Abstract
Despite evidence of a substantial genetic component, the genetic factors that underlie longevity in humans remain to be identified. Previous genome-wide linkage and association studies have not found strong evidence for the contribution of common variants besides the APOE gene, suggesting the role of rare variants in human longevity. To discover rare variants that might contribute to longevity, we selected 988 candidate genes and performed a pilot study to identify novel non-synonymous variants in 6 Ashkenazi Jewish centenarians older than 105. Our candidate genes act in pathways implicated in aging and longevity, including neurodegeneration, cognitive function, lipid metabolism, DNA repair, and genome maintenance. By implementing custom-designed Agilent SureSelect target capture and next-generation sequencing, we discovered a total of 89 novel non-synonymous SNPs (nsSNPs) and validated 51 nsSNPs by iPLEX MassArray assays. Genotyping analysis of these novel SNPs in 410 Ashkenazi Jewish controls and 390 centenarians showed significant enrichment (5.3 fold, p=0.02) of the p.Y318C variant in PMS2 and significant depletion (7.5 fold, p=0.04) of the p.V465A variant in GABRR3 in centenarians compared to controls. Our study presents the potential of targeted next-generation sequencing for discovery of rare but functional genetic variation which may lead to exceptional longevity in humans.
Keywords: Centenarian, Human longevity, Functional variant, Candidate genes, Target capture and next-generation sequencing
1. Introduction
Longevity is known to have a genetic component. The heritability of average life expectancy has been estimated to be 15%-25% (Murabito et al., 2012). Family studies on centenarians, those who are 100 years of age or above, suggest that the relationship between genetics and longevity is stronger in the oldest-old adults (Perls et al., 2002; Robine and Allard, 1998). For example, male and female siblings of centenarians have 17 and 8 times, respectively, greater probability of surviving to age 100 than the siblings of people who have an average life expectancy (Perls et al., 2002). Furthermore, offspring of centenarians are healthier with a lower prevalence of age-related diseases such as Alzheimer's disease (AD), cardiovascular disease, cancer, and type 2 diabetes, as compared to age- and gender- matched controls. Therefore, centenarians have been utilized as a model system for genetics studies of human longevity, with extreme age used as a phenotype to discover genetic variations predisposing people to longevity. Genetic studies of centenarians are based on the premise that they may lead to the identification of alleles that are either particularly enriched in these populations due to positive effects on extreme longevity or under-represented due to negative impact on human health (Tazearslan et al., 2012).
Genome-wide analyses have been performed to identify genomic regions linked to longevity followed by identification of the genetic variation causal to the trait. Both the genome-wide linkage studies (GWLS) in family-based designs and genome-wide association studies (GWAS) in population-based designs confirmed the role of APOE alleles in human longevity. These studies suggest that centenarians are likely to harbor rare protective variants with strong effects (Deelen et al., 2011; Passtoors et al., 2012). Considering the frequency of centenarians is only ∼1/5,000 individuals in the population (Andersen et al., 2012), these alleles may not be detectable in general populations. Indeed, a comprehensive candidate approach demonstrated the presence of rare protective alleles enriched in centenarians (Suh et al., 2008; Tazearslan et al., 2011).
Rapid technological advances in next generation sequencing has opened up the door to discover all possible genetic variations in the entire genome, including not only common alleles with small effects typically detected by GWAS, but also rare, causal variants with large effects in population-based studies. Currently whole genome sequencing results from one female and one male supercentenarian have been reported (Sebastiani et al., 2011). The bioinformatics challenge in analysis of whole genome sequencing data and high cost can be overcome by taking a candidate approach for comprehensive sequencing analysis of hundreds to thousands of genes implicated in aging and longevity pathways. Here we used a target capture and next-generation sequencing to discover rare but potentially causal variants in 988 candidate genes in 6 Ashkenazi Jewish centenarians older than 105 years of age. Candidate genes from neurodegeneration/cognitive function, lipid metabolism, DNA damage repair and genome maintenance were selected because they are involved in pathways which have demonstrated roles in aging and longevity.
There is increasing evidence that longevity genes protect against cognitive decline, dementia and neurodegeneration including AD (Barzilai et al., 2006; Christensen et al., 2006; Sanders et al., 2010), consistent with the epidemiologic finding that dementia is absent or markedly delayed amongst centenarians (age of onset > 93 years) (Hitt et al., 1999). For example, a functional longevity allele (p.I405V) in the cholesteryl ester transfer protein (CETP) gene is significantly associated with slower memory decline and lower risk for dementia and AD (Sanders et al., 2010). In addition, centenarians are enriched with the AD-protective APOE ε2 allele, while they are depleted of AD-predisposing APOE ε4 allele (Christensen et al., 2006). Remarkably, a recent GWAS has demonstrated that SNPs associated with longevity are most significantly enriched in genes related to AD, dementia and tauopathies (Sebastiani et al., 2012). Furthermore, genetic variations associated with longevity have been associated with beneficial lipid profiles. The APOE protein functions as a lipid transporter and the APOE ε2 allele was associated with a beneficial lipid profile and lower risk of coronary heart disease (Bennet et al., 2007). Longevity associated variation (p.I405V) in the lipid transfer protein, CETP was associated with larger particle size of LDL and HDL (Barzilai et al., 2003) and lipoprotein particle size was significantly increased among centenarians and their offsprings compared to controls (Barzilai et al., 2003; Vaarhorst et al., 2011). The frequency of homozygotes for the -641C allele in the apolipoprotein-C3 (APOC3) promoter was higher in centenarians and their offspring when compared to controls (Atzmon et al., 2006). Taken together, we selected genes involved in pathways implicated in neurodegeneration, cognitive function, and lipid metabolism as our candidate genes.
Accumulation of DNA damage and mutations has been considered an important causal factor in age-related deterioration and death (Garinis et al., 2008; Hoeijmakers, 2009). This concept is in keeping with the proposed evolutionary basis of aging, i.e., the declining force of natural selection with age, which does not predict maximization of cellular maintenance and repair (Gavrilov and Gavrilova, 2002). DNA damage continuously occurs by both endogenous and exogenous sources such as reactive oxygen species (ROS) produced as byproducts of metabolism, UV light and ionizing radiation. DNA damage is known to cause aging-related cellular degeneration and diseases. The insulin/insulin-like growth factor I (IGF-I) signaling pathway modulates DNA damage and stress response, and is well established as a conserved pathway of aging (Kenyon, 2010; Tazearslan et al., 2012). Mutations in insulin/IGF-1 signaling have been shown to delay aging and extend life span in model organisms (Bartke, 2011). In humans, genetic variations in this pathway are implicated in longevity through genetic association studies (Pawlikowska et al., 2009; Suh et al., 2008; van Heemst et al., 2005). DNA repair pathways counteract the deleterious effects of DNA damage, maintaining genomic stability. There is increasing evidence that genome maintenance is a major longevity assurance pathway (Vijg and Suh, 2005) and defects in this pathway lead to a shorter life span and premature aging in humans and mice (Garinis et al., 2008; Martin, 2005). Thus, for our study we included genes involved in major DNA repair pathways, including nucleotide excision repair, base excision repair, homologous recombination, nonhomologous end joining, mismatch repair, and telomere maintenance, as well as genes involved in regulation of these pathways such as ubiquitylation (Thomson and Guerra-Rebollo, 2010).
From our candidate aging and longevity pathways, we selected a total of 988 candidate genes spanning 5.69 Mb for target capture and next generation sequencing analysis (see Supplementary Table S1 for entire gene list). We report discovery and validation of novel non-synonymous variants discovered in 6 centenarians and further genotyping and association analysis of these variants in 410 controls and 390 centenarians of Ashkenazi Jewish descent.
2. Materials and Methods
2.1. Population and sample collection
In total, 800 human samples were used in this study: 410 Ashkenazi Jewish controls and 390 Ashkenazi Jewish centenarians that were previously collected as part of a longevity study by Dr. Nir Barzilai of the Albert Einstein College of Medicine (Barzilai et al., 2003). All blood samples were rapidly processed to obtain DNA at the General Clinical Research Center of the Albert Einstein College of Medicine. Informed written consent was obtained in accordance with the policy of the Committee on Clinical Investigations of the Albert Einstein College of Medicine. A centenarian is defined as a healthy individual living independently at 95 years of age or older and a control is defined as an individual without a family history of unusual longevity; parents of controls survived to the age of 85 years or less. The control group consisted largely of spouses of the offspring of centenarians. For target capture and next-generation sequencing of our candidate genes, we used genomic DNA extracted from immortalized B-lymphocytes.
2.2. Target selection and generation of customized target capture
For sequencing, we selected 988 genes implicated in the pathways of aging and longevity in humans. The rationale for selection and their criteria is described in the section 3.1 under Results. The complete list of the 988 candidate genes is provided in the Supplementary Table S1. We designed a customized Agilent SureSelect in-solution target capture to enrich for our candidate genes. For each selected gene locus, we included 2 kb upstream region of the transcription start site, all exons, and 20 bp of each exon-intron junction. We used the Agilent eArray (http://earray.chem.agilent.com/earray/) to design and assess coverage across the target genomic regions of bait libraries. Design efficiency from eArray was 99%, calculated as the covered fraction of target bases in the oligo design (6,229,389 bp), out of the total submitted target regions (6,291,136 bp). The final customized target capture contained 5,692,804 bp covering the target regions of the 988 candidate genes.
2.3. Library preparation, target enrichment and sequencing
We isolated DNA from immortalized B-lymphocytes established from 6 centenarians and generated libraries for Illumina sequencing. Each library was indexed with a unique barcode, which allowed for target capture, enrichment of pooled libraries and multiplex sequencing. The library preparation and enrichment methods were performed according to the Agilent Illumina Paired-End Sequencing Platform Library Prep Protocol, with the following modifications: (1) genomic DNA was sheared with Covaris system for 250bp; (2) Illumina adapters were replaced with custom-made, indexed adapters supplied by IDT DNA (see Supplementary Table S2 for adapter sequence information); (3) size-selection and DNA elution were performed by precast E-gel (Invitrogen) Size Select 2% with E-gel precast gel cassette and fragments with desired size (300bp ∼ 350bp) were eluted with 25 μl water; (4) for the pre-capture enrichment PCR, DNA was amplified in 8 cycles; (5) post-enriched DNA libraries were combined in equal amounts to a total quantity of 1 μg; (6) hybridization mixture was incubated for 24 hours at 65°C to minimize evaporation. Target-enriched libraries were sent to Axeq Technologies and sequenced paired-end on the Illumina HiSeq2000 with cluster kit v2 according to the manufacturer's protocol. Reads generated were 101 bp in length.
2.4. Data analysis
Sequencing reads were aligned to the human genome, revision hg19 (Lander et al., 2001), using BWA, version 0.5.9 (Li and Durbin, 2009), with setting “-q 17” provided when running the “aln” command. Following alignment, we utilized Picard [http://picard.sourceforge.net] to detect potential PCR duplicates and to calculate on-target statistics. Subsequent processing was performed using the Genome Analysis Toolkit (GATK) (McKenna et al., 2010). To call variants, we first preprocessed the data, locally realigning the reads around known and suspected indel events, then recalibrating the Illumina base quality scores to more closely reflect actual mismatch rates. We then performed multi-sample genotyping and filtered the results by fitting a Gaussian mixture model to the 7-dimensional point cloud formed by the joint distributions of various statistical annotations calculated for each SNP (DePristo et al., 2011). The model was trained to assign log-odds likelihood probabilities on the basis of sets of known SNPs from public databases, expanding the set retained until a sensitivity threshold of 99% of accessible, known SNPs was reached. Recommended settings were given during all steps, as detailed in the GATK software manual (http://www.broadinstitute.org/gatk/gatkdocs/). SNP databases used were the 1000 Genomes Project (Genomes Project, 2010), dbSNP build 132 (Sherry et al., 2001), and HapMap3 r3 (Altshuler et al., 2010). Functional annotations were provided by ANNOVAR (Liu et al., 2011; Wang et al., 2010). Correlation between SNP calls obtained here and those publicly released was determined with the CompOverlap evaluation module of the VariantEval tool from the GATK.
2.5. Genotyping
To validate novel nsSNPs discovered in 6 centenarians, we designed iPLEX MassArray assays (Sequenom) using the web-based assay design suite program on mysequenom website (http://www.mysequenom.com). From a total of 89 novel nsSNPs, 67 nsSNPs were designed for iPLEX assays in 4 separate multiplexing groups based on the high quality of peaks visualized in the MassARRAY® Typer software after performing MassArray. Using the iPLEX assays, genotyping was performed in 410 controls and 390 centenarians in the Genomics Shared Facility at Albert Einstein College of Medicine.
2.6. Statistical analysis
The association analyses were performed using SNP & Variation suite version 7.6.11 (Golden helix). For a basic allelic test, the genotypes dd, Dd, and DD were resolved into pairs of alleles d and d, D and d, or D and D. Both elements of each subject's genotype are considered to correspond to the same value of the dependent variable. The associations with these individual alleles are then tested. For a genotypic test, the genotypes dd, Dd, and DD were tested without regard to any “order”, allelic count, or allelic pairing they might have. Statistical differences among groups were assessed by the Fisher's exact test, for both allelotype and genotype distribution comparisons. A two-tailed p-value of < 0.05 was considered significant.
3. Results
3.1. Discovery of novel nsSNPs in 988 candidate genes from 6 centenarians
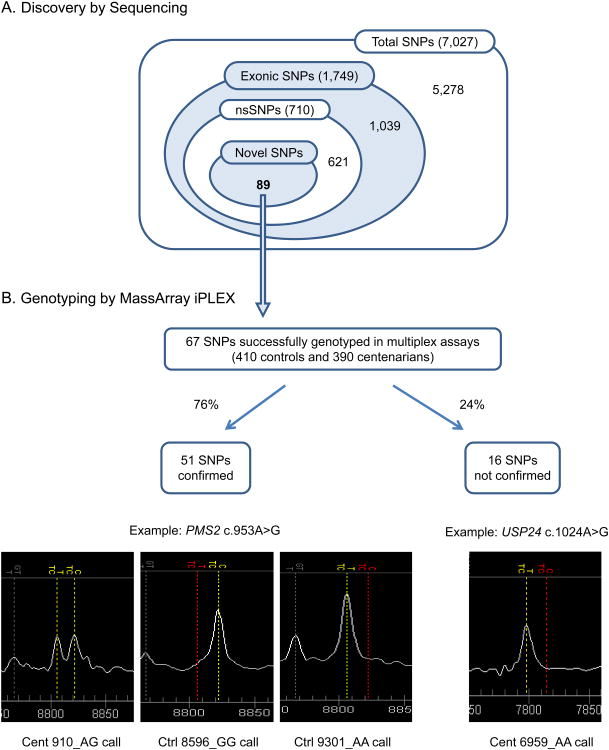
Analysis of the results from target capture and next-generation sequencing of 6 centenarians (Table 1) identified a total of 7,027 genetic variations (excluding variations annotated as ncRNAs) in our candidate genes, among which 1,749 variations were located in exonic regions including 710 nsSNPs. In this study, we focused on novel nsSNPs that have not been discovered in other studies as they may exert strong effects leading to the rare phenotype of exceptional longevity in humans by directly affecting protein structures and/or functions. We identified 89 out of 710 nsSNPs as novel after comparing our sequence results to the dbSNP 132 which contained results from 1000 genomes at the time of analysis (Figure 1A). Since novel nsSNPs are likely rare variants, which are subject to sequencing errors, we first validated if they were true variants and further tested whether these novel nsSNPs were enriched in centenarians as compared to controls by genotyping and association analysis.
Table 1. Demographic information of AJ Centenarian study subjects for sequencing analysis.
| Gender | Age of recruitment |
|---|---|
| M | 105 |
| M | 106 |
| M | 106 |
| F | 107 |
| F | 108 |
| F | 109 |
“M” indicates male, “F” indicates female.
Fig. 1.
Schematic overview of results from sequencing and genotyping analysis. (A) Discovery of SNPs by target capture and next-generation sequencing in 6 AJ centenarians. Total numbers of SNPs discovered in each category are indicated. (B) Genotyping and validation analysis of novel nsSNPs by MassArray iPLEX assays. Genotyping was performed for 410 controls and 390 centenarians. Total numbers of assayable, confirmed, and not confirmed nsSNPs are indicated. c.953A>G in PMS2 and c.1024A>G in USP24 represent specific examples of confirmed nsSNP and not confirmed calls, respectively.
3.2. Validation of novel nsSNPs discovered in 6 centenarians
To validate the 89 novel nsSNPs discovered in our candidate genes from 6 centenarians, we designed iPLEX genotyping assays which allowed us to detect 67 nsSNPs in 4 multiplexed assays (Figure 1A). A total of 51 nsSNPs (76%) were confirmed by genotyping. Sixteen nsSNPs (24%) were considered not confirmed because the genotyping assays could not detect heterozygote calls in the same individuals that the novel nsSNPs were initially discovered and in a larger number of controls (n=410) and centenarians (n=390) (Figure 1B).
3.3. Genotyping and association analysis of novel nsSNPs in 410 controls and 390 centenarians
Using the established iPLEX assays, we performed genotyping analysis in 410 controls and 390 centenarians. The result from allelotypic analysis is shown in Table 2 with alleles ranked according to nominal p-values by Fisher's exact test. We found a total of 13 nsSNPs that were detected only in centenarians but not in controls. These include 11 nsSNPs (c.1469G>T in PCDH10, c.10501A>G in DNAH1, c.768G>A in SHC1, c.484G>A in GRIA3, c.1343T>C in CHRNG, c.41G>C in IL1A, c.991G>A in OPRK1, c.1453C>T in PCDH12, c.1024A>G in USP24, c.634T>G in CHEK2 and c.109A>G in GRIK2) that were detected only in the initial discovery panel of 6 centenarians (minor allele frequency (MAF)=0.1%) and 2 nsSNPs (c.1172C>T in ZMIZ1 and c.118C>T in PLCD4) for which additional centenarians were also found to be carriers. Most of the novel nsSNPs were rare with MAF less than 1%, and thus significance of rare alleles could not be estimated with the current sample size. Notably, MAF of the c.1394T>C in GABRR3 was significantly higher in controls as compared to centenarians (1.25% vs. 0.13%, p=0.01). MAFs of some of the novel nsSNPs reached up to 5%, e.g. c.1021G>A in FOXO3, but none of them showed significant differences in frequencies between controls and centenarians. Genotypic analysis identified two novel nsSNPs (c.953A>G in PMS2 and c.1394T>C in GABRR3) that were significantly different between controls and centenarians (Table 3). Heterozygote genotype frequency for the c.953A>G variant in PMS2 was 0.5% in controls as compared to 2.64% in centenarians (5.3 fold difference, p=0.02), while that of c.1394T>C variant in GABRR3 was higher in controls than in centenarians (2.02% and 0.27%, p=0.04). No other genotypes showed significant differences in frequencies between controls and centenarians.
Table 2. Allelotypic analysis of novel nsSNPs in 410 controls and 390 centenarians.
| Gene | Transcript | Nucleotide changea | Amino acid change | Minor allele count (%)b | p-valuec | |
|---|---|---|---|---|---|---|
|
| ||||||
| Controls | Centenarians | |||||
| GABRR3 | NM_001105580 | c.1394T>C | p.V465A | 10 (1.26) | 1 (0.13) | 0.01* |
| EPHB2 | NM_017449 | c.1106G>A | p.R369Q | 26 (3.23) | 38 (4.97) | 0.10 |
| PMS2 | NM_000535 | c.953A>G | p.Y318C | 4 (0.50) | 10 (1.32) | 0.11 |
| ZMIZ1 | NM_020338 | c.1172C>T | p.P391L | 0 (0.00) | 3 (0.41) | 0.11 |
| GRM1 | NM_001114329 | c.2581G>A | p.G861S | 14 (1.76) | 22 (2.96) | 0.13 |
| RAD52 | NM_34424 | c.1186C>T | p.R396C | 14 (1.76) | 20 (2.70) | 0.23 |
| PLCD4 | NM_032726 | c.118C>T | p.L40F | 0 (0.00) | 2 (0.26) | 0.23 |
| CDC25A | NM_001789 | c.391A>G | p.I131V | 10 (1.25) | 16 (2.12) | 0.24 |
| AFTPH | NM_203437 | c.1796G>A | p.R599Q | 12 (1.48) | 6 (0.79) | 0.24 |
| RELN | NM_173054 | c.9340A>G | p.I3114V | 9 (1.13) | 4 (0.54) | 0.27 |
| PCDH11X | NM_032968 | c.3140G>A | p.G1047E | 5 (0.62) | 9 (1.19) | 0.29 |
| FOXO3 | NM_001455 | c.1021G>A | p.A341T | 44 (5.54) | 32 (4.29) | 0.29 |
| UBE3C | NM_014671 | c.1406G>A | p.R469Q | 1 (0.13) | 3 (0.41) | 0.36 |
| SENP6 | NM_001100409 | c.1447G>C | p.D483H | 1 (0.13) | 3 (0.40) | 0.36 |
| OPRM1 | NM_001008505 | c.1256A>G | p.Y419C | 13 (1.61) | 18 (2.37) | 0.36 |
| ATM | NM_000051 | c.4388T>G | p.F1463C | 21 (2.63) | 26 (3.45) | 0.38 |
| ANAPC5 | NM_016237 | c.2027C>T | p.A676V | 9 (1.13) | 13 (1.75) | 0.39 |
| MLH3 | NM_014381 | c.713A>C | p.Y238S | 11 (1.39) | 14 (1.94) | 0.43 |
| PPIL2 | NM_148175 | c.655G>A | p.E219K | 12 (1.51) | 16 (2.15) | 0.45 |
| FANCI | NM_018193 | c.1963G>A | p.G655R | 10 (1.26) | 6 (0.80) | 0.46 |
| PCDH10 | NM_032961 | c.1469G>T | p.R490L | 0 (0.00) | 1 (0.16) | 0.47 |
| DNAH1 | NM_015512 | c.10501A>G | p.I3501V | 0 (0.00) | 1 (0.14) | 0.48 |
| SHC1 | NM_003029 | c 768G>A | p.M256I | 0 (0.00) | 1 (0.13) | 0.48 |
| GRIA3 | NM_000828 | c.484G>A | p.V162M | 0 (0.00) | 1 (0.13) | 0.48 |
| CHRNG | NM_005199 | c.1343T>C | p.I448T | 0 (0.00) | 1 (0.13) | 0.48 |
| IL1A | NM_000575 | c.41G>C | p.C14S | 0 (0.00) | 1 (0.13) | 0.48 |
| OPRK1 | NM_000912 | c.991G>A | p.A331T | 0 (0.00) | 1 (0.13) | 0.49 |
| PCDH12 | NM_016580 | c.1453C>T | p.H485Y | 0 (0.00) | 1 (0.13) | 0.49 |
| USP24 | NM_015306 | c.1024A>G | p.I342V | 0 (0.00) | 1 (0.13) | 0.49 |
| CHEK2 | NM_001005735 | c.634T>G | p.F212V | 0 (0.00) | 1 (0.13) | 0.49 |
| GRIK2 | NM_175768 | c.109A>G | p.R37G | 0 (0.00) | 1 (0.13) | 0.49 |
| LTF | NM_002343 | c.1841C>T | p.P614L | 16 (2.00) | 19 (2.60) | 0.49 |
| FARP1 | NM_005766 | c.692C>T | p.P231L | 17 (2.09) | 20 (2.64) | 0.51 |
| THBS1 | NM_003246 | c.583G>A | p.A195T | 14 (1.86) | 16 (2.30) | 0.58 |
| POLD1 | NM_002691 | c.2275G>A | p.V759I | 14 (1.77) | 17 (2.28) | 0.59 |
| HERC1 | NM_003922 | c.1130G>T | p.W377L | 1 (0.13) | 2 (0.27) | 0.61 |
| PCDH15 | NM_001142769 | c.4669A>G | p.K1557E | 3 (0.38) | 1 (0.13) | 0.63 |
| POLI | NM 007195 | c.1169C>G | p.P390R | 10 (1.26) | 7 (0.94) | 0.63 |
| MAPK8IP3 | NM_015133 | c.1772G>A | p.R591H | 29 (3.64) | 23 (3.14) | 0.67 |
| NF1 | NM_001042492 | c.7595C>T | p.A2532V | 8 (0.99) | 6 (0.80) | 0.79 |
| HIF1A | NM_181054 | c.2080G>A | p.A694T | 13 (1.63) | 11 (1.48) | 0.84 |
| SIRT2 | NM_012237 | c.347G>A | p.R116H | 29 (3.58) | 26 (3.46) | 1.00 |
| ANAPC7 | NM_001137664 | c.96G>C | p.Q32H | 25 (3.09) | 23 (3.04) | 1.00 |
| ABCA1 | NM_005502 | c.3053A>G | p.D1018G | 8 (0.99) | 8 (1.06) | 1.00 |
| GOLM1 | NM_177937 | c.179G>A | p.R60K | 7 (0.88) | 6 (0.81) | 1.00 |
| MNAT1 | NM_001177963 | c.521T>C | p.I174T | 4 (0.50) | 4 (0.53) | 1.00 |
| MLH3 | NM_014381 | c.3145G>A | p.D1049N | 1 (0.13) | 1 (0.13) | 1.00 |
| POLE | NM_006231 | c.2131T>C | p.S711P | 2 (0.25) | 1 (0.13) | 1.00 |
| RAD23A | NM_005053 | c.434G>A | p.G145D | 1 (0.13) | 1 (0.14) | 1.00 |
| F13A1 | NM_000129 | c.1861G>T | p.A621S | 2 (0.26) | 2 (0.28) | 1.00 |
| FARP1 | NM_005766 | c.2200A>G | p.M734V | 10 (1.25) | 9 (1.23) | 1.00 |
cDNA change,
% minor allele frequency,
Fisher's exact test,
p <0.05.
Table 3. Genotypic analysis of novel nsSNPs in 410 controls and 390 centenarians.
| Gene | Transcript | Nucleotide changea | Amino acid change | Homb count (%)c | Hetd count (%) | p-valuee | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Controls | Centenarians | Controls | Centenarians | |||||
| PMS2 | NM_000535 | c.953A>G | p.Y318C | 1 (0.25) | 0 (0.00) | 2 (0.50) | 10 (2.64) | 0.02* |
| GABRR3 | NM_001105580 | c.1394T>C | p.V465A | 1 (0.25) | 0 (0.00) | 8 (2.02) | 1 (0.27) | 0.04* |
| ZMIZ1 | NM 020338 | c.1172C>T | p.P391L | 0 (0.00) | 0 (0.00) | 0 (0.00) | 3 (0.82) | 0.11 |
| EPHB2 | NM_017449 | c.1106G>A | p.R369Q | 0 (0.00) | 2 (0.52) | 26 (6.45) | 34 (8.90) | 0.14 |
| GRM1 | NM_001114329 | c.2581G>A | p.G861S | 0 (0.00) | 2 (0.54) | 14 (3.52) | 18 (4.85) | 0.23 |
| PLCD4 | NM_032726 | c.118C>T | p.L40F | 0 (0.00) | 0 (0.00) | 0 (0.00) | 2 (0.53) | 0.23 |
| AFTPH | NM_203437 | c.1796G>A | p.R599Q | 0 (0.00) | 0 (0.00) | 12 (2.96) | 6 (1.58) | 0.24 |
| RELN | NM_173054 | c.9340A>G | p.I3114V | 0 (0.00) | 0 (0.00) | 9 (2.26) | 4 (1.08) | 0.27 |
| PCDH11X | NM_032968 | c.3140G>A | p.G1047E | 0 (0.00) | 0 (0.00) | 5 (1.23) | 9 (2.37) | 0.28 |
| RAD52 | NM_134424 | c.1186C>T | p.R396C | 0 (0.00) | 1 (0.27) | 14 (3.53) | 18 (4.85) | 0.32 |
| CDC25A | NM_001789 | c.391A>G | p.I131V | 0 (0.00) | 2 (0.53) | 10 (2.50) | 12 (3.18) | 0.36 |
| UBE3C | NM_014671 | c.1406G>A | p.R469Q | 0 (0.00) | 0 (0.00) | 1 (0.25) | 3 (0.81) | 0.36 |
| OPRM1 | NM_001008505 | c.1256A>G | p.Y419C | 0 (0.00) | 0 (0.00) | 13 (3.23) | 18 (4.75) | 0.36 |
| SENP6 | NM_001100409 | c.1447G>C | p.D483H | 0 (0.00) | 0 (0.00) | 1 (0.25) | 3 (0.79) | 0.36 |
| ANAPC5 | NM_016237 | c.2027C>T | p.A676V | 0 (0.00) | 0 (0.00) | 9 (2.27) | 13 (3.50) | 0.39 |
| ANAPC7 | NM_001137664 | c.96G>C | p.Q32H | 1 (0.25) | 3 (0.79) | 23 (5.68) | 17 (4.50) | 0.41 |
| MLH3 | NM_014381 | c.3145G>A | p.D1049N | 0 (0.00) | 0 (0.00) | 11 (2.77) | 14 (3.88) | 0.42 |
| FANCI | NM_018193 | c.1963G>A | p.G655R | 0 (0.00) | 0 (0.00) | 10 (2.51) | 6 (1.60) | 0.45 |
| FOXO3 | NM_001455 | c.1021G>A | p.A341T | 4 (1.01) | 1 (0.27) | 36 (9.07) | 30 (8.04) | 0.46 |
| PCDH10 | NM_032961 | c.1469G>T | p.R490L | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.33) | 0.47 |
| DNAH1 | NM_015512 | c.10501A>G | p.I3501V | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.27) | 0.48 |
| SHC1 | NM_003029 | c.768G>A | p.M256I | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.27) | 0.48 |
| GRIA3 | NM_000828 | c.484G>A | p.V162M | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.26) | 0.48 |
| CHRNG | NM_005199 | c.1343T>C | p.I448T | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.26) | 0.48 |
| IL1A | NM_000575 | c.41G>C | p.C14S | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.26) | 0.48 |
| OPRK1 | NM_000912 | c.991G>A | p.A331T | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.26) | 0.49 |
| PCDH12 | NM_016580 | c.1453C>T | p.H485Y | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.26) | 0.49 |
| USP24 | NM_015306 | c.1024A>G | p.I342V | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.26) | 0.49 |
| CHEK2 | NM_001005735 | c.634T>G | p.F212V | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.27) | 0.49 |
| GRIK2 | NM_175768 | c.109A>G | p.R37G | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.26) | 0.49 |
| LTF | NM_002343 | c.1841C>T | p.P614L | 0 (0.00) | 0 (0.00) | 16 (3.99) | 19 (5.19) | 0.49 |
| ATM | NM_000051 | c.4388T>G | p.F1463C | 0 (0.00) | 1 (0.27) | 21 (5.26) | 24 (6.37) | 0.49 |
| FARP1 | NM_005766 | c.692C>T | p.P231L | 0 (0.00) | 0 (0.00) | 17 (4.19) | 20 (5.28) | 0.50 |
| PPIL2 | NM_148175 | c.655G>A | p.E219K | 0 (0.00) | 1 (0.27) | 12 (3.02) | 14 (3.76) | 0.55 |
| THBS1 | NM_003246 | c.583G>A | p.A195T | 2 (0.53) | 1 (0.29) | 10 (2.65) | 14 (4.02) | 0.56 |
| POLD1 | NM_002691 | c.2275G>A | p.V759I | 0 (0.00) | 0 (0.00) | 14 (3.54) | 17 (4.57) | 0.58 |
| HERC1 | NM_003922 | c.1130G>T | p.W377L | 0 (0.00) | 0 (0.00) | 1 (0.25) | 2 (0.53) | 0.61 |
| PCDH15 | NM_001142769 | c.4669A>G | p.K1557E | 0 (0.00) | 0 (0.00) | 3 (0.75) | 1 (0.27) | 0.62 |
| POLI | NM_007195 | c.1169C>G | p.P390R | 0 (0.00) | 0 (0.00) | 10 (2.51) | 7 (1.87) | 0.63 |
| MAPK8IP3 | NM_015133 | c.1772G>A | p.R591H | 3 (0.75) | 1 (0.27) | 23 (5.78) | 21 (5.74) | 0.79 |
| NF1 | NM_001042492 | c.7595C>T | p.A2532V | 0 (0.00) | 0 (0.00) | 8 (1.98) | 6 (1.59) | 0.79 |
| HIF1A | NM_181054 | c.2080G>A | p.A694T | 0 (0.00) | 0 (0.00) | 13 (3.26) | 11 (2.96) | 0.84 |
| SIRT2 | NM_012237 | c.347G>A | p.R116H | 2 (0.49) | 1 (0.27) | 25 (6.17) | 24 (6.38) | 1.00 |
| POLE | NM_006231 | c.2131T>C | p.S711P | 0 (0.00) | 0 (0.00) | 2 (0.50) | 1 (0.27) | 1.00 |
| MNAT1 | NM_001177963 | c.521T>C | p.I174T | 0 (0.00) | 0 (0.00) | 4 (0.99) | 4 (1.06) | 1.00 |
| RAD23A | NM_005053 | c.434G>A | p.G145D | 0 (0.00) | 0 (0.00) | 1 (0.26) | 1 (0.27) | 1.00 |
| MLH3 | NM_014381 | c.3145G>A | p.D1049N | 0 (0.00) | 0 (0.00) | 1 (0.25) | 1 (0.26) | 1.00 |
| GOLM1 | NM_177937 | c.179G>A | p.R60K | 0 (0.00) | 0 (0.00) | 7 (1.75) | 6 (1.61) | 1.00 |
| ABCA1 | NM_005502 | c.3053A>G | p.D1018G | 0 (0.00) | 0 (0.00) | 8 (1.98) | 8 (2.12) | 1.00 |
| F13A1 | NM_000129 | c.1861G>T | p.A621S | 0 (0.00) | 0 (0.00) | 2 (0.52) | 2 (0.57) | 1.00 |
| FARP1 | NM_005766 | c.2200A>G | p.M734V | 0 (0.00) | 0 (0.00) | 10 (2.51) | 9 (2.45) | 1.00 |
cDNA change,
Homozygote genotype of minor alleles,
% minor allele frequency,
Heterozygote genotype,
Fisher's exact test,
p <0.05.
4. Discussion
This is a pilot study to explore the hypothesis that individuals with exceptional longevity such as centenarians harbor individually rare but collectively more frequent functional variants in major pathways of aging and longevity as compared to controls (Suh et al., 2008; Tazearslan et al., 2012). We capitalized on recent advances in next-generation sequencing technologies to identify novel, potentially causal variants in 988 candidate genes involved in major pathways of aging and longevity from 6 Ashkenazi Jewish centenarians who were 105 years or older at the time of sample collection. No such scale of a study has been reported thus far in genetics studies of human longevity. In this study, we focused on 89 novel nsSNPs discovered from 6 centenarians as potentially functional rare variants, among which 67 nsSNPs were assayable for genotyping and 51 nsSNPs were validated to be real variants. Our genotyping analysis of these variants in 410 AJ controls and 390 AJ centenarians indicated that indeed most of the novel nsSNPs are rare in frequency (Table 2 and 3). Although we had a limited power to detect significant association of rare variants (MAF < 1.5%) with this sample size, we found that a novel nsSNP in PMS2 (p.Y318C) was significantly enriched in centenarians (5.3 fold difference, p=0.02), while a novel nsSNP in GABRR3 (p.V465A) was significantly depleted in centenarians (7.5 fold difference, p=0.04), as compared to controls (Table 3).
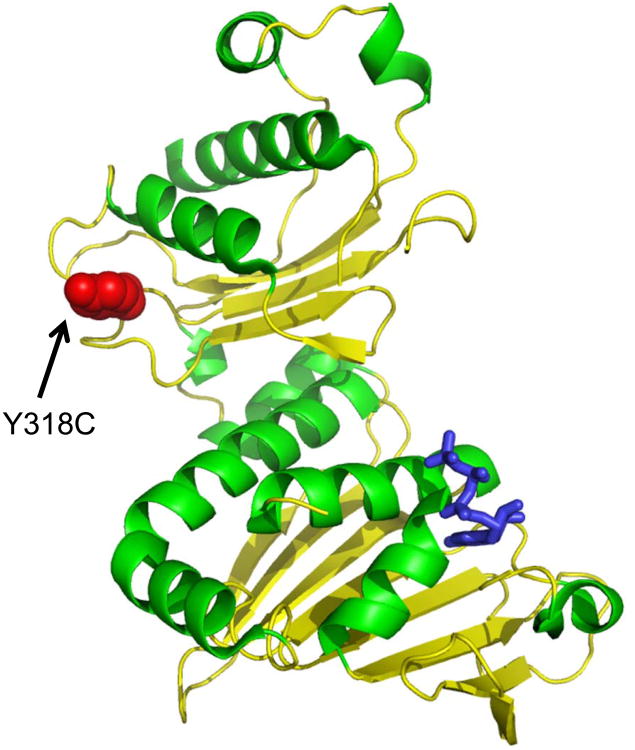
PMS2 is a component of the post-replicative DNA mismatch repair (MMR) system. It heterodimerizes with MLH1 to form MUTL and introduces single-strand breaks near the mismatch and generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. It is also implicated in DNA damage signaling which induces cell cycle arrest and can lead to apoptosis in the case of major DNA damage (Kadyrov et al., 2006; Sacho et al., 2008). It is well reported that defects in this gene are involved in hereditary non-polyposis colorectal cancer type 4 (HNPCC4) (Gill et al., 2005). Mice defective in this gene (Pms2) have about a 100-fold elevated mutation frequency in all tissues, but they did not appear to age more rapidly than normal (Narayanan et al., 1997). However, there are recent reports showing that PMS2 deficiency prolonged the mean lifespan and median survival of telomerase-deficient mice concomitant with rescue of degenerative pathologies (Siegl-Cachedenier et al., 2007). PMS2 amino acid change (p.Y318C) was predicted to be possibly damaging or deleterious according to the in silico tools, SIFT (Kumar et al., 2009) and PolyPhen2 (Adzhubei et al., 2010), which predict the possible impact of an amino acid substitution on the structure or function of a protein. In addition, a protein structural modeling of the PMS2 nsSNP suggests that the side chain of Y318 (Figure 2, red spheres) is exposed and could be mediating interactions with other factors. Interestingly, none of the reported mutations from HNPCC patients is near this region, suggesting that the centenarian specific nsSNP may have a gain of function property beneficial to long lifespan.
Fig. 2.
Protein structural modeling of the PMS2 nsSNP (p.Y318C) associated with longevity. The location of mutated amino acid in the PMS2 is indicated as red spheres in the crystallographic structure of PMS2 (Guarne et al., 2001). Arrows indicate the location of the novel nsSNP identified in centenarians. Blue indicates the bound AMPPNP, marking the ATPase active site.
GABRR3 encodes the rho3 subunit of GABAA-rho subclass receptor which is composed of 5 subunits. The 5 subunits can be made up with any 3 kinds of subunit (rho 1, 2, 3) as a homo or hetero pentamer complex. GABAA-rho receptor is a kind of ionotropic GABA receptor which responds to the inhibitory neurotransmitter, gamma-aminobutyric acid (GABA). About 10% of neurons in the hippocampus are inhibitory and most of them are GABAergic. It was reported that there was an aging related change in GABA receptor composition and that the GABA receptor could be relevant to AD pathogenesis (Rissman and Mobley, 2011). Our discovery on significant depletion of GABRR3 (p.V465A) genotype in centenarians as compared to controls suggests its potential role in human aging and longevity. GABRR3 amino acid change (p.V465A) was predicted to be benign according to the SIFT and PolyPhen2. However, the variation occurs at residue 465 of a 467 amino acid integral membrane protein. Transmembrane-predicting programs suggest that this residue will be right at the end of the last transmembrane helix. It is conceivable that V to A substitution might introduce minor flexibility in this region by leaving a hydrophobic pocket to fill. If the C-terminus interacts with a factor like a PDZ domain, which recognizes C-terminal sequence motifs, it could have an impact on GABBR3 function. Further functional analysis will be needed to uncover molecular mechanisms by which these variants lead to changes in protein function and contribute to longevity.
We also identified 13 novel nsSNPs that were detected only in centenarians but not in controls (Table 2), although none of them reached statistical significance due to the small sample size. In particular, we could detect additional centenarians harboring the p.P391L variant in ZMIZ1 or the p.L40F variant in PLCD4. Both SIFT and PolyPhen2 predicts those two amino acid substitutions to be “damaging” or “deleterious”. Additionally, there were 11 variants that were detected only in the discovery panel of 6 centenarians. Interestingly, all but the p.I342V variant in USP24 were predicted to be functional by the SIFT or PolyPhen2 analysis. These variants could be good candidates for further genotyping analysis in a larger sample size and for functional analysis to assess the relevance of the genetic variations to longevity.
We identified a total of 7,027 genetic variations (excluding variations annotated as ncRNAs) in our 988 candidate genes from 6 centenarians, including 710 nsSNPs. In this study, we followed up on 89 novel nsSNPs (1.3% of total SNPs and 12.5% of total nsSNPs), among which 67 nsSNPs (9.4% of total nsSNPs) were assayed for a validation study and genotyping analysis by iPLEX MassArray method. As expected, these novel nsSNPs were found to be rare in our populations (Table 2). Nevertheless, we could validate 51 novel nsSNPs (76%) to be true positives. We concluded that our customized target capture and next-generation sequencing can reproducibly detect genetic variation in candidate genes, licensing for a large scale genetic discovery study to identify all possible variations including rare variants in a cost-effective manner. Since our targets contain regulatory regions including 2kb proximal promoter, exon-intron junctions, and 5′ and 3′ UTRs, we hope to discover functional regulatory variants (Wray, 2007) of our candidate genes associated with longevity and to investigate the molecular mechanisms underlying the association in future studies.
Supplementary Material
Highlights.
Aim to study the role of rare functional variants in human longevity
Implementation of target capture and next-generation sequencing
Comprehensive analysis of 988 candidate genes for discovery of novel nsSNPs
Identification of GABRR3 and PMS2 as potential longevity-associated genes
Acknowledgments
We would like to thank Genomics Shared Facility at Albert Einstein College of Medicine for their assistance with the iPLEX MassArray assay. This work was funded by NIH grant AG024391, AG027734, and AG17242. YS is the recipient of a Glenn Award for Research in Biological Mechanisms of Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Sebastiani P, Dworkis DA, Feldman L, Perls TT. Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J Gerontol A Biol Sci Med Sci. 2012;67:395–405. doi: 10.1093/gerona/glr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G, Rincon M, Schechter CB, Shuldiner AR, Lipton RB, Bergman A, Barzilai N. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS biology. 2006;4:e113. doi: 10.1371/journal.pbio.0040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A. Single-gene mutations and healthy ageing in mammals. Philos Trans R Soc Lond B Biol Sci. 2011;366:28–34. doi: 10.1098/rstb.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Derby CA, Bauman JM, Lipton RB. A genotype of exceptional longevity is associated with preservation of cognitive function. Neurology. 2006;67:2170–2175. doi: 10.1212/01.wnl.0000249116.50854.65. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, Cheng S, Shuldiner AR. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, Keavney B, Collins R, Wiman B, de Faire U, Danesh J. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van der Breggen R, Suchiman HE, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van Heemst D, de Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov LA, Gavrilova NS. Evolutionary theories of aging and longevity. ScientificWorldJournal. 2002;2:339–356. doi: 10.1100/tsw.2002.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project, C. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Lindor NM, Burgart LJ, Smalley R, Leontovich O, French AJ, Goldberg RM, Sargent DJ, Jass JR, Hopper JL, Jenkins MA, Young J, Barker MA, Walsh MD, Ruszkiewicz AR, Thibodeau SN. Isolated loss of PMS2 expression in colorectal cancers: frequency, patient age, and familial aggregation. Clin Cancer Res. 2005;11:6466–6471. doi: 10.1158/1078-0432.CCR-05-0661. [DOI] [PubMed] [Google Scholar]
- Guarne A, Junop MS, Yang W. Structure and function of the N-terminal 40 kDa fragment of human PMS2: a monomeric GHL ATPase. EMBO J. 2001;20:5521–5531. doi: 10.1093/emboj/20.19.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitt R, Young-Xu Y, Silver M, Perls T. Centenarians: the older you get, the healthier you have been. Lancet. 1999;354:652. doi: 10.1016/S0140-6736(99)01987-X. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM. Genetic engineering of mice to test the oxidative damage theory of aging. Ann N Y Acad Sci. 2005;1055:26–34. doi: 10.1196/annals.1323.005. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murabito JM, Yuan R, Lunetta KL. The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long-lived individuals. J Gerontol A Biol Sci Med Sci. 2012;67:470–479. doi: 10.1093/gerona/gls089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan L, Fritzell JA, Baker SM, Liskay RM, Glazer PM. Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2. Proc Natl Acad Sci U S A. 1997;94:3122–3127. doi: 10.1073/pnas.94.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passtoors WM, Beekman M, Deelen J, van der Breggen R, Maier AB, Guigas B, Derhovanessian E, van Heemst D, de Craen AJ, Gunn DA, Pawelec G, Slagboom PE. Gene expression analysis of mTOR pathway: association with human longevity. Aging Cell. 2012 doi: 10.1111/acel.12015. in press. [DOI] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perls TT, Wilmoth J, Levenson R, Drinkwater M, Cohen M, Bogan H, Joyce E, Brewster S, Kunkel L, Puca A. Life-long sustained mortality advantage of siblings of centenarians. Proc Natl Acad Sci U S A. 2002;99:8442–8447. doi: 10.1073/pnas.122587599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman RA, Mobley WC. Implications for treatment: GABAA receptors in aging, Down syndrome and Alzheimer's disease. J Neurochem. 2011;117:613–622. doi: 10.1111/j.1471-4159.2011.07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine JM, Allard M. The oldest human. Science. 1998;279:1834–1835. doi: 10.1126/science.279.5358.1831h. [DOI] [PubMed] [Google Scholar]
- Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol Cell. 2008;29:112–121. doi: 10.1016/j.molcel.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AE, Wang C, Katz M, Derby CA, Barzilai N, Ozelius L, Lipton RB. Association of a functional polymorphism in the cholesteryl ester transfer protein (CETP) gene with memory decline and incidence of dementia. JAMA. 2010;303:150–158. doi: 10.1001/jama.2009.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Riva A, Montano M, Pham P, Torkamani A, Scherba E, Benson G, Milton JN, Baldwin CT, Andersen S, Schork NJ, Steinberg MH, Perls TT. Whole genome sequences of a male and female supercentenarian, ages greater than 114 years. Front Genet. 2011;2:90. doi: 10.3389/fgene.2011.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Hoh J, Perls TT. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl-Cachedenier I, Munoz P, Flores JM, Klatt P, Blasco MA. Deficient mismatch repair improves organismal fitness and survival of mice with dysfunctional telomeres. Genes Dev. 2007;21:2234–2247. doi: 10.1101/gad.430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci U S A. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazearslan C, Cho M, Suh Y. Discovery of functional gene variants associated with human longevity: opportunities and challenges. J Gerontol A Biol Sci Med Sci. 2012;67:376–383. doi: 10.1093/gerona/glr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazearslan C, Huang J, Barzilai N, Suh Y. Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging Cell. 2011;10:551–554. doi: 10.1111/j.1474-9726.2011.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson TM, Guerra-Rebollo M. Ubiquitin and SUMO signalling in DNA repair. Biochem Soc Trans. 2010;38:116–131. doi: 10.1042/BST0380116. [DOI] [PubMed] [Google Scholar]
- Vaarhorst AA, Beekman M, Suchiman EH, van Heemst D, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE, Heijmans BT. Lipid metabolism in long-lived families: the Leiden Longevity Study. Age (Dordr) 2011;33:219–227. doi: 10.1007/s11357-010-9172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D, Beekman M, Mooijaart SP, Heijmans BT, Brandt BW, Zwaan BJ, Slagboom PE, Westendorp RG. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4:79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- Vijg J, Suh Y. Genetics of longevity and aging. Annu Rev Med. 2005;56:193–212. doi: 10.1146/annurev.med.56.082103.104617. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.