Chem. 2008;283:30101–30111. doi: 10.1074/jbc.M804613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scala E., Ariani F., Mari F., Caselli R., Pescucci C., Longo I., Meloni I., Giachino D., Bruttini M., Hayek G., Zappella M., Renieri A. CDKL5/STK9 is mutated in Rett syndrome variant with infantile spasms. J. Med. Genet. 2005;42:103–107. doi: 10.1136/jmg.2004.026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholzen T., Gerdes J. The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Schwab M. MYCN in neuronal tumours. Cancer Lett. 2004;204:179–187. doi: 10.1016/S0304-3835(03)00454-3. [DOI] [PubMed] [Google Scholar]
- 40.Singh J., Kaur G. Transcriptional regulation of polysialylated neural cell adhesion molecule expression by NMDA receptor activation in retinoic acid-differentiated SH-SY5Y neuroblastoma cultures. Brain Res. 2007;1154:8–21. doi: 10.1016/j.brainres.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 41.Tao J. Van, Esch H., Hagedorn-Greiwe M., Hoffmann K., Moser B., Raynaud M., Sperner J., Fryns J.P., Schwinger E., Gecz J., Ropers H.H., Kalscheuer V.M. Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5/STK9) gene are associated with severe neurodevelopmental retardation. Am. J. Hum. Genet. 2004;75:1149–1154. doi: 10.1086/426460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavyev Asher Y.J., Scaglia F. Molecular bases and clinical spectrum of early infantile epileptic encephalopathies. Eur. J. Med. Genet. 2012;55:299–306. doi: 10.1016/j.ejmg.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 43.van Bokhoven H., van Celli J., Reeuwijk J., Rinne T., van Glaudemans B., Beusekom E., Rieu P., Newbury-Ecob R.A., Chiang C., Brunner H.G. MYCN haploinsufficiency is associated with reduced brain size and intestinal atresias in Feingold syndrome. Nat. Genet. 2005;37:465–467. doi: 10.1038/ng1546. [DOI] [PubMed] [Google Scholar]
- 44.Weaving L.S., Christodoulou J., Williamson S.L., Friend K.L., McKenzie O.L., Archer H., Evans J., Clarke A., Pelka G.J., Tam P.P., Watson C., Lahooti H., Ellaway C.J., Bennetts B., Leonard H., Gecz J. Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am. J. Hum. Genet. 2004;75:1079–1093. doi: 10.1086/426462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinmann A.S., Farnham P.J. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 46.Weiss W.A., Aldape K., Mohapatra G., Feuerstein B.G., Bishop J.M. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williamson S.L., Giudici L., Kilstrup-Nielsen C., Gold W., Pelka G.J., Tam P.P., Grimm A., Prodi D., Landsberger N., Christodoulou J. A novel transcript of cyclin-dependent kinase-like 5 (CDKL5) has an alternative C-terminus and is the predominant transcript in brain. Hum. Genet. 2011;131:187–200. doi: 10.1007/s00439-011-1058-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in the present study.