Abstract
The highly pathogenic filoviruses, Marburg and Ebola virus, belong to the nonsegmented negative-sense RNA viruses of the order Mononegavirales. The mode of replication and transcription is similar for these viruses. On one hand, the negative-sense RNA genome serves as a template for replication, to generate progeny genomes, and, on the other hand, for transcription, to produce mRNAs. Despite the similarities in the replication/transcription strategy, filoviruses have evolved structural and functional properties that are unique among the nonsegmented negative-sense RNA viruses. Moreover, there are also striking differences in the replication and transcription mechanisms of Marburg and Ebola virus. This includes nucleocapsid formation, the structure of the genomic replication promoter, the protein requirement for transcription and the use of mRNA editing. In this article, the current knowledge of the replication and transcription strategy of Marburg and Ebola virus is reviewed, with focus on the observed differences.
Keywords: ebola virus, hemorrhagic fever, marburg virus, nonsegmented negative-sense RNA viruses, nucleocapsid complex, replication, reverse genetics, transcription
Marburg (MARV) and Ebola virus (EBOV) cause severe hemorrhagic fever in humans and nonhuman primates. With mortality rates as high as 90%, these viruses represent some of the most deadly human pathogens [1]. Owing to their high pathogenicity and the lack of either vaccines or effective therapies, MARV and EBOV are classified as biosafety level four agents.
MARV and EBOV comprise the family Filoviridae that, together with the paramyxo-, rhabdo- and borna-viruses, belong to the order Mononegavirales. All viruses within this order are enveloped and contain a nonsegmented negative-strand (NNS) RNA genome. The family Filoviridae is divided into two genera: Ebolavirus, with four distinct species (Zaire, Sudan, Côte d’Ivoire and Reston ebolavirus), and Marburgvirus, with the single species Lake Victoria marburgvirus [2].
Genome organization & viral proteins
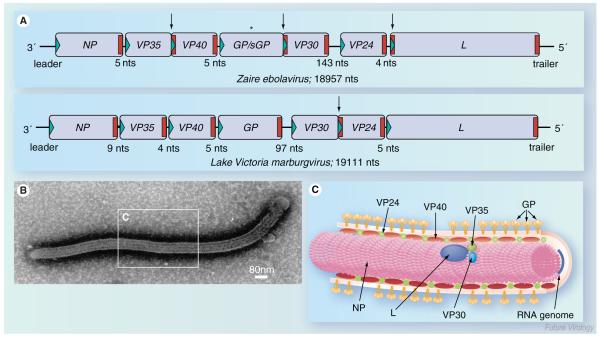
The genome organization of the different filoviruses is rather similar. Seven viral genes are arranged tandemly on the negative-sense RNA genome, which is approximately 19 kb in length (Figure 1A). The genes contain the respective open reading frame (ORF) flanked by unusually long nontranslated sequences, ranging from 57 to 684 nucleotides. Among the NNS RNA viruses, this feature has only been observed for filoviruses and Nipah and Hendra viruses [3]. Thus far, the function of these long untranslated regions is not known. At the beginning and end of each gene there are highly conserved transcription start and stop signals. The genes are either separated by short intergenic regions spanning 4–7 nonconserved nucleotides or they overlap. There is only one longer intergenic region located between the GP and VP30 gene in the case of MARV and between the VP30 and VP24 gene in the case of the different EBOV species. The genome ends consist of short nontranscribed regions containing cis-acting signals important for replication, transcription initiation and encapsidation of the genomic RNA [4–7].
Figure 1. Filovirus genome organization and structure.
(A) Schematic diagram of Ebola virus (EBOV) and Marburg virus (MARV) genomes. The genes are depicted as boxes and nontranscribed regions as black bars (leader, trailer and intergenic regions). Transcription start signals are depicted as green triangles and stop signals as red bars. Gene overlaps are marked by arrows. The mRNA editing site within the EBOV GP gene is indicated by an asterisk. The length of the intergenic regions is shown below the scheme. Note that the EBOV VP24 gene contains two transcription stop signals. (B) Electron micrograph of an EBOV particle. (C) Schematic presentation of EBOV structure. The RNA genome is encapsidated by nucleocapsid proteins NP, VP35, VP30 and L. VP40 and VP24 are matrix proteins. GP trimers are inserted into the viral membrane.
GP: Glycoprotein; L: RNA-dependent RNA polymerase; NP: Nucleoprotein; VP: Viral protein.
Filoviruses encode seven structural proteins (Figure 1B & C). While five of these proteins, the nucleoprotein (NP), the viral proteins (VP) 35 and 40, the glycoprotein (GP) and the RNA-dependent RNA polymerase (L) are counterparts of proteins found in all NNS RNA viruses, VP24 appears to be unique to filoviruses, and a protein similar to VP30 is only described for the pneumoviruses [8].
The only surface protein, GP, is inserted into the viral membrane and mediates receptor binding and fusion during virus entry [9–11]. VP40 is the counterpart of the matrix proteins (M) of other NNS RNA viruses. It is a peripheral membrane protein located on the inner side of the membrane and plays a key role in the budding of viral particles (see below) [10,12]. The viral protein VP24 is presumed to be a minor matrix protein involved in nucleocapsid formation and assembly [13–17]. Since VP24 plays a crucial role in host tropism determination and is able to counteract the type I interferon response, it is considered to be an important virulence factor [18–20]. The four remaining proteins are tightly associated with the viral RNA genome, forming the nucleocapsid [21]. These are the NP, the polymerase cofactor VP35, the transcription activator VP30, and the major component of the RNA-dependent RNA polymerase, L (Figure 1C). The nucleocapsid proteins play a dual role in the viral replication cycle; they are structural components of the nucleocapsid complex and are, therefore, involved in viral morphogenesis and catalyze the replication and transcription of the RNA genome [22–25].
Additional functions have been determined for VP35, which has been shown to act as a type I interferon antagonist by interfering with retinoic acid inducible gene I-dependent activation of the interferon regulatory factor 3 and to block the activation of the double-stranded RNA-dependent protein kinase [26–28].
EBOV genomes code for an additional protein, the soluble GP (sGP). As with GP, sGP is encoded by the fourth gene but is translated from nonedited mRNA species [29]. sGP is not incorporated into viral particles but is secreted from infected cells. The function of the protein is not fully understood. A putative role of sGP in immune modulation has been discussed controversially [30–32]. Recently, it has been shown to act as an anti-inflammatory factor by protecting the endothelial cell barrier function during infection [32].
Filovirus replication & transcription
Filovirus replication and transcription take place in the cytoplasm of the infected cells. The viruses encode their own RNA-dependent RNA polymerase, which recognizes the encapsidated negative-strand RNA genome as a template for replication and transcription. This replication and transcription strategy resembles that of other NNS RNA viruses [33]. However, there are some features unique to filoviruses that will be described in this review.
Nucleocapsid assembly
The driving force for RNA encapsidation is NP. MARV NP forms large helical structures resembling the nucleocapsids found in mature virions [22,34]. For EBOV, the minimal protein requirements for the formation of nucleocapsid-like structures are NP, VP35 and VP24 [17,35]. Recently, Noda and colleagues have proposed a model for EBOV nucleocapsid formation and subsequent budding [23]. According to this model, NP forms helical tubes which differ from nucleocapsid-like structures. VP35 and VP24 interact with the pre-formed NP-derived helices, resulting in the formation of nucleocapsid-like structures [36]. Both transport of the nucleocapsids to the plasma membrane and incorporation into virions are mediated by the interaction of NP with VP40 [23,37]. In addition, Johnson and colleagues have shown that EBOV VP35 interacts with the matrix protein VP40 and EBOV minigenomes, thus mediating the packaging of the minigenome RNA into virus-like particles [38].
Filovirus replication
Protein requirements
Replication of the RNA genome begins with the synthesis of a positive-sense replicative intermediate, the antigenome, which is the reverse complement of the RNA genome. The antigenome, in turn, serves as a template for the generation of new genomes. Both the genomes and the antigenomes are encapsidated by the nucleocapsid proteins (Figure 2) [24,25].
Figure 2. Hypothetical model of filovirus replication and transcription.
Transcription results in the synthesis of seven monocistronic mRNAs that are capped and polyadenylated. The mRNAs are not encapsidated. For MARV transcription, NP, VP35 and L are sufficient. By contrast, EBOV transcription is dependent on the transcription activator VP30. Hypothetical model for EBOV transcription: (1) Transcription initiation: it is proposed that the polymerase consisting of L and VP35 initially binds to a single promoter site within the leader region and initiates transcription at the transcription start site of the first gene, the NP gene. Immediately after transcription initiation, the first 23 nucleotides of the nascent mRNA form a stable secondary structure (Figure 4A). In the absence of VP30, this RNA structure hampers movement of the polymerase along the RNA template and, consequently, the transcription complex pauses. (2) Transcription antitermination: the transcription block caused by the RNA secondary structure is abrogated by VP30 by a mechanism that is as yet unknown. It may be speculated that VP30 resolves the RNA structure or directs additional cofactors to the folded RNA. (3) Elongation and transcription reinitiation: transcription elongation and reinitiation at the following genes takes place independently of VP30, although each transcription start signal is involved in the formation of an RNA stem-loop structure [51]. Please note that the transcription start signal of MARV NP mRNA may also be involved in secondary structure formation. Nevertheless, MARV transcription occurs independently of VP30. mRNA editing has been observed only for EBOV GP mRNA [64,65].
Replication starts with the synthesis of a full-length positive-sense antigenome. The antigenome, in turn, serves as a template for the generation of progeny genomes. Both the genome and the antigenome are tightly encapsidated by the nucleocapsid proteins. NP, VP35 and L are sufficient to mediate MARV and EBOV replication. Since the antigenomic replication promoter is considered to be stronger than the genomic promoter, production of genomic RNA occurs more efficiently, as indicated by the blue arrow. Replication is performed by three proteins, NP, VP35 and L. Blue: negative-sense RNA; Red: positive-sense RNA.
EBOV: Ebola virus; GP: Glycoprotein; L: RNA-dependent RNA polymerase; MARV: Marburg virus; NP: Nucleoprotein; VP: Viral protein.
The replication and transcription processes of MARV and EBOV (species Zaire ebolavirus and Reston ebolavirus) have been investigated using reconstituted replication and transcription systems based on the simultaneous expression of the nucleocapsid protein genes and a virus-specific minireplicon in transfected cells (Box 1 & Figure 3) [24,25,39,40]. With the use of these systems it was demonstrated that three of the four nucleocapsid proteins were sufficient to support MARV and EBOV replication: NP, VP35 and L. For paramyxo- and rhabdovirus replication, similar minimum protein requirements have been described, consisting of the NP, the phosphoprotein (P) and the large protein (L) [33,41]. Thus, it is evident that the filoviral VP35 is the counterpart of the phosphoprotein of other NNS RNA viruses. The active polymerase complex is presumably composed of VP35 and L [25]. VP35 forms homo-oligomers, and interacts with L and NP [42–44]. While homo-oligomerization mediated by an aminoterminally located coiled-coil motif is a prerequisite for L binding, and, therefore, for an active polymerase complex, destruction of the oligomerization domain does not affect binding to NP-derived inclusion bodies [43]. In addition, triple complexes consisting of NP, VP35 and L have been identified, indicating that VP35 directs L to the encapsidated RNA genomes by acting as a linker protein between NP and L [42]. The L proteins of NNS RNA viruses are thought to contain all catalytic functions required for replication and transcription, such as polymerization, polyadenylation and methylation [45].
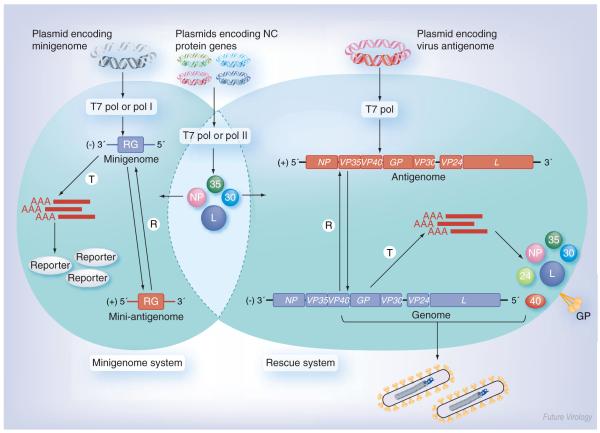
Figure 3. Schematic diagram of filovirus reverse genetics systems.
Minigenome system (left): cells are transfected with plasmids encoding the nucleocapsid proteins along with a plasmid encoding a virus-specific minigenome. The minigenome consists of a RG flanked by the 3′ and 5′ ends of the viral genome containing the signals for replication, encapsidation and transcription. The minigenome is T and R by the nucleocapsid proteins, leading to RG expression (reporter). Rescue system (right): here, cells are transfected with plasmids encoding the nucleocapsid proteins, along with a plasmid encoding a full-length copy of the positive-sense viral antigenome. The antigenome is first R by the nucleocapsid proteins resulting in the generation of a negative-sense genome. The genome serves as a template for replication and transcription, leading to viral mRNA production and subsequent protein synthesis. Finally, the genomes are packaged by the viral proteins and released into mature infectious virus particles. The nucleocapsid protein genes, minigenomes and full-length antigenomes can be cloned under the control of different promoters (T7 pol, pol I and pol II). Negative-sense RNA is indicated in blue, positive-sense RNA in red.
GP: Glycoprotein; L: RNA-dependent RNA polymerase; NP: Nucleoprotein; pol I: RNA polymerase I; pol II: RNA polymerase II; R: Replicated; RG: Reporter gene; T7 pol: T7 RNA polymerase; T: Transcribed; VP: Viral protein.
Genomic replication promoter
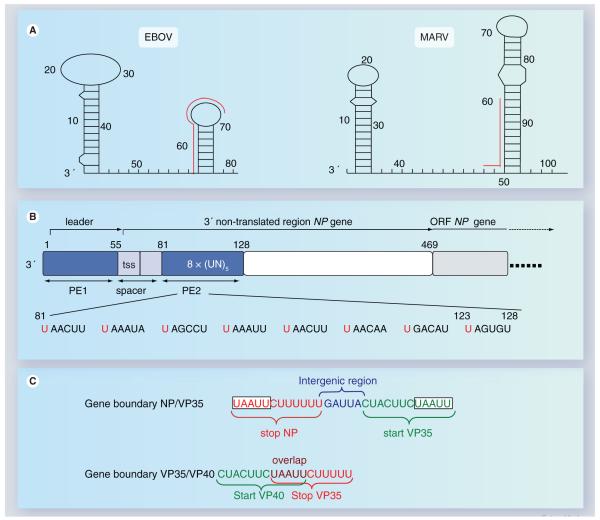
The 3′ termini of filovirus genomes and anti-genomes contain the replication promoters for plus- and minus-sense RNA synthesis. Defective interfering particles generated by serial passaging of Zaire ebolavirus revealed a minimal promoter length of 155 and 176 nucleotides for the genomic and antigenomic RNA 3′ termini, respectively [46]. Since 13–23 nucleotides of the 3′ and 5′ termini of the filovirus genomes show a high degree of complementarity, base-pairing of the respective nucleotides could lead to the formation of a panhandle structure. Notably, the leader and trailer regions contain a second stretch of nucleotides complementary to the very end of the genome, approximately 34–40 nucleotides from either terminus, allowing the formation of an alternative internal hairpin loop [47]. Indeed, chemical modification assays performed with in vitro transcribed EBOV-specific RNA suggested the formation of an internal stem-loop structure within the leader region (Figure 4A) [48]. However, it remains to be elucidated if RNA secondary structure formation of the encapsidated genome really occurs and if it is necessary for replication [47].
Figure 4. Cis-acting elements on filovirus genomes.
(A) RNA secondary structure formation at the 3′ terminus of MARV and EBOV genomes. The first predicted RNA secondary structure is located within the leader region of each genome, the second RNA stem-loop structure is formed by the respective first transcription start site and downstream-located sequences. The red lines running along the side of the stem-loops identify the transcription start signals. Predicted panhandle structures formed by complementary regions of the 3′ and 5′ ends are not shown. Nucleotide numbers refer to the genomic RNA of Zaire ebolavirus, strain Mayinga (accession number AF086833) and Lake Victoria marburgvirus, strain Musoke (accession number DQ217792), respectively. (B) Structure of the Zaire ebolavirus genomic replication promoter. The promoter is located at the 3′ end of the genome. It is bipartite consisting of PE 1 and 2. PE 1 and 2 are depicted as blue boxes. PE 1 and 2 are separated by a spacer (light blue) that contains the transcription start signal of the NP gene. PE 1 spans the leader region, PE 2 is located within the nontranslated region of the NP gene and consists of a stretch of eight UN5 hexamers. The ORF of the NP gene is depicted in light gray. The nucleotide sequence of PE 2 is shown below the scheme. Conserved hexameric U residues are indicated in red. (C) Two typical gene boundaries within the EBOV genome. Transcription start signals are shown in green, transcription stop signals in red. Intergenic region is shown in blue, gene overlap in purple. A conserved pentamer, which is part of the transcription start signals as well as of the transcription stop signals, is boxed.
EBOV: Ebola virus; MARV: Marburgvirus; NP: Nucleoprotein; ORF: Open-reading frame; PE: Promoter element; tss: Transcription start signal; VP: Viral protein.
Concerning their promoter structure, NNS RNA viruses can be broadly divided into two groups. The genomic replication promoters of Rhabdoviridae and Pneumovirinae are short monopartite elements located entirely within the leader [33,49]. By contrast, the genomic replication promoters of the Paramyxovirinae subfamily are bipartite, consisting of a first promoter element located within the leader and a second promoter element contained within the nontranslated region of the first gene. The second promoter element consists of three consecutive hexamers [50]. While the total genome length of rhabdo- and pneumo-viruses does not appear to influence replication efficiency, the genome length of the Paramyxovirinae must be a multiple of six to allow for efficient replication [50]. A genome length rule, such as the ‘rule of six’, was neither found for filovirus genomes nor for naturally occurring EBOV-specific defective interfering particles [46]. Nevertheless, the Zaire ebola-virus genomic replication promoter is a bipartite structure consisting of a first promoter element spanning the leader region (nucleotides 1–55), and a second promoter element comprising nucleotides 81–128 within the nontranslated region of the first gene, the NP gene (Figure 4B). The two promoter elements are separated by a spacer region consisting of the transcription start signal of the NP gene and a downstream located region involved in secondary structure formation and transcription initiation (see later). While proper spacing of the two promoter elements is critical for replication, the spacer sequence itself is not important. The structure of the second promoter element is unique among the NNS RNA viruses and consists of a stretch of eight consecutive UN5 hexamers, where N is any nucleotide (Figure 4B). Only nucleotide insertions or deletions of six or a multiple of six in the spacer region are tolerated, indicating that the genomic replication promoter obeys the rule of six whereas the total genome length does not [48].
Filovirus transcription
Transcription start & stop signals
During transcription, the negative-strand RNA genome is transcribed into seven monocistronic mRNA species. These mRNAs are capped and polyadenylated [51,52]. It is believed that for all NNS RNA viruses, the polymerase gains access to the viral genes through a single polymerase binding site at the 3′ end of the genome. Once bound, the polymerase complex proceeds along the RNA template by stopping and reinitiating at each gene junction and thereby transcribing the individual genes sequentially in their 3′ to 5′ order (Figure 2) [33]. Owing to the release of the polymerase complex from the RNA template, reinitiation at downstream located genes is attenuated. Accordingly, the first gene, NP, is transcribed at the highest levels, while the last gene, L, is transcribed at the lowest [53].
Filovirus genes are flanked by highly conserved transcription start and stop signals. The transcription start signals comprise of a stretch of 12 nucleotides and vary slightly between MARV (3′ CUUCUUA/GUAAUU) and EBOV (3′ CUC/ACUUCUAAUU). Analysis of the 5′ ends of the viral mRNAs revealed that transcription starts precisely with the first nucleotide of the respective transcription start signal [52]. The consensus stop signal contains the sequence 3′ UAAUUC followed by a stretch of five to six uridine residues that serves as a template for the addition of the poly-A tail [4,5]. With analogy to other NNS RNA viruses, it is believed that polyadenylation of the mRNAs occurs through the mechanism of transcriptional stuttering [33]. A highly conserved pentamer, 3′ UAAUU, is located at the 5′end of each start site and the 3′ end of each stop signal (Figure 4C). Notably, the same pentamer can be shared by the transcription stop signal of the downstream and the start signal of the upstream-located gene, leading to gene overlaps whose structure is unique among the NNS RNA viruses (Figure 4C). Genes that do not overlap are separated by short intergenic regions varying in length and nucleotide composition (Figure 1A).
A unique feature of filovirus transcription start signals is that they are predicted to form stable RNA secondary structures [52,54]. The function of most predicted RNA structures remains to be determined. However, the secondary structure of the start site of the EBOV NP gene at least was found to play an important role during viral transcription (see later).
Protein requirements
Typically, transcription of the NNS RNA virus is performed by NP, P and L [41]. The homologous filovirus proteins, namely NP, VP35 and L, were sufficient to support the transcription of MARV minigenomes in a reconstituted replication/transcription system [24]. By contrast, transcription of EBOV minigenomes was strongly dependent on the presence of the fourth nucleocapsid protein VP30 (Figure 2) [25,39,40]. In addition, rescue of full-length EBOV clones was only possible when VP30 was added as the helper plasmid [29,55,56].
EBOV transcription factor VP30
Besides its function as a transcription activator, EBOV VP30 is involved in nucleocapsid assembly by binding to NP-derived inclusion bodies [21]. VP30 is a P that contains a zinc-binding domain and forms hexamers [57–60]. Phosphorylation of VP30 is presumed to be an important regulatory factor by determining whether the protein is involved in transcription activation or assembly [57]. The aminoterminal part of the protein contains an unconventional Cys3–His zinc-binding domain that was also found in MARV VP30 and in the M2–1 proteins of pneumoviruses [58,61]. Destruction of the zinc-binding domain led to the loss of its function as a transcription factor, but VP30 was still able to bind NP inclusions [58]. Recently, the crystal structure of the C-terminal domain of EBOV VP30 has been determined [60]. The C-terminal part of the protein mediates the formation of dimers, which assemble to form hexamers by a leucine-rich oligomerization domain located in the N-terminal part of the protein. The hexamers are incorporated into virions [59,60]. Hexamerization is essential for transcription activation but dispensable for interacting with NP inclusion bodies. Interestingly, the C-terminal part of the protein is sufficient for the incorporation of nucleocapsid-associated VP30 into virions, whereas the N-terminal part of the protein is not required for this process [59].
The precise mechanism of VP30-regulated transcription is not yet fully understood. It is known that VP30 regulates a very early step of transcription, most likely transcription initiation or early antitermination. Interestingly, VP30-dependent transcription is regulated by a RNA hairpin loop formed by the promoter–proximal transcription start signal of the NP gene and a downstream located sequence (Figure 4A). When the formation of the RNA secondary structure was impaired, transcription took place independently of VP30. Although each EBOV-specific transcription start signal is predicted to form a RNA hairpin loop, VP30 is not required for transcription reinitiation at the internally located gene start sites [51]. A hypothetical model of VP30-dependent transcription is depicted in Figure 2.
Among the other members of the order Mononegavirales, a comparable process has not yet been described. Whether MARV VP30 is involved in transcription activation remains elusive. Although transcription took place independently of VP30 in a reconstituted MARV minigenome system, a full-length MARV clone could only be recovered when MARV VP30 was added as a helper plasmid [62]. In addition, RNA interference targeting MARV VP30 led to the down-regulation of the intracellular levels of all other viral proteins in infected cells, thereby suggesting that VP30 plays an essential role in transcription/replication [63]. However, in contrast to EBOV, a MARV VP30 mutant containing a destroyed zinc-binding motif was able to mediate the rescue of a full-length MARV clone [62]. Thus, it appears that MARV VP30 is essential for the viral life cycle but probably not involved in transcription activation.
Besides filoviruses, the only other NNS RNA viruses possessing a fourth nucleocapsid protein are the pneumoviruses. The fourth nucleocapsid protein of respiratory syncytial virus, M2–1, acts in a later stage of transcription as a processivity factor [49].
mRNA editing
There are two overlapping ORFs within the EBOV GP gene and neither of these are able to encode the virion-associated GP. By contrast, sGP is directly encoded by the first ORF. During transcription, an additional nontemplated adenosine residue is inserted into the nascent mRNA chain within a stretch of seven adenosines via the mRNA editing mechanism. Approximately 20% of GP-specific mRNA is edited [29,64,65]. Due to the insertion of the adenosine residue, the reading frame is shifted, leading to the expression of the membrane-anchored GP. High levels of GP were produced in cells infected with a recombinant EBOV clone encoding GP by a continuous ORF, whereas sGP was not expressed. Interestingly, the mutant was significantly more cytopathogenic than the wild-type virus, indicating that GP mRNA editing is an important biological mechanism for the control and down-regulation of the expression of the cytotoxic GP [29,66].
Future perspective
Replication and transcription of the RNA genome are crucial processes in the filovirus life-cycle. Since filoviruses encode their own replication/transcription machinery, the components involved in replication/transcription; cis-acting elements on the genomes and VPs mediating replication/transcription, are ideal targets for antiviral strategies. Indeed, initial approaches to use viral genes and proteins involved in replication/transcription as targets for antiviral compounds were quite promising [59,63,67–70]. Thus, a deeper understanding of the way in which filoviruses replicate and transcribe their genomes will lead to potential benefits in controlling filovirus infections. The reverse genetics systems now available for MARV and EBOV are especially powerful tools in the development of antiviral therapies and vaccines.
Another focus of future filovirus research will be the correlation of viral replication and antiviral response. It is already known for the polymerase cofactor VP35 that it interferes with the innate immune system [26–28]. Within the same vein, bioinformatic analysis has revealed that filovirus species differing in their replication efficiency also differ in their ability to evade the cellular antiviral response [71].
Conclusion
In this review, the replication and transcription strategy of MARV and EBOV has been described. Although in many aspects, filoviruses follow the general mechanisms reported for NNS RNA virus replication/transcription, they have evolved individual molecular mechanisms that differ from all other NNS RNA viruses. These unique features include the structure of the genomic replication promoter, the structure of overlapping genes, the need for an additional nucleocapsid protein, VP30, as a transcription activation factor, and the use of mRNA editing to regulate the expression of the cytotoxic EBOV GP.
Box 1. Tools to investigate filovirus replication and transcription.
For a long time, research on Marburg (MARV) and Ebola virus (EBOV) replication was hampered by the requirement for biosafety level four containment facilities. Then, in 1998 and 1999, respectively, the first transfection-based filovirus-specific minigenome systems were described, allowing for the investigation of the replication and transcription strategy of these dangerous viruses under lower biosafety conditions [24,25]. The central element of these systems is a virus-specific minigenome consisting of the 3′ and 5′ ends of the filoviral genome containing the cis-acting signals required for replication/transcription initiation and RNA packaging. In this construct, all viral genes are replaced by a reporter gene. The minigenome was cloned under the control of the T7 RNA polymerase promoter and used to transfect cells expressing the T7 RNA polymerase. The same cells were transfected with plasmids containing the viral nucleocapsid protein genes under the control of the T7 RNA polymerase promoter. Recognition and subsequent replication and transcription of the minigenome by the nucleocapsid proteins could be determined by the detection of the replicated and transcribed RNA species or by reporter gene assays (Figure 3). Meanwhile, alternative minigenome systems exist. Groseth and colleagues have described an RNA polymerase I- and II-driven minigenome system in which the minigenome is supplied by RNA polymerase I and the supporting nucleocapsid proteins by RNA polymerase II (Figure 3) [40].
In order to generate infectious EBOV-like particles containing a minigenome RNA, the plasmid-based minigenome system was extended with the inclusion of the viral genes needed for assembly [16,72].
Based on the minigenome system, rescue systems have been established for MARV and EBOV, allowing the recovery of complete recombinant viruses entirely from cDNA (Figure 3) [29,55,62,73].
Excecutive summary.
Genome organization & viral proteins
The nonsegmented negative-sense RNA genome of Marburg (MARV) and Ebola viruses (EBOV) are 19 kb in length and contain seven genes (Figure 1A).
At the 3′ and 5′ ends of the genome are short extragenic regions containing signals required for replication, transcription initiation and packaging of the RNA genome.
Genes are flanked by highly conserved transcription start and stop signals.
Genes are either separated by short intergenic regions or they overlap (Figure 4C).
Filoviruses encode seven structural proteins (Figure 1B).
The only surface protein, glycoprotein (GP) binds to the cellular receptor and mediates fusion. Viral protein (VP)-40 is the major matrix protein and is required for the transport of the nucleocapsids to the plasma membrane and for virus budding. VP24 is presumed to be a minor matrix protein involved in nucleocapsid formation and assembly. It is considered to be a virulence factor.
In contrast to most other Mononegavirales, filoviruses possess four nucleocapsid proteins. These are nucleoprotein (NP), polymerase cofactor VP35, transcription activator VP30 and RNA-dependent RNA polymerase (L).
The nucleocapsid proteins are tightly associated with the RNA genome.
Filovirus replication
Replication of the RNA genome starts with the synthesis of a positive-sense reverse complement of the genome, the antigenome. The antigenome, in turn, serves as a template for the generation of new genomes (Figure 2).
The minimal protein requirement for filovirus replication is NP, VP35 and L.
The structure of the genomic replication promoter of EBOV is unique among nonsegmented negative-sense RNA viruses (Figure 4B).
Filovirus transcription
During transcription, the negative-strand RNA genome is transcribed into seven monocistronic mRNA species. These mRNAs are capped and polyadenylated by the viral RNA-dependent RNA polymerase (Figure 2).
A unique feature of filovirus transcription start signals is that they are predicted to form stable RNA secondary structures.
NP, VP35 and L are sufficient to support the transcription of MARV minigenomes. By contrast, the minimal protein requirement for EBOV transcription is NP, VP35, L and VP30.
The EBOV transcription factor, VP30, regulates a very early step of transcription, most likely transcription initiation or early antitermination.
VP30-dependent transcription is regulated by an RNA hair-pin loop formed by the transcription start signal of the first gene, the NP gene.
EBOV GP mRNA is subjected to mRNA editing to regulate the expression of cytotoxic GP. mRNA editing has not been observed with MARV transcription.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Hensley LE, Jones SM, Feldmann H, Jahrling PB, Geisbert TW. Ebola and Marburg viruses: pathogenesis and development of countermeasures. Curr. Mol. Med. 2005;5:761–772. doi: 10.2174/156652405774962344. [DOI] [PubMed] [Google Scholar]
- 2.Feldmann H, Geisbert TW, Jahrling PB, et al. In: Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Elsevier/Academic Press; San Diego, USA: 2005. pp. 645–653. [Google Scholar]
- 3.Wang L, Harcourt BH, Yu M, et al. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 2001;3:279–287. doi: 10.1016/s1286-4579(01)01381-8. [DOI] [PubMed] [Google Scholar]
- 4.Towner JS, Khristova ML, Sealy TK, et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J. Virol. 2006;80:6497–6516. doi: 10.1128/JVI.00069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez A, Rollin PE. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res. 2005;113:16–25. doi: 10.1016/j.virusres.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Groseth A, Ströher U, Theriault S, Feldmann H. Molecular characterization of an isolate from the 1989/90 epizootic of Ebola virus Reston among macaques imported into the United States. Virus Res. 2002;87:155–163. doi: 10.1016/s0168-1702(02)00087-4. [DOI] [PubMed] [Google Scholar]
- 7.Ikegami T, Calaor AB, Miranda ME, et al. Genome structure of Ebola virus subtype Reston: differences among Ebola subtypes. Brief report. Arch. Virol. 2001;146:2021–2027. doi: 10.1007/s007050170049. [DOI] [PubMed] [Google Scholar]
- 8.Biacchesi S, Skiadopoulos MH, Boivin G, et al. Genetic diversity between human metapneumovirus subgroups. Virology. 2003;315:1–9. doi: 10.1016/s0042-6822(03)00528-2. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn JH, Radoshitzky SR, Guth AC, et al. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 2006;281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 10.Jasenosky LD, Kawaoka Y. Filovirus budding. Virus Res. 2004;106:181–188. doi: 10.1016/j.virusres.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann H, Volchkov VE, Volchkova VA, Ströher U, Klenk HD. Biosynthesis and role of filoviral glycoproteins. J. Gen. Virol. 2001;82:2839–2848. doi: 10.1099/0022-1317-82-12-2839. [DOI] [PubMed] [Google Scholar]
- 12.Hartlieb B, Weissenhorn W. Filovirus assembly and budding. Virology. 2006;344:64–70. doi: 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Bamberg S, Kolesnikova L, Möller P, Klenk HD, Becker S. VP24 of Marburg virus influences formation of infectious particles. J. Virol. 2005;79:13421–13433. doi: 10.1128/JVI.79.21.13421-13433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Z, Boshra H, Sunyer JO, Zwiers SH, Paragas J, Harty RN. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 2003;77:1793–1800. doi: 10.1128/JVI.77.3.1793-1800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Licata JM, Johnson RF, Han Z, Harty RN. Contribution of ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 2004;78:7344–7351. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoenen T, Groseth A, Kolesnikova L, et al. Infection of naive target cells with virus-like particles: implications for the function of ebola virus VP24. J. Virol. 2006;80:7260–7264. doi: 10.1128/JVI.00051-06. • Describes the establishment of an infectious virus-like particle system for Ebola virus (EBOV) in naive cells.
- 17.Noda T, Aoyama K, Sagara H, Kida H, Kawaoka Y. Nucleocapsid-like structures of Ebola virus reconstructed using electron tomography. J. Vet. Med. Sci. 2005;67:325–328. doi: 10.1292/jvms.67.325. [DOI] [PubMed] [Google Scholar]
- 18.Volchkov VE, Chepurnov AA, Volchkova VA, Ternovoj VA, Klenk HD. Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology. 2000;277:147–155. doi: 10.1006/viro.2000.0572. [DOI] [PubMed] [Google Scholar]
- 19.Reid SP, Leung LW, Hartman AL, et al. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. • First report that a filovirus protein interferes with interferon signaling.
- 20.Ebihara H, Takada A, Kobasa D, et al. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog. 2006;2:e73. doi: 10.1371/journal.ppat.0020073. • Identifies virus proteins involved in host tropism determination by using recombinant EBOV.
- 21.Mühlberger E, Becker S. Marburg virus replication and nucleocapsid formation: different jobs, same players. In: Bogner E, Holzenburg A, editors. Structure-Function Relationships of Human Pathogenic Viruses. Kluwer Academic/Plenum Publishers; London, UK: 2002. pp. 89–97. [Google Scholar]
- 22.Kolesnikova L, Mühlberger E, Ryabchikova E, Becker S. Ultrastructural organization of recombinant Marburg virus nucleoprotein: comparison with Marburg virus inclusions. J. Virol. 2000;74:3899–3904. doi: 10.1128/jvi.74.8.3899-3904.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noda T, Ebihara H, Muramoto Y, et al. Assembly and budding of Ebolavirus. PLoS Pathog. 2006;2:e99. doi: 10.1371/journal.ppat.0020099. • Proposes a nice model for EBOV virion formation.
- 24.Mühlberger E, Lötfering B, Klenk HD, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35 and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. • First description of a Marburg virus (MARV) minigenome system.
- 25.Mühlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J. Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. • First description of an EBOV minigenome system.
- 26.Cardenas WB, Loo YM, Gale M, Jr, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits α/β interferon production induced by RIG-I signaling. J. Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. • Determines cellular target for EBOV interferon antagonist VP35.
- 27.Hartman AL, Dover JE, Towner JS, Nichol ST. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J. Virol. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Z, Cerveny M, Yan Z, He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA dependent protein kinase PKR. J. Virol. 2006;81:182–193. doi: 10.1128/JVI.01006-06. • Shows that EBOV VP35 blocks antiviral proteins.
- 29.Volchkov VE, Volchkova VA, Mühlberger E, et al. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science. 2001;291:1965–1969. doi: 10.1126/science.1057269. • First description of an EBOV rescue system.
- 30.Kindzelskii AL, Yang Z, Nabel GJ, Todd RF, 3rd, Petty HR. Ebola virus secretory glycoprotein (sGP) diminishes Fc RIIIB-to-CR3 proximity on neutrophils. J. Immunol. 2000;164:953–958. doi: 10.4049/jimmunol.164.2.953. [DOI] [PubMed] [Google Scholar]
- 31.Sui J, Marasco WA. Evidence against Ebola virus sGP binding to human neutrophils by a specific receptor. Virology. 2002;303:9–14. doi: 10.1006/viro.2002.1715. [DOI] [PubMed] [Google Scholar]
- 32.Wahl-Jensen V, Kurz SK, Hazelton PR, et al. Role of Ebola virus secreted glycoproteins and virus-like particles in activation of human macrophages. J. Virol. 2005;79:2413–2419. doi: 10.1128/JVI.79.4.2413-2419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 34.Mavrakis M, Kolesnikova L, Schoehn G, Becker S, Ruigrok RW. Morphology of Marburg virus NP-RNA. Virology. 2002;296:300–307. doi: 10.1006/viro.2002.1433. [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and post-translational modification of nucleoprotein. Mol. Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe S, Noda T, Kawaoka Y. Functional mapping of the nucleoprotein of Ebola virus. J. Virol. 2006;80:3743–3751. doi: 10.1128/JVI.80.8.3743-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noda T, Watanabe S, Sagara H, Kawaoka Y. Mapping of theVP40-binding regions of the Nucleoprotein of Ebola virus. J. Virol. 2007 doi: 10.1128/JVI.02183-06. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J. Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boehmann Y, Enterlein S, Randolf A, Mühlberger E. A reconstituted replication and transcription system for Ebola virus Reston and comparison with Ebola virus Zaire. Virology. 2005;332:406–417. doi: 10.1016/j.virol.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 40.Groseth A, Feldmann H, Theriault S, Mehmetoglu G, Flick R. RNA polymerase I-driven minigenome system for Ebola viruses. J. Virol. 2005;79:4425–4433. doi: 10.1128/JVI.79.7.4425-4433.2005. • Description of an RNA polymerase I-, II-driven EBOV minigenome system.
- 41.Conzelmann KK. Reverse genetics of mononegavirales. Curr. Top. Microbiol. Immunol. 2004;283:1–41. doi: 10.1007/978-3-662-06099-5_1. [DOI] [PubMed] [Google Scholar]
- 42.Becker S, Rinne C, Hofsäss U, Klenk HD, Mühlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology. 1998;249:406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 43.Möller P, Pariente N, Klenk HD, Becker S. Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J. Virol. 2005;79:14876–14886. doi: 10.1128/JVI.79.23.14876-14886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid SP, Cardenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 46.Calain P, Monroe MC, Nichol ST. Ebola virus defective interfering particles and persistent infection. Virology. 1999;262:114–128. doi: 10.1006/viro.1999.9915. [DOI] [PubMed] [Google Scholar]
- 47.Crary SM, Towner JS, Honig JE, Shoemaker TR, Nichol ST. Analysis of the role of predicted RNA secondary structures in Ebola virus replication. Virology. 2003;306:210–218. doi: 10.1016/s0042-6822(02)00014-4. [DOI] [PubMed] [Google Scholar]
- 48.Weik M, Enterlein S, Schlenz K, Mühlberger E. The Ebola virus genomic replication promoter is bipartite and follows the rule of six. J. Virol. 2005;79:10660–10671. doi: 10.1128/JVI.79.16.10660-10671.2005. • Describes the unique structure of the EBOV promoter.
- 49.Cowton VM, McGivern DR, Fearns R. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J. Gen. Virol. 2006;87:1805–1821. doi: 10.1099/vir.0.81786-0. [DOI] [PubMed] [Google Scholar]
- 50.Kolakofsky D, Roux L, Garcin D, Ruigrok RW. Paramyxovirus mRNA editing, the ‘rule of six’ and error catastrophe: a hypothesis. J. Gen. Virol. 2005;86:1869–1877. doi: 10.1099/vir.0.80986-0. [DOI] [PubMed] [Google Scholar]
- 51.Weik M, Modrof J, Klenk HD, Becker S, Mühlberger E. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J. Virol. 2002;76:8532–8539. doi: 10.1128/JVI.76.17.8532-8539.2002. • Describes the exceptional EBOV transcription strategy.
- 52.Mühlberger E, Trommer S, Funke C, Volchkov V, Klenk HD, Becker S. Termini of all mRNA species of Marburg virus: sequence and secondary structure. Virology. 1996;223:376–380. doi: 10.1006/viro.1996.0490. [DOI] [PubMed] [Google Scholar]
- 53.Mühlberger E. Genome organization, replication, and transcription of filoviruses. In: Feldmann HaK, HD, editors. Ebola and Marburg viruses: Molecular and Cellular Biology. Horizon Scientific Press; Wymondham, Norfolk, UK: 2004. pp. 1–12. [Google Scholar]
- 54.Sanchez A, Kiley MP, Holloway BP, Auperin DD. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 55.Neumann G, Feldmann H, Watanabe S, Lukashevich I, Kawaoka Y. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 2002;76:406–410. doi: 10.1128/JVI.76.1.406-410.2002. • Description of an EBOV rescue system based on RNA polymerase II helper plasmids expressing the nucleocapsid proteins.
- 56.Theriault S, Groseth A, Neumann G, Kawaoka Y, Feldmann H. Rescue of Ebola virus from cDNA using heterologous support proteins. Virus Res. 2004;106:43–50. doi: 10.1016/j.virusres.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Modrof J, Mühlberger E, Klenk HD, Becker S. Phosphorylation of VP30 impairs ebola virus transcription. J. Biol. Chem. 2002;277:33099–33104. doi: 10.1074/jbc.M203775200. [DOI] [PubMed] [Google Scholar]
- 58.Modrof J, Becker S, Mühlberger E. Ebola virus transcription activator VP30 is a zinc-binding protein. J. Virol. 2003;77:3334–3338. doi: 10.1128/JVI.77.5.3334-3338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hartlieb B, Modrof J, Mühlberger E, Klenk HD, Becker S. Oligomerization of Ebola virus VP30 is essential for viral transcription and can be inhibited by a synthetic peptide. J. Biol. Chem. 2003;278:41830–41836. doi: 10.1074/jbc.M307036200. [DOI] [PubMed] [Google Scholar]
- 60.Hartlieb B, Muziol T, Weissenhorn W, Becker S. Crystal structure of the C-terminal domain of Ebola virus VP30 reveals a role in transcription and nucleocapsid association. Proc. Natl Acad. Sci. USA. 2007;104:624–629. doi: 10.1073/pnas.0606730104. • First data on the crystal structure of EBOV VP30.
- 61.Hardy RW, Wertz GW. The Cys(3)-His(1) motif of the respiratory syncytialvirus M2–1 protein is essential for protein function. J. Virol. 2000;74:5880–5885. doi: 10.1128/jvi.74.13.5880-5885.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Enterlein S, Volchkov V, Weik M, et al. Rescue of recombinant Marburg virus from cDNA is dependent on nucleocapsid protein VP30. J. Virol. 2006;80:1038–1043. doi: 10.1128/JVI.80.2.1038-1043.2006. • First description of a MARV rescue system.
- 63.Fowler T, Bamberg S, Möller P, et al. Inhibition of Marburg virus protein expression and viral release by RNA interference. J. Gen. Virol. 2005;86:1181–1188. doi: 10.1099/vir.0.80622-0. [DOI] [PubMed] [Google Scholar]
- 64.Volchkov VE, Becker S, Volchkova VA, et al. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. • First reports about EBOV glycoprotein (GP) mRNA editing.
- 65.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl Acad. Sci. U S A. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. • First reports about EBOV GP mRNA editing.
- 66.Alazard-Dany N, Volchkova V, Reynard O, et al. Ebola virus glycoprotein GP is not cytotoxic when expressed constitutively at a moderate level. J. Gen. Virol. 2006;87:1247–1257. doi: 10.1099/vir.0.81361-0. [DOI] [PubMed] [Google Scholar]
- 67.Geisbert TW, Hensley LE, Kagan E, et al. Postexposure protection of guinea pigs against a lethal ebola virus challenge is conferred by RNA interference. J. Infect. Dis. 2006;193:1650–1657. doi: 10.1086/504267. • Protection of guinea pigs from a lethal EBOV challenge by small interefering RNAs targeting the RNA-dependent RNA polymerase gene.
- 68.Warfield KL, Swenson DL, Olinger GG, et al. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2006;2:e1. doi: 10.1371/journal.ppat.0020001. • Describes the antiviral effect of antisense oligomers targeting EBOV genes in monkeys.
- 69.Enterlein S, Warfield KL, Swenson DL, et al. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob. Agents Chemother. 2006;50:984–993. doi: 10.1128/AAC.50.3.984-993.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Müller S, Möller P, Bick MJ, et al. Inhibition of filovirus replication by the zinc finger antiviral protein. J. Virol. 2006;81(5):2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kash JC, Mühlberger E, Carter V, et al. Global suppression of the host antiviral response by Ebola- and Marburg viruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol. 2006;80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. • Describes the correlation of replication efficiency and the ability to suppress the antiviral response.
- 72.Watanabe S, Watanabe T, Noda T, et al. Production of novel ebola virus-like particles from cDNAs: an alternative to ebola virus generation by reverse genetics. J. Virol. 2004;78:999–1005. doi: 10.1128/JVI.78.2.999-1005.2004. • First description of an infectious virus-like particle system for EBOV.
- 73.Towner JS, Paragas J, Dover JE, et al. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]