Abstract
Here we report an integrated analysis that leverages data from treatment of genetic mouse models of prostate cancer along with clinical data from patients to elucidate new mechanisms of castration resistance. We show that castration counteracts tumor progression in a Pten-loss driven mouse model of prostate cancer through the induction of apoptosis and proliferation block. Conversely, this response is bypassed upon deletion of either Trp53 or Lrf together with Pten, leading to the development of castration resistant prostate cancer (CRPC). Mechanistically, the integrated acquisition of data from mouse models and patients identifies the expression patterns of XAF1-XIAP/SRD5A1 as a predictive and actionable signature for CRPC. Importantly, we show that combined inhibition of XIAP, SRD5A1, and AR pathways overcomes castration resistance. Thus, our co-clinical approach facilitates stratification of patients and the development of tailored and innovative therapeutic treatments.
We conceived the co-clinical platform as an innovative approach to improve the design, speed and outcomes of rational and personalized clinical treatments1. The power of this approach lies in harnessing data gained from pre-clinical studies performed in several genetic mouse models and combining it with the concurrent investigation of therapies, either conventional or experimental, in human patients.
One goal of this approach is to expedite the identification of key genetic and molecular determinants that dictate resistance to a specific therapy in order to facilitate the stratification of human candidates for enrollment in treatment with different therapies, optimize combination treatments and accelerate new drug approval.
In this study we have implemented this paradigm in an investigation of androgen deprivation therapy (ADT), which is a widely-prescribed standard-of-care therapy for prostate cancer. Although almost invariably effective in the short term, ADT is consistently followed by the development of castration resistant prostate cancer (CRPC) in the majority of patients2. Given the diverse genetic alterations reported to drive prostate cancer initiation and progression, we hypothesized that these same genetic alterations also govern the tumor response to ADT, and may be utilized to develop novel combinational treatments.
Testing this hypothesis with the co-clinical approach led to the discovery of new pathways underlying ADT resistance, as well as new potential modalities for overcoming this serious therapeutic hurdle.
RESULTS
Prostate cancer genetic dictates androgen deprivation response
PTEN, LRF, and TP53 loss or mutation has been associated with prostate cancer progression in human and mouse models (Supplementary Table 1)4-12. We therefore enrolled prostate conditional Pten null mice (Ptenflox/flox;Probasin-Cre), Pten and Lrf double null mice (Ptenflox/flox;Lrfflox/flox;Probasin-Cre), and Pten and p53 double null mice (Ptenflox/flox;p53flox/flox;Probasin-Cre) to test the hypothesis that advanced prostate cancer with distinct genetic makeups respond differentially to ADT. For each genetically-distinct tumor type, we generated cohorts of 15 mutant mice that were castrated at the age at which they had developed prostate cancer (high grade-prostatic epithelial neoplasia (HG-PIN) characterized by focal invasive carcinoma) as previously reported4,5,13. Age-matched non-castrated cohorts were used as controls. All cohorts were monitored for 3 months post-castration.
Groups of 5 mice from each cohort (castrated and non-castrated) were sacrificed at 1, 2 and 3 months post-castration for histopathological analysis (see Fig. 1a for timeline of the analysis). Mice from the 3-month post-castration group were also imaged by magnetic resonance imaging (MRI) for tumor volume assessment on a monthly schedule (Fig. 1a).
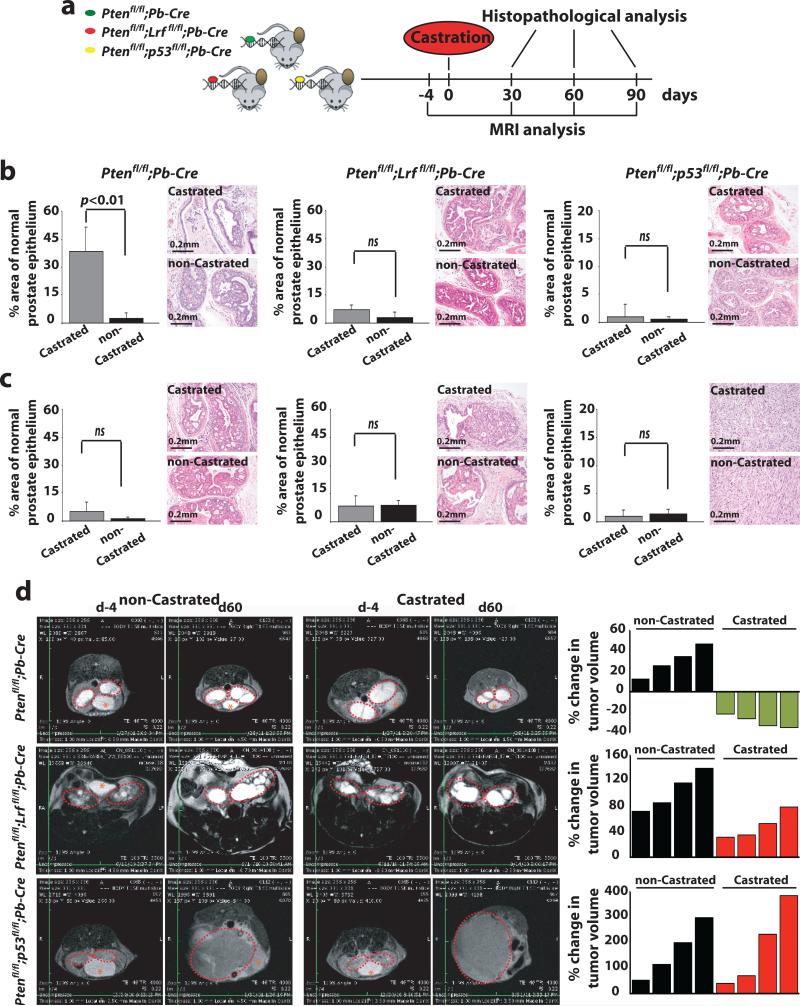
Figure 1. Prostate tumors characterized by distinct genetic alterations differentially respond to castration.
(a) Timeline of analysis of Ptenflox/flox;Probasin(Pb)-Cre, Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre, and Ptenflox/flox;p53flox/flox;Probasin(Pb)-Cre noncastrated (n=15) and castrated (n=15) cohorts of mice. (b-c) Quantification of percentage area of normal epithelium in the prostate of Ptenflox/flox;Probasin(Pb)-Cre (left), Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre (middle), and Ptenflox/flox;p53flox/flox;Probasin(Pb)-Cre (right) castrated and non-castrated mice at 60 (b) and 90 (c) days post-castration. Representative images of H&E staining of DLP from castrated and non-castrated mice of the indicated genotypes are shown. (d) Representative MRI images of prostate cancers (Anterior Lobe AP) in castrated and non-castrated mice of indicated genotype 4 days before and 60 days post-castration. Tumor volume (area outlined in red) was quantified as described in the experimental procedures. Asterisk represents the location of the bladder.
Histopathological analysis of Ptenflox/flox;Probasin-Cre mice at 1 and 2 months post-castration identified the presence of a significant amount of normal epithelium in castrated mice, indicative of a clear response to castration, when compared to the levels of normal epithelium in the non-castrated control group [1 month: PI 0 = 25% vs 0 < 5%, (p < 0.01) in Supplementary Fig. 1a left panel and 2 months: PI 0 = 40% vs. 0 < 5%, (p < 0.01) Fig. 1b left panels and Supplementary Fig. 1a]. Importantly, and in agreement with a previous report14, examination of the DLP from castrated Ptenflox/flox;Probasin-Cre mice at 3 months post-castration demonstrated a strong reduction of normal epithelium, indicating onset of acquired resistance to ADT (Fig. 1c left panel and Supplementary Fig. 1a). In sharp contrast to this finding, we did not observe any response in DLP from either castrated Ptenflox/flox;Lrfflox/flox;Probasin-Cre (Fig. 1b-c center panels and Supplementary Fig. 1a) or Ptenflox/flox;p53flox/flox;Probasin-Cre mutants (Fig. 1b-c right panels and Supplementary Fig. 1a). These results indicate that loss of either p53 or Lrf in a Pten-null genetic context confers a de novo resistance to androgen deprivation in the DLP tumors.
Interestingly, and in line with recent publications15, examination of the ventral lobe of the mouse prostate (VP) from castrated Ptenflox/flox;Probasin-Cre mice identified, an intrinsic propensity of the tumors derived from this lobe to be resistant to ADT (PI 0 = 0%; Supplementary Fig. 1b).
The anterior lobe of the mouse prostate (AP) of Ptenflox/flox;Probasin-Cre, Ptenflox/flox;Lrfflox/flox;Probasin-Cre, Ptenflox/flox;p53flox/flox;Probasin-Cre mice are characterized by a marked cystic dilation of prostate ducts5, a feature preventing accurate pathological evaluation of the extent and severity of tumor lesions. Therefore, to assess ADT responses in this lobe, we quantitated the tumor volume of AP lesions by MRI (Fig. 1d). In line with the results described for the DLP above, we observed that castration of Ptenflox/flox;Probasin-Cre led to reduced tumor volumes (Fig. 1d upper panels), while no regression was observed in castrated Ptenflox/flox;Lrfflox/flox;Probasin-Cre (Fig. 1d middle panels) and Ptenflox/flox;p53flox/flox;Probasin-Cre mice (Fig. 1d lower panels).
In accordance with the results obtained through physical castration, Ptenflox/flox;Probasin-Cre mice displayed sensitivity (as previously described)16 while Ptenflox/flox;Lrfflox/flox;Probasin-Cre and Ptenflox/flox;p53flox/flox;Probasin-Cre mice displayed resistance to chemical castration (bicalutamide), thereby validating that resistance to ADT is tumor-intrinsic and not dependent on the specific treatment (Supplementary Fig. 1c-f).
Together, these data demonstrate that Pten-null prostate cancer is initially sensitive to androgen deprivation, yet invariably acquires castration-resistance. In contrast, our analysis clearly demonstrates that both Pten;Lrf and Pten;p53 double null prostate cancers display complete resistance to castration.
Genetic determinants of ADT response in human patients
We next determined whether the differential response to castration we observed in the different mouse models would allow us to predict responses to ADT in human prostate cancer (Fig. 2a). To this end, we constructed a tissue microarray (TMA) designed to interrogate ADT response biomarkers by immunohistochemistry (IHC). The TMA included 84 biopsied specimens from prostatectomized human primary prostate cancer (Gleason Score 6/7) that had been treated with neo-adjuvant ADT and showed variable response to the treatment, as measured by the ratio in circulating prostate specific antigen (PSA) levels before and after ADT, proliferation rate and percentage of cells with nuclear AR localization (Fig. 2b, Supplementary Fig. 2a-c, Supplementary Table 2 and Supplementary Note). We also performed an additional analysis to validate our stratification criteria. It has been previously reported that levels of 10 ng/ml of PSA at diagnosis are frequently predictive of worse prognosis when compared with patients with levels below 10 ng/ml [reviewed in Ref.17,18]. 31 of our patients (out of 84) had a PSA value higher than 10 ng/ml before treatment with ADT in neo-adjuvant setting (Supplementary Fig. 2c). After 3 months on ADT, patients have now been classified as either good (16 patients) or poor responders (15 patients) based on the ratio of PSA levels after and before treatment (Supplementary Fig. 2c). Importantly, all the patients that retained levels of PSA > 10 ng/ml post-ADT belonged to the category of poor responder (11 out of 15) and none was found among the 16 good responders (p=1.612E-05; Supplementary Fig. 2c). Finally, we obtained and analyzed the follow-up information of 36 of the 84 patients originally enrolled in this study. Notably, only 2 out of 23 patients classified as “Good Responders” have developed resistance to ADT, while a consistent 6 out of 13 belong to the category of the “Poor Responders” (p=0.01; Supplementary Fig. 2d). These data further suggest the notion that the category that we have identified as poor responder represents the group of patients most likely to develop tumor recurrence and castration-resistant distant metastasis [reviewed in Ref.17,18].
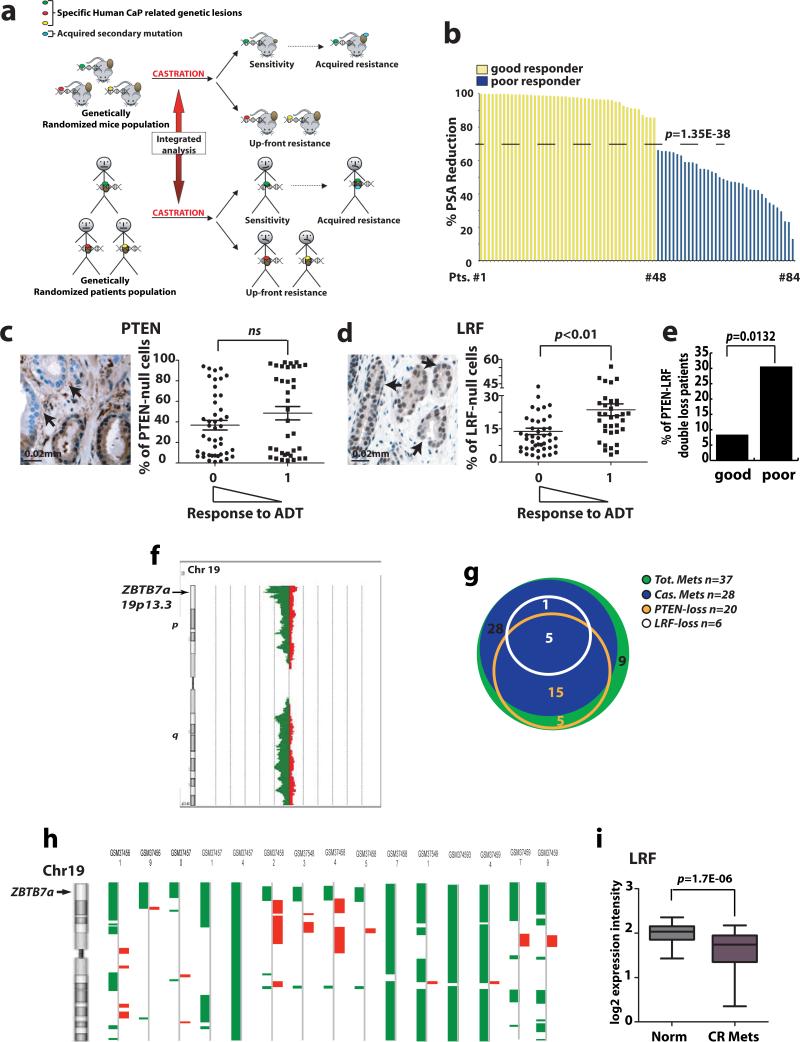
Figure 2. Analysis of human prostate cancer unveils the relevance of LRF loss in the response to androgen deprivation.
(a) Cross-species integrated genetic screenings (b) Histogram showing the percentage of PSA-reduction following treatment used to stratify patients as good responders (yellow) or poor responders (blue) to ADT. Mean value of percentage of PSA reduction distribution is shown (dashed line). (c-d) PTEN and LRF IHC in human prostate cancer TMAs. The percentage of tumor cells with loss of PTEN and LRF were measured. Patients were considered good responders (0) or poor responders (1) to ADT according to the percentage of PSA reduction. Arrows highlight PTEN and LRF null cells. (e) Percentage of patients that experienced combined loss of PTEN and LRF in the category of good (8.7%) and poor responders (30.5%). This distribution resulted statistically significant p=0.0132 in a Fisher test. (f) Schematic representation of the deletion of the p arm on chromosome 19 (green) in human CRPC. ZBTB7a genetic locus is indicated (arrow). (g) Diagram showing the distribution of PTEN-loss (orange) and LRF-loss (white) genetic alterations in castrated (blue) and noncastrated (green) metastasis. (h) CGH array data for 58 castration resistant prostate cancer metastatic samples shows genetic loss of the telomeric region of chromosome 19 containing the Zbtb7a locus. Data were processed by Partek Genomic Suite 6.4 Segmentation Algorithm (deletions are shown in green on the left side, amplification are showed in red on the right side). (i) Expression profile analysis of human castration resistant metastases shows statistically significant down-regulation of LRF expression.
As mentioned, TP53 loss or mutation has been associated with tumor progression in a subset of human patients (Supplementary Table 1)6-11. Taking into account that p53 loss cannot be evaluated by IHC, as the protein levels are usually extremely low, on our TMA we could only assess mutational events, which would result in p53 overexpression. Mutation in p53 has been reported to correlate with progression to higher Gleason Scores (4 + 3 or 4 + 4) and CRPC6, while the patients in our TMA had lower Gleason Scores (mainly 3 + 3 and 3 + 4 as detailed in the Supplementary Table 2). In agreement with these findings, in these earlier prostate cancer lesions from our cohort we could not identify any sample with a homogeneously elevated p53 immunohistochemical staining. We therefore interrogated a pre-existing array-based comparative genomic hybridization (aCGH) database made up of 37 human prostate cancer metastases (28 out of 37 were castration resistant)11. 43% of these castration resistant metastatic patients (12 of 28) presented heterozygous loss and 7% homozygous loss (2 out of 28) of p53, thereby confirming the relevance of loss in this tumor suppressor gene in CRPC.
We next focused our attention on PTEN and LRF and performed IHC on our low-Gleason TMAs (Fig. 2c-d). In this analysis, we observed no difference in the mean PTEN loss in those patients who responded poorly or favorably to ADT (Fig. 2c). In contrast, patient tumors harboring a significantly larger mean number of LRF-null cells stratified as poor responders to ADT (Fig. 2d). Additionally, the number of patients that displayed both PTEN- and LRF-protein loss was 15 out of 84 total samples, 4 in the category of good responders (4/48 patients, 8.7%) and 11 in the category of poor responders (11/36 patients, 30.5%) (Fig. 2e, p=0.0132), suggesting that concomitant loss of PTEN and LRF stratifies for poor responsiveness to castration.
In order to mirror in humans the data obtained in genetic mouse models, we sought to assess in prostate cancer patients whether concomitant genetic loss of LRF with PTEN may be a stronger indicator of CRPC development than loss of PTEN only. To this end, we interrogated again the aCGH database made up of 37 human prostate cancer metastases (28 out of 37 were castration resistant)11. We identified 336 genes undergoing genetic losses solely in castration-resistant specimens characterized by concomitant loss of PTEN in at least 80% of cases (Supplementary Fig. 2e and Supplementary Table 3). Intriguingly, 110 of these genes were located on the telomeric p arm of chromosome 19 (Supplementary Fig. 2e) including LRF gene (Zbtb7a) (Fig. 2f). This suggests that in humans, as predicted by our mouse study, there is a functional association between loss of PTEN and genetic deletion of LRF for CRPC development (Fig. 2g and Supplementary Fig. 2f).
The relevance of LRF loss in CRPC was further confirmed by the analysis of two other large pre-existing aCGH datasets made up of 58 and 50 human castration resistant metastatic prostate cancer samples19,20. LRF was found to be genetically lost in 26% of cases (15 out of 58), and in 33% of cases (15 out of 50) respectively19,20. In particular, 10 castration resistant metastatic specimens out of 15 (67%) in the first dataset19 and 8 out of 15 (53%) in the second dataset20 showed specific deletion of the telomeric p arm of chromosome 19 which contains the LRF gene (Zbtb7a) (Fig. 2h, and Supplementary Fig. 2g). Of note, in the dataset recently published by Grasso and colleagues20, 2 out of 3 described castration resistant bone marrow metastases show deletion of the telomeric p arm of chromosome 19 (WA48, and WA51), while the remaining one (WA23) is characterized by a somatic missense mutation (K60R) in the BTB/POZ domain of LRF, a fundamental region for its transcriptional repressive activity21.
Finally, a similar result was also obtained by the analysis of the expression profile data set from Tomlins et al.22, where we observed a significant reduction in LRF expression in castration resistant metastases (20) from several organs (Fig. 2i), thereby further corroborating a role for LRF loss in resistance to ADT.
Together, these data show that prostate cancers characterized by distinct genetic background differentially respond to ADT in mice as well as human patients, and that LRF represents an unexpected player in the response of prostate cancer to androgen deprivation.
Biological responses to androgen deprivation
Castration typically results in a major decrease in prostatic mass, with the key molecular hallmarks including decreased proliferation and increased apoptosis23. Accordingly, we examined whether the absence of androgen differentially affects the proliferative index of these tumor types. To this end, we performed IHC on the VP, DLP, and AP of castrated mice utilizing the cellular proliferation marker Ki67. In Ptenflox/flox;Probasin-Cre mice we observed at one month post-castration a significant reduction in the overall proliferative index (Fig. 3a and 3b for quantifications, left panels). In contrast, we observed no difference in the proliferative index in any prostate lobes from either Ptenflox/flox;Lrfflox/flox;Probasin-Cre mice (Fig. 3a and 3b for quantifications, center panels), or Ptenflox/flox;p53flox/flox;Probasin-Cre (Fig. 3a and 3b for quantifications, right panels) at one month post-castration compared to non-castrated controls. These findings suggest that Lrf or p53 deletion in Pten-null prostate epithelium confers an ability to proliferate in conditions of androgen depletion. Notably, no difference in Ki67 staining was observed in the VP of the same mice (Supplementary Fig. 3a).
Figure 3. Differential responses to castration in different genetic backgrounds.
(a-b) IHC staining of DLP and AP from castrated and non-castrated mice at 30 days post-castration. Upper panel: Ki-67 staining on prostate sections from castrated and noncastrated mice of indicated genotype. Lower panel: quantification of Ki-67 staining. Quantification has been performed as described in the methods. For each genotype, castrated (n=3, grey bar) and non-castrated (n=3, black bar) mice were analyzed. Percentage of Ki67 positive cells in DLP and AP prostate lobes was evaluated on a total of 15,000 cells per lobe. Data are presented as mean ± standard deviation. (c) Western blot analysis on VP, DLP and AP from castrated Ptenflox/flox;Probasin(Pb)-Cre, Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre and Ptenflox/flox;p53flox/flox;Probasin(Pb)-Cre mice. Western blot analyses were performed on sample lysates collected from mice sacrificed 4 days after castration. (d) IHC staining of AR on DLP and AP from castrated and noncastrated mice sacrificed 30 days after castration. (e) Quantification of cells with nuclear AR localization in castrated Ptenflox/flox;Probasin(Pb)-Cre (light grey bar), Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre (dark grey bar) and Ptenflox/flox;p53flox/flox;Probasin(Pb)-Cre (black bar) prostate tumors. Number of cells with nuclear AR staining in DLP and AP prostate lobes was evaluated in n=3 castrated mice/genotype on a total of 2,500 cells for each prostate lobe. Data are presented as mean ± standard deviation.
We next measured the induction of apoptosis in the prostatic epithelium of castrated Ptenflox/flox;Probasin-Cre, Ptenflox/flox;Lrfflox/flox;Probasin-Cre, and Ptenflox/flox;p53flox/flox;Probasin-Cre mice. The induction of an early apoptotic response was observed in all lobes of the prostate and in all genotypes, as analyzed by measuring elevation of caspase-3 cleavage (Fig. 3c). In contrast, Parp cleavage and pyknotic nuclei (Supplementary Fig. 3b), feature of terminal apoptosis, were observed in all prostate lobes of castrated Ptenflox/flox;Probasin-Cre mice (Fig. 3c), but were completely absent in all lobes of Ptenflox/flox;Lrfflox/flox;Probasin-Cre, and Ptenflox/flox;p53flox/flox;Probasin-Cre castrated mice, indicating apoptotic inhibition in these tumors.
We next investigated the effects of androgen withdrawal on androgen receptor (AR) localization in our different prostate cancer genotypes. AR was predominantly translocated to the cytoplasm in all genotypes shortly after castration, as expected in the absence of androgen24 (Fig. 3d for DLP and AP, Supplementary Fig. 3c for VP). However, quantitative IHC analysis of prostates from castrated Ptenflox/flox;Lrfflox/flox;Probasin-Cre, and Ptenflox/flox;p53flox/flox;Probasin-Cre mice identified a modest but significantly greater number of cells with nuclear AR compared to the prostates of castrated Ptenflox/flox;Probasin-Cre mice (Fig. 3e and relevant quantification), suggesting that cells switch to or harbor pre-existing alternative pathways of AR activation in Pten;Lrf or Pten;p53 double null CRPC (see below).
In summary, these data suggest that Pten;Lrf or Pten;p53 double null tumor prostatic cells, in contrast to Pten-null only, posses the ability to sustain androgen-independent proliferation, while escaping the engagement of an apoptotic response.
XAF1 and SRD5A1 are key biomarkers for ADT response in mouse and human
Next we sought to identify the molecular mechanisms underlying the differential sensitivity displayed by Ptenflox/flox;Probasin-Cre vs. Ptenflox/flox;Lrfflox/flox;Probasin-Cre and Ptenflox/flox;p53flox/flox;Probasin-Cre double null prostate tumor cells when challenged by androgen deprivation. For this purpose, we interrogated microarray gene expression profiles from mouse prostate specimens4. Strikingly, we observed a significant down-regulation (p=1.07E-12) of X-linked inhibitor of apoptosis protein-associated factor-1 (Xaf1; Fig. 4a) in Ptenflox/flox;Lrfflox/flox;Probasin-Cre mice compared to Ptenflox/flox;Probasin-Cre. XAF1 was first identified as a protein that binds and opposes the anti-caspase activity of X-linked inhibitor of apoptosis (XIAP), thereby promoting apoptosis25. A growing body of evidence has also demonstrated strong involvement of the XAF1-XIAP pathway in cell cycle regulation through yet unknown mechanisms26-32. The down-regulation of Xaf1 was confirmed by qRT-PCR, IHC and Western blot in specimens from Ptenflox/flox;Lrfflox/flox;Probasin-Cre mice (Fig. 4b-d). Since XAF1 has been recently reported to be a p53-responsive gene33, we assessed Xaf1 levels in Ptenflox/flox;p53flox/flox;Probasin-Cre mice. As expected, Xaf1 was significantly down-regulated in Ptenflox/flox;p53flox/flox;Probasin-Cre prostates as measured by qRT-PCR, IHC and Western blot (Fig. 4b-d). Interestingly, IHC of Xaf1 in DLP from castrated Ptenflox/flox;Probasin-Cre mice analyzed 90 days post-castration showed reduced levels of staining-positive cells compared to ones analyzed 30 days post-castration, supporting the concept of Xaf1 as a good biomarker for castration resistance (Supplementary Fig. 4a).
Figure 4. Deregulation of XAF1 and SRD5A1 levels dictates prostate cancer progression and castration resistance in mouse and human.
(a) Analysis of the expression profiles of Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre versus Ptenflox/flox;Probasin(Pb)-Cre prostates shows a significant down-regulation of Xaf1 and a concomitant up-regulation of Srd5a1. (b) Quantitative RT-PCR of Xaf1 in VP (n=3, black bars), DLP (n=3, dark grey bars) and AP (n=3, light grey bars) samples collected from Ptenflox/flox;Probasin(Pb)-Cre, Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre, and Ptenflox/flox;p53flox/flox;Probasin(Pb)-Cre mice. Data are presented as mean ± standard deviation. (c) IHC with anti-Xaf1 of the same samples in b. (d) Western blot analysis on the same samples in b. (e) Quantitative RT-PCR for Srd5a1 in VP (n=3, black bars), DLP (n=3, dark grey bars) and AP (n=3, light grey bars) samples collected from WT, Ptenflox/flox;Probasin(Pb)-Cre, Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre and Ptenflox/flox;p53flox/flox;Probasin(Pb)-Cre mouse prostates. Data are presented as mean ± standard deviation. (f) XAF1 staining and relative quantification in human prostate cancer TMA. (g) LRF and XAF1 down regulation and SRD5A1 induction in castration resistant prostate cancer xenografts (orange bars) versus castration sensitive (black bars). (h-l) XAF1, XIAP, and SRD5A1 expression levels in primary and metastatic prostate cancer compared to normal prostatic epithelium.
Further interrogation of expression profiles of Ptenflox/flox;Lrfflox/flox;Probasin-Cre murine prostates identified a significant up-regulation of 3-oxo-5-alpha-steroid 4-dehydrogenase 1 (Srd5a1) gene compared to Ptenflox/flox;Probasin-Cre mice (Fig. 4a). Srd5a1 and Srd5a2 enzymes catalyze the conversion of testosterone into dihydrotestosterone (DHT), a more potent and stable form of testosterone, and Srd5a1 is known to be frequently up-regulated in human CRPC35. Production of DHT represents an important alternative mechanism that allows activation of AR at castration-levels of androgen35. qRT-PCR on the prostates from Ptenflox/flox;Lrfflox/flox;Probasin-Cre andPtenflox/flox;p53flox/flox;Probasin-Cre mice confirmed Srd5a1 up-regulation compared to Ptenflox/flox;Probasin-Cre prostate cancer (Fig. 4e), while no changes in the level of Srd5a2 transcript were observed (Supplementary Fig. 5a). Interestingly, qRT-PCR analysis in DLP from castration resistant Ptenflox/flox;Probasin-Cre mice analyzed 90 days post-castration also showed higher levels of Srd5a1 compared to the castration sensitive Ptenflox/flox;Probasin-Cre prostates analyzed 30 days post-castration, whereas only a mild increase was observed in the level of Srd5a2 (Supplementary Fig. 4b). Notably, while Western blot analysis of AR in DLP from castrated Ptenflox/flox;Probasin-Cre mice analyzed 90 days post-castration showed no differences in the levels of the protein compared to ones analyzed 30 days post-castration (Supplementary Fig. 4a), IHC analysis showed different localization of AR in the two different contexts. In the castration sensitive setting (30 days after castration) AR, as expected, was mainly localized in the cytoplasm. In contrast, AR was found to be nuclear 90 days post castration in large areas of the Ptenflox/flox;Probasin-Cre mutant tumor, as frequently observed in human prostate cancers that have acquired resistance to ADT34, and Western blot analysis showed higher levels of the AR transcriptional target Probasin (Supplementary Fig. 4a).
In order to translate the relevance of biomarkers found in the mouse to the human context, we assessed the status of XAF1 and SRD5A1 pathways in human prostate cancer. For this, we performed IHC with anti-XAF1 on the custom TMA mentioned above (Fig. 4f), and quantitated the number of cells that had down-regulation of XAF1 (Fig 4f, right panel). In this analysis, we observed that XAF1 protein levels are lower in human prostate cancer and XAF1 staining is significantly lost in those patients that respond poorly to ADT (Fig. 4f).
Furthermore, we analyzed the expression of XAF1/XIAP and SRD5A1 mRNA in several human prostate cancer expression arrays (Fig. 4g-l)11,36-39. Importantly, we observed that the expression levels of XAF1 are significantly decreased, in contrast to XIAP and SRD5A1, which were significantly increased, in the patients with the most aggressive prostate cancer, as characterized by Gleason Score, recurrence and metastasis (Fig. 4g-l). This data, combined with the results obtained in an androgen deprivation setting, suggests that down-regulation of XAF1 and up-regulation of SRD5A1 in high Gleason Score prostate cancer and metastasis might be predictive of poor sensitivity to ADT.
XIAP inhibition sensitizes castration resistant prostate cancers to ADT
Our findings suggest a potential therapeutic modality based on the combination of ADT with a XIAP inhibitor for CRPC. Therefore, we treated the AR-mutant/DHT-sensitive LnCaP C4-2 and the AR-null/DHT-insensitive DU145 and PC3 cell lines with the plant-derived XIAP inhibitor embelin32 and ADT [charcoal/dextran treated steroid-free FBS (SFM)] as single agents and in combination (Supplementary Fig. 6a-d). While single treatments with SFM or embelin at a concentration of 5 μM for 4 days did not induce any apoptosis, the combination of embelin and SFM triggered a clear increase in the apoptotic response in LnCaP C4-2 cells, as measured by PARP cleavage (Supplementary Fig. 6a). Notably, the combination of embelin with SFM and MDV3100 (an experimental androgen receptor antagonist drug developed for the treatment of castration-resistant prostate cancer currently in Phase III clinical trial) strikingly increased the apoptotic response in LnCaP C4-2 castration resistant cell line (Supplementary Fig. 6a). Likewise XIAP silencing induced apoptosis in SFM conditions (Supplementary Fig. 6b), confirming the fundamental role of XAF1/XIAP pathway in human CRPC. As expected, the AR-null DU145 and PC3 prostate cancer cell lines displayed de novo resistance to the SFM-plus-embelin combination treatment since SFM (mimicking ADT) is unable to trigger any apoptotic response in these cell lines (Supplementary Fig. 6c-d). Conversely, in these cells ionizing radiation (IR) synergizes with embelin by triggering apoptosis40.
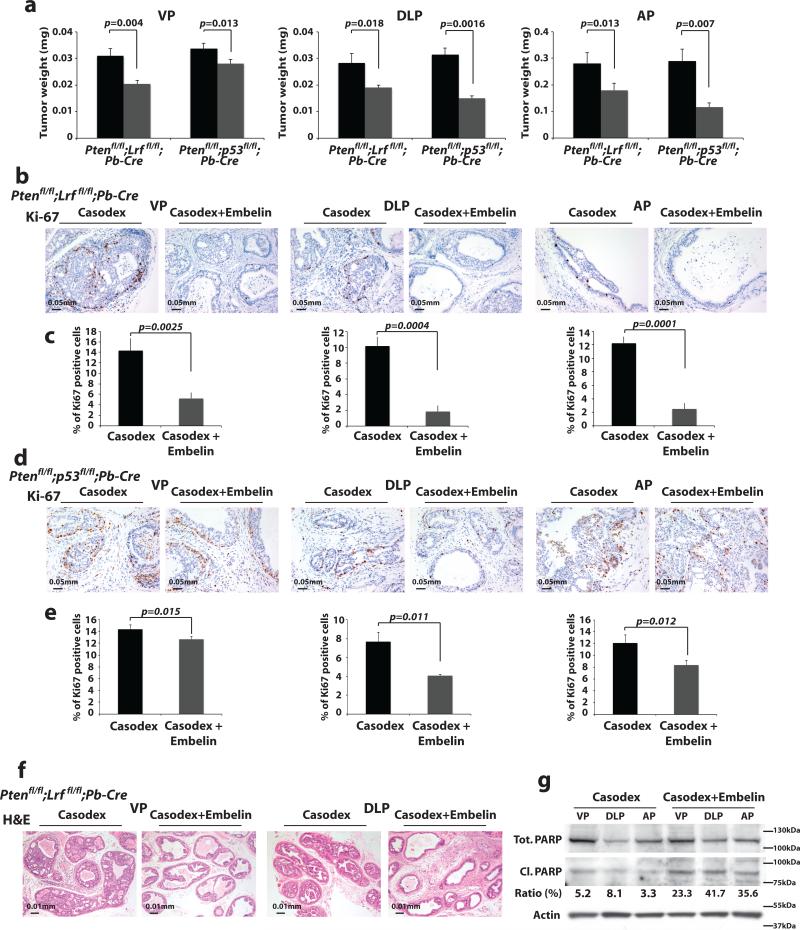
To corroborate our in vitro results in vivo, we next enrolled our castration-resistant Ptenflox/flox;Lrfflox/flox;Probasin-Cre and Ptenflox/flox;p53flox/flox;Probasin-Cre mouse models in treatment programs with embelin and ADT. Specifically, we treated 4 month-old Ptenflox/flox;Lrfflox/flox;Probasin-Cre and 3 month-old Ptenflox/flox;p53flox/flox;Probasin-Cremice with bicalutamide (Casodex) or embelin alone and in combination. Remarkably, while both Ptenflox/flox;Lrfflox/flox;Probasin-Cre and Ptenflox/flox;p53flox/flox;Probasin-Cre prostate cancers exhibited resistance to bicalutamide or embelin treatment as single agents (Supplementary Fig. 7a-b), all prostate lobes showed a marked tumor regression when treated with bicalutamide in combination with embelin (Fig. 5a and Supplementary Fig. 7a-b). Specifically, we found that treatment ofPtenflox/flox;Lrfflox/flox;Probasin-Cre and Ptenflox/flox;p53flox/flox;Probasin-Cre prostate cancers with bicalutamide plus embelin was characterized by a profound reduction in the proliferation rate (Fig. 5b-e), which in the case of Ptenflox/flox;Lrfflox/flox;Probasin-Cre prostate cancers was also accompanied by the rescue of the apoptotic response (Fig. 5f-g and Supplementary Fig. 7c-d).
Figure 5. Synergistic effect of ADT and embelin, in mouse CRPC.
(a) Weight of VP, DLP, and AP of Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre and Ptenflox/flox;p53flox/flox;Probasin(Pb)-Cre mice treated with bicalutamide (Casodex) (n=8) (black bars) or bicalutamide plus embelin (n=8) (grey bars) for 4 weeks. Data are presented as mean ± standard deviation. (b-e) Proliferation rate analysis as percentage of Ki67 positive cells in VP, DLP, and AP of the samples described in a. Data are presented as mean ± standard deviation. (f-g) H&E and apoptosis analysis through Western blot quantification of Parp cleavage versus total Parp in VP, DLP, and AP of the Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre prostate cancers described in a.
These findings clearly demonstrate that castration-resistant prostate tumors characterized by the impairment of the XAF1/XIAP pathway become sensitive to ADT when treated with the XIAP inhibitor embelin.
Combination of ADT, embelin and dutasteride for the treatment of CRPC
We hypothesized that management of CRPC could further benefit from the addition of an SRD5A1 inhibitor to the combination therapy with embelin and ADT. Importantly, the SRD5A1 inhibitor dutasteride is currently utilized in clinical trials in combination with ADT for the treatment of advanced prostate cancer (see URLs)41-43.
At first, therefore, we treated the VCaP, LnCaP and 22RV1 human prostate cell lines with embelin, dutasteride and ADT as single agents and in triple combination. Strikingly, the triple combination of embelin plus dutasteride and SFM induced an even more potent apoptotic response compared to each of the double combinations, embelin plus SFM and dutasteride plus SFM (Fig. 6a-d and Supplementary 6e-f).
Figure 6. Genetic and molecular assessments in mice and patients dictate new experimental treatment to overcome CRPC.
(a-d) Apoptosis analysis in human prostate cancer cell line (VCaP) treated with ADT, XIAP inhibitor (embelin), or SRD5A1 inhibitor (dutasteride), as single agent or in combination. Quantifications of PARP cleavage versus total PARP after 2 days of treatment are shown. (e) Weight of VP, DLP, and AP of castration resistant Ptenflox/flox;Lrfflox/flox;Probasin(Pb)-Cre and Ptenflox/flox;p53flox/flox;Probasin(Pb)-Cre mice treated with bicalutamide (Casodex) (n=4/genotype, black bars), bicalutamide plus embelin (n=4/genotype, dark grey bars), or bicalutamide plus embelin plus dutasteride (n=4/genotype, light grey bars) for 4 weeks. Data are presented as mean ± standard deviation.
We next treated castration resistant 4 month-old Ptenflox/flox;Lrfflox/flox;Probasin-Cre and 3 month-old Ptenflox/flox;p53flox/flox;Probasin-Cre mice with ADT in combination with embelin and dutasteride. Both Pten;Lrf and Pten;p53 double-null prostate cancers exhibited a further significant decrease in tumor burden after the triple treatment with bicalutamide, embelin, and dutasteride when compared to the treatment with the dual combination bicalutamide plus embelin (Fig. 6e).
These findings clearly demonstrate that castration-resistant prostate tumors characterized by the impairment of the XAF1/XIAP and SRD5A1 pathways are sensitized to ADT when treated with embelin and dutasteride, thereby rendering this triple combination an effective novel therapeutic approach to the treatment of XAF1/XIAP/SRD5A1 genetically stratified CRPC patients.
DISCUSSION
Our data support the hypothesis that the response to ADT is dictated by the genetic make-up of prostate cancer. Our findings argue that genetic mouse models can be extremely valuable in tailoring the administration of standard-of-care treatments on the basis of genetic and molecular stratification.
We find that Pten-loss driven prostate cancer exhibits an overall reduction in tumor volume in response to castration, indicating that this genetic mouse model is initially sensitive to androgen deprivation. Nevertheless, the mice demonstrate a rapid recurrence of prostate cancer at 3 months post-castration. CRPC development in Pten-null prostate cancer appears to be accompanied by concomitant down-regulation of Xaf1, up-regulation of Srd5a1, and re-localization of AR in the nucleus (Supplementary Fig. 4a).
This model of “acquired” resistance is consistent with the initial response patterns and invariable latent resistance observed in human prostate cancer subjects. Importantly, we have now shown that concomitant Pten and either Lrf or p53 deletions abrogate the response of Pten-null tumors to ADT.
Lrf- and p53-loss associated castration resistant prostate cancers exhibit low levels of Xaf1, a phenomenon which potentially explains the lack of both apoptosis induction and proliferation arrest. The importance of this pathway is further highlighted by the observation that XIAP is highly up-regulated during prostate cancer progression and CRPC both in human44 and mice (Supplementary Fig. 5b). Accordingly, we have shown that the XIAP inhibitor embelin, when combined with ADT, potently suppresses tumor growth in vitro, confirming what previously reported45, and in vivo in our castration resistant genetic mouse models.
We also found that Pten;Lrf and Pten;p53 double null CRPCs, as well as Pten null CRPC, are characterized by the up-regulation of Srd5a135, and, importantly, that dutasteride (SRD5A1 inhibitor) further increases prostate tumor response to embelin plus ADT.
We think our integrated mouse and human analysis has identified potential new therapeutic strategies. Patients with prostate cancer characterized by XAF1-XIAP/SRD5A1 deregulations are likely to quickly develop CRPC, thus ADT in such patients should be combined with embelin and dutasteride (Supplementary Fig. 7e).
In sum, our study demonstrates the validity of a cross-species integrated genetic approach to the evaluation of prostate cancer therapies. A particularly powerful aspect of the approach is the combined access it affords to data regarding genetic and molecular differences between tumors whether in human patients or mouse models. Importantly, this study has led to the identification of critical pathways and genetic alterations dictating responses to standard-of-care therapeutic modalities, as well as novel “druggable” biomarkers for overcoming resistance to such treatments. This approach can be customized for any tumor types when appropriate genetic mouse models are available, and for testing of experimental targeted therapies1,46. Thus the applications of an integrated approach to the field of human cancer therapy are clearly significant, and potentially transformative.
URLs
ClinicalTrial.gov: http://www.clinicaltrials.gov
Taylor et al., 2010: http://www.cbioportal.org/public-portal/index.do?cancer_study_id=mskcc_prad
Grasso et al., 2012: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35988
Tomlins et al., 2007: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6099
Liu et al., 2009: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14996
Bioconductor simpleaffy package: http://www.bioconductor.org/
METHODS
Pten, p53 and Lrf mouse models
Previously generated Ptenflox/flox, p53flox/flox and Lrfflox/flox mice5,13,47 were crossed with the Probasin-Cre mice48 for the deletion of Pten, p53 and Lrf specifically in the prostate epithelium. The pathological index (PI) we used was as follows: 0 (normal tissue), 1 (glands with a monolayer of epithelial cells with evident cellular neoplastic features), 2 (HG-PIN characterized by glands with more than 50% of cell-free lumen) to 3 (HG-PIN characterized by glands with less than 50% of cell-free lumen). Number of animals per genotype per experiment was estimated accordingly to the error variance. Mice were randomized and all the mouse work performed blindly to the genotype of the mice and in accordance with our IACUC approved protocol. For genotyping, tail DNA was subjected to PCR following the protocols previously described5,13,47.
Western blot, immunohistochemistry and proliferation analysis on human and mouse prostate tissue
Cell lysates were prepared with RIPA buffer [1 X PBS, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Roche)] and cleaned by centrifugation. The following antibodies were used for Western blot analysis: mouse anti-cleaved Parp D214 (mouse specific), rabbit anti-Parp 46D11, rabbit anti-cleaved Caspase 3 (Asp175), rabbit anti-Gapdh 14C10, rabbit anti-XIAP 3B6 (Cell signaling), rabbit anti-AR PG-21 (Millipore), rabbit anti-Xaf1 SAB2900401 (Sigma), IMG-379 (Imgenex), goat anti-Probasin sc17126 (Santa Cruz). For immunohistochemistry, prostate tissues were fixed in 10% neutral-buffered formalin (Sigma) overnight, subsequently washed once with PBS, transferred into 50% ethanol and stored in 70% ethanol. Prostate lobes were embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E) in accordance with standard procedures. Sections were stained with the following antibodies: rabbit anti-Ki-67 (Novacastra), rabbit anti-AR (Millipore), rabbit anti-Xaf1 (Sigma), rabbit anti cleaved Caspase3 (Cell Signaling). Proliferative cells were identified by positive Ki-67 staining. A total of 5000 cells were counted from 50 different fields for each section for each lobe. The analysis was repeated on three different mice for each genotype.
MRI analysis
Mouse prostate images were acquired on an ASPECT Model M2 1T tabletop MRI scanner (ASPECT Magnet Technologies Ltd., Netanya, Israel). The M2 is a permanent magnet with zero magnetic fringe field. All mice were placed in the supplied 35mm mouse RF coil, calibrated to a RF frequency of between 43-45 MHz which is used for both transmission and reception. Prior to acquisition, mice were anesthetized with a mixture of isoflurane and oxygen via an external vaporizer at a percentage of 2.0 and 1.5%, respectively. Percentages were adjusted as needed during MR acquisition, as respiration was monitored using a small animal physiological monitoring system (BIOPAC Systems, Inc., Aero Camino Goleta, California). Scout images were acquired using a GRE steady state sequence to acquire images in all three orthogonal planes/orientations. For prostate imaging, mice were positioned in the center of the magnet with bladder used as the anatomical reference for the center point. Axial T2 weighted SE images were first acquired with the following parameters: FOV 40 × 40 mm, 1/0 mm thickness/gap, TR/TE of 4600/40 ms, 256 × 256 matrix, 2 NEX, and a dwell time of 30 μs. Subsequent to the T2 weighted scan, a T1 SE sequence was also acquired by simply changing the TR/TE values to 625/10 ms and decreasing the dwell to 16μs. The NRG Console GUI performs online reconstruction which then allows data to be converted into DICOM files and archived for export into any image analysis software. Tumor volume quantification was performed as previously described49.
Quantitative RT-PCR
Total RNA was prepared from mice frozen tissues using the Trizol method (Invitrogen). cDNA was obtained with iScriptTM cDNA Synthesis Kit (Bio-Rad). Triplicate samples for quantitative PCR were run in the Lightcycler 480 (Roche) using the SYBR Green I Master (Roche). Each value was adjusted by using Gapdh levels as reference (see Supplementary Table 4 for primer sequences).
Human mRNA profiling and aCGH analysis
CGH array data for 37 metastatic samples, yet pre-processed with RAE algorithm, were downloaded by GEO database (see URLs)11. CGH array data for 50 metastatic samples, pre-processed with RAE algorithm, were downloaded from GEO database (see URLs)20. Data for 13 EPI and 20 Metastatic patient samples analyzed on a spotted cDNA platform were obtained from GEO database (see URLs)22. 58 metastatic samples data were downloaded from GEO database and analyzed by Affymetrix Genome-Wide human single nucleotide polymorphism Array 6.0 (see URLs)19. MAS5 algorithm, as implemented in Bioconductor simpleaffy package (see URLs), was used to normalize raw data and Affymetrix annotation na31 to annotate probe-set ID to the corresponding Entrez Gene ID. Differential expression and statistical significance have been evaluated by Bioconductor limma package (see URLs). P-value lower than 0.05 were considered significant.
Patient samples
This study was performed after approval of the Institutional Review Board of the Dana-Farber Cancer Institute and Brigham and Women's Hospital, Boston, MA. Archival paraffin-embedded tissue blocks of prostate cancer from patients were retrieved from the Departments of Pathology from Beth Israel Deaconess Medical Center, Boston, and from the University of Ancona, Italy.
Tissue microarray construction, immunohistochemistry and image analysis
Among men in the study cohort, we obtained archival formalin-fixed, paraffin-embedded radical prostatectomy samples (n=87; all study subjects). The study pathologists (R.L., M.L.) reviewed H&E slides of each FFPE tissue block to select areas of tumor and benign tissue for construction of a tissue microarray (TMA). The TMA was constructed by taking three 0.6-mm cores of tumor tissue and two 0.6-mm cores of benign tissue per case from a formalin-fixed, primary tumor nodule and transferring each core to a recipient block. The tumor and benign specimens from all 87 cases were included on two different TMAs. We characterized the expression profile of PTEN, and LRF in tumors included on the TMAs by immunohistochemical evaluation of protein expression for each, using a BioGenex i6000 automated staining platform (BioGenex Laboratories Inc., Fremont CA). TMA sections were cut at 4μm, and subsequently deparaffinized in xylene, followed by a graded alcohol rehydration. Antigen retrieval was performed by microwaving the tissue in citrate buffer (LRF), or EDTA buffer (PTEN) for 5 minutes. Antisera for PTEN (Cell Signaling Technologies, Beverly, MA) and LRF (Novus Biologicals, Littleton, CO) were then applied (PTEN: 1/200 overnight; LRF: 1/500 for 1 hour). Primary antibody detection was performed using a polymer-HRP detection kit (Biogenix) with visualization achieved using a commercially available chromogenic DAB kit (Vector Laboratories Inc., Burlingame, CA). Finally, sections were counterstained with hematoxylin, and dehydrated in a graded series of alcohols prior to the application of a coverslip. TMA slides stained with either PTEN or LRF were scanned using the CRi Vectra 200-slide scanning platform (v1.4.1, CRi/Caliper, Hopkinton, MA) following a standard bright-field scanning protocol. For each tumor core represented on the TMA, the system acquired images at 20nm intervals and combined them into a stack file that represents one image. To detect both DAB and hematoxylin, a maximum likelihood method was used to extract each spectrum, which represents each element of the IHC profile. The analysis of each core image stack was performed using InForm (v1.3, CRi/Caliper). To do this, a training set comprising two classes of tissue was created: ‘tumor’ and ‘other’. Representative areas for each class were marked on 16-20 images from the TMA. Using the hematoxylin and DAB spectra, InForm was trained to differentiate between the two classes. Within the training set, InForm's ability to correctly differentiate tumor from other was determined by a trained pathologist (R.T.L), and the process was repeated until further training of InForm correctly identified all tumor tissue. IHC images were then analyzed using InForm's ‘object-based’ algorithm. The multispectral imaging profile obtained with the Vectra scanner allows the software to segment the nucleus and cytoplasm using the unmixed spectra of the nuclear counterstain and the DAB immunohistochemical stain used for each biomarker. To quantify the nuclear staining for each biomarker, the following parameters were employed: auto scale and fill hole were enabled, and the area was set to 0.15. For percentage analyses, a nuclear threshold for each biomarker was set as follows: PTEN was 0.288; LRF was 0.08. For cytoplasmic staining, the following parameters were set: the inner distance to nuclei was 0, the outer distance to nuclei was 7, the minimum size (pixel) was 1, and the threshold was 0.56. For percentage analyses, a cytoplasmic threshold for each biomarker was set as follows: PTEN was 0.066, LRF was 0.056. For each core, the DAB intensity of each biomarker for each cell is the determined by InForm. With a threshold applied, InForm calculated the percentage of cells that were positive for each biomarker in the nucleus, in the cytoplasm, and in both compartments. TMA cores that were difficult to classify due to technical limitations (complications with tissue presentation or core loss) were removed from all analyses. For the current analysis, of the 87 cases represented on the two TMA, three were removed from the analysis due to insufficient representation on the TMA (core loss). For all data metrics obtained, tissue samples were reviewed by a trained pathologist (R.T. L. and M.L.) to ensure appropriate assignment of scores.
In vitro treatments
VCaP, LnCaP, LnCaP C4-2, 22Rv1, PC3, and DU145 human prostate cancer cells were purchased from the ATCC, cultured in DMEM or RPMI medium (as indicated by the ATCC) supplemented with 10% FBS and tested for mycoplasma contamination every month. For androgen deprivation and drug treatments, the regular medium plus 10% FBS was replaced with regular medium supplemented with 10% charcoal/dextran treated FBS (Hyclone) (Steroid Free Medium) and 10 μM dutasteride, and/or 10 μM MDV3100, and/or 5μM embelin for 2 or 4 days. For siRNA experiments, cells were transfected with XIAP siRNA (obtained from Dharmacon) and 72 hrs after transfection, the regular medium plus 10% FBS was replaced with regular medium supplemented with 10% charcoal/dextran treated FBS (Hyclone) (Steroid Free Medium). For western blotting, cells were lysed in Ripa buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, and 0.1% SDS) supplemented with protease inhibitors (Complete; Roche), briefly sonicated, and resolved on SDS-PAGE gels. The proteins were transferred to nitrocellulose membranes and detected by chemiluminescence after incubation with antibodies against PARP (1:1000, Cell Signaling), XIAP (1:1000, Cell Signaling), AR (1:1000, Millipore), Actin (1:4000, Sigma), GAPDH (Cell Signaling), and LRF (13E9).
In vivo treatments
Chemical castration studies in mice were conducted with bicalutamide (Casodex) at a dose of 10 mg/kg by oral gavage, daily for 5 days a week for 4 weeks. Tumor-bearing mice were treated with a combination of bicalutamide (10 mg/kg) plus XIAP inhibitor embelin (60 mg/kg) in 0.1% carboxymethyl cellulose (CMC) (as vehicle) by oral gavage, daily for 5 days a week for 4 weeks. Triple treatment experiments were conducted with bicalutamide (10 mg/kg) plus XIAP inhibitor embelin (60 mg/kg) in 0.1% carboxymethyl cellulose (CMC) (as vehicle) by oral gavage, 3 days a week and bicalutamide (10 mg/kg) plus Srd5a1 inhibitor dutasteride (XY mg/kg) in 0.1% carboxymethyl cellulose (CMC) (as vehicle) by oral gavage, 2 days a week for 4 weeks. Mice were euthanized and tumors were dissected, weighed, and processed for histopathology and molecular analyses at the conclusion of these studies.
Statistical Analysis
Results are expressed as mean ± SD. Comparisons between groups were assessed using Student's t-Test analysis. P ≤ 0.05 was considered significant.
Supplementary Material
Editorial Summary (PF).
Pier Paolo Pandolfi and colleagues report that compound loss of Pten with Lrf or Trp53 leads to de novo resistance to androgen deprivation therapy in prostate cancer. Integrative analysis of mouse and patient data in a co-clinical approach identified XIAP and SRD5A1 inhibitors as potential therapies for castration-resistant prostate cancer.
ACKNOWLEDGMENTS
We would like to thank current members of the Pandolfi lab for critical discussion and Thomas Garvey for insightful editing. We are grateful to the Preclinical Murine Pharmacogenetics Facility at BIDMC and Dana-Farber/Harvard Cancer Center for expert support in all the aspects related to the work in mice. Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant # NIH 5 P30 CA06516. We also thank Dr. Abate-Shen for kindly providing the Probasin antibody, and the Small Animal Imaging Facility at BIDMC for the MRI imaging work. Additionally, we are grateful to Giuseppe Fedele and Xiaoqiu Wu from Loda's lab and the Center for Molecular Oncologic Pathology Dana-Farber/Brigham and Women's for their technical support in the generation, staining and analysis of the TMA. U.A. has been supported by a fellowship from the Italian Association for Cancer Research (AIRC) under grant IG-9408. AL has been supported in part by a fellowship from the Istituto Toscano Tumori (ITT, Italy). This work has been supported through the MMHCC/NCI grant (RC2 CA147940-01) to P.P.P.
Footnotes
AUTHOR CONTRIBUTIONS
A.L., C.N., and P.P.P. designed, realized and analyzed the experiments. U.A., W.L.G., R.M., M.B, E.C.S., R.L., A.P., L.C.W., G.B., C.C., W.L.G., and R.M., conducted the human genetic and molecular analysis. K.A.W. performed the H&E and IHC on mouse prostate samples. M.T.E, L.S., J.G.C., and G.W., helped with the experiments. M.L. and S.S. reviewed all mouse pathology. A.L., C.N., and P.P.P. wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
REFERENCES
- 1.Nardella C, Lunardi A, Patnaik A, Cantley LC, Pandolfi PP. The APL paradigm and the “co-clinical trial” project. Cancer Discov. 2011;1:108–16. doi: 10.1158/2159-8290.CD-11-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catalona WJ. Management of cancer of the prostate. N Engl J Med. 1994;331:996–1004. doi: 10.1056/NEJM199410133311507. [DOI] [PubMed] [Google Scholar]
- 3.Molina A, Belldegrun A. Novel therapeutic strategies for castration resistant prostate cancer: inhibition of persistent androgen production and androgen receptor mediated signaling. J Urol. 2011;185:787–94. doi: 10.1016/j.juro.2010.10.042. [DOI] [PubMed] [Google Scholar]
- 4.Wang G. Lrf suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet. 2013 doi: 10.1038/ng.2654. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlomm T, et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21:1371–8. doi: 10.1038/modpathol.2008.104. [DOI] [PubMed] [Google Scholar]
- 7.Tepper CG, et al. Profiling of gene expression changes caused by p53 gain-of-function mutant alleles in prostate cancer cells. Prostate. 2005;65:375–89. doi: 10.1002/pros.20308. [DOI] [PubMed] [Google Scholar]
- 8.Markert EK, Mizuno H, Vazquez A, Levine AJ. Molecular classification of prostate cancer using curated expression signatures. Proc Natl Acad Sci U S A. 2011;108:21276–81. doi: 10.1073/pnas.1117029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abou-Kheir WG, Hynes PG, Martin PL, Pierce R, Kelly K. Characterizing the contribution of stem/progenitor cells to tumorigenesis in the Pten-/-TP53-/- prostate cancer model. Stem Cells. 2010;28:2129–40. doi: 10.1002/stem.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108:17087–92. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pourmand G, et al. Role of PTEN gene in progression of prostate cancer. Urol J. 2007;4:95–100. [PubMed] [Google Scholar]
- 13.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–21. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 15.Carver BS, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, et al. Inhibition of tumor growth progression by antiandrogens and mTOR inhibitor in a Pten-deficient mouse model of prostate cancer. Cancer Res. 2009;69:7466–72. doi: 10.1158/0008-5472.CAN-08-4385. [DOI] [PubMed] [Google Scholar]
- 17.Kent EC, Hussain MH. The patient with hormone-refractory prostate cancer: determining who, when, and how to treat. Urology. 2003;62(Suppl 1):134–40. doi: 10.1016/j.urology.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Kent EC, Hussain MH. Neoadjuvant Therapy for Prostate Cancer: An Oncologist's Perspective. Rev Urol. 2003;5(Suppl 3):S28–37. [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SU, Maeda T. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol Rev. 2012;247:107–19. doi: 10.1111/j.1600-065X.2012.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlins SA, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 23.Baretton GB, Klenk U, Diebold J, Schmeller N, Lohrs U. Proliferation- and apoptosis-associated factors in advanced prostatic carcinomas before and after androgen deprivation therapy: prognostic significance of p21/WAF1/CIP1 expression. Br J Cancer. 1999;80:546–55. doi: 10.1038/sj.bjc.6690390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–15. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Liston P, et al. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol. 2001;3:128–33. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- 26.Huang J, et al. XAF1 as a prognostic biomarker and therapeutic target in pancreatic cancer. Cancer Sci. 2010;101:559–67. doi: 10.1111/j.1349-7006.2009.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu SP, et al. Restoration of XAF1 expression induces apoptosis and inhibits tumor growth in gastric cancer. Int J Cancer. 2009;125:688–97. doi: 10.1002/ijc.24282. [DOI] [PubMed] [Google Scholar]
- 28.Jiang C, Yi XP, Shen H, Li YX. Targeting X-linked inhibitor of apoptosis protein inhibits pancreatic cancer cell growth through p-Akt depletion. World J Gastroenterol. 2012;18:2956–65. doi: 10.3748/wjg.v18.i23.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Y, et al. Adenovirus-mediated down-regulation of X-linked inhibitor of apoptosis protein inhibits colon cancer. Mol Cancer Ther. 2009;8:2762–70. doi: 10.1158/1535-7163.MCT-09-0509. [DOI] [PubMed] [Google Scholar]
- 30.Qiao L, et al. Gene expression profile in colon cancer cells with respect to XIAP expression status. Int J Colorectal Dis. 2009;24:245–60. doi: 10.1007/s00384-008-0566-1. [DOI] [PubMed] [Google Scholar]
- 31.Dubrez-Daloz L, Dupoux A, Cartier J. IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle. 2008;7:1036–46. doi: 10.4161/cc.7.8.5783. [DOI] [PubMed] [Google Scholar]
- 32.Nikolovska-Coleska Z, et al. Discovery of embelin as a cell-permeable, small- molecular weight inhibitor of XIAP through structure-based computational screening of a traditional herbal medicine three-dimensional structure database. J Med Chem. 2004;47:2430–40. doi: 10.1021/jm030420+. [DOI] [PubMed] [Google Scholar]
- 33.Zou B, et al. XIAP-associated factor 1 (XAF1), a novel target of p53, enhances p53-mediated apoptosis via post-translational modification. Mol Carcinog. 2012;51:422–32. doi: 10.1002/mc.20807. [DOI] [PubMed] [Google Scholar]
- 34.Yamaoka M, Hara T, Kusaka M. Overcoming persistent dependency on androgen signaling after progression to castration-resistant prostate cancer. Clin Cancer Res. 2010;16:4319–24. doi: 10.1158/1078-0432.CCR-10-0255. [DOI] [PubMed] [Google Scholar]
- 35.Chang KH, et al. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2011;108:13728–33. doi: 10.1073/pnas.1107898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo JH, et al. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 37.Liu P, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–9. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 38.LaTulippe E, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–506. [PubMed] [Google Scholar]
- 39.Stephenson AJ, et al. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer. 2005;104:290–8. doi: 10.1002/cncr.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai Y, et al. Natural IAP inhibitor Embelin enhances therapeutic efficacy of ionizing radiation in prostate cancer. Am J Cancer Res. 2011;1:128–43. [PMC free article] [PubMed] [Google Scholar]
- 41.Taplin ME, et al. Phase II study of androgen synthesis inhibition with ketoconazole, hydrocortisone, and dutasteride in asymptomatic castration- resistant prostate cancer. Clin Cancer Res. 2009;15:7099–105. doi: 10.1158/1078-0432.CCR-09-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sartor O, Gomella LG, Gagnier P, Melich K, Dann R. Dutasteride and bicalutamide in patients with hormone-refractory prostate cancer: the Therapy Assessed by Rising PSA (TARP) study rationale and design. Can J Urol. 2009;16:4806–12. [PubMed] [Google Scholar]
- 43.Sartor O, et al. Activity of dutasteride plus ketoconazole in castration-refractory prostate cancer after progression on ketoconazole alone. Clin Genitourin Cancer. 2009;7:E90–2. doi: 10.3816/CGC.2009.n.030. [DOI] [PubMed] [Google Scholar]
- 44.Varambally S, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Danquah M, Duke CB, 3rd, Patil R, Miller DD, Mahato RI. Combination Therapy of Antiandrogen and XIAP Inhibitor for Treating Advanced Prostate Cancer. Pharm Res. 2012;29:2079–91. doi: 10.1007/s11095-012-0737-1. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–7. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maeda T, et al. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev Cell. 2009;17:527–40. doi: 10.1016/j.devcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–9. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 49.Nastiuk KL, et al. In vivo MRI volumetric measurement of prostate regression and growth in mice. BMC Urol. 2007;7:12. doi: 10.1186/1471-2490-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.