Abstract
Inflammation is a protective response of organisms to pathogens, irritation or injury. Primary inflammatory sensors activate an array of signaling pathways that ultimately converge upon a few transcription factors such as AP1, NFκB and STATs that in turn stimulate expression of inflammatory genes to ultimately eradicate infection and repair the damage. A disturbed balance between activation and inhibition of inflammatory pathways can set the stage for chronic inflammation which is increasingly recognized as a key pathogenic component of autoimmune, metabolic, cardiovascular and neurodegenerative disorders. Nuclear Receptors (NRs) are a large family of transcription factors many of which are known for their potent anti-inflammatory actions. Activated by small lipophilic ligands, NRs interact with a wide range of transcription factors, cofactors and chromatin-modifying enzymes, assembling numerous cell- and tissue-specific DNA-protein transcriptional regulatory complexes with diverse activities. Here we discuss established and emerging roles and mechanisms by which NRs and, in particular, the glucocorticoid receptor (GR) repress genes encoding cytokines, chemokines and other pro-inflammatory mediators.
Keywords: Inflammation, transcriptional repression, nuclear receptors, glucocorticoid receptor
Inflammation is the response of the host innate immune system to infection, irritation or injury that involves the production of cytokines and chemokines, changes in vascular permeability, activation and mobilization of specialized immune cells to the affected area and, ultimately, elimination of the infectious agents and the repair of damaged tissue. Inflammation is a broad and relatively non-specific response usually initiated upon the recognition of conserved microbial components by pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), Rig-I-like receptors (RLRs) and Nod-like receptors (NLRs). TLRs reside either on the cell surface or endosomal membranes and recognize a wide range of structures such as lipopolysaccharides (LPS), lipopeptides, proteins and single- and double-stranded nucleic acids. TLRs trigger an array of downstream signaling events that converge upon the transcription factors Nuclear Factor (NF) κB, activator protein-1 (AP-1), interferon response factors (IRF) and signal transducer and activator of transcription (STAT) (Takeuchi and Akira 2010) that induce the transcription of key immune modulators such as TNFα, interleukins (IL)-1α, -1β, -6, and chemokines CXCL10, CCL2 among others. The production of cytokines and chemokines, particularly by resident macrophages and dendritic cells at the site of inflammation, enables the influx and activation of neutrophils, which release additional inflammatory mediators such as oxygen-independent antimicrobial effectors (e.g., defensins, cathepsins, lactoferrin) and reactive oxygen and nitrogen species ultimately aimed at clearing the infection and initiating tissue repair (Nathan 2006).
Activators of inflammatory gene expression programs
Since their discovery over 25 years ago, the NFκB/Rel transcription factors have been established as primary regulators of innate immune responses. The members of this family share the Rel homology domain that participates in DNA binding and protein-protein interactions. Five mammalian members have been identified: p65 (RelA), RelB, cRel, p50 and p52. In addition to the Rel domain p65, RelB and cRel each contain the N-terminal transactivation domain (TAD). The NFκB subunits form either homo- or heterodimers with different transcriptional activities; the p65:p50 heterodimer (Fig. 1A) is the most potent activator whereas the p50:50 and p50:52 dimers that lack TAD often confer repression (Saccani, et al. 2003; Smale 2012; Yan, et al. 2012). In unstimulated cells, the majority of NFκB is sequestered in an inactive cytoplasmic complex with the Inhibitor of κB (IκB) proteins. Induction of TLR pathways activates the IκB kinases (IKK), which in turn phosphorylate IκB thereby marking it for ubiquitn-mediated proteasomal degradation (Hayden and Ghosh 2012). Free NFκB translocates into the nucleus and binds specific recognition (κB) sites containing the consensus sequence, 5’-GGGRNWYYCC-3’ (Fig. 1A) (Kunsch, et al. 1992; Siggers, et al. 2011). DNA bound NFκB recruits additional cofactors with chromatin remodeling and histone modifying activities that promote formation of a transcriptionally permissive chromatin structure (Zhong, et al. 2002).
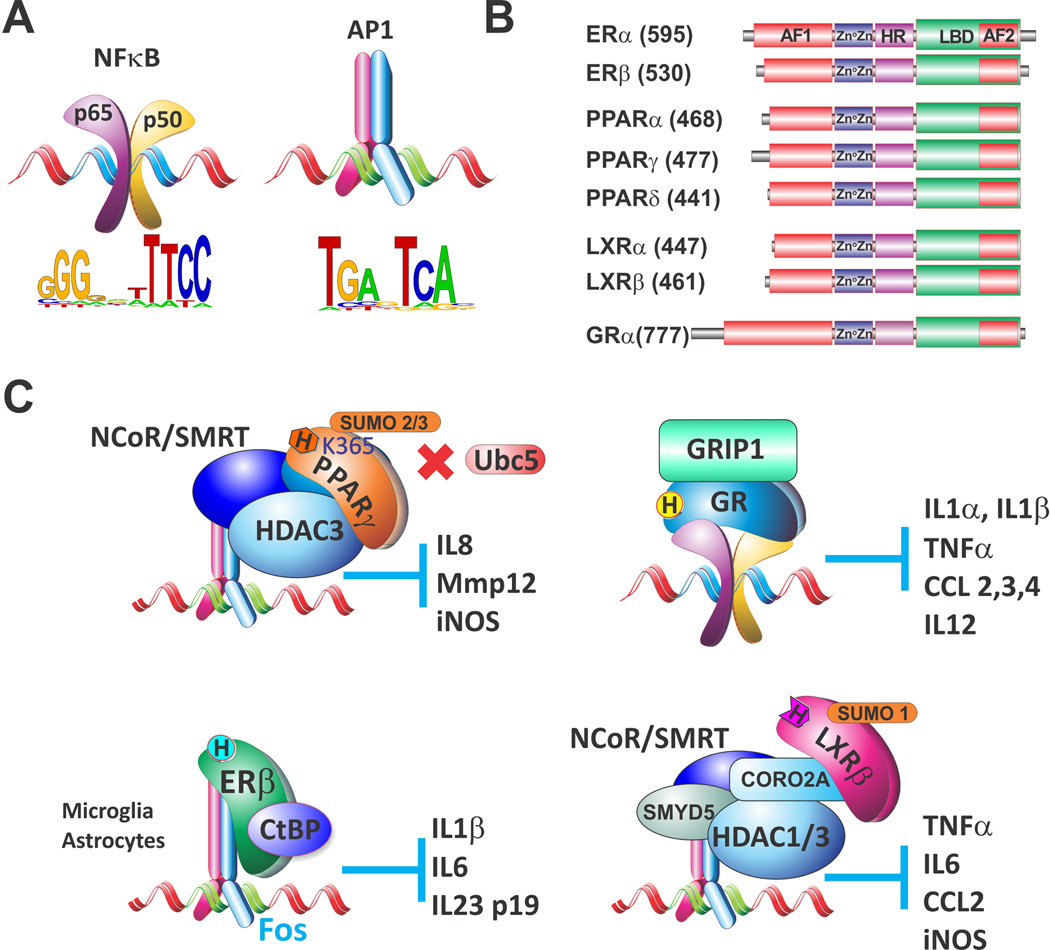
Fig. 1.
Nuclear receptor ‘transrepression’ complexes. Detailed descriptions appear in the text
In addition to activating NFκB TLR ligation also results in phosphorylation of downstream signaling adapters such as TRAF6, leading to sequential activation of TAK1 followed by MAP kinases JNK and p38 that phosphorylate serine/threonine residues on cJun and cFos, commonly referred to as AP-1 proteins (Johnson and Lapadat 2002). AP-1 transcription factors belong to a large family that share a conserved bZIP domain responsible for DNA binding and dimerization. AP-1 proteins (cJun, cFos, Fra-1, Fra-2, FosB, JunB and JunD) homo- and heterodimerize (Chinenov and Kerppola 2001) and bind to AP-1 (TGA(C/G)TCA) or CRE (TGACGTCA) elements (Fig. 1A) in target promoter regions often adjacent to NFκB, IRF, PU.1 or nuclear factor of activated T-cells (NFAT) to coordinately induce transcription (Chinenov and Kerppola 2001; Pascual and Glass 2006).
Rapid activation of NFκB, AP-1 and IRFs amplifies the initial response to TLR ligands by activating the transcription of multiple cytokine genes including IL-6, IFN α and β that bind their cognate cell surface receptors and initiate the Janus kinase (JAK)-STAT signaling cascade (Flammer and Rogatsky 2011; O'Shea, et al. 2013; Yu, et al. 2009). Ligated cytokine receptors change their conformation and bring associated JAK tyrosine kinases in close proximity inducing their self-phosphorylation. Activated JAKs phosphorylate specific tyrosine residues of the cytokine receptors thus creating recognition sites for the STAT family of transcription factors. There are at least seven eukaryotic STAT proteins that upon receptor binding undergo tyrosine phosphorylation which induces conformational rearrangement and dimerization via SH2 domains which recognizes phosphotyrosines (Mao, et al. 2005; Ota, et al. 2004). Most STAT proteins (1, 3, 4, 5 and 6) form homodimers, whereas STAT1 and 2 form heterodimers that further interact with IRF9 to form a heterotrimeric ISGF3 complex (Flammer and Rogatsky 2011). STATs have also been proposed to exist as inactive dimers in the absence of cytokine signals. Activated STAT dimers translocate into the nucleus, bind DNA and induce the transcription of cytokine genes, thereby ensuring secondary amplification of the inflammatory signal (Shuai and Liu 2003).
Nuclear receptors and control of inflammation
Although effective at rapidly stopping infection, most innate immune effectors are relatively non-specific and do not distinguish well between the host and pathogens which can lead to inflammation-associated tissue damage. Indeed, excessive cytokine production, sometimes referred to as “cytokine storm” (Ferrara, et al. 1993), results in pathologies of varying severity from fever, hypoglycemia and weight loss to organ damage and, in extreme circumstances, death (Clark, et al. 1987; Spriggs, et al. 1988; Tracey, et al. 1986). Furthermore, a wide range of autoimmune diseases including rheumatoid arthritis, psoriasis, inflammatory bowel disease and many others have been associated with augmented cytokine production and signaling (Palucka, et al. 2005). Hence, numerous local and systemic regulatory pathways have evolved to control inflammation and promote postinflammatory tissue repair and remodeling (Lee and Mazmanian 2010; Medzhitov 2008). Locally, innate immune and surrounding cells in the affected tissue produce antiinflammatory cytokines and suppressor proteins, such as IL-10, TGFβ and SOCS family members, initiate degradation of cytokine mRNAs, and induce the synthesis and release of small inhibitory molecules such as neuropeptides, lipoxins, prostaglandins and eicosanoids (Medzhitov and Horng 2009). Systemically, circulating cytokines either directly or via stimulation of the vagus nerve initiate a hormonal stress response (Webster, et al. 2002) and production of steroid molecules such as glucocorticoids described in subsequent sections.
Many anti-inflammatory steroids are recognized by the nuclear receptor (NR) superfamily of transcription factors. NR are modular proteins containing the C4-Zn-finger DNA binding and Ligand binding domains (DBD and LBD) linked by a short disordered (or flexible) hinge region in the center of the molecule (Fig. 1B). The highly variable N-terminal regulatory domain encompasses the activation function-1 (AF1) whereas the C-terminal domain including a portion of LBD forms ligand-dependent activation function-2 (AF2). NRs that participate in regulation of inflammation may reside in the nucleus or translocate from the cytoplasm into the nucleus upon ligand binding. Once in the nucleus, NRs either bind directly to specific DNA sequences to induce or repress target genes or become ‘tethered’ to DNA by unrelated DNA-bound transcription factors via protein:protein interactions and typically repress associated genes in ‘trans’ (a.k.a. transrepression). This process is mechanistically and functionally distinct from repression by the same receptor when bound to DNA (‘cis’) and has been well characterized particularly for estrogen receptors (ER), peroxisome proliferator-activated receptors (PPAR), liver-X receptors (LXR) and glucocorticoid receptor (GR) in the context of inflammation (Fig. 1B). Below, we will provide a few examples of the roles of ER, PPAR and LXR in the regulation of inflammation via tethering followed by a more detailed discussion of GR-mediated transrepression.
Estrogen receptors
Estrogens exhibit either pro- or anti-inflammatory activities, depending on the cell type, immune stimulus, hormonal concentration and relative amounts of the predominant ER subtype (α or β) (Cutolo, et al. 2010; Straub 2007). Tethering of either ERα or ERβ to other transcription factors has been proposed as a major mechanism mediating the ‘ERE-independent’ effects of estrogen (Heldring, et al. 2007). In particular, ER interacts in vivo and in vitro with multiple AP-1 family members including cFos, cJun, JunD, JunB, small Maf proteins, ATF2 and CREB1 (Eferl and Wagner 2003; Heldring, et al. 2011; Yang-Yen, et al. 1990), and genome-wide studies demonstrate extensive co-localization of ER and AP-1 binding sites in estradiol-stimulated MCF7 breast cancer cells (Carroll, et al. 2006; Kininis, et al. 2007). More recently, genome wide ChIP-chip experiments in HeLa cells with reintroduced ERα (Heldring et al. 2011) have shown a strong enrichment of ERα at AP-1 sites following estradiol treatment. Liganded ERα recruitment promoted activation of AP-1-driven native genes and enhanced promoter occupancy by cFos and CREB1 suggesting that ERα stabilizes relatively weak DNA binding by AP-1 by an unknown mechanism. Conversely, ER binding to NFκB repressed transiently transfected NFκB-driven IL-6 reporters in estradiol-treated HeLa cells (Ray, et al. 1994). Similarly, in microglia and astrocytes, two cell types involved in the innate immune responses in the brain, liganded ERβ isoform repressed the endogenous production of pro-inflammatory cytokines IL-1β, IL-6 and IL-23 p19. The molecular mechanism of repression involved initial ERβ tethering to phosphorylated cFos followed by ERβ-mediated recruitment of CtBP co-repressor (Fig. 1C). Consequently, treatment with ERβ-specific ligands conferred neuroprotection against experimentally induced autoimmune encephalomyelitis (EAE), a murine model for multiple sclerosis (MS) (Saijo, et al. 2011). Thus, the progression of MS, and possibly other autoimmune disorders of the brain, could potentially be linked to a dysregulated ERβ transrepression pathway (Gosselin and Rivest 2011).
Peroxisome proliferator-activated receptors
PPAR isoforms α, γ and δ (Fig. 1B) have been implicated in regulating inflammation in numerous physiological contexts ranging from atherosclerosis to encephalitis (Li and Yang 2011; Schnegg, et al. 2012) PPARs regulate inflammatory responses through multiple mechanisms including both activation of genes encoding anti-inflammatory mediators, such as IκB (Delerive, et al. 2002), soluble IL-1 receptor antagonist (Stienstra, et al. 2007) and TGFβ, and repression of inflammatory genes via tethering to AP-1, NFκB and STAT1 (Ricote, et al. 1998; Wahli and Michalik 2012). In macrophages, PPAR agonists induce a number of genes related to glucose and lipid metabolism and repress a large subset of genes induced by inflammatory stimuli (Welch, et al. 2003) suggesting that PPARs integrate the two processes making these receptors highly relevant in metabolic diseases including type-II diabetes and atherosclerosis (Varga, et al. 2011). Indeed, in human aortic smooth muscle cells, PPARα inhibits production of IL-6, a key cytokine involved in the pathogenesis of atherosclerosis, by directly tethering to p65 and cJun (Delerive, et al. 1999). Interestingly, PPARα was also proposed to attenuate inflammation indirectly, by potentiating the anti-inflammatory activities of glucocorticoids. Although the mechanistic details are scarce, in a proposed model, PPARα interferes with GR DNA binding at palindromic GREs thereby increasing the pool of free GR available to be tethered to DNA by NFκB or AP-1 (Bougarne, et al. 2009). Co-treatment of L929sA mouse fibrosarcoma cells with the synthetic glucocorticoid, dexamethasone (Dex) and PPARα ligand GW647 resulted in greater suppression of NFκB- and AP-1-dependent inflammatory genes than Dex alone.
PPARγ transrepression of inflammatory genes has been proposed to be regulated by receptor SUMOylation. In this model, AP-1 and NFκB at the IL-8, Mmp12 and iNOS promoters are basally bound by NCoR/SMRT:HDAC3 co-repressor complexes (Hoberg, et al. 2004; Ogawa, et al. 2004). In the presence of ligand, PPARγ undergoes SUMOylation at the conserved Lys-365, which targets PPARγ to promoter-bound NCoR complexes and prevents LPS-induced recruitment of ubiquitin ligase Ubc5 to NCoR (Fig. 1C). This in turn precludes proteasomal degradation and clearance of the co-repressor complex, thereby attenuating LPS-induced transcription (Pascual, et al. 2005). It has been proposed that the NCoR-dependent repression by PPARγ is protective against inflammation-induced insulin resistance associated with obesity and type-II diabetes (Olefsky and Glass 2010).
Liver X receptors
LXR α and β that recognize oxysterols are central to the regulation of cholesterol and lipid metabolism in hepatocytes. In macrophages, LXRs repress a number of atherogenic inflammatory mediators including TNFα, IL-6, CCL2 and iNOS (Pourcet, et al. 2011; Zelcer and Tontonoz 2006). Moreover, inflammatory gene repression by LXRs is not a macrophage-specific phenomenon and has been observed in other cell types such as astrocytes (Lee, et al. 2009) and hepatocytes (Venteclef, et al. 2010). Thus, similar to PPARs, LXRs integrate the maintenance of lipid homeostasis with inhibition of inflammation (Kidani and Bensinger 2012). Despite a significant overlap between inflammatory genes repressed by LXRs and PPARs, a large subset of genes was sensitive to only one receptor (Ogawa, et al. 2005). Furthermore, although the repression of inflammatory genes by both LXR and PPAR involves SUMOylation, each receptor relies on a different E3 ligase (HDAC4 and PIAS-1, respectively) that conjugates distinct SUMO proteins (SUMO-2/3 and SUMO-1, respectively) and are functionally independent of each other (Ghisletti, et al. 2007). Similar to PPARs, SUMOylated LXRs repress inflammatory genes by preventing NCoR and SMRT (Fig. 1C) co-repressor clearance from promoter regions of LPS-stimulated genes in macrophages (Ghisletti, et al. 2009). Resident NCoR/SMRT complexes recruit several chromatin modifying enzymes with “repressing” activities, including HDAC1, HDAC3 and histone methyltransferase SMYD5 that trimethylates H4 K20 (Gao, et al. 2005; Stender, et al. 2012). NCoR promoter clearance is mediated by CORO2A, an actin-binding subunit of the NCoR complex that also harbors a conserved SUMO-interaction motif. The SUMOylated LXR binds CORO2A and prevents the actin-dependent NCoR release that is required for transcription activation (Huang, et al. 2011).
Glucocorticoid-mediated suppression of inflammation
Of the NR superfamily, GR is the most widely known target of anti-inflammatory therapy. Indeed, since the Nobel Prize winning discovery of their anti-inflammatory actions (Hench, et al. 1950; Reichstein 1951), glucocorticoids have remained among the most commonly prescribed drugs to treat a multitude of autoimmune and inflammatory diseases including rheumatoid arthritis, asthma, lupus, inflammatory bowel disease, and multiple sclerosis.
The increase in serum level of glucocorticoids during the immune response has been first noted in the mid-1970s (Besedovsky, et al. 1975) and was linked to the increased serum concentration of cytokines such as IL-1 (Besedovsky, et al. 1986; Woloski, et al. 1985). The level of circulating glucocorticoids is globally controlled by the hypothalamic-pituitary axis (HPA) whereby in response to various stresses, corticotropin-releasing hormone (CRH) produced in paraventricular nucleus of the hypothalamus activates the production of adrenocorticotropic hormone (ACTH) by the anterior lobe of the pituitary ultimately triggering the synthesis and release of glucocorticoids by the adrenal cortex (Silverman and Sternberg 2012; Webster et al. 2002). The regulation of the CRH and POMC genes encoding CRH and ACTH, respectively, is complex and integrates multiple signaling pathways (Kageyama, et al. 2011; Kariagina, et al. 2004). IL1, IL6, TNFα and LIF can each stimulate transcription of the CRH and POMC genes in cell culture directly (Katahira, et al. 1998; Liu, et al. 2008; Wei, et al. 2002). In vivo, however, the contribution of direct activation via cytokines in circulation vs. the hypothalamic stimulation through afferent neuronal circuitry remains to be fully elucidated (Watts and Sanchez-Watts 2002). In a classic example of a feed-back loop regulation, glucocorticoids inhibit POMC transcription and ACTH production (Bilodeau, et al. 2006; Gagner and Drouin 1985). The effect of glucocorticoids on CRH expression is more complex: although glucocorticoids inhibit the transcription of CRH in pituitary cell cultures (Adler, et al. 1988) in placenta, the effect is opposite (Robinson, et al. 1988).
Unliganded GR resides in the cytoplasm as a part of a large aporeceptor complex with chaperon proteins including heat shock protein (HSP) 70, HSP90, HSP40 co-chaperones p23 and Hop, and immunophilin FKBP51. Upon glucocorticoid binding, structural changes in GR result in the complex rearrangement and exchange of FKBP51 for FKBP2, binding to a motor-protein dynein that translocates GR to the nucleus where it activates or represses target gene transcription (Echeverria and Picard 2010). GR can directly bind specific palindromic glucocorticoid response elements (GREs) that contain consensus AGAACA half-sites arranged as an inverted palindrome separated by 0–3 bp providing a binding site for the GR homodimer. Liganded GR then recruits various co-factors, such as the NCoA/p160 family of co-factors (SRC1, SRC2/GRIP1/TIF2 and SRC3/AIB1/ACTR/Rac3), which in turn recruit histone modifiers including histone acetyltransferases CBP/p300; histone methyltransferases CARM1, G9a (York and O'Malley 2010) and ATP-dependent chromatin remodellers, e.g., BRG1 (Burd, et al. 2012; Trotter and Archer 2008). Several GR-induced genes encode anti-inflammatory molecules such as Dusp1 (Imasato, et al. 2002; Vandevyver, et al. 2012), Tsc22d3 (GILZ) (Cannarile, et al. 2009; Mittelstadt and Ashwell 2001) and IκB (Scheinman, et al. 1995a). The role of these factors in inflammation is beyond the scope of this review and has been extensively discussed elsewhere (Clark 2007). Recently, GR binding at GREs with atypical spacers of 1–2 nucleotides was shown to result in transcriptional repression (Surjit, et al. 2011). Although the mechanism and biological relevance of this phenomenon remains to be explored, the recent crystal structure of GR DBD bound to such a site revealed an unusual binding orientation that precluded GR dimerization (Hudson, et al. 2013) suggesting that the repressive activity of GR at nGREs relies on monomeric GR.
At “tethering” GREs GR does not directly bind DNA but instead interacts with other DNA-bound factors and modifies their transcriptional activity. Indeed, GR interacts with several AP-1 family members such as cJun, cFos (Konig, et al. 1992), JunD, JunB and CREB1 (Matsumoto, et al. 2012), NFκB subunit p65 (Nissen and Yamamoto 2000), STAT3 (Langlais, et al. 2012), SMAD3 (Li, et al. 2003), p53 (Sengupta, et al. 2000; Sengupta and Wasylyk 2001) and C/EBPα (Rudiger, et al. 2002) and represses transcription of their target genes. The initial analysis of the repression function of GR mutants with disrupted dimerization interface suggested that at tethering sites GR dimerization may not be required (Reichardt, et al. 2001), however, later studies indicated that the importance of dimeric GR needs to be analyzed on a case by case basis for activated and repressed GR targets alike (Chen, et al. 2013; Rogatsky, et al. 2003). This review will focus on the mechanisms and physiological consequences of GR-mediated transrepression.
GR interactions with AP-1
The first evidence of GR-mediated antagonism of AP-1 activity emerged two decades ago when GR activation was shown to inhibit transcription from transfected AP-1 reporters (Jonat, et al. 1990; Yang-Yen et al. 1990). Subsequently, these in vitro observations were extended to endogenous genes demonstrating by nuclear run-on assays that GR repressed transcription of AP-1-driven collagenase I (Mmp1) gene in human skin fibroblasts treated with Dex. This glucocorticoid-dependent repression did not interfere with AP-1 DNA binding in vitro based on electrophoretic mobility shift assays (EMSA), and did not involve chromatin remodeling as evidenced by the lack of any Dex-induced changes in genomic dimethyl sulfate footprinting at the AP-1 binding sites (Konig et al. 1992). Moreover, mutations affecting GR DNA binding had no effect on its ability to repress AP-1 (Heck, et al. 1994). Later, the advent of ChIP assays enabled a direct quantitative assessment of GR recruitment to genomic AP-1 sites associated with the Mmp13 and IL-8 genes along with cJun and cFos (Kassel, et al. 2004; Rogatsky, et al. 2001) thus solidifying the model for GR tethering to AP-1 and inhibiting its activity. Furthermore, the analysis of overrepresented binding sites within GR binding peaks identified by ChIP-seq in mammary epithelial cells revealed an enrichment of AP-1 motifs. Interestingly, over 90% of AP-1 binding sites, including those coinciding with GR sites, have been associated with constitutively “open” chromatin as shown by DNase-seq assays. Thus, most of the GR binding sites enriched for AP-1 display constitutive GC-independent DNase I hypersensitivity (Biddie, et al. 2011; John, et al. 2011). GR binding to the majority of these sites required intact AP-1 binding activity and was disrupted by cFos DNA binding-deficient mutants when used instead of the WT cFos (Biddie et al. 2011). Taken together, these findings demonstrate the ubiquitous nature and functional importance of GR tethering to AP-1 in modulating its activity.
GR interactions with NFκB
Resembling antagonism of AP-1, initial studies with NFκB showed that GR inhibited NFκB-dependent expression of reporter constructs driven by the IL-6 and ICAM1 proximal promoter regions, or tandem κB sites (Caldenhoven, et al. 1995; Ray and Prefontaine 1994; Scheinman, et al. 1995b). Interactions between GR and p65, p50 and Rel proteins have been further confirmed by in vitro pull-down and co-immunoprecipitation experiments (Ray and Prefontaine 1994; Scheinman et al. 1995b) Mutation analysis identified the GR LBD and DBD as being crucial for repression (Caldenhoven et al. 1995) and mapped the GR:p65 binding interface to the second zinc finger in GR DBD (Liden, et al. 1997; Nissen and Yamamoto 2000) and Rel homology domain (Wissink, et al. 1997). Curiously, a single point mutation in the second zinc finger of the rat GR (Arg488Gln) disrupts GR-mediated repression of NFκB reporters without affecting GR:p65 interactions (Bladh, et al. 2005). Using Bioluminescence Resonance Energy Transfer (BRET), interactions between GR DBD and NFκB have been observed in living cells (Garside, et al. 2004). GR recruitment to the IL-8, ICAM1, Mmp13, and IL-1 α and β κB sites did not alter p65 occupancy (Chinenov, et al. 2012; Kassel et al. 2004; Nissen and Yamamoto 2000) further corroborating the “tethering” model for GR inhibition of NFκB.
The physiological importance of GR-mediated repression in regulating p65-driven inflammation has been highlighted in several genome-wide studies. Microarray expression profiling of macrophages treated with LPS for 6 h in the absence or presence of Dex, revealed over 500 LPS-induced genes of which more than half were repressed by glucocorticoids at least two-fold (Ogawa et al. 2005). In our recent RNA-seq study, a 1-h LPS treatment upregulated over 350 genes >2 fold of which 150 were repressed by Dex, including key inflammatory mediators such as TNFα, IP10 and IL-1 a and β (Chinenov et al. 2012). In another study, co-treatment of HeLa cells with TNFα (a potent inducer of NFκB-driven transcription) and a synthetic glucocorticoid triamcinolone acetonide (TA) for 4 h resulted in the induction of 1033 novel GR binding sites (~12% of total GR binding) that were not induced by TA alone (Rao, et al. 2011). 572 of these “gained” sites overlapped with p65 binding sites induced by TNFα alone. Binding motif analysis revealed that NFκB and AP-1 elements, and not palindromic GREs, were overrepresented among the gained GR binding sites, consistent with the GR recruitment to DNA via tethering by NFκB or AP-1 (Rao et al. 2011). Furthermore, gained GR binding sites were associated with genes encoding cytokines, cytokine receptor families, MAP kinases and mediators of TLR signaling and apoptosis.
GR interactions with STAT3
GR interactions with several STAT family members, including STAT 3 and 5, result in either synergistic induction of STAT-bound genes or inhibition of the GR-bound genes ((Rogatsky and Ivashkiv 2006) and references therein). For example, the first evidence of STAT5:GR interaction was reported for the β-casein gene which lacks a known GRE but contains a STAT5 binding site. This interaction was shown to cooperatively induce β-casein expression relative to STAT5 alone (Stocklin, et al. 1996; Stoecklin, et al. 1997). GR and STAT5 have also been shown to induce the TLR2 gene in TNFα + Dex treated A549 cells, although whether this induction is due to direct protein:protein interactions remains to be determined (Hermoso, et al. 2004). STAT3:GR interaction results in an increased activation of the STAT3 target gene α2-macroglobin (Lerner, et al. 2003). Surprisingly, GR interaction with STAT3 was also reported to result in superactivation of GR targets with STAT3 operating as a GR co-activator at a GRE. Upon co-treatment of rat hepatoma H4IIE cells with IL-6 and Dex which stimulate STAT3 and GR binding, respectively, the two proteins could be co-precipitated. Moreover, in the EMSA experiment an IL-6+Dex-induced protein complex bound to a probe with a STAT3 binding site, and this could be inhibited by anti-GR and super-shifted by anti-STAT3 antibodies suggesting the presence of both proteins in the same DNA-bound complex (Zhang, et al. 1997). Interestingly, recent genome-scale studies have uncovered a widespread GR tethering to STAT3 which results in the negative regulation STAT3 target genes. Microarray-based expression profiling showed that Dex together with the STAT3-activating cytokine LIF, modulates the transcription of over 700 genes (Langlais et al. 2012). Moreover, cluster analysis of ChIP-seq data demonstrated that GR was recruited to over 300 STAT3-bound sites. Notably, the genes in this cluster were repressed by liganded GR in a LIF-dependent manner (Langlais et al. 2012). These results are consistent with a model whereby GR tethers to STAT3 and represses associated genes by as yet unknown mechanism.
Mechanisms of GR-mediated repression
Although the physiological relevance of GR-mediated repression of AP-1- and NFκB-induced transcription is well established, the underlying molecular mechanisms remain to be determined. Conceivably, a single unifying mechanism does not exist rather, distinct processes contribute to repression in a gene-, cell type- and tissue-specific manner with the regulatory outcome ultimately determined by the relative concentrations, activities and repertoire of the context-specific components. Several models of GR action have been proposed over the years and are discussed below.
GR competition for limited co-activators
Regulators of different families rely on a defined repertoire of co-regulators to control the basal transcriptional machinery and chromatin dynamics. Thus, cofactors utilized by both GR and AP-1 or NFκB may in principle become limiting such that activation of GR would sequester them away from AP-1 and NFκB thereby attenuating transcription of genes dependent on these regulators. One example of such shared co-regulators are the histone acetyltransferases CBP/p300, which are used by NRs as well as p65 and Jun. Indeed, introduction of CBP was shown to alleviate GR-mediated repression of AP-1- and NFκB-driven synthetic reporters (Kamei, et al. 1996; Sheppard, et al. 1998). This model was later debated when similar CBP ‘rescue’ experiments had no effect on GR-mediated repression of IL-6 reporters driven by either p65 or Jun (De Bosscher, et al. 2001; De Bosscher, et al. 2000). This suggests that perhaps, competition for co-activators as the basis of GR-mediated repression might be gene-specific and requires validation on a genome-wide scale.
Consistent with the mutually exclusive cofactor utilization mechanism, in macrophages, sequestration of the p160 protein GRIP1 by GR may account for glucocorticoid inhibition of transcription regulated by members of the IRF family (reviewed in (Flammer and Rogatsky 2011)). IRF3 and IRF9 are responsible for anti-viral defenses by activating transcription in response to TLR3 ligands and type I interferons, respectively. GRIP1 was found to interact with several IRF proteins including IRF3 and 9 (Bhandare, et al. 2011; Flammer, et al. 2010; Reily, et al. 2006) in a glucocorticoid-sensitive fashion. In macrophages, knockdown of GRIP1 or Dex treatment both markedly attenuated transcription of IRF3 and IRF9 target genes, whereas GR antagonists that interfere with GR:GRIP1 complex formation reversed Dex-mediated inhibition. These findings were consistent with the role of GRIP1 as a rate-limiting co-activator for IRF proteins, which is sequestered away by the agonist-bound GR. Interestingly, this effect was determined by the total amount of cellular GRIP1: in fibroblasts that, unlike macrophages, express GRIP1 at high levels, IFN-induced transcription was refractory to Dex (Flammer et al. 2010), illustrating how a specific cellular context influences the regulatory outcome.
Contextual “gain of function”
Reportedly, NRs utilize over 300 different co-regulators (Lonard, et al. 2007) that form a combinatorial interaction network participating in gene-, cell- and signal-specific complexes with varying, sometimes opposite, activities. Surprisingly, unlike LXR and PPAR, GR transrepression does not seem to involve the NCoR complex and, at least in macrophages, is unaffected by NCoR depletion (Ogawa et al. 2005). Alternative cofactors have been implicated in GR-mediated transrepression, including GRIP1 and TRIP6.
Similar to several other GR cofactors (Bittencourt, et al. 2012), GRIP1 was first identified as a co-activator of the p160 family with a typical multidomain structure encompassing a centrally located NR interacting domain, the N-terminal bHLH-PAS domain and several C-terminal domains involved in protein:protein interactions. In the context of palindromic GREs, GR-bound GRIP1 activates transcription by recruiting several co-activators such as CBP/p300 and CARM1 (York and O'Malley 2010). GRIP1 also interacts with several nonreceptor transcription activators including MEF2C and MyoD (Chen, et al. 2000; Wu, et al. 2005) and, as discussed above, several IRF proteins. The first indication for an additional role of GRIP1 in GR signaling was its ability to potentiate glucocorticoid repression of the human Mmp13 gene via the AP-1 response element in a GR agonist- and GR recruitment-dependent manner (Rogatsky et al. 2001). GRIP1 repression activity was associated with a unique Repression Domain (RD), located immediately downstream of the NR interacting domain. RD does not have any discernible sequence or structural motifs suggestive of function and is absent in the other p160 proteins (Rogatsky, et al. 2002). Consistently, other members of p160 family (SRC1 and SRC3) fail to potentiate GR-mediated repression of AP-1- or NFκB-dependent transcription. In mouse macrophages, GRIP1 is recruited to NFκB binding sites concomitantly with GR upon co-treatment with Dex and LPS. Furthermore, in macrophages derived from mice conditionally lacking GRIP1 in hematopoietic cells, genes encoding inflammatory mediators such as TNFα, IL-1β and CCL4 were less susceptible to GR-mediated repression than those in wild-type macrophages (Chinenov et al. 2012). Remarkably, GRIP1 depletion attenuated GR-mediated repression of nearly half of the LPS-induced, Dex-repressed genes including a large number of mediators of inflammation (TNFα, IL-1 α and β, CCL2, 3 and 4, LIF, IP10 and IFNβ) thus illustrating a broad role of GRIP1 in controlling the inflammatory transcriptome in macrophages. Furthermore, the GRIP1-deficient mice were more sensitive to a systemic inflammatory challenge in vivo succumbing to endotoxin shock due to augmented levels of serum inflammatory cytokines relative to wild-type mice (Chinenov et al. 2012).
The molecular mechanism of the GRIP1 co-repressor function awaits elucidation. GRIP1 contains several protein:protein interaction domains that recruit transcriptionally repressive chromatin-modifying enzymes such as methyltransferases G9a and Suv4–20h1 (Chinenov, et al. 2008; Lee, et al. 2006) that methylate H3 K9 and H4 K20, respectively. Curiously, in the context of LXR-mediated repression of TLR4 target genes, H4 K20 is trimethylated by methyltransferase SMYD5 associated with the LXR/NCoR complex (Stender et al. 2012). Whether Suv4–20h1 plays a similar role in GR-mediated repression is unknown.
TRIP6 is another multifunctional protein whose role in transcription was overlooked at the time of the initial functional characterization (Wang, et al. 1999). TRIP6 is a LIM domain-containing protein from the zyxin family that was first discovered in focal adhesion contacts and later shown to shuttle between focal adhesion contacts and the nucleus (Nix and Beckerle 1997). In addition to TRIP6, several members of this family including paxillin and Hic-5 interact with GR and other NRs and typically act as co-activators (reviewed in (Hervy, et al. 2006). TRIP6 was first identified as a thyroid receptor binding partner activator recruited to palindromic GREs in a ligand-dependent manner (Diefenbacher, et al. 2010). Based on electrophoretic mobility TRIP6 exist as three different isoforms of which the fastest (nTRIP6) is localized to the nucleus (Kassel et al. 2004). Whether these isoforms are RNA splice variants or distinct protein species generated through posttranslational modifications is unknown. Interestingly, following AP-1 and NFκB stimulation, nTRIP6 is also recruited to AP-1 and κB sites (via interactions with cFos and NFκB) and potentiates transcription of endogenous Mmp13 and IL-8 genes. Notably, upon glucocorticoid treatment GR was recruited to the same binding sites, likely forming a complex with AP-1:nTRIP6 and NFκB:nTRIP6, which correlated with the repression of Mmp13 and IL-8. Indeed, TRIP6 depletion attenuated GR recruitment to these promoters and subsequent repression without affecting either AP-1 or NFκB loading (Diefenbacher, et al. 2008; Kassel et al. 2004). Similarly, the TRIP6 mutants interacting with cFos and p65 but not with GR, activated AP-1 and NFκB-driven reporters but failed to potentiate repression by GR (Kassel et al. 2004).
Transcription Initiation vs Elongation
Activation of transcription includes two sequential steps: initiation and elongation. Transcription initiation involves the assembly of RNA polymerase2 (Pol2) and general transcription factors at target promoters into the pre-initiation complex (PIC) which coincides with the phosphorylation of the Ser-5 residue of the heptapeptide repeats in the Pol2 C-terminal domain (CTD) by the cyclin H/CDK7 CAK kinase. The subsequent transition into productive elongation involves phosphorylation of the Ser-2 of the Pol2 CTD repeats by the Positive Transcription Elongation Factor (PTEF)-b complex (Nechaev and Adelman 2010). In principle, GR can target either or both steps to inhibit activation of transcription by AP-1 or NFκB.
For example, it was shown that GR represses the IL-8 and ICAM-1 genes in TNFα-stimulated A549 cells by inhibiting transcription elongation. Indeed, at the promoters of these genes, glucocorticoids did not affect TNFα-induced Pol2 recruitment and Ser-5 phosphorylation, thus implying that initiation occurred normally. However, the recruitment of PTEF-b to the IL-8 promoter, and hence, Ser-2 phosphorylation was blocked by Dex (Luecke and Yamamoto 2005; Nissen and Yamamoto 2000) resulting in the early elongation arrest of the initiated Pol2.
Recruitment of HDACs
Another potential target for GR is chromatin itself. Indeed, GR could inhibit transcription by changing either directly or via its co-regulators chromatin structure at the target promoters, making them inaccessible to the basal transcriptional machinery. For example, in A549 cells, glucocorticoids inhibit the IL-1β stimulated, p65-dependent expression of the GM-CSF gene which correlates with reduced histone H4 acetylation at the GM-CSF κB site. The potential enzyme responsible, HDAC2, was identified as a GR:p65 associated protein using co-immunoprecipitation assays. Both the inhibition and depletion of HDAC2 resulted in elevated H4 acetylation of the GM-CSF promoter and increased GM-CSF production in the presence of Dex, implicating HDAC2 in GR-mediated inhibition of the GM-CSF gene (Ito, et al. 2000; Ito, et al. 2001; Ito, et al. 2006). Interestingly, GR itself is subject to hormone-dependent acetylation at Lys-494 and 495 in its DBD, and HDAC2 was able to reverse this process. Furthermore, HDAC2 depletion both inhibited GR deacetylation and also prevented GR:p65 interaction without affecting GR DNA binding (Ito et al. 2006). Thus, the requirement for HDAC2 in repression in this system might be due in part to its permissive effects on GR recruitment to the NFκB complexes rather than changes in H4 acetylation.
Summary and perspectives
NR ligands constitute 13% of unique FDA approved drugs (Overington, et al. 2006). Among them, potent anti-inflammatory properties of glucocorticoids ensured the widespread use of these compounds to treat a variety of immune and inflammatory disorders. 4000–5000 papers related to the use or mechanisms of glucocorticoid actions are published annually. Despite such intense scrutiny, how glucocorticoids inhibit inflammation remains a hotly debated topic. Numerous conflicting mechanisms are oftentimes based on data obtained in reductionist model systems that yield monofactorial readouts of a few “key” genes. Failed attempts to generalize these findings or translate in vitro results to more complex systems suggest that the discovery of the universal mechanism of anti-inflammatory actions of glucocorticoids may be an unattainable goal for not only technical but for broader biological reasons. Perhaps, much like the diverse inflammatory pathologies themselves, the anti-inflammatory approaches should be a disease- or condition-specific. Inflammatory disorders exhibit strikingly different profiles of pro- and anti-inflammatory gene expression and involve different cell types. For example, although rheumatoid arthritis and systemic lupus erythematosus both have a strong inflammatory component, the pathogenesis of the former is driven by TNFα (Feldmann 2002)whereas the latter is associated with dysregulated IFNα (Bennett, et al. 2003). Strikingly, in MS, another inflammatory autoimmune disease, IFNα is anti-inflammatory and is used therapeutically (Guarda, et al. 2011).The recognition of disease-specific pathogenic inflammatory cell types adds another layer of complexity to the design of anti-inflammatory strategies. Although “common” glucocorticoid-regulated genes certainly exist, system-specific regulatory events may drastically affect the kinetics of the response, relative activities of these genes and their products as well as their relevance to the pathogenesis of a specific disorder. Thus, it is difficult to predict whether “large” change in few genes observed in a given cell type would produce desired outcome in a unique disease settings. Conversely, minor, seemingly insignificant changes might initiate cooperative, cascading responses with profound biological consequences. Finally, data obtained in mouse models of human disease should be interpreted with caution given fundamental inter-species differences and well known strain-to-strain variations. Therefore, system-wide analyses in disease-relevant model systems and efforts to validate murine data in human are required to understand and ultimately manipulate GR regulatory circuits.
Highlights.
Nuclear receptors inhibit inflammatory gene transcription via distinct mechanisms
GR colocalizes with other regulators and represses their targets genome-wide
GR utilizes cofactors as context-specific coactivators or corepressors
Acknowledgements
We thank K. Zarember (NIH-NIAID) for helpful comments on the manuscript. This work was supported by grants from the National Institutes of Health (to I.R.) and from American Heart Association Grant 11SDG5160006 (to Y.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler GK, Smas CM, Majzoub JA. Expression and dexamethasone regulation of the human corticotropin-releasing hormone gene in a mouse anterior pituitary cell line. J Biol Chem. 1988;263:5846–5852. [PubMed] [Google Scholar]
- Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besedovsky H, del Rey A, Sorkin E, Dinarello CA. Immunoregulatory feedback between interleukin-1 and glucocorticoid hormones. Science. 1986;233:652–654. doi: 10.1126/science.3014662. [DOI] [PubMed] [Google Scholar]
- Besedovsky H, Sorkin E, Keller M, Muller J. Changes in blood hormone levels during the immune response. Proc Soc Exp Biol Med. 1975;150:466–470. doi: 10.3181/00379727-150-39057. [DOI] [PubMed] [Google Scholar]
- Bhandare R, Damera G, Banerjee A, Flammer JR, Keslacy S, Rogatsky I, Panettieri RA, Amrani Y, Tliba O. Glucocorticoid receptor interacting protein-1 restores glucocorticoid responsiveness in steroid-resistant airway structural cells. Am J Respir Cell Mol Biol. 2011;42:9–15. doi: 10.1165/rcmb.2009-0239RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Vallette-Kasic S, Gauthier Y, Figarella-Branger D, Brue T, Berthelet F, Lacroix A, Batista D, Stratakis C, Hanson J, et al. Role of Brg1 and HDAC2 in GR trans-repression of the pituitary POMC gene and misexpression in Cushing disease. Genes Dev. 2006;20:2871–2886. doi: 10.1101/gad.1444606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittencourt D, Wu DY, Jeong KW, Gerke DS, Herviou L, Ianculescu I, Chodankar R, Siegmund KD, Stallcup MR. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2012;109:19673–19678. doi: 10.1073/pnas.1211803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladh LG, Liden J, Dahlman-Wright K, Reimers M, Nilsson S, Okret S. Identification of endogenous glucocorticoid repressed genes differentially regulated by a glucocorticoid receptor mutant able to separate between nuclear factor-kappaB and activator protein-1 repression. Mol Pharmacol. 2005;67:815–826. doi: 10.1124/mol.104.005801. [DOI] [PubMed] [Google Scholar]
- Bougarne N, Paumelle R, Caron S, Hennuyer N, Mansouri R, Gervois P, Staels B, Haegeman G, De Bosscher K. PPARalpha blocks glucocorticoid receptor alpha-mediated transactivation but cooperates with the activated glucocorticoid receptor alpha for transrepression on NF-kappaB. Proc Natl Acad Sci U S A. 2009;106:7397–7402. doi: 10.1073/pnas.0806742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CJ, Ward JM, Crusselle-Davis VJ, Kissling GE, Phadke D, Shah RR, Archer TK. Analysis of chromatin dynamics during glucocorticoid receptor activation. Mol Cell Biol. 2012;32:1805–1817. doi: 10.1128/MCB.06206-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldenhoven E, Liden J, Wissink S, Van de Stolpe A, Raaijmakers J, Koenderman L, Okret S, Gustafsson JA, Van der Saag PT. Negative cross-talk between RelA and the glucocorticoid receptor: a possible mechanism for the antiinflammatory action of glucocorticoids. Mol Endocrinol. 1995;9:401–412. doi: 10.1210/mend.9.4.7659084. [DOI] [PubMed] [Google Scholar]
- Cannarile L, Cuzzocrea S, Santucci L, Agostini M, Mazzon E, Esposito E, Muia C, Coppo M, Di Paola R, Riccardi C. Glucocorticoid-induced leucine zipper is protective in Th1-mediated models of colitis. Gastroenterology. 2009;136:530–541. doi: 10.1053/j.gastro.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chen SH, Masuno K, Cooper SB, Yamamoto KR. Incoherent feed-forward regulatory logic underpinning glucocorticoid receptor action. Proc Natl Acad Sci U S A. 2013;110:1964–1969. doi: 10.1073/pnas.1216108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y, Gupte R, Dobrovolna J, Flammer JR, Liu B, Michelassi FE, Rogatsky I. Role of transcriptional coregulator GRIP1 in the anti-inflammatory actions of glucocorticoids. Proc Natl Acad Sci U S A. 2012;109:11776–11781. doi: 10.1073/pnas.1206059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Sacta MA, Cruz AR, Rogatsky I. GRIP1-associated SET-domain methyltransferase in glucocorticoid receptor target gene expression. Proc Natl Acad Sci U S A. 2008;105:20185–20190. doi: 10.1073/pnas.0810863105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AR. Anti-inflammatory functions of glucocorticoid-induced genes. Mol Cell Endocrinol. 2007;275:79–97. doi: 10.1016/j.mce.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Clark IA, Cowden WB, Butcher GA, Hunt NH. Possible roles of tumor necrosis factor in the pathology of malaria. Am J Pathol. 1987;129:192–199. [PMC free article] [PubMed] [Google Scholar]
- Cutolo M, Brizzolara R, Atzeni F, Capellino S, Straub RH, Puttini PC. The immunomodulatory effects of estrogens: clinical relevance in immune-mediated rheumatic diseases. Ann N Y Acad Sci. 2010;1193:36–42. doi: 10.1111/j.1749-6632.2009.05383.x. [DOI] [PubMed] [Google Scholar]
- De Bosscher K, Vanden Berghe W, Haegeman G. Glucocorticoid repression of AP-1 is not mediated by competition for nuclear coactivators. Mol Endocrinol. 2001;15:219–227. doi: 10.1210/mend.15.2.0591. [DOI] [PubMed] [Google Scholar]
- De Bosscher K, Vanden Berghe W, Vermeulen L, Plaisance S, Boone E, Haegeman G. Glucocorticoids repress NF-kappaB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc Natl Acad Sci U S A. 2000;97:3919–3924. doi: 10.1073/pnas.97.8.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delerive P, De Bosscher K, Besnard S, Vanden Berghe W, Peters JM, Gonzalez FJ, Fruchart JC, Tedgui A, Haegeman G, Staels B. Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-kappa B and AP-1. J Biol Chem. 1999;274:32048–32054. doi: 10.1074/jbc.274.45.32048. [DOI] [PubMed] [Google Scholar]
- Delerive P, De Bosscher K, Vanden Berghe W, Fruchart JC, Haegeman G, Staels B. DNA binding-independent induction of IkappaBalpha gene transcription by PPARalpha. Mol Endocrinol. 2002;16:1029–1039. doi: 10.1210/mend.16.5.0826. [DOI] [PubMed] [Google Scholar]
- Diefenbacher M, Sekula S, Heilbock C, Maier JV, Litfin M, van Dam H, Castellazzi M, Herrlich P, Kassel O. Restriction to Fos family members of Trip6-dependent coactivation and glucocorticoid receptor-dependent transrepression of activator protein-1. Mol Endocrinol. 2008;22:1767–1780. doi: 10.1210/me.2007-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbacher ME, Litfin M, Herrlich P, Kassel O. The nuclear isoform of the LIM domain protein Trip6 integrates activating and repressing signals at the promoter-bound glucocorticoid receptor. Mol Cell Endocrinol. 2010;320:58–66. doi: 10.1016/j.mce.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Echeverria PC, Picard D. Molecular chaperones, essential partners of steroid hormone receptors for activity and mobility. Biochim Biophys Acta. 2010;1803:641–649. doi: 10.1016/j.bbamcr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Feldmann M. Development of anti-TNF therapy for rheumatoid arthritis. Nat Rev Immunol. 2002;2:364–371. doi: 10.1038/nri802. [DOI] [PubMed] [Google Scholar]
- Ferrara JL, Abhyankar S, Gilliland DG. Cytokine storm of graft-versus-host disease: a critical effector role for interleukin-1. Transplant Proc. 1993;25:1216–1217. [PubMed] [Google Scholar]
- Flammer JR, Dobrovolna J, Kennedy MA, Chinenov Y, Glass CK, Ivashkiv LB, Rogatsky I. The type I interferon signaling pathway is a target for glucocorticoid inhibition. Mol Cell Biol. 2010;30:4564–4574. doi: 10.1128/MCB.00146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flammer JR, Rogatsky I. Minireview: Glucocorticoids in autoimmunity: unexpected targets and mechanisms. Mol Endocrinol. 2011;25:1075–1086. doi: 10.1210/me.2011-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagner JP, Drouin J. Opposite regulation of pro-opiomelanocortin gene transcription by glucocorticoids and CRH. Mol Cell Endocrinol. 1985;40:25–32. doi: 10.1016/0303-7207(85)90154-6. [DOI] [PubMed] [Google Scholar]
- Gao Z, Chiao P, Zhang X, Lazar MA, Seto E, Young HA, Ye J. Coactivators and corepressors of NF-kappaB in IkappaB alpha gene promoter. J Biol Chem. 2005;280:21091–21098. doi: 10.1074/jbc.M500754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garside H, Stevens A, Farrow S, Normand C, Houle B, Berry A, Maschera B, Ray D. Glucocorticoid ligands specify different interactions with NF-kappaB by allosteric effects on the glucocorticoid receptor DNA binding domain. J Biol Chem. 2004;279:50050–50059. doi: 10.1074/jbc.M407309200. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Rivest S. Estrogen receptor transrepresses brain inflammation. Cell. 2011;145:495–497. doi: 10.1016/j.cell.2011.04.018. [DOI] [PubMed] [Google Scholar]
- Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, Farlik M, Decker T, Du Pasquier RA, Romero P, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck S, Kullmann M, Gast A, Ponta H, Rahmsdorf HJ, Herrlich P, Cato AC. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994;13:4087–4095. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N, Isaacs GD, Diehl AG, Sun M, Cheung E, Ranish JA, Kraus WL. Multiple sequence-specific DNA-binding proteins mediate estrogen receptor signaling through a tethering pathway. Mol Endocrinol. 2011;25:564–574. doi: 10.1210/me.2010-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, et al. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Hench PS, Kendall EC, Slocumb CH, Polley HF. Effects of cortisone acetate and pituitary ACTH on rheumatoid arthritis, rheumatic fever and certain other conditions. Arch Intern Med (Chic) 1950;85:545–666. doi: 10.1001/archinte.1950.00230100002001. [DOI] [PubMed] [Google Scholar]
- Hermoso MA, Matsuguchi T, Smoak K, Cidlowski JA. Glucocorticoids and tumor necrosis factor alpha cooperatively regulate toll-like receptor 2 gene expression. Mol Cell Biol. 2004;24:4743–4756. doi: 10.1128/MCB.24.11.4743-4756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr Opin Cell Biol. 2006;18:524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Hoberg JE, Yeung F, Mayo MW. SMRT derepression by the IkappaB kinase alpha: a prerequisite to NF-kappaB transcription and survival. Mol Cell. 2004;16:245–255. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, et al. Coronin 2A mediates actin-dependent derepression of inflammatory response genes. Nature. 2011;470:414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol. 2013;20:53–58. doi: 10.1038/nsmb.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imasato A, Desbois-Mouthon C, Han J, Kai H, Cato AC, Akira S, Li JD. Inhibition of p38 MAPK by glucocorticoids via induction of MAPK phosphatase-1 enhances nontypeable Haemophilus influenzae-induced expression of toll-like receptor 2. J Biol Chem. 2002;277:47444–47450. doi: 10.1074/jbc.M208140200. [DOI] [PubMed] [Google Scholar]
- Ito K, Barnes PJ, Adcock IM. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol Cell Biol. 2000;20:6891–6903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Jazrawi E, Cosio B, Barnes PJ, Adcock IM. p65-activated histone acetyltransferase activity is repressed by glucocorticoids: mifepristone fails to recruit HDAC2 to the p65-HAT complex. J Biol Chem. 2001;276:30208–30215. doi: 10.1074/jbc.M103604200. [DOI] [PubMed] [Google Scholar]
- Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, Adcock IM. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-kappaB suppression. J Exp Med. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, Herrlich P. Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- Kageyama K, Tamasawa N, Suda T. Signal transduction in the hypothalamic corticotropin-releasing factor system and its clinical implications. Stress. 2011;14:357–367. doi: 10.3109/10253890.2010.536279. [DOI] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kariagina A, Romanenko D, Ren SG, Chesnokova V. Hypothalamic-pituitary cytokine network. Endocrinology. 2004;145:104–112. doi: 10.1210/en.2003-0669. [DOI] [PubMed] [Google Scholar]
- Kassel O, Schneider S, Heilbock C, Litfin M, Gottlicher M, Herrlich P. A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1- and NF-kappaB-regulated promoters. Genes Dev. 2004;18:2518–2528. doi: 10.1101/gad.322404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira M, Iwasaki Y, Aoki Y, Oiso Y, Saito H. Cytokine regulation of the rat proopiomelanocortin gene expression in AtT-20 cells. Endocrinology. 1998;139:2414–2422. doi: 10.1210/endo.139.5.6005. [DOI] [PubMed] [Google Scholar]
- Kidani Y, Bensinger SJ. Liver X receptor and peroxisome proliferator-activated receptor as integrators of lipid homeostasis and immunity. Immunol Rev. 2012;249:72–83. doi: 10.1111/j.1600-065X.2012.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27:5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig H, Ponta H, Rahmsdorf HJ, Herrlich P. Interference between pathway-specific transcription factors: glucocorticoids antagonize phorbol ester-induced AP-1 activity without altering AP-1 site occupation in vivo. EMBO J. 1992;11:2241–2246. doi: 10.1002/j.1460-2075.1992.tb05283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunsch C, Ruben SM, Rosen CA. Selection of optimal kappa B/Rel DNA-binding motifs: interaction of both subunits of NF-kappa B with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell. 2012;47:38–49. doi: 10.1016/j.molcel.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem. 2006;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Park SM, Kim OS, Lee CS, Woo JH, Park SJ, Joe EH, Jou I. Differential SUMOylation of LXRalpha and LXRbeta mediates transrepression of STAT1 inflammatory signaling in IFN-gamma-stimulated brain astrocytes. Mol Cell. 2009;35:806–817. doi: 10.1016/j.molcel.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner L, Henriksen MA, Zhang X, Darnell JE., Jr STAT3-dependent enhanceosome assembly and disassembly: synergy with GR for full transcriptional increase of the alpha 2-macroglobulin gene. Genes Dev. 2003;17:2564–2577. doi: 10.1101/gad.1135003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang S, Gelehrter TD. Identification of glucocorticoid receptor domains involved in transrepression of transforming growth factor-beta action. J Biol Chem. 2003;278:41779–41788. doi: 10.1074/jbc.M305350200. [DOI] [PubMed] [Google Scholar]
- Li MD, Yang X. A Retrospective on Nuclear Receptor Regulation of Inflammation: Lessons from GR and PPARs. PPAR Res. 2011;2011:742785. doi: 10.1155/2011/742785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liden J, Delaunay F, Rafter I, Gustafsson J, Okret S. A new function for the C-terminal zinc finger of the glucocorticoid receptor. Repression of RelA transactivation. J Biol Chem. 1997;272:21467–21472. doi: 10.1074/jbc.272.34.21467. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kamitakahara A, Kim AJ, Aguilera G. Cyclic adenosine 3’,5’-monophosphate responsive element binding protein phosphorylation is required but not sufficient for activation of corticotropin-releasing hormone transcription. Endocrinology. 2008;149:3512–3520. doi: 10.1210/en.2008-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, Lanz RB, O'Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, McMurray JS, Demeler B, Darnell JE, Jr, Chen X. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Kuriwaka-Kido R, Kondo T, Endo I, Kido S. Regulation of osteoblast differentiation by interleukin-11 via AP-1 and Smad signaling. Endocr J. 2012;59:91–101. doi: 10.1507/endocrj.ej11-0219. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Mittelstadt PR, Ashwell JD. Inhibition of AP-1 by the glucocorticoid-inducible protein GILZ. J Biol Chem. 2001;276:29603–29610. doi: 10.1074/jbc.M101522200. [DOI] [PubMed] [Google Scholar]
- Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2010;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci U S A. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol. 2004;5:208–215. doi: 10.1038/ni1032. [DOI] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci U S A. 2005;102:3372–3377. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G, Glass CK. Nuclear receptors versus inflammation: mechanisms of transrepression. Trends Endocrinol Metab. 2006;17:321–327. doi: 10.1016/j.tem.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Pourcet B, Feig JE, Vengrenyuk Y, Hobbs AJ, Kepka-Lenhart D, Garabedian MJ, Morris SM, Jr, Fisher EA, Pineda-Torra I. LXRalpha regulates macrophage arginase 1 through PU.1 and interferon regulatory factor 8. Circ Res. 2011;109:492–501. doi: 10.1161/CIRCRESAHA.111.241810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NA, McCalman MT, Moulos P, Francoijs KJ, Chatziioannou A, Kolisis FN, Alexis MN, Mitsiou DJ, Stunnenberg HG. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011;21:1404–1416. doi: 10.1101/gr.118042.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Prefontaine KE. Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc Natl Acad Sci U S A. 1994;91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Prefontaine KE, Ray P. Down-modulation of interleukin-6 gene expression by 17 beta-estradiol in the absence of high affinity DNA binding by the estrogen receptor. J Biol Chem. 1994;269:12940–12946. [PubMed] [Google Scholar]
- Reichardt HM, Tuckermann JP, Gottlicher M, Vujic M, Weih F, Angel P, Herrlich P, Schutz G. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J. 2001;20:7168–7173. doi: 10.1093/emboj/20.24.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichstein T. [Chemistry of adrenal cortex hormones] Bull Schweiz Akad Med Wiss. 1951;7:359–370. [PubMed] [Google Scholar]
- Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. EMBO J. 2006;25:108–117. doi: 10.1038/sj.emboj.7600919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- Robinson BG, Emanuel RL, Frim DM, Majzoub JA. Glucocorticoid stimulates expression of corticotropin-releasing hormone gene in human placenta. Proc Natl Acad Sci U S A. 1988;85:5244–5248. doi: 10.1073/pnas.85.14.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Ivashkiv LB. Glucocorticoid modulation of cytokine signaling. Tissue Antigens. 2006;68:1–12. doi: 10.1111/j.1399-0039.2006.00599.x. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci U S A. 2002;99:16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq CM, Darimont BD, Garabedian MJ, Yamamoto KR. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci U S A. 2003;100:13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Zarember KA, Yamamoto KR. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 2001;20:6071–6083. doi: 10.1093/emboj/20.21.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger JJ, Roth M, Bihl MP, Cornelius BC, Johnson M, Ziesche R, Block LH. Interaction of C/EBPalpha and the glucocorticoid receptor in vivo and in nontransformed human cells. FASEB J. 2002;16:177–184. doi: 10.1096/fj.01-0226com. [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. Modulation of NF-kappaB activity by exchange of dimers. Mol Cell. 2003;11:1563–1574. doi: 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Saijo K, Collier JG, Li AC, Katzenellenbogen JA, Glass CK. An ADIOL-ERbeta-CtBP transrepression pathway negatively regulates microglia-mediated inflammation. Cell. 2011;145:584–595. doi: 10.1016/j.cell.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinman RI, Cogswell PC, Lofquist AK, Baldwin AS., Jr Role of transcriptional activation of I kappa B alpha in mediation of immunosuppression by glucocorticoids. Science. 1995a;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- Scheinman RI, Gualberto A, Jewell CM, Cidlowski JA, Baldwin AS., Jr Characterization of mechanisms involved in transrepression of NF-kappa B by activated glucocorticoid receptors. Mol Cell Biol. 1995b;15:943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnegg CI, Kooshki M, Hsu FC, Sui G, Robbins ME. PPARdelta prevents radiation-induced proinflammatory responses in microglia via transrepression of NF-kappaB and inhibition of the PKCalpha/MEK1/2/ERK1/2/AP-1 pathway. Free Radic Biol Med. 2012;52:1734–1743. doi: 10.1016/j.freeradbiomed.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Vonesch JL, Waltzinger C, Zheng H, Wasylyk B. Negative cross-talk between p53 and the glucocorticoid receptor and its role in neuroblastoma cells. EMBO J. 2000;19:6051–6064. doi: 10.1093/emboj/19.22.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Wasylyk B. Ligand-dependent interaction of the glucocorticoid receptor with p53 enhances their degradation by Hdm2. Genes Dev. 2001;15:2367–2380. doi: 10.1101/gad.202201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard KA, Phelps KM, Williams AJ, Thanos D, Glass CK, Rosenfeld MG, Gerritsen ME, Collins T. Nuclear integration of glucocorticoid receptor and nuclear factor-kappaB signaling by CREB-binding protein and steroid receptor coactivator-1. J Biol Chem. 1998;273:29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-kappaB family DNA binding. Nat Immunol. 2011;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman MN, Sternberg EM. Glucocorticoid regulation of inflammation and its functional correlates: from HPA axis to glucocorticoid receptor dysfunction. Ann N Y Acad Sci. 2012;1261:55–63. doi: 10.1111/j.1749-6632.2012.06633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunol Rev. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- Spriggs DR, Sherman ML, Michie H, Arthur KA, Imamura K, Wilmore D, Frei E, 3rd, Kufe DW. Recombinant human tumor necrosis factor administered as a 24-hour intravenous infusion. A phase I and pharmacologic study. J Natl Cancer Inst. 1988;80:1039–1044. doi: 10.1093/jnci/80.13.1039. [DOI] [PubMed] [Google Scholar]
- Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R, Mandard S, Tan NS, Wahli W, Trautwein C, Richardson TA, Lichtenauer-Kaligis E, Kersten S, Muller M. The Interleukin-1 receptor antagonist is a direct target gene of PPARalpha in liver. J Hepatol. 2007;46:869–877. doi: 10.1016/j.jhep.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- Stoecklin E, Wissler M, Moriggl R, Groner B. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol Cell Biol. 1997;17:6708–6716. doi: 10.1128/mcb.17.11.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RH. The complex role of estrogens in inflammation. Endocr Rev. 2007;28:521–574. doi: 10.1210/er.2007-0001. [DOI] [PubMed] [Google Scholar]
- Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tracey KJ, Beutler B, Lowry SF, Merryweather J, Wolpe S, Milsark IW, Hariri RJ, Fahey TJ, 3rd, Zentella A, Albert JD, et al. Shock and tissue injury induced by recombinant human cachectin. Science. 1986;234:470–474. doi: 10.1126/science.3764421. [DOI] [PubMed] [Google Scholar]
- Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandevyver S, Dejager L, Van Bogaert T, Kleyman A, Liu Y, Tuckermann J, Libert C. Glucocorticoid receptor dimerization induces MKP1 to protect against TNF-induced inflammation. J Clin Invest. 2012;122:2130–2140. doi: 10.1172/JCI60006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga T, Czimmerer Z, Nagy L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim Biophys Acta. 2011;1812:1007–1022. doi: 10.1016/j.bbadis.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteclef N, Jakobsson T, Ehrlund A, Damdimopoulos A, Mikkonen L, Ellis E, Nilsson LM, Parini P, Janne OA, Gustafsson JA, et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes Dev. 2010;24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab. 2012;23:351–363. doi: 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dooher JE, Koedood Zhao M, Gilmore TD. Characterization of mouse Trip6: a putative intracellular signaling protein. Gene. 1999;234:403–409. doi: 10.1016/s0378-1119(99)00168-7. [DOI] [PubMed] [Google Scholar]
- Watts AG, Sanchez-Watts G. Interactions between heterotypic stressors and corticosterone reveal integrative mechanisms for controlling corticotropin-releasing hormone gene expression in the rat paraventricular nucleus. J Neurosci. 2002;22:6282–6289. doi: 10.1523/JNEUROSCI.22-14-06282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster JI, Tonelli L, Sternberg EM. Neuroendocrine regulation of immunity. Annu Rev Immunol. 2002;20:125–163. doi: 10.1146/annurev.immunol.20.082401.104914. [DOI] [PubMed] [Google Scholar]
- Wei R, Phillips TM, Sternberg EM. Specific up-regulation of CRH or AVP secretion by acetylcholine or lipopolysaccharide in inflammatory susceptible Lewis rat fetal hypothalamic cells. J Neuroimmunol. 2002;131:31–40. doi: 10.1016/s0165-5728(02)00251-5. [DOI] [PubMed] [Google Scholar]
- Welch JS, Ricote M, Akiyama TE, Gonzalez FJ, Glass CK. PPARgamma and PPARdelta negatively regulate specific subsets of lipopolysaccharide and IFN-gamma target genes in macrophages. Proc Natl Acad Sci U S A. 2003;100:6712–6717. doi: 10.1073/pnas.1031789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink S, van Heerde EC, Schmitz ML, Kalkhoven E, van der Burg B, Baeuerle PA, van der Saag PT. Distinct domains of the RelA NF-kappaB subunit are required for negative cross-talk and direct interaction with the glucocorticoid receptor. J Biol Chem. 1997;272:22278–22284. doi: 10.1074/jbc.272.35.22278. [DOI] [PubMed] [Google Scholar]
- Woloski BM, Smith EM, Meyer WJ, Fuller GM, 3rd, Blalock JE. Corticotropin-releasing activity of monokines. Science. 1985;230:1035–1037. doi: 10.1126/science.2997929. [DOI] [PubMed] [Google Scholar]
- Wu HY, Hamamori Y, Xu J, Chang SC, Saluna T, Chang MF, O’Malley BW, Kedes L. Nuclear hormone receptor coregulator GRIP1 suppresses, whereas SRC1A and p/CIP coactivate, by domain-specific binding of MyoD. J Biol Chem. 2005;280:3129–3137. doi: 10.1074/jbc.M412560200. [DOI] [PubMed] [Google Scholar]
- Yan Q, Carmody RJ, Qu Z, Ruan Q, Jager J, Mullican SE, Lazar MA, Chen YH. Nuclear factor-kappaB binding motifs specify Toll-like receptor-induced gene repression through an inducible repressosome. Proc Natl Acad Sci U S A. 2012;109:14140–14145. doi: 10.1073/pnas.1119842109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- York B, O'Malley BW. Steroid receptor coactivator (SRC) family: masters of systems biology. J Biol Chem. 2010;285:38743–38750. doi: 10.1074/jbc.R110.193367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones S, Hagood JS, Fuentes NL, Fuller GM. STAT3 acts as a co-activator of glucocorticoid receptor signaling. J Biol Chem. 1997;272:30607–30610. doi: 10.1074/jbc.272.49.30607. [DOI] [PubMed] [Google Scholar]
- Zhao MK, Wang Y, Murphy K, Yi J, Beckerle MC, Gilmore TD. LIM domain-containing protein trip6 can act as a coactivator for the v-Rel transcription factor. Gene Expr. 1999;8:207–217. [PMC free article] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]