SUMMARY
Allosteric interactions provide precise spatiotemporal control over signaling proteins, but how allosteric activators and their targets co-evolve is poorly understood. Here, we trace the evolution of two allosteric activator motifs within the yeast scaffold protein Ste5 that specifically target the mating MAP kinase Fus3. One activator (Ste5-VWA) provides pathway insulation and dates to the divergence of Fus3 from its paralog, Kss1; a second activator (Ste5-FBD) that tunes mating behavior is, in contrast, not conserved in most lineages. Surprisingly, both Ste5 activator motifs could regulate MAP kinases that diverged from Fus3 prior to the emergence of Ste5, suggesting that Ste5 activators arose by exploiting latent regulatory features already present in the MAPK ancestor. The magnitude of this latent allosteric potential drifts widely among pre-Ste5 MAP kinases, providing a pool of hidden phenotypic diversity that, when revealed by new activators, could lead to functional divergence and the evolution of distinct signaling behaviors.
INTRODUCTION
Eukaryotic signaling proteins display highly diverse and divergent allosteric regulation. Although any one genome might contain many evolutionarily related signaling molecules, such as protein kinases, individual family members usually display divergent substrate specificity and unique allosteric regulation by various partner proteins. By controlling when and where signaling proteins are activated, these allosteric regulatory interactions play a central role in determining the specific “wiring” of the molecular networks that control cellular behavior (Fig. 1A).
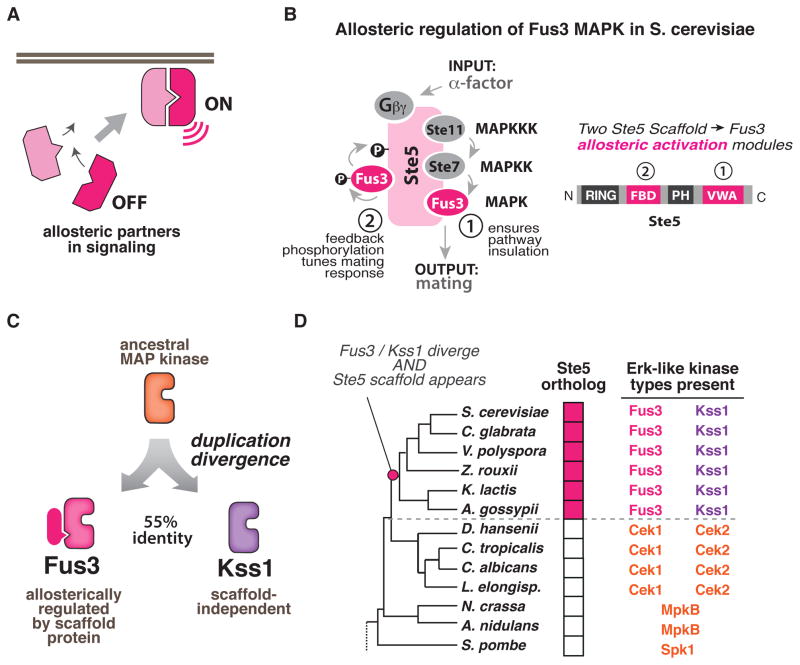
Figure 1. Fungal Erk kinase signaling repertoires provide a model system for biochemically interrogating the evolution of novel and divergent allosteric activation mechanisms.
(A) Allosteric interactions between signaling partners control when and where signaling molecules are activated in cells. (B) In the S. cer. mating MAP kinase pathway, two unique allosteric activities of the Ste5 scaffold regulate the MAP kinase Fus3 but not its paralog Kss1: (1) a VWA domain in Ste5 (Ste5-VWA) that is required to allosterically prime Fus3 for phosphorylation by the upstream MAPKK Ste7; and (2) a Fus3-binding domain (FBD) which stimulates Fus3 autophosphorylation as part of a negative feedback loop that shapes the morphological response of cells to mating pheromone. (C) Fus3 and Kss1 are Erk-like kinases that are 55% identical and arose from a duplication of an ancestral MAP kinase. (D) Abbreviated phylogeny of fungal species from Ascomycota with the signaling repertoire (number and types of ERK-like kinases present; presence or absence of Ste5 scaffold) indicated for each species (see also Fig. S1A–C).
Despite their importance, little is known about how these complex allosteric regulatory partnerships in signaling networks evolve. The molecular complexity of these systems represents a challenge for evolution: allosteric activators and the target proteins that they act on must seemingly acquire their complementary regulatory properties simultaneously for these systems to be functional and provide a selective advantage. These allosteric activators must also be specific enough to ensure that they do not inadvertently target homologous signaling components in the cell. The viable paths by which such multicomponent regulatory systems can evolve are therefore unclear.
In other complex systems, many new features appear to evolve by taking advantage of pre-existing or latent behavior: an active site that catalyzes a particular reaction can, with increased promiscuity, perform similar reactions on other substrates; a binding pocket that favors binding of one nuclear hormone can be adapted accommodate a yet-to-be-evolved hormone with somewhat similar structural features (Aharoni et al., 2005; Baker et al., 2012; Bridgham et al., 2006; O’Brien and Herschlag, 1999; Khersonsky and Tawfik, 2010; Wise et al., 2005). While such latent capacities provide clear toeholds for new enzymatic activities or ligand binding capacities, these changes represent a shift in an already well-established and constitutive molecular activity. It is thus unclear the extent to which these evolutionary models apply to allosteric systems in which new protein partnerships must develop that are unrelated to any existing form of regulation and that must produce complex structural reorganization. Computational and protein engineering studies suggest that certain features of protein structure and dynamics may endow proteins with some latent capacity for allosteric regulation (Lee et al., 2008; Reynolds et al., 2011). Whether natural systems have harnessed such latent features to produce new allosteric regulation during evolution, however, has not been established.
Comparative studies that track the appearance of specific molecular properties across related species were instrumental in uncovering the role of latent protein features in the evolution of other systems and have provided great insights into how new enzymatic activities, receptor/ligand pairs, and transcriptional circuits evolve (Afriat et al., 2006; Booth et al., 2010; Gerlt and Babbitt, 2001; O’Brien and Herschlag, 2001; Roodveldt and Tawfik, 2005; Taylor Ringia et al., 2004; Thornton, 2001). However, applying these approaches to multi-component allosteric regulation of signaling proteins has been hindered by a lack of model systems that can be biochemically interrogated over species spanning a considerable window of evolutionary time.
The budding yeast MAP kinase network presents a unique model system with which to take a comparative approach to understand how complex multi-component allosteric regulation might have evolved. Prior biochemical studies have shown that, in S. cerevisiae, the function of the mating-pathway specific MAP kinase Fus3 requires its allosteric activation by the scaffold protein Ste5 (Fig. 1B). This scaffold-mediated allosteric activation ensures that Fus3 is only activated in signaling complexes that are organized in response to pheromone stimulation, thus preventing inappropriate cross-talk in which distinct MAP kinase mediated pathways trigger mating (Zalatan et al., 2012). Interestingly, the closely related starvation-responsive MAP kinase, Kss1, functions independent of Ste5 regulation, despite the fact that Fus3 and Kss1 are 55 % identical, are both targets of the MAPKK Ste7, and likely arose from duplication of the same Erk-like MAP kinase ancestor (Fig. 1C) (Madhani and Fink, 1998).
Given their common MAPK ancestor, how did Fus3 become dependent on allosteric regulation while Kss1 did not? The availability of a large number of sequenced fungal genomes provides an opportunity to gain insights into this evolutionary question by exploring the regulatory properties of orthologs from scaffold and MAP kinase species throughout the fungal tree. Comparison of Erk-like MAP kinase sequences from across the Ascomycota fungi (to which S. cer. belongs) indicates that these kinases are highly divergent and fall into distinct classes that are associated with specific fungal lineages (Fig. S1A–C). Interestingly, only those species that have both a Fus3 and Kss1 ortholog also have a Ste5 scaffold ortholog (Fig. 1D; detailed in supplement). It is unclear how both a potent allosteric activator (Ste5) and its regulated target (Fus3) could simultaneously evolve as a two-part complementary system. However, because we have access to signaling repertoires from species that clearly diverged from the S. cer. lineage prior to the appearance of Fus3, Kss1 and Ste5, we have the potential to uncover the mechanism by which this allosteric partnership evolved.
Here we expressed and purified Erk-like MAP kinases and Ste5 orthologs (if present) from 13 diverse fungal species that span from S. pombe to S. cer. (~1 billion years of divergence—comparable to the divergence between sea squirt and human). Using an in vitro reconstituted system, we determined the ability of these orthologs to cross-activate one another, even for species that do not contain a Ste5 protein. These quantitative data allowed us to determine when specific kinase and scaffold biochemical features arose during evolution and to formulate a model for the evolution of the allosteric regulatory schemes observed in S. cer.
First, we find that the Ste5 allosteric interaction required for Fus3 activation by the MAPKK Ste7 (Ste5-VWA) is a conserved scaffold feature of all Fus3/Kss1 containing species, while a second allosteric region in Ste5 (Ste5-FBD) that tunes the ultrasensitiviy of the mating response is, in general, not conserved outside of S. cer. This is consistent with a model in which a core function of the Ste5 scaffold protein has been to functionally insulate Fus3 and Kss1 since their divergence, but also suggests that Ste5/Fus3 interactions might continue to evolve to meet specific organismal needs. Second, and surprisingly, we find that the Ste5 scaffold can allosterically activate orthologous MAP kinases from species that diverged prior to the evolution of Ste5, i.e. kinases that are likely to never have co-existed with the Ste5 scaffold. This result suggests that the Ste5 allosteric interactions evolved by tapping into latent, pre-existing dynamic properties of the MAP kinase. The magnitude of this latent allostery appears to drift significantly within the pre-Ste5 MAP kinases—some orthologs are primed for Fus3-like regulation (strong allosteric response) while others are primed for Kss1-like regulation (inability to respond). We propose that hidden diversity in these latent allosteric properties provides a toehold that new partner molecules can exploit to develop novel, component-specific allosteric regulatory relationships, simplifying the evolutionary paths to allosteric controls that shape pathway behavior and distinguish functional identity.
RESULTS
The S. cer. Ste5 scaffold protein allosterically activates the Fus3 MAP kinase via two mechanisms
In prior work we identified two modes by which the Ste5 scaffold protein allosterically activates the Fus3 MAP kinase in budding yeast S. cer. (Fig. 1B). The first allosteric interaction involves a Von-Wildebrand Type A (VWA) domain in the Ste5 scaffold protein that is required to allosterically unlock Fus3 to allow for its dual phosphorylation and activation by the upstream MAP kinase kinase (MAPKK), Ste7 (Fig. 2A). This VWA allosteric co-activation is essential for the transmission of the mating signal but has no influence on activation of the paralagous starvation-specific MAP kinase Kss1, which is also a substrate for the MAPKK Ste7 (Good et al., 2009). In the resting Ste5 molecule, Ste5-VWA activity is autoinhibited by other domains in Ste5. This autoinhibition prevents Fus3 from being activated until mating inputs relieve this inhibition, providing insulation from alternative inputs that activate the upstream MAPKK Ste7, such as starvation (Zalatan et al., 2012).
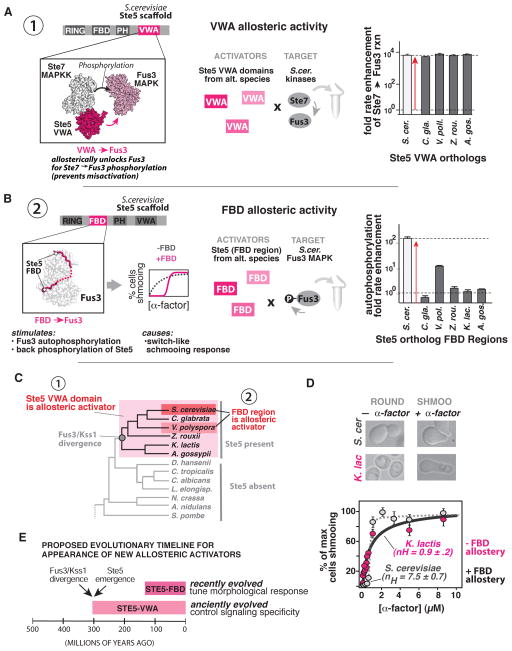
Figure 2. Tracking the emergence of MAPK allosteric activating domains within the Ste5 scaffold protein.
(A) In S. cer., Ste5-VWA is required to allosterically unlock Fus3 for phosphorylation by Ste7; Ste7 cannot effectively phosphorylate Fus3 in the absence of this domain. Fold rate enhancements (mean ± SEM) for Ste7 catalyzed phosphorylation of S. cer. Fus3 in the presence of saturating amounts of the indicated Ste5-VWA ortholog are shown (see also titration curves in Fig. S2A). (B) In S. cer., the Ste5-FBD is a linear motif between the Ste5 RING and PH domains that binds Fus3 and stimulates its autophosphorylation activity as part of a mechanism that results in a switch-like morphological dose response profile to α-factor. Rate-enhancements for S. cer. Fus3 autophosphorylation (mean ± SEM) provided by addition of 25 μM of the indicated Ste5-FBD are shown (see also titration curves in Fig. S3B). (C) Phylogeny of Ascomycota indicating the appearance of Ste5-VWA and Ste5-FBD scaffold activities as inferred from (A) and (B). (D) Morphological response to α-factor (mean ± SEM for % of maximum cells shmooing, n ≥ 500 cells) of S. cer. (grey dots; dashed line) and K. lactis (pink dots; solid line). Data were fit to a hill-equation to extract the parameter nH. (E) Timeline indicating the proposed appearance of Ste5 allosteric activators relative to the appearance of the Ste5 and the Fus3/Kss1 divergence (dating estimates from (Dujon, 2006; Taylor and Berbee, 2006)).
The second allosteric interaction involves a linear motif in Ste5 called the “Fus3 binding domain” (Ste5-FBD), which binds Fus3 and allosterically activates autophosphorylation of the MAP kinase on its activation loop tyrosine (Fig. 2B) (Bhattacharyya et al., 2006). This partially activated form of Fus3 back-phosphorylates Ste5 to down-regulate mating pathway output and reshapes the morphological response of cells to α-factor (“shmooing”) to be switch-like (ultrasensitive) instead of graded (Malleshaiah et al., 2010). The FBD allosteric activation is not essential for mating signaling, but instead appears to fine-tune the quantitative aspects of the mating response.
The Ste5-VWA allosteric interaction dates back to Fus3/Kss1 divergence, while the Ste5-FBD allosteric interaction is a recent innovation that tunes mating behavior in a few specific lineages
We first examined when Ste5-VWA allosteric activity appeared relative to the emergence of Fus3 and Kss1 kinases families. We purified Ste5-VWA domain orthologs from diverse fungal species that contain the Ste5 scaffold, and determined if they could allosterically co-activate S. cer. Fus3 phosphorylation by the S. cer. Ste7 MAPKK (henceforth, Ste7) (Fig. 2A and Fig. S2A). As observed previously, phosphorylation of S. cer. Fus3 by Ste7 is very slow in the absence of S. cer. Ste5-VWA (kcat = 6.0±0.4 x10−7 s−1) and the addition of saturating S. cer. Ste5-VWA stimulates this rate by greater than 3 orders of magnitude (6250 ± 610 fold). When saturating amounts of other Ste5-VWA orthologs were provided instead, rate enhancements were nearly identical to that of S. cer. Ste5-VWA. The most parsimonious interpretation of these data is that the Ste5-VWA domain possessed potent allosteric activity towards Fus3 in the last common ancestor of these species (Fig. 2C). Consistent with this, chimeric S. cer. Ste5 molecules in which the native VWA domain was replaced with the VWA domain from other Ste5 orthologs were able to support robust mating in vivo (Fig. S2E).
Is the Ste5-VWA domain of other orthologs subject to autoinhibition, as in S. cer.? A simple diagnostic for Ste5 autoinhibition is that full-length Ste5 provides a smaller rate enhancement for Fus3 phosphorylation than the isolated VWA domain (Zalatan et al., 2012). Thus we compared the rate enhancement provided by the longest Ste5 construct we could express for each ortholog to that of the corresponding isolated VWA domain (Fig. S2A, S2C). As in S. cer., every ortholog we examined was less effective than the isolated VWA domain in enhancing the Ste7 → Fus3 reaction. The extent of this autoinhibition ranged from values comparable to the S. cer. inhibition (~10-fold) to values that were as much as 80-fold inhibited. Additional experiments indicate that the molecular mechanism of this inhibition is likely the same as in S. cer. Ste5 (Fig. S2D), and thus the simplest explanation for these data is that this mechanism to control Ste5 VWA allosteric activation was a conserved Ste5 feature present in the last common ancestor of these species.
We next examined the evolutionary history of the S. cer. Ste5-FBD allosteric regulatory interaction. Orthologous Ste5-FBD sequences (detailed in supplement) were purified and assayed for the ability to stimulate S. cer. Fus3 autophosphorylation (Fig. 2B and Fig. S3A–B). As observed previously, S. cer. Ste5-FBD potently stimulated the rate of S. cer. Fus3 autophosphorylation (149.7 ± 13.5 fold rate enhancement). In contrast, the FBD region from all but one Ste5 ortholog failed to provide a detectable rate enhancement for S. cer. Fus3 autophosphorylation. The one exception was the FBD sequence from V. pol., which provided an intermediate effect (12.8 ± 0.3 fold rate enhancement). One possible explanation for the lack of allosteric activity we observed for most Ste5-FBD sequences is that perhaps each FBD motif is optimized for its corresponding Fus3 ortholog. However, no differences were observed when the Fus3 ortholog from the same source species was used as a target instead of S. cer. Fus3 (Fig. S3D). Thus, unlike Ste5-VWA regulation, the Ste5-FBD regulation found in S. cer. is not conserved in every organism that contains Fus3, Kss1, and Ste5 (Fig. 2C).
Most likely, the FBD interaction evolved as a recent lineage specific feature to tune the mating behavior of S. cer. It has previously been shown that mutating the Ste5-FBD in S. cer. converts a switch-like (ultrasensitive) shmooing response to α-factor into a graded (linear) response (Malleshaiah et al., 2010). This model would predict that species lacking an active FBD motif would show a linear shmooing response. To test this model, we quantitatively examined the morphological responses of K. lactis – a species that lacks an active FBD motif but retains an active VWA domain (Fig. 2D). As predicted, the morphological dose response of K. lactis to α-factor was graded (nH = 0.9 +/− 0.2) in comparison to the switch-like response observed in S. cer. (nH = 7.7 +/− 0.7). The fact that K. lactis cultures must undergo prolonged phosphate starvation to be mating competent (Booth et al., 2010; Tuch et al., 2008) may complicate a direct comparison of these two profiles. Nonetheless, together with our biochemical analyses, these data suggest that the Ste5-FBD interaction arose well after the divergence of Fus3 and Kss1 as a mechanism to fine tune quantitative mating responses. We note, however, that we cannot definitively rule out repeated loss of the Ste5-FBD from multiple lineages as an alternative explanation of these data.
Together, the simplest evolutionary model for these data is that a potent but tightly regulated Ste5-VWA activity was present in the last common ancestor of the species that contain both Fus3 and Kss1 MAP kinase types, while the Ste5-FBD activity was likely layered on top of the core conserved Ste5 activities to reshape the morphological response to mating pheromone in only certain species (Fig. 2E). This suggests that a core function of the Ste5 scaffold protein has been to functionally insulate Fus3 and Kss1 since their divergence but also suggests that Ste5 scaffold interactions with the Fus3 kinase might continue to evolve to meet specific organismal signaling needs.
Latent Allostery: Ste5 allosteric activator domains can stimulate MAP kinases that diverged prior to the evolution Ste5
We then turned to the converse question of understanding how the Fus3 MAPK acquired the necessary features to serve as a target of these two Ste5 allosteric interactions. Here we reversed our in vivo cross-reaction components and tested the extent to which Fus3 and Kss1 orthologs from other species could be regulated by the S. cer. Ste5 scaffold activities. Starting with the S. cer. Ste5-VWA domain (Fig. 3A), we found that all Fus3 orthologs were strongly allosterically regulated by the VWA domain: they were poor substrates for Ste7 in the absence of S. cer. Ste5-VWA (kcat < 5×10−6 s−1) but the addition of S. cer. Ste5-VWA enhanced phosphorylation of each MAPK by greater than 2000 fold. This strong allosteric activation was identical when other Ste5-VWA orthologs were used in place of S. cer. Ste5-VWA (Fig. S2B).
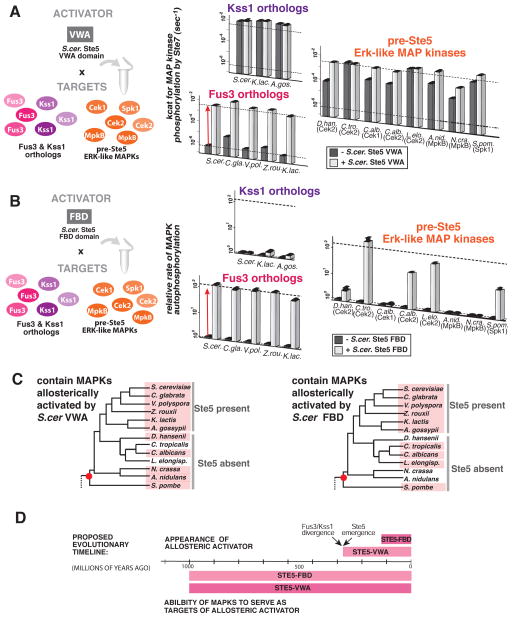
Figure 3. Ste5 scaffold domains can allosterically activate Erk-like MAP kinases that diverged prior to the evolution of Ste5.
(A) Rate constants (mean ± SEM) for Ste7 catalyzed phosphorylation of the indicated MAP kinase substrate in the presence (white) or absence (grey) of saturating amounts of S. cer. Ste5-VWA for the indicated Fus3 orthologs, Kss1 orthologs or pre-Ste5 Erk-like MAP kinases. (B) Relative rates (mean ± SEM) for MAP kinase autophosphorylation (normalized to the no Ste5-FBD rate) in the presence (white) or absence (grey) of saturating amounts of S. cer. Ste5-FBD for the indicated Fus3 orthologs, Kss1 orthologs or pre-Ste5 Erk-like MAP kinases. (C) Phylogeny of Ascomycota fungi with species that contains an Erk-like kinase that can be regulated by the indicated Ste5 activator domain marked in pink. The red dot represents the last common ancestor that likely contained an Erk-like kinase that could be regulated by the indicated Ste5 activator. (D) Proposed timeline (date estimates from (Dujon, 2006; Taylor and Berbee, 2006)) indicating when the capacity for regulation by a Ste5 activator appeared in the fungal Erk-like MAP kinase family relative to the appearance of Ste5 scaffold activator domains that target that capacity for regulation.
In contrast, all of the Kss1 orthologs we tested were not targets for VWA activation — these MAPKs were ideal substrates for Ste7 (kcat > 1×10−3s−1) in the absence of any other molecules, and were unaffected by the addition of S. cer. Ste5-VWA (rate-enhancement < 1.5 fold). From these data, we infer that Fus3 and Kss1 likely possessed their divergent responses to Ste5-VWA regulation in the last common ancestor of the species that contain these kinases. The ability of Fus3 orthologs to be activated by the VWA domain, thus, appears to be tightly conserved after the functional divergence of the Fus3 and Kss1 MAPKs.
We then tested whether Erk-like kinases from species that diverged from S. cer. prior to the evolution of Ste5—henceforth referred to as “pre-Ste5” Erk-like kinases— had the capacity to be regulated by the modern S. cer Ste5 VWA domain (Fig. 3A). Unlike S. cer Fus3, these pre-Ste5 kinases were intrinsically good substrates for Ste7 catalyzed phosphorylation in vitro (kcat >7×10−5 s−1). However, addition of S. cer. Ste5-VWA surprisingly stimulated phosphorylation of many of these kinases by as much as a 42-fold rate enhancement. Thus, these pre-Ste5 MAP kinases are similar to Fus3 in that they have a modest capability to serve as a target for allosterically activation by the Ste5 VWA domain, despite the fact that the species from which they come lack Ste5.
We then analogously examined when the ability to serve as a target for the Ste5-FBD interaction arose within the MAP kinase family (Fig. 3B). Although only the S. cer Ste5 ortholog possessed potent Ste5-FBD activity, we surprisingly found that the Fus3 orthologs from nearly every species that we examined were targets for FBD activation – like S. cer Fus3, they all displayed a FBD enhanced rate autophosphorylation of greater than 100-fold. In contrast, S. cer. Ste5-FBD did not significantly enhance the rate of autophosphorylation of the Kss1 orthologs we tested. We conclude that Fus3 was primed for regulation by a S. cer. Ste5-FBD mechanism in the common ancestor of these species, even before the FBD activity had evolved in the Ste5 scaffold; little change to the kinase was necessary for the S. cer. Ste5-FBD to be able to influence the rate of autophosphorylation.
We then tested whether S. cer. Ste5 FBD could enhance the rate of autophosphorylation of the pre-Ste5 Erk-like kinases (Fig. 3B). We observed a broad range of capacities for regulation by S. cer. Ste5-FBD. Several kinases were not allosterically affected by the S. cer. Ste5-FBD (D. han. Cek2, C. alb. Cek1, A. nid. MpkB, N. cra. MpkB) even though these kinases readily bound to S. cer. Ste5-FBD (Fig. S4A). Some kinases, however, showed intermediate effects (S. pom. Spk1, C. alb. Cek2); and still others showed allosteric responses that approached or even exceeded the enhancement in autophosphorylation that is seen for S. cer. Fus3 (C. tro. Cek2, L. elo. Cek2). Thus, many of the pre-Ste5 MAPKs display the ability to serve as a target for both VWA and FBD mediated allosteric activation.
These findings suggest that both the VWA and FBD allosteric interactions evolved by tapping into latent allosteric features that pre-existed within this family of kinases. Because the pre-Ste5 Erk-like kinases — including the Spk1 kinase from S. pombe, which is the most distantly related to S. cer. Fus3 — broadly show modest regulation by both Ste5-VWA and Ste5-FBD, the most parsimonious explanation of these data is that some capacity for both of these forms of allosteric regulation was likely present in the ancestral kinase of all the orthologs we inspected (Fig. 3C). Although it is formally possible that there exist other alternative allosteric regulators that capitalize on these modest features in the pre-Ste5 lineages, several lines of reasoning argue against this. First, we tested several likely candidate proteins present in these organisms for such activity and found no evidence in support of this (Fig. S4B, Fig. S4D–E). Second, the extensive variation in the latent allosteric features of the pre-Ste5 MAPKs—including the absence of these features in particular orthologs—suggests that these features are not under selective pressure. That is, these particular allosteric regulations of the MAPK substrate have not been fixed in all of the pre-Ste5 branches of the Ascomycota to the extent that they have been fixed in the post-Ste5 species, casting doubt on the existence of other critical allosteric regulators that are using the latent allosteric features. Third, given that the Ste5-VWA regulation is functionally required for pathway specificity—i.e. discriminating between the Fus3 and Kss1 kinases—it is unclear why such allosteric effectors would exist in lineages that contain only a single Erk-like kinase. Finally, the observed allosteric effects on the pre-Ste5 kinases are in most cases relatively small — all of the kinases were adequate MAPKK substrates in the absence of any additional Ste5 regulation. Thus, it is unlikely that these species functionally require such allosteric effectors. As such, we favor a model in which the capacity for the allosteric regulation we observed was already present in the ancestral kinases, providing a toe-hold for the emergence of new forms allosteric regulation.
Drift in latent allostery produces evolutionary related kinases that are primed for divergent responses to new allosteric activators
A model of latent allostery within the MAP kinase family provides a simple framework for how new allosteric regulators such as the Ste5-FBD and Ste5-VWA domain may have evolved. However, it also raises an important question in terms of divergent regulation: how then is it that Fus3 orthologs are targets for this allosteric regulation, while Kss1 orthologs are not?
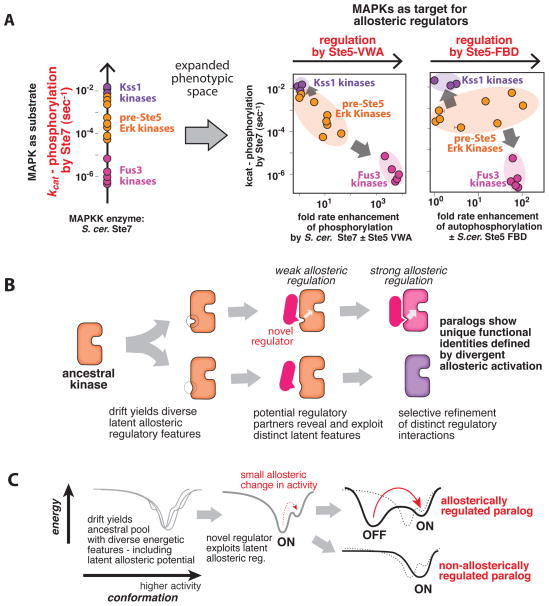
To gain insights into this question, we examined the diversity in the distribution of properties observed for the pre-Ste5 Erk-like kinases (Fig. 4A). When only considered as substrates for the MAPKK Ste7, pre-Ste5 Erk-like kinases generally cluster together and appear to be similar quality substrates in the absence of any scaffold coactivator. However, the latent capacity for allosteric regulation in each of these substrates results in additional dimensions of MAPK phenotypic diversity beyond their basic properties as substrates for phosphorylation by the MAPKK Ste7. This diversity is easily visualized by plotting each of the kinases on phenotypic morphospace plots in which one dimension is the rate of Ste7 → Fus3 phosphorylation—the apparent kinase diversity in the absence of any allosteric activators—and a second dimension is the allosteric enhancement of either of the two Ste5 allosteric interactions—the hidden phenotypic diversity that is only revealed upon interaction with scaffold effectors (Fig. 4A—see also Fig. S4C). For both Ste5-VWA and Ste5-FBD activities, the highly divergent regulation of Fus3 and Kss1 orthologs places them in opposite regions of this space, while most of the pre-Ste5 Erk-like kinases are ‘hybrids’ that, as a set, occupy a region of space in between Fus3 and Kss1. Importantly, these plots reveal that kinases that may appear close together in the one-dimensional perspective as substrates for MAPKK phosphorylation can be far apart along these hidden allosteric dimensions. Thus, drift in these hidden phenotypic properties (latent allostery) results in a distribution of family members, with some much closer to Fus3 in behavior, and others much closer to Kss1.
Figure 4. Drift in latent protein allostery provides a path for evolution of divergent regulatory phenotypes within seemingly equivalent kinase paralogs.
(A) Morphospace visualizations of MAP kinase biochemical diversity found in this study, either only as substrates for phosphorylation by the upstream MAPKK, or taking into account the additional hidden dimensions of diversity in substrates upon interaction with the Ste5 scaffold. Circles in the plots correspond to individual MAP kinases (Fus3-type kinases: pink; Kss1-type kinases: purple; pre-Ste5 Erk-like kinases: orange) that we examined and indicate their associated properties. See also Fig. S4C. (B) Evolutionary model for novel and divergent regulation by exploitation of latent allosteric diversity. An ancestral kinase (orange) with some capacity for allosteric regulation is duplicated. In the absence of an effector, drift yields paralagous kinases with distinct latent regulatory features. Potential regulatory partners can reveal and exploit differences in these distinct latent features, providing a foothold for selection to refine and optimize the targets and effectors by coevolution to produce paralagous kinases with divergent allosteric responses to an effector molecule. (C) The above model illustrated in terms of the conformational energy landscape of the proteins.
These findings suggest a simple and general mechanism for the evolution of novel and divergent allosteric regulation of paralogous signaling components such as the Kss1 and Fus3 MAP kinases (Fig. 4B and Fig. 4C). Neutral drift in a latent capacity for allosteric regulation produces paralagous variants that are primed for divergent responses to regulation. Appearance of a new interaction partner with weak activity against this latent allosteric feature ‘reveals’ the pre-existing diversity and provides a toehold for Darwinian processes to exploit these differences and drive these kinases into divergent regulatory modes by selection, as was observed for the divergent responses of Fus3 and Kss1 to the Ste5-VWA domain. Such selection events have the potential to fix other latent allosteric properties within the newly selected lineage owing to founder-effects or hitchhiking, which could explain why all of the Fus3 and Kss1 orthologs we tested also display divergent responses to Ste5-FBD regulation even before this activity evolved.
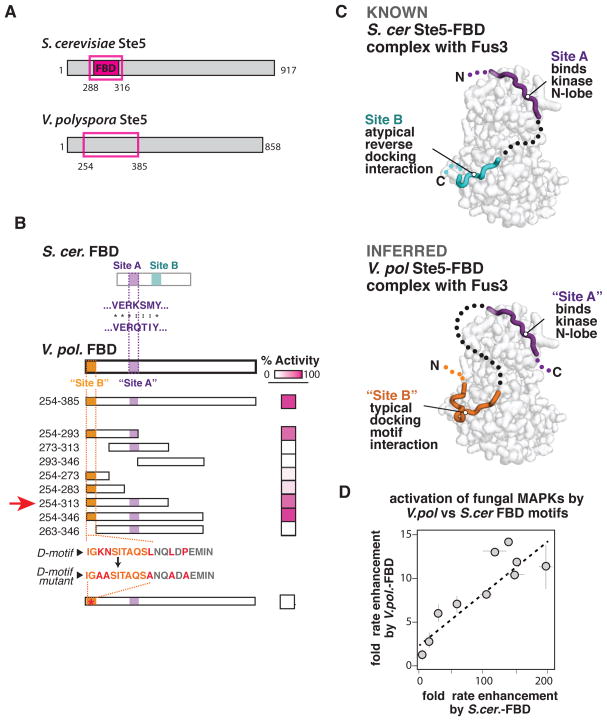
Dissection of the V. pol. Ste5-FBD with intermediate allosteric activity reveals alternative paths for coopting the same latent regulatory features
Our data demonstrate that a significant capacity for allosteric regulation is present in kinases prior to the evolution of the effectors that provide that regulation in S. cer. How, at a molecular level, does evolution discover activators that can tap into these hidden allosteric features and coopt this pre-existing capacity for new regulation? Our biochemical screen of Ste5-FBD motifs, identified a sequence with intermediate allosteric activity from V. polyspora (V. pol. Ste5-FBD) that gives us an opportunity to biochemically dissect how this FBD-mediated allosteric activity may have arisen (Fig. 5A) (Addressing this question for Ste5-VWA domain regulation is difficult because of the lack of any forms of Ste5 that show intermediate VWA activities).
Figure 5. Dissection of the V. pol. Ste5-FBD motif with intermediate allosteric activity reveals alternative paths for coopting the same latent regulatory features.
(A) Position of minimal S. cer. Ste5-FBD sequence mapped previously (Bhattacharyya et al., 2006) and initial region of V. pol. Ste5 that showed FBD activity that serves as the starting point for further analysis. (B) Truncation mapping of the V. pol. Ste5-FBD fragment, showing the relative activity of an indicated truncation or fragment (see also Fig. S5). This analysis identifies two sites that are required for activity. One site (“Site A”) is similar to a sequence required for activity in the S. cer. Ste5-FBD fragment. A second site (“Site B”) resembles a traditional MAPK docking peptide sequence. (C) Comparison of the known structure of the S. cer. Ste5-FBD•Fus3 complex to the inferred structure based on homology and the truncation mapping from (B). The two kinases appear to use similar “Site A” sequences to bind the N-lobe of the kinase but use distinct mechanisms to engage the docking groove of the MAP kinase C-lobe. (D) A plot indicating the magnitude (mean ± SEM) of the S. cer. FBD effect and V. pol. FBD effect; each point corresponds to an individual kinases we inspected. The tight linear relationship between these effects suggests that both the S. cer. and V. pol. Ste5-FBD sequences target the same allosteric features and diversity present in the MAP kinases we inspected.
We wanted to determine whether the V. pol. Ste5-FBD functions through a related mechanism to that used by the S. cer. Ste5-FBD. We previously showed that the S. cer. Ste5-FBD sequence binds to Fus3 in a bipartite manner to allosterically activate the Fus3 kinase: an “A-site” motif binds to the N-lobe of the kinase, while a second “B-site” motif binds to a canonical docking groove on the C-lobe of the MAP kinase (albeit in a non-canonical reverse C-to-N terminal orientation); linking these two binding sites is thought to constrain the two kinase domain into a more active conformation that promotes autophosphorylation (Fig. 5C) (Bhattacharyya et al., 2006). Inspection of the V. pol. Ste5-FBD sequence reveals a sequence that resembles the “A-site” motif of S. cer. Ste5-FBD, but there is no obvious sequence that resembles the “B-site” motif.
To better understand the mechanism of the V. pol. Ste5-FBD interaction, we used deletion analysis to map the regions of this sequence that were required for its allosteric activity (Fig. 5B and Fig. S5A). Like S. cer. Ste5-FBD, we found that two distinct regions were required for activity. One of these regions contained the motif that resembles the “A-site” of the S. cer. Ste5-FBD, suggesting that both V. pol. Ste5-FBD and S. cer. Ste5-FBD use this “A-site” sequence to engage the N-lobe of Fus3. Unlike in the S. cer. Ste5-FBD, however, the second region of V. pol. Ste5-FBD required for allosteric activity was on the opposite side of the “A site” (N-terminal to it, i.e. the opposite orientation relative to the S. cer. Ste5-FBD). This second required region in the V. pol. Ste5-FBD motif fits the consensus MAPK docking motif ([R/K]1-2-X2-6-Φ–x-Φ–x-Φ) that is used by many signaling partners to interact with MAPKs (Reményi et al., 2005; Tanoue et al., 2000). Consistent with this, mutation of the residues within this motif that would disrupt a MAPK docking interaction completely abolished the allosteric activity of V. pol. Ste5-FBD (Fig. 5B).
From these data, we infer a model for how V. pol. Ste5-FBD interacts with Fus3 to exert allosteric influence (Fig. 5C). At low resolution, both the S. cer. and V. pol. FBD mechanisms appear very similar – they both bind at the same two sites on the MAPK, potentially constraining the kinase N and C lobes relative to one another in a manner than increases autophosphorylation. Nonetheless, while the S. cer. Ste5-FBD binds Fus3 with an “A-site”-”B-site” bipartite polypeptide, the V. pol. Ste5-FBD appears to binds to Fus3 with a “docking motif”- “A-site” bipartite polypeptide (where the docking motif functionally replaces the B-site motif). In both cases, functionally analogous motifs that bind the C-lobe docking groove, cooperate with binding of the “A-site” motif to the N-lobe of the kinase to achieve allosteric activation. We postulate that this distinct but analogous bipartite binding represents a case of convergent evolution – both bipartite peptides can constrain the kinase lobes required to stimulate autophosphorylation, albeit to different degrees (Fig. S5B).
Is the V. pol. Ste5-FBD motif, despite its detailed differences, tapping into the same latent allosteric features present in the fungal MAPK family as those exploited by the S. cer. Ste5-FBD? If so, then we predict that the effects of the V. pol. Ste5-FBD motif on diverse members of the Erk-like fungal kinase family should mirror those observed for the S. cer Ste5-FBD motif. Indeed, we observe a linear relationship across fungal species between the degree to which the V. pol. Ste5-FBD and the S. cer Ste5-FBD motifs can activate individual MAPK family members (Fig. 5E; see also Fig. S3D). Thus, even this weak activator appears to reveal the same latent potential for allosteric regulation that is present in many fungal MAP kinases, including the pre-Ste5 Erk-like kinases. During the course of evolution, once a weak effector like the V. pol. Ste5-FBD uncovers these latent kinase regulatory features, Darwinian processes can proceed to optimize this allosteric regulation. Because different yeast species occupy distinct environments and exhibit different mating preferences (Booth, 2010), the outcomes of these Darwinian processes will differ depending on local selective pressures and organismal niche: the activity can be optimized to increase potency (as in S. cer. Ste5-FBD), it can be maintained as a weak effector (as in V. pol. Ste5-FBD), or it can be turned over to a state in which Ste5-FBD regulation is lost (as observed in C. gla.—see Fig. 2B).
DISCUSSION
Our analysis of kinase and scaffold properties from across Ascomycota fungi allowed us to determine when particular Ste5 scaffold allosteric activator functions most likely arose, as well as when the capacity of a MAP kinase to serve as a target for such allosteric regulation arose. From this analysis, we made the surprising observation that many kinases from species that diverged from S. cer. prior to the evolution of the Ste5 scaffold can still be regulated by the allosteric motifs within Ste5. These findings suggest that a latent capacity for allosteric regulation was present within this MAP kinase family long before the evolution of effectors that target this allostery for regulation.
Exploitation of an existing latent capacity to derive a new molecular regulatory relationship is similar to proposed models for the evolution of new catalytic activities by catalytic promiscuity (O’Brien and Herschlag, 1999; Khersonsky and Tawfik, 2010) and new hormone receptor signaling responses by molecular exploitation (Bridgham et al., 2006). In this case, however, the latent allosteric kinase features we have described in this study are not obviously similar to some pre-existing regulatory interaction, but represent new regulatory connections that can redirect and reshape information flow in cell signaling pathways.
The dynamic protein kinase structure as a source of latent and diverse allosteric behaviors
We postulate that the dynamic nature of the protein kinase structure itself may provide the latent allosteric potential and diversity observed in these fungal MAP kinases. Indeed, both Ste5-VWA domain and Ste5-FBD motif are thought to allosterically activate the Fus3 MAP kinase by altering the kinase flexibility (Bhattacharyya et al., 2006; Good et al., 2009), and such flexibility is an innate but variable feature of protein kinases (Huse and Kuriyan, 2002). Thus, rather than requiring that evolution create unprecedented structural features to produce allosteric innovation, the ruggedness of the MAP kinase conformational landscape itself may provide toeholds for weak, but specific, activators that act by selecting and stabilizing particular kinase conformations (Ma et. al., 1999) (Fig. 4C). These relationships can then be strengthened by evolutionary mechanisms that tune and modulate the stability of certain states of the molecule or widen the difference in activity between alternative states. These findings are consistent with patterns observed in many other members of the protein kinase family: all kinases appear to require the proper assembly of the same core catalytic and structural elements in order to adopt an active state; but different kinases adopt a wide array of distinct inactive conformations, each of which requires a different set of inputs to stabilize the conserved active conformation (Huse and Kuriyan, 2002; Kornev et al., 2006). More generally, modes of flexibility intrinsic to particular protein folds may provide the starting point for future regulatory evolution (Halabi et al., 2009).
The hidden regulatory diversity that we find in the fungal MAP kinases may be a more general feature of many protein kinases as well as other dynamically regulated macromolecules. In fact, many drugs may act as effectors that uncover this regulatory potential. Indeed, such hidden conformational toe-holds serve as the basis of action of the Abl-specific tyrosine kinase inhibitor Gleevac, which stabilizes a inactive conformation that is uniquely accessible to that kinase (Schindler et al., 2000). Similarly some small molecules have been found to allosterically activate regulatory proteins, despite the lack of a clear physiologic analog that normally targets that site (Hardy et al., 2004).
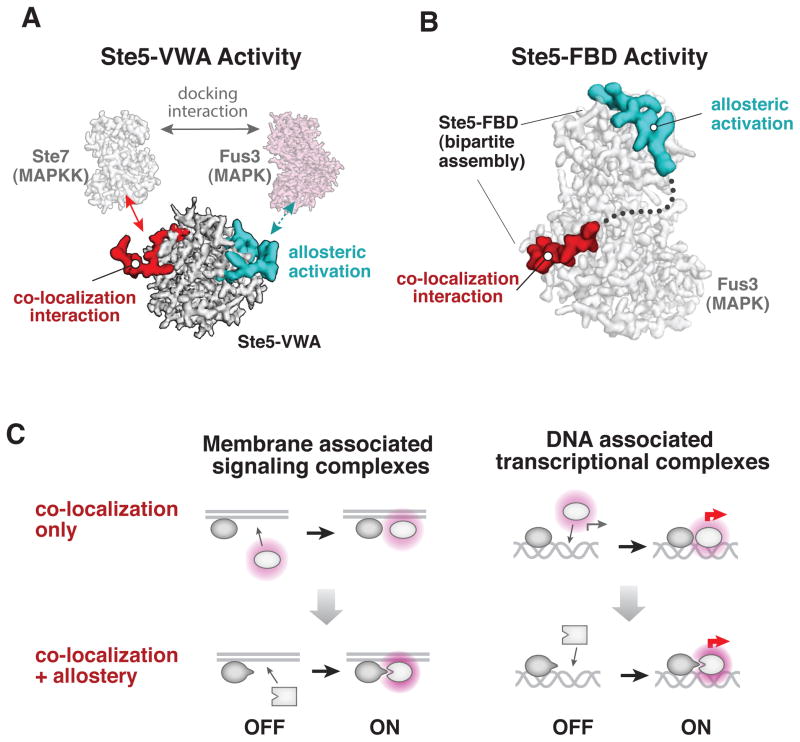
Co-localization may facilitate the evolution of new allosteric regulation
The latent allosteric properties in MAP kinases we have described must be ‘revealed’ by an effector in order for selection to be possible. How do such primitive allosteric regulators evolve? The mechanism of the Ste5 VWA and FBD motifs suggests that co-localization may facilitate this process, as both interactions involve the interplay between allosteric interactions and co-localization interactions. The VWA domain allosterically regulates Fus3 in the context of a higher order molecular complex (a Ste5 Fus3 Ste7 ternary complex) that is assembled by non-allosteric co-localization interactions that are sufficient for tight complex formation (Fig. 6A). Similarly, binding of the Ste5-FBD A-site motif to the Fus3 kinase depends on a second-site interaction with the docking groove of MAP kinase which is sufficient for complex formation on its own (Fig. 6B). A simple model is that co-localization of the future allosteric target and regulator was an early step in the evolution of these allosteric relationships. Such co-localization establishes effective concentrations of the components in the milimolar range in which fleeting and weak interactions occur more readily (Kuriyan and Eisenberg, 2007; Reynolds et al., 2011), thus enhancing the likelihood of uncovering a weak interaction that reveals a latent allosteric feature in a target.
Figure 6. The role of co-localization in the evolution of new allosteric regulation.
(A) Colocalization interactions that are distinct from the essential allosteric surface of the Ste5-VWA assemble a Ste5•Fus3•Ste7 ternary complex that is essential for the Ste5-VWA to allosterically activate Fus3. These colocalization interactions are sufficient for tight complex assembly on their own. (B) Binding of the Ste5-FBD A-site motif to the Fus3 MAP kinase, which is essential for allosteric activation of autophosphorylation, requires a second-site interaction with the docking groove of MAP kinase. This docking interaction is, by itself, a non-allosteric colocalization interaction that can be sufficient for complex formation. (C) Co-localization-based activation mechanisms—whether on scaffolds, membranes, or DNA—can facilitate the evolution of allosteric interactions between the co-localized components and yield the tighter, precise spatiotemporal control of activation that is observed in modern pathways.
This evolutionary ‘co-localization first’ strategy is similar to the novel ‘tethering’ approach used for developing small molecule allosteric effectors, in which a library of disulfide-containing small molecules is localized to a particular cysteine residue on the drug target (Erlanson et al., 2004) allowing for the identification of weak effectors that bind to surprising new allosteric protein sites (Hardy and Wells, 2004). The synergy between evolutionary and engineering approaches suggests that genetically encoded libraries that ‘tether’ a variable protein or RNA library to a target might offer an effective in vivo screening approach for identifying new allosteric effectors.
Finally, these observations indicate that the co-localization of signaling components on scaffolds or at the membrane may play a more active role in the evolution of new signaling pathways and behaviors than previously appreciated, by producing local environments in which hidden allosteric diversity and the ruggedness of conformational landscapes are revealed by high effective concentrations and potential new effector interactions. Indeed, many primitive signaling pathways may have initially simply consisted of components that became co-localized upon stimulation with an input signal (Fig. 6C). These assemblies, however, might then provide a context that would facilitate the evolution of allosteric regulation, as described here, that yielded the diverse forms of precision control that we observe in modern pathways. An analogous progression of regulatory evolution is suggested to take place among DNA binding factors that are tethered at a promoter (Baker et al., 2012; Tuch et al., 2008b).
EXPERIMENTAL PROCEDURES
Identification, sequence analysis, and cloning of kinase and scaffold orthologs
S. cer. Fus3, Kss1 and Ste5 sequences were used to query the fungal orthogroups database to identify orthologous sequences in the Ascomycota (additional details in supplemental methods) which were subsequently cloned from gDNA or synthesized directly. Phylogenetic analysis of these sequences is detailed in supplementary materials. A complete list of all constructs used in this study is in Table S1.
Protein purification
MAP kinases, Ste5 scaffold fragments, and the SR13 Fab antibody were expressed in BL21(T1R) E. coli cells. The S. cer. Kss1 ortholog and the constitutively active form of the MEK Ste7 (Ste7EE) were expressed from S. frugiperda (SF9) cells. Proteins were purified similarly as described previously (Good et al., 2009; Reményi et al., 2005; Zalatan et al., 2012) with minor modification as detailed in supplemental methods.
In vitro kinase activity assays
Initial rates for Ste7 catalyzed phosphorylation of a MAPK as well as MAPK autophosphorylation were measured by quantitative western blotting as described and detailed in supplemental methods. Under saturating conditions, VWA reactions contained 50 nM of MBP-Ste7EE, 5 μM MAPK substrate, and (if present) 5 μM Ste5-VWA ortholog; saturating FBD reactions contained 10 μM MAPK and, if present 25 μM of a Ste5-FBD sequence.
Morphological dose response to α-factor
Morpohological responses to α-factor were performed for S. cer. (strain W303) and K. lac. (strain yLB17a (Booth et al., 2010)) as described previously (Malleshaiah et al., 2010). For K. lac., the response was measured after 6 hours of growth in SCD media lacking phosphate to ensure cells were mating competent (detailed in supplement). The percentage of cells shmooing at a given concentration of pheromone was determined by microscopy and the resulting dose response curves were fit to a Hill-equation to extract the hill-coefficient parameter nH.
Supplementary Material
HIGHLIGHTS.
Mapped evolutionary timeline of allosteric interactions regulating yeast MAPK Fus3.
Pre-Ste5 MAPKs show latent allosteric regulation by Ste5 scaffold protein.
Drift creates pool of kinases primed for divergent responses to new regulation.
Novel allosteric activators evolve by coopting latent features pre-existing in target protein.
Acknowledgments
S.M.C. is supported by a National Science Foundation Graduate Research Fellowship. This work was supported by NIH grants RO1 GM55040, PN2 EY016546, P50 6M081879 (W.A.L.), and the Howard Hughes Medical Institute (W.A.L.). We thank R. Almeida, P. Dumesic, J. Fraser, R. Gordley, A. Johnson, I. Nocadel, L. Pack, S. Peisajovich, T. Sorrels, J. Walter, A. Weeks, P. Wei, E. Woods and J. Zalatan for helpful discussions. S. Rajan, S.S. Sidhu, T. Sorrels and J. Zalatan provided strains and reagents.
Footnotes
Supplemental Information includes Extended Experimental Procedures, 5 supplemental figures, and 1 supplemental table.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afriat L, Roodveldt C, Manco G, Tawfik DS. The Latent Promiscuity of Newly Identified Microbial Lactonases Is Linked to a Recently Diverged Phosphotriesterase. Biochemistry. 2006;45:13677–13686. doi: 10.1021/bi061268r. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Gaidukov L, Khersonsky O, Gould SM, Roodveldt C, Tawfik DS. The ‘evolvability’ of promiscuous protein functions. Nature Genetics. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Baker CR, Booth LN, Sorrells TR, Johnson AD. Protein Modularity, Cooperative Binding, and Hybrid Regulatory States Underlie Transcriptional Network Diversification. Cell. 2012;151:80–95. doi: 10.1016/j.cell.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 Scaffold Allosterically Modulates Signaling Output of the Yeast Mating Pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- Booth LN, Tuch BB, Johnson AD. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature. 2010;468:959–963. doi: 10.1038/nature09560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of Hormone-Receptor Complexity by Molecular Exploitation. Science. 2006;312:97–101. doi: 10.1681/01.asn.0000926836.46869.e5. [DOI] [PubMed] [Google Scholar]
- Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends in Genetics. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Wells JA, Braisted AC. TETHERING: Fragment-Based Drug Discovery. Annual Review of Biophysics and Biomolecular Structure. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- Good M, Tang G, Singleton J, Reményi A, Lim WA. The Ste5 Scaffold Directs Mating Signaling by Catalytically Unlocking the Fus3 MAP Kinase for Activation. Cell. 2009;136:1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein Sectors: Evolutionary Units of Three-Dimensional Structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Current Opinion in Structural Biology. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The Conformational Plasticity of Protein Kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS. Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective. Annual Review of Biochemistry. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Kornev AP, Haste NM, Taylor SS, Eyck LFT. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. PNAS. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. The riddle of MAP kinase signaling specificity. Trends in Genetics. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- Malleshaiah MK, Shahrezaei V, Swain PS, Michnick SW. The scaffold protein Ste5 directly controls a switch-like mating decision in yeast. Nature. 2010;465:101–105. doi: 10.1038/nature08946. [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Herschlag D. Functional Interrelationships in the Alkaline Phosphatase Superfamily: Phosphodiesterase Activity of Escherichia coli Alkaline Phosphatase. Biochemistry. 1999;40:5691–5699. doi: 10.1021/bi0028892. [DOI] [PubMed] [Google Scholar]
- Reményi A, Good MC, Bhattacharyya RP, Lim WA. The Role of Docking Interactions in Mediating Signaling Input, Output, and Discrimination in the Yeast MAPK Network. Molecular Cell. 2005;20:951–962. doi: 10.1016/j.molcel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Reynolds KA, McLaughlin RN, Ranganathan R. Hot Spots for Allosteric Regulation on Protein Surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodveldt C, Tawfik DS. Shared promiscuous activities and evolutionary features in various members of the amidohydrolase superfamily. Biochemistry. 2005;44:12728–12736. doi: 10.1021/bi051021e. [DOI] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural Mechanism for STI-571 Inhibition of Abelson Tyrosine Kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nature Cell Biology. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Berbee ML. Dating divergences in the Fungal Tree of Life: review and new analyses. Mycologia. 2006;98:838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- Taylor Ringia EA, Garrett JB, Thoden JB, Holden HM, Rayment I, Gerlt JA. Evolution of enzymatic activity in the enolase superfamily: functional studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis. Biochemistry. 2004;43:224–229. doi: 10.1021/bi035815+. [DOI] [PubMed] [Google Scholar]
- Thornton JW, Need E, Crews D. Resurrecting the Ancestral Steroid Receptor: Ancient Origin of Estrogen Signaling. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The Evolution of Combinatorial Gene Regulation in Fungi. PLoS Biol. 2008a;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Li H, Johnson AD. Evolution of Eukaryotic Transcription Circuits. Science. 2008b;319:1797–1799. doi: 10.1126/science.1152398. [DOI] [PubMed] [Google Scholar]
- Wise EL, Yew WS, Akana J, Gerlt JA, Rayment I. Evolution of enzymatic activities in the orotidine 5′-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase. Biochemistry. 2005;44:1816–1823. doi: 10.1021/bi0478143. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. Conformational Control of the Ste5 Scaffold Protein Insulates Against MAP Kinase Misactivation. Science. 2012;337:1218–1222. doi: 10.1126/science.1220683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.