Abstract
Maturation of the nickel-containing urease of Klebsiella aerogenes is facilitated by the UreD, UreF, and UreG accessory proteins along with the UreE metallo-chaperone. A fusion of the maltose binding protein and UreD (MBP-UreD) was co-isolated with UreF and UreG in a soluble complex possessing a (MBP-UreD:UreF:UreG)2 quaternary structure. Within this complex a UreF:UreF interaction was identified by chemical cross-linking of the amino termini of its two UreF protomers, as shown by mass spectrometry of tryptic peptides. A pre-activation complex was formed by the interaction of (MBP-UreD:UreF:UreG)2 and urease. Mass spectrometry of intact protein species revealed a pathway for synthesis of the urease pre-activation complex in which individual hetero-trimer units of the (MBP-UreD:UreF:UreG)2 complex bind to urease. Together, these data provide important new insights into the structures of protein complexes associated with urease activation.
Introduction
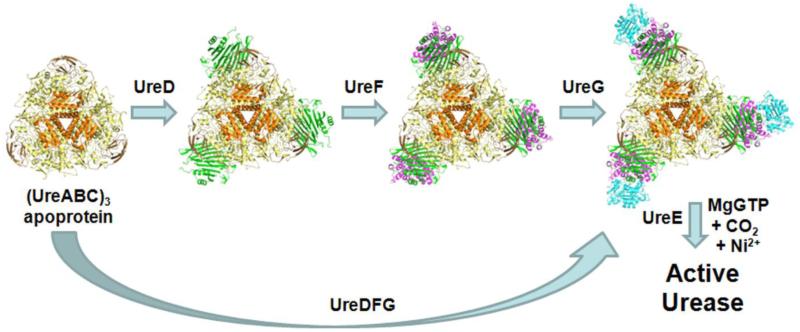
Urease, an enzyme that hydrolyzes urea to form ammonia and carbonic acid [1, 2], plays important roles in nitrogen cycling and pathogenesis [3, 4]. Like most bacterial ureases, the Klebsiella aerogenes protein contains three subunits (UreA, UreB, and UreC) arranged in a (UreABC)3 architecture with a dinuclear nickel active site in each of its UreC subunits [5]. The enzyme is synthesized by an elaborate biosynthetic pathway that requires four accessory proteins (UreD, UreE, UreF, and UreG) along with carbon dioxide, GTP, and the metal ions [1, 2, 6]. A working hypothesis for K. aerogenes urease activation (Figure 1) shows the (UreABC)3 apoprotein [7] forming sequential complexes with UreD [8], UreF [9], and UreG [10] or binding a pre-formed complex of UreD, UreF, and UreG (UreDFG) [11]. The resulting pre-activation urease complex carbamylates an active site lysine side chain to form a metal-binding ligand [12] and acquires nickel ions from the UreE metallo-chaperone [13] in a process driven by GTP hydrolysis [14]. Completion of this process is accompanied by dissociation of the accessory proteins.
Figure 1.
Working model of urease activation. The urease apoprotein (orange UreA, sand UreB, and yellow UreC) sequentially binds UreD (green), UreF (magenta), and UreG (cyan) or binds a complex of these three accessory proteins to form a pre-activation complex. Synthesis of active enzyme requires carbamylation of an active site lysine by CO2, GTP hydrolysis by UreG, and Ni2+ transfer from the UreE metallochaperone. Structures of the docked complexes are derived from computational models [47].
The protein complexes in the urease activation pathway have been studied to varying extents, but many questions about their structures and properties remain to be answered. Chemical cross-linking of (UreABC)3:UreD followed by proteolysis and matrix-assisted laser-desorption ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS) indicates that UreD binds to UreB and UreC [15]. Small angle x-ray scattering (SAXS) data from this species are consistent with the UreD density being located at the vertices of the triangular urease apoprotein [16]. Corresponding cross-linking/proteolysis/MALDI-TOF-MS and SAXS analyses of the (UreABC)3:UreD:UreF species show a UreF linkage to UreB and yield similar positional results. Moreover, cross-linking and computational flexibility studies provide evidence that UreB is loosely associated with UreC in this complex and can move in a hinge-like motion that potentially enhances access to the active site [15, 16]. (UreABC)3:UreD, (UreABC)3:UreD:UreF, and (UreABC)3:UreDFG exhibit multiple bands on native gels, and analysis of these bands on denaturing gels reveals peptide staining intensities consistent with varied numbers of UreD, UreF, and UreG bound to the (UreABC)3 apoprotein. The structure of K. aerogenes urease apoprotein is known [17], but no urease:accessory protein complexes have been structurally elucidated. By contrast, the urease-free structures are known for Helicobacter pylori UreF [18] and the complex of UreF with UreH (the UreD homologue in H. pylori) [19], while UreG homology models have been generated by using the related HypB dimeric structure from Methanocaldococcus jannaschii [20].
Whereas the sequential protein binding urease activation model (Figure 1, top) depicts individual protomers of the accessory proteins binding to the urease apoprotein vertices, evidence from homologous systems suggests that the pre-formed accessory protein complex may be dimeric. For example, H. pylori UreF and UreH:UreF each possess an overall dimeric structure (i.e., (UreF)2, (UreH:UreF)2) [18, 19]. Furthermore, UreG is dimeric as purified from some microorganisms [21, 22], although the K. aerogenes protein is monomeric [11, 23]. It is plausible that distinct quaternary structures of the urease accessory proteins are found in K. aerogenes and H. pylori. Alternatively, it is possible that K. aerogenes also produces a dimeric accessory protein complex, but it dissociates to the monomeric form when it binds to urease. In addition, it is feasible that the complexes obtained by sequential binding of accessory proteins are distinct from those generated by binding of UreDFG. To resolve these questions, we utilize chemical cross-linking/proteolysis/MS methods to characterize a soluble UreDFG complex from K. aerogenes, we demonstrate that the accessory protein complex binds to urease, and we use MS tuned to retain fragile non-covalent contacts to examine the intact structures of various urease complexes. Our results provide substantial support for the protein complex assembly model (Figure 1 lower path) and present new insights into the stability of various urease complexes.
Experimental
Preparation of Protein Samples
Urease holoenzyme was produced in E. coli BL21-Gold(DE3) cells containing pKK17 (the pKK223-3 vector with the complete ureDABCEFG gene cluster) that were grown in lysogeny broth (LB) with 300 μg ml−1 ampicillin plus 1 mM NiCl2 and induced at an optical density (600 nm) of ~0.4 with 0.1 mM isopropyl β-thiogalactopyranoside. The protein was purified as previously described [24]. The (UreABC)3 apoprotein was purified in the same manner from the same E. coli cells that were grown in LB medium without added NiCl2.
The Strep-tag II version of (UreABC)3:UreDFG was formed in cells transformed with pKKG containing ureDABCEFG where ureG is fused to the codons for Strep-tag II [25]. This complex was purified by use of a Strep-tactin column followed by Superdex 200 gel filtration chromatography, as previously described [25].
A hetero-trimeric MBP-UreDFG species, containing a maltose binding protein (MBP) fusion of UreD (MBP-UreD) along with UreF and UreG, was obtained from E. coli BL21-Gold(DE3) cells co-expressing pEC005 (with ureFG cloned into pACT3) and pEC002 (with ureD cloned into pASK-IBA3plus), as previously described [24, 26]. The complex was isolated by sequential amylose, DEAE-Sepharose, and Superdex 200 column chromatography of cell-free extracts. A version of this species containing the K165A variant of UreF was purified by similar approaches using cells with pEC005-UreF-K165A (a modified pEC005) and pEC002 [25]. MBP-UreDFG or its K165A UreF version was mixed with excess urease apoprotein or holoenzyme, incubated 60 min, and separated by chromatography on amylose resin to remove excess urease.
General Properties of MBP-UreDFG
The native molecular mass of MBP-UreDFG was assessed by Superdex 200 gel filtration chromatography in 20 mM Tris-HCl buffer (pH 7.5) containing 1 mM EDTA, 1 mM β-mercaptoethanol, and 25 mM NaCl and using thyroglobulin, γ-globulin, ovalbumin, and myoglobin (Bio-Rad) as molecular mass markers. The apparent molecular masses of denatured components in various samples were obtained by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12% running gels) [27], followed by staining with Coomassie brilliant blue. Integration of band intensities was assessed by gel scanning (Alpha Innotech Corp., using AlphaEase FC software).
Chemical Cross-Linking, Proteolysis, and MALDI-TOF-MS of MBP-UreDFG
MBP-UreDFG and its K165A UreF derivative were further characterized by a chemical cross-linking approach. The samples (3.75 μM of the heterotrimer in 20 μl) were incubated with 160-fold excess (600 μM) BS3 (Thermo Fisher Scientific) in 25 mM HEPES (4(-2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4) buffer containing 1 mM EDTA, 1 mM dithiothreitol and 1% glycerol for one h, then quenched with 15 μl of 1 M glycine.
Untreated and BS3 treated samples were subjected to SDS-PAGE (12% gel) and the protein bands of interest were excised with a clean razor blade and chopped into ~1-mm3 pieces. The gel fragments were washed with 100 μl of 100 mM NH4HCO3 buffer (pH 8.4) for 5 min, dehydrated at room temperature in 50 μl of 100% acetonitrile, and rehydrated in 50 μl of 10 mM dithiothreitol in 100 mM NH4HCO3 at 56°C for one h. Dehydration and rehydration was repeated, with final rehydration in 50 μl of 55 mM iodoacetamide in 100 mM NH4HCO3. The mixtures were incubated at room temperature and in the dark for one h. Samples were repeatedly washed with 50 μl of 100 mM NH4HCO3, dehydrated, rehydrated in 20 μl 50 mM NH4HCO3 containing 15 ng/μl of sequencing grade trypsin (Promega), and incubated overnight at 37°C. Supernatants from the trypsin digest were saved, and the gel pieces were sonicated three times with 50 μl of 60% acetonitrile/1% trifluoroacetic acid in an ice water bath for 5 min followed by centrifugation. The solutions were pooled with the initial supernatant, dried to ~2 μl in a Speed Vac, and dissolved in 20 μl of 2% acetonitrile/0.1% trifluoroacetic acid. The final samples were desalted using C-18 pipette tip columns (Agilent Technologies).
MALDI-TOF MS was performed by using a Shimadzu Axima plus instrument equipped with a nitrogen laser operating at 337 nm. Samples (5 μl) were mixed with 5 μl of a saturated α-cyano-4-hydroxycinnamic acid solution (Sigma) in 50% acetonitrile containing 0.1% trifluoroacetic acid. Aliquots (1 μl) were spotted on a MALDI plate and allowed to dry in a hood. Spectra were obtained in linear positive mode and externally calibrated with bradykinin fragment (1-7, RPPGFSP, 756.86 Da), angiotensin I (1296.48 Da), and human ß-chain insulin (3495.95 Da). The mass values at the centroid peak positions are reported, i.e. the average value across the isotopic peaks.
MS Analysis of Intact Protein Complexes
Samples containing UreABC, MBP-UreDFG and its K165A UreF version, UreABC:MBP-UreDFG and its K165A UreF version, and UreABC:UreDFG (where UreG is fused to Strep-tag II) with protein concentrations of ~0.46 mg/mL were buffer exchanged into 200 mM ammonium acetate buffer using Micro Bio-Spin 6 columns (Bio-Rad, Hercules, CA) just prior to MS analysis. A quadrupole-ion mobility (IM)-TOF MS instrument (Synapt G2 HDMS; Waters, Milford, MA) was used to collect the intact protein complex data. Protein ions were typically generated by nano-electrospray ionization (nESI) using a capillary voltage of 1.5-1.7 kV. Ions traveled through a mass-selective quadrupole and entered an ion trap constructed from a traveling wave ion guide, pressurized at 3×10−2 mbar of argon. Ions were then released from the trap cell into the IM separator pressurized with 2.7 mbar of N2. Typical settings for the traveling waves within the IM drift region were 150 m/s wave velocity and a wave height of 15 V [28]. Ions underwent m/z separation in a TOF mass analyzer, which was maintained at a vacuum of 1.7×10−6 mbar, and operated using a mass range of 500 to 25,000 m/z. All other instrument parameters were tuned to preserve fragile non-covalent protein interactions [29-31].
Tandem MS experiments were carried out to increase the confidence of the protein stoichiometries and compositions assigned from intact mass alone. Ions of interest were generated in the same fashion as above and then a single m/z was selected in the quadrupole mass filter. Charge states selected for such dissociation experiments varied from complex to complex, but typically those signals that presented the least overlap with other protein complex ions having similar m/z and those with the greatest intensity values were chosen for tandem MS. Following isolation, ions were given excess kinetic energy upon accelerating into the ion trap region prior to the IM drift cell by increasing the acceleration voltage to 200V. These data are presented with minimal smoothing and no background subtraction.
Results
Characterization of the MBP-UreDFG Complex
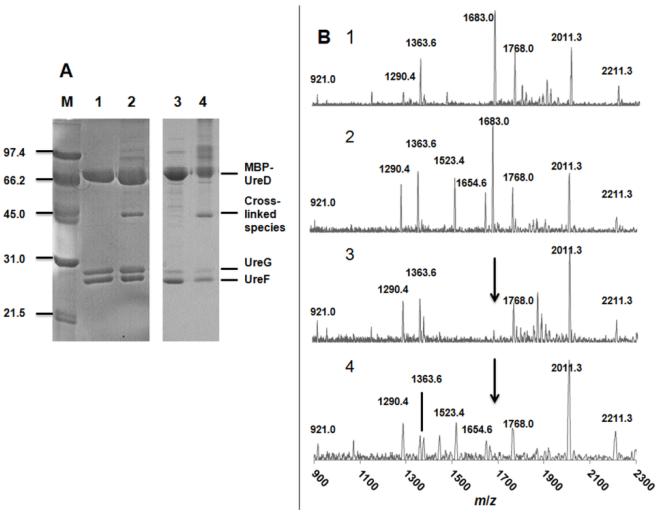
Of the protein complexes illustrated in Figure 1, the least well studied is UreDFG due to its insolubility [11]. To overcome this hurdle, the maltose binding protein (MBP) fusion with UreD (MBP-UreD, known to be functional for urease activation in the cell) was used to obtain a soluble hetero-trimeric complex abbreviated here as MBP-UreDFG [24, 26]. As an initial step in characterizing this species, the sample was digested with thrombin to remove the MBP tag; however, the resulting species was unstable, so further investigations focused on the properties of the intact MBP-UreDFG complex. SDS-PAGE analysis of purified sample (Figure 2A, lane 1) revealed the expected three bands (72,892, 25,222, and 21,944 Da, respectively, according to their sequences; note that UreG migrates anomalously during SDS-PAGE and is positioned above UreF), and division of the integrated band intensities by their relative masses indicated the components were present at nearly equivalent ratios. Gel filtration analysis provided an apparent relative mass of ~275 kDa (data not shown), consistent with a (MBP-UreDFG)2 quaternary structure (predicted sequence mass of 240,116 Da). Furthermore, there was no evidence of an equilibrium involving a single heterotrimeric species or of dissociation into the individual protein subunits.
Figure 2.
MALDI-TOF MS analysis of chemically cross-linked MBP-UreDFG. (A) SDS-PAGE analysis of samples. Lanes: M, molecular mass markers (in kDa); 1, MBP-UreDFG (19.2 μg); 2, MBP-UreDFG (19.2 μg) treated with BS3 (0.9 μg); 3, MBP-UreD:UreF(K165A):UreG (9.0 μg); 4, MBP-UreD:UreF(K165A):UreG (9.0 μg) treated with BS3 (6.9 μg). Note that UreG migrates anomalously slowly for its known mass. (B) MALDI-TOF MS of in-gel trypsin digests of bands from panel A. Spectra: 1, UreF; 2, ~45 kDa band from cross-linked MBP-UreDFG; 3, UreF(K165A); 4, ~45 kDa band of cross-linked MBP-UreD:UreF(K165A):UreG. Theoretical digest fragments of UreF include m/z 921.0 (residues 214-220), 1290.4 (70-81, carbamidomethylated), 1363.6 (181-191, carbamidomethylated), 1683.0 (166-180, absent in samples for K165A UreF), 1768.0 (109-122, carbamidomethylated), and 2010.3 (192-213). The fragment at m/z 2211.3 is from trypsin. Cross-links of UreF peptides 2-7 plus 2-7 and peptides 2-7 plus 1-7 have predicted m/z 1523.4 and 1654.6, respectively.
To further characterize the (MBP-UreDFG)2 complex, purified sample was subjected to chemical cross-linking with the amine-specific reagent bis(sulfosuccinimidyl)suberate (BS3) followed by trypsin proteolysis and MALDI-TOF-MS. SDS-PAGE analysis of the complex treated with BS3 demonstrated the formation of a species (~44 kDa) that was not present in the untreated sample (Figure 2A, lane 2). A mixture of additional less intense bands corresponding to intact protein masses greater than MBP-UreD were detected, but not further characterized. The apparent molecular mass of the ~44-kDa species could be explained by UreF-UreF, UreF-UreG, or UreG-UreG cross-links. Digestion of this band with trypsin and MALDI-TOF-MS analysis of the resulting peptides identified the species as arising solely from UreF, demonstrating a UreF-UreF cross-link. The MALDI-TOF-MS spectrum of tryptic peptides derived from untreated UreF was compared to that for BS3-treated sample (Figure 2B, spectra 1 and 2). Both samples contained a fragment corresponding to residues 166-180 (m/z = 1683; LVPFGQQAAQQLILR) arising from cleavage after K165, the only lysine in this protein; thus, cross-linking did not involve this residue. The BS3-treated sample gave rise to two new tryptic peptides that were both consistent with cross-linking that involves the amino-termini (with partial removal of the initial Met); a peak near m/z of 1523 corresponded to two peptides containing residues 2-7 (m/z = 692; STAEQR) and the linker region (m/z = 140) while that at m/z of 1655 linked residues 1-7 (m/z = 823) to residues 2-7. Confirmation of the amino-terminal linkages was obtained by similar studies with a variant complex containing K165A UreF, thus lacking free lysine residues. A ~44 kDa band derived from BS3 cross-linking again was observed by SDS-PAGE (Figure 2A, lanes 3 and 4). MALDI-TOF-MS analysis (Figure 2B, spectra 3 and 4) again revealed the presence of fragments with m/z of 1523 and 1655, but a fragment with m/z of 1683 was absent, consistent with a lack of proteolytic cleavage after the altered residue. These results demonstrate a close juxtaposition of the amino termini of two UreF peptides in (MBP-UreDFG)2.
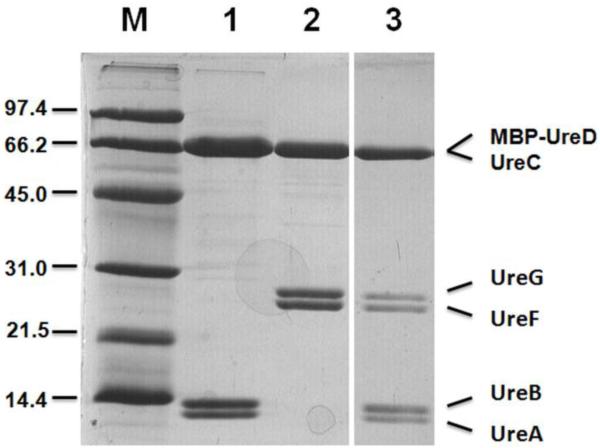
The addition of excess urease holoenzyme or apoprotein to (MBP-UreDFG)2 or its UreF(K165A) derivative led to the formation of pre-activation complexes that were separated from free urease by amylose affinity chromatography (Figure 3). This procedure allowed for further structural studies of the pre-activation complex.
Figure 3.
Interaction of MBP-UreDFG with urease. Lanes: M, molecular mass markers (in kDa); 1, Urease apoprotein (8.8 μg); 2, MBP-UreDFG (4.7 μg); 3, 1/20th of the elution fraction (10 mM maltose in 20 mM Tris buffer, pH 7.8, containing 85 mM NaCl and 1 mM EDTA) from an amylose resin after applying a mixture of urease apoprotein and MBP-UreDFG (207 μg and 41.6 μg, respectively; incubated 1 h, in the same buffer), washing with maltose-free buffer, and eluting with buffer containing maltose.
MS Analysis of Intact Urease Complexes
To examine the composition and connectivity of protein complexes along the urease activation pathway in more detail, MS measurements were conducted on numerous urease-related samples under conditions designed to retain fragile, non-covalent interactions in the gas-phase [30, 32-37]. Through the proper tuning of the nESI process, as well as the correct managing of both pressure and focusing potentials within the instrument, MS enables the measurement of heterogeneous multiprotein assemblies at low micromolar concentrations with unprecedented precision and accuracy [38]. Errors in this mass measurement arise primarily from buffer material that adheres to the surface of protein ions created by gentle nESI, resulting in expected positive deviations between experimentally determined and expected molecular masses of <3% [33, 39]. Once sufficient signal intensity is achieved, tandem MS approaches, involving collision induced dissociation (CID), can be employed on multiprotein complex ions [40-42]. The fragment ions produced typically take the form of highly-charged monomeric subunits and charge-reduced assemblies stripped of the aforementioned monomers [43]. If these data are combined with ion mobility (IM) spectrometry, which enables the separation of protein complex ions based on their orientationally averaged size and shape, then closely-related assemblies can be individually analyzed and identified [29, 44, 45]. While the data shown here were acquired in an IM-MS mode, and in many cases the IM data were used to guide our MS analysis and differentiate overlapping charge state distributions resulting from assemblies of similar intact mass, only the resulting MS data and its analysis are discussed here. Limited IM-MS data comparing the stability differences observed between holo-enzyme and apo-protein complexes are available in Supplementary Material, with complexes containing holo-enzymes demonstrating superior stability in our measurements (Figure S1). Also illustrated in the Supplementary Material are example IM-MS data (Figure S2) enabling a more-detailed analysis of our tandem MS datasets.
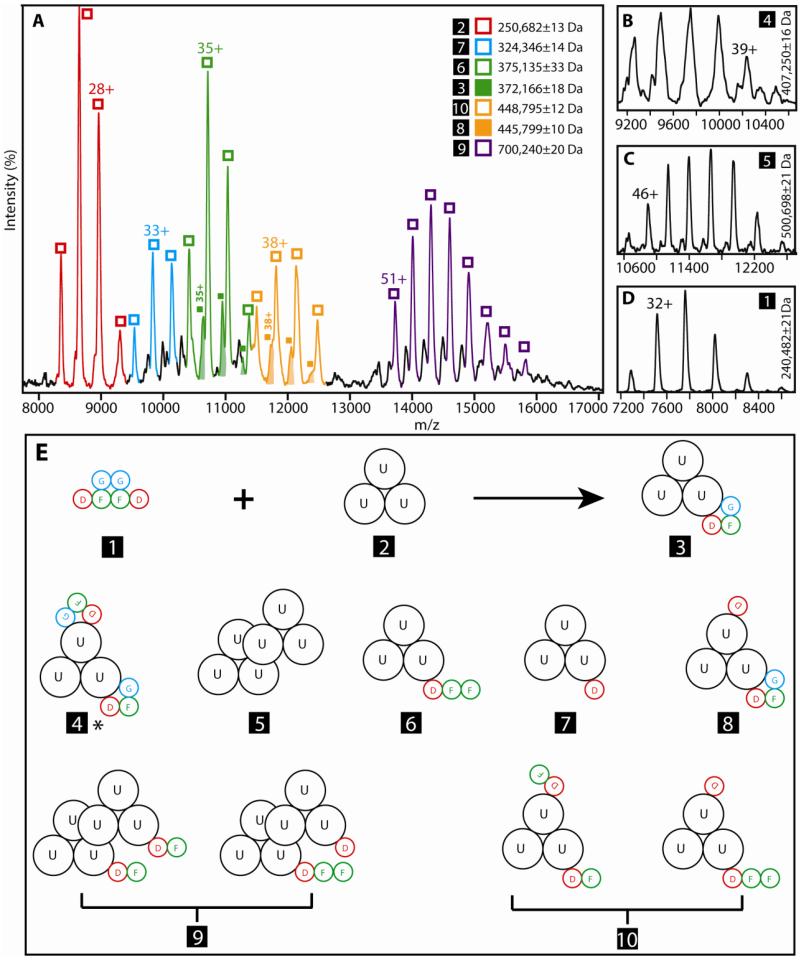
To test if the accessory protein complex binds to urease as the dimeric species or whether one heterotrimer binds while the other dissociates, these components were incubated under conditions where urease was in three-fold excess and the products were analyzed by MS (Figures 4A). Figure 4E serves as a framework for describing the results. MBP-UreDFG (species No. 1) in a 200 mM ammonium acetate solution yielded the well-resolved charge state series in the mass spectrum of Figure 4D. This species had an intact mass corresponding to the expected value for (MBP-UreDFG)2 (240,482 ±21 Da measured and 239,856 Da expected). No indication of a single heterotrimer, free UreF, or UreF:UreF dimer was detected (see also Figure S3). The observation of this overall dimeric species corroborates the dimerization suggested by our gel filtration analysis and the MALDI-MS data that had indicated UreF-UreF cross-linking (Figure 2). Significantly, individual heterotrimeric units of this dimer bind to the core urease assembly (UreA:UreB:UreC)3 (Figure 4A, No. 2) to create a (UreABC)3:MBP-UreDFG assembly (Figure 4A, No. 3). A hypothetical pathway for synthesis of this species is illustrated in Figure S4.
Figure 4.
Intact complexes of UreDFG and urease as probed through ESI-MS. (A), (B), (C) and (D) are mass spectra recorded for various stabilized urease complexes discussed in the text. (E) Topological diagrams of the complex stoichiometries identified by ESI-MS, where structures are indicated by numbers 1-10. MBP-UreD, UreF, and UreG are abbreviated as D, F, and G, except for complex No. 4 which was isolated directly from cells, lacks MBP-tagged UreD, and has UreF containing Strep-tag II (labeled 4*). Depicted topologies are congruent with the experimental observations and other structural information known for these assemblies. High confidence data from this work strongly indicates that complexes 1 and 2 form primarily complex 3 in vitro. Additional protein complexes are also observed, but likely result from the dissociation of higher-order complexes, and not from intermediates in the assembly of urease activation complexes. Complexes No. 9 and No. 10 each have at least two possible topologies that match the ESI-MS data (as shown).
Figure 4A also provides evidence of other protein complexes that presumably represent incompletely synthesized, partially decomposed, or aggregated assemblies. These include (but are not limited to): the (UreABC)3 central complex bound to MBP-UreD:UreF:UreF (No. 6), to one copy of MBP-UreD (No. 7), or to two copies of MBP-UreD:UreF (No. 10, alternatively depicted as one copy of MBP-UreD and one copy of MBP-UreD:UreF:UreF), and complex No. 3 with an additional MBP-UreD (No. 8). For a different sample purified directly from the cells (rather than generated by mixing purified proteins in solution) we detect a complex with two copies of UreDFG (lacking MBP and with UreG fused to Strep-tag II) bound to urease (No. 4*, Fig 4B). The addition of a second (UreDFG)2 to the complex marked as No. 3, indicated as a second transient species in brackets in Figure S4, is likely needed to satisfy the formation of several of these assemblies. Alternatively, complexes No. 7 and 8, as well as some possible topologies of complexes No. 9 and 10 can be formed through the binding of excess MBP-UreD to smaller assemblies. MS data reveal additional complexes which result from the oligomerization of (UreABC)3 to form ((UreABC)3)2 (Figure 4A, No. 5) and that same complex bound to two copies of MBP-UreD:UreF (No. 9, alternatively depicted as one MBP-UreD and one MBP-UreD:UreF:UreF). Due to the low working concentration of the samples used in our MS analysis of these intact complexes (<10 μM), it is unlikely that these larger oligomers can be described as artifacts of the nESI process.
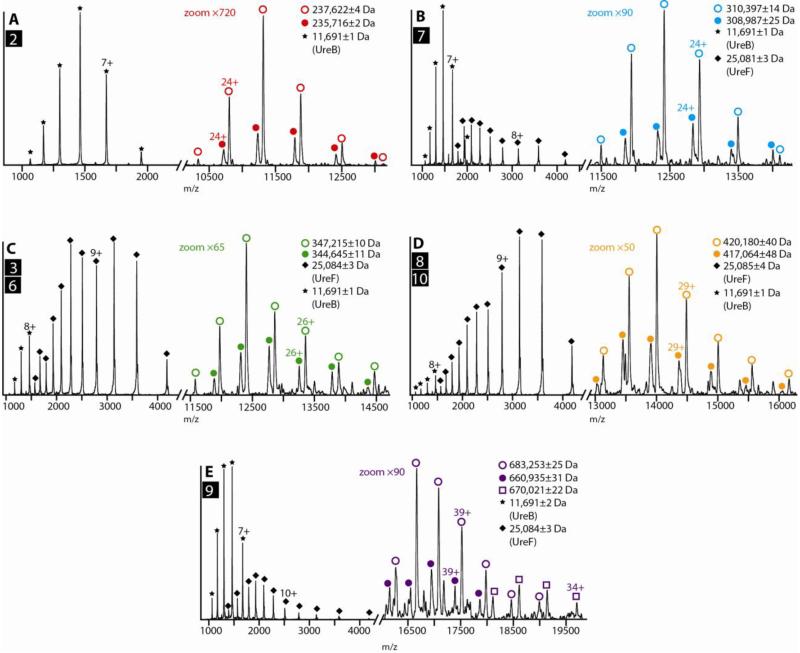
All of the identifications discussed above are supported with tandem MS data, where CID generates fragment ions that enable the high-confidence assignment of the protein complex composition and stoichiometry. Figure 5 shows selected tandem MS spectra from our dataset, using the same numerical identifiers as used in Figure 4 to refer to the assemblies originally isolated for gas-phase dissociation. In all cases shown, multiple stripped protein populations are observed and rationalized based on the monomer components known to exist within each sample and the intact mass of the isolated protein complex ion (Table S1 in Supplementary Material). In the cases of Figures 5B, C, D and E, for example, two overlapping protein complex ion populations are co-isolated in each dataset due to their similar intact m/z values (Figure 4A), and produce fragment ion populations that allow for their discrete identification. In contrast, fragment ions recorded in Figure 5A correspond to complex No. 2 and a previously undetected putative truncated form present at low relative abundance. We also observe signals in our tandem MS data corresponding to multiple dissociated monomers, dominated by signals from UreB and UreF at low m/z in Figures 5B, C, D and E. For example, the ejection energies for UreF and UreB from complex No. 9 are similar, resulting in two prominent stripped protein ion populations (indicated by open purple circles and squares) in Figure 5E. In contrast, UreF and UreB are ejected at significantly different collision energies for complex No. 3 versus No. 6 or complex No. 8 versus No. 10, thus aiding our interpretation of the recorded CID patterns (Figure 5C and D).
Figure 5.
Protein complex identification through precise mass measurements enabled by tandem MS. Tandem MS CID spectra recorded for the urease-related species that appear in Figure 4B, acquired at a trap collision voltage of 200 V (A, B, C, D and E). CID spectra are typically dominated by signals for ejected UreB and UreF monomers at low m/z, indicated by star and diamond notations respectively, while the signals corresponding to stripped protein complex ions at high m/z are magnified (magnification factors indicated on each spectrum) and marked by color-coded circles. Detailed annotations are given in Supplementary Material as Table S1.
Surprisingly, Figure 5B contains signals corresponding to both UreB and UreF, when the complex we intended to isolate (No. 7) contains only the former as a constituent. Using IM-MS data to guide our interpretation, however, we discovered that the 33+ charge state of complex No. 7 and the 34+ charge state of (UreABC)3:MBP-UreD:UreF lacking one UreB subunit coexist in our dataset and possess m/z values that cannot be distinguished by our quadruple MS (Figure S2). The latter complex contains UreF, and thus accounts for the appearance of those signals in Figure 5B.
Overall, the specific datasets shown clearly assign the stoichiometries and compositions of the complexes indicated in Figure 4E. By summing the fragment ion population masses observed in Figure 5 we obtain intact complex masses that closely agree with the sequence masses for the protein constituents indicated (see Table S1). Nevertheless, protein complex topology is not established in all cases. For example, in addition to the two versions shown for species No. 9 one could alternatively position both MBP-UreD:UreF units (or the MBP-UreD and MPB-UreD:UreF:UreF units) on the same urease trimer; however, a urease:MBP-UreD:UreF:UreF:MBP-UreD:urease arrangement is excluded by the CID results. Similarly, two versions are shown for species No. 10, whereas the CID data preclude a single MBP-UreD:UreF:UreF:MBP-UreD unit bound to the urease. In other cases where many possible protein complex topologies occupy the same molecular mass, results from multiple samples under different conditions inform our assignments. A hypothetical assembly pathway that is most consistent with the MS data is depicted in Figure S4.
Discussion
The soluble (MBP-UreDFG)2 complex derived from the K. aerogenes proteins is an overall dimer, compatible with the previously reported (UreF)2 and (UreH:UreF)2 structures of the H. pylori proteins [18, 19]. Furthermore, the chemical cross-linking results demonstrate a close juxtaposition (BS3 forms a crosslink of 11.4 Å) of the amino termini of the two UreF protomers within this complex. This result also is consistent with the published structural results; the first 25 residues of H. pylori UreF are unstructured, but the shorter K. aerogenes protein lacks these residues and its dimer is predicted to have the two amino termini on the same protein face with the first four residues of each protomer likely to have substantial flexibility. The (MBP-UreDFG)2 complex binds to the urease apoprotein and, unexpectedly, to the urease holoenzyme. The dimeric structure of (MBP-UreDFG)2 raised the question of whether the intact species binds to urease or if it dissociates to form a heterotrimeric species upon binding.
Our intact protein complex MS results display a number of features related to urease structure and stability that confirm both our chemical cross-linking and SDS-PAGE data, as well as previously reported findings. First, the loss of UreB from urease (Figure 5A) and several of its complexes (e.g., Figure 5B-5E) may relate to its proposed weak interaction with UreC, providing access to the nascent active site [16]. Second, even though UreF can be cross-linked to UreB in the (UreABC):UreD:UreF complex [15], previous data have indicated UreF to be relatively weakly bound to the urease core complex [24]; these findings suggest that UreF uses UreD as a scaffold to bind to urease rather than having a significant region of direct interface. This weak binding is observed both in the stoichiometry of the complexes recorded in our intact MS dataset that are most-likely formed through UreF dissociation, specifically the conversion between complexes No. 6 to 7 and No. 4 to 8 (Figure 4). In addition, our tandem MS data reveal several instances of the preferential ejection of UreF over other proteins upon collisional activation in the gas phase, specifically in the cases of complexes No. 3, 6, 8, 9, and 10, despite the constant presence of the much smaller UreB, the ejection of which is favored in (UreABC)3 CID. Furthermore, our intact protein complex MS data strongly indicate the formation of a UreF dimer within (MBP-UreDFG)2, primarily through the observation of complex No. 6, which is also consistent with our MALDI-MS cross-linking data (Figure 2). In general, MS data obtained with intact protein complexes also agree well with our gel filtration analysis, in that the stoichiometry of the MBP-UreDFG complex determined through both techniques is identical.
In addition to these aspects of commonality, intact protein complex MS analysis reveals unique aspects of urease-related structures and enzyme activation that were previously unknown. We find substantial evidence for protein complexes that are likely off-pathway in the formation of activated urease. Complexes such as No. 6, 7, 8, and 10 all represent assemblies that could possibly result from the decay of the core urease complex bound to one or two MBP-UreDFG. In addition, complexes No. 5 and 9 appear to result from the dimerization of either (UreABC)3 or (UreABC)3:MBP-UreD:UreF, and are also likely to represent off-pathway assemblies. Of potential interest, the hexameric architecture of urease subunits in these species is reminiscent of jack bean urease, which is a hexameric enzyme [46]. To account for all of the subcomplexes and decay products observed in our intact protein complex MS data, we invoked sequential (MBP-UreDFG)2 additions to the core urease complex to generate complexes No. 3 and 4 (Figure S4), both of which are likely to lie on the activation pathway for the enzyme. We presume that incubation of (MBP-UreDFG)2 in great excess over (UreABC)3 would lead to addition of a third MBP-UreDFG unit to the enzyme; however, our experiment was designed to test for the presence of No. 3 rather than larger complexes that would less clearly address the question. Each of the pre-activation complexes appears to undergo dissociation under our analytical conditions to produce other partially assembled complexes that are unlikely to contribute to urease activation. Significantly, however, (MBP-UreDFG)2 does not dissociate to yield MBP-UreD:UreF:UreG, free UreF, or UreF:UreF dimer (Figure S3), suggesting that these species are not available and do not add to urease during our in vitro incubation studies. The observed assemblies are present over relatively long time-scales (hours/days), suggesting that the complexes identified in Figure 4 are all relatively stable forms of these proteins in vitro.
Conclusion
We present here unique data on the urease activation complexes formed between the (UreABC)3 core assembly and a complex comprised of UreD, UreF, and UreG. Our MS data of the intact urease assemblies involved in enzyme activation are supported by measurements involving SDS-PAGE, gel filtration, chemical cross-linking, and IM separations. First, we detect clear evidence that a (UreDFG)2 complex with juxtaposed UreF subunits is the primary biological unit generated by UreD, UreF and UreG under the conditions used for our experiments. Secondly, we detect the stepwise addition of UreDFG units to the core (UreABC)3 assembly upon exposure to (UreDFG)2 in solution, or from complexes isolated directly from cells. These (UreABC)3(UreDFG) and (UreABC)3(UreDFG)2 assemblies are on route to the putative, fully-assembled pre-activation complex shown in Figure 1 through the addition of UreDFG units. This result, therefore, lends credence to a urease activation mechanism in which UreDFG binds to urease apoprotein (Figure 1, bottom track), but does not preclude the sequential protein binding pathway (Figure 1, top). Importantly, our findings are inconsistent with the repeatedly seen UreF:UreF interaction remaining intact. Finally, we also observe evidence for many off-pathway complexes that appear in high abundance in our intact MS measurements, suggesting either the relative fragility of the pre-activation urease complexes probed, or the likely complexity of the activation process. Future work in this area will likely capitalize on the data shown here to further probe the structures and stabilities of urease activation complexes, and eventually construct a complete map of their assembly in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Dan Jones and the Mass Spectrometry and Metabolomics Core facility at Michigan State University for assistance with MALDI-TOF MS, along with Dr. Jones and Dr. Lee Macomber for helpful discussions. We also thank Joseph Eschweiler (UM) for aiding in the collection of some of the intact MS data on the urease complexes reported here. This project was funded by the National Institutes of Health awards to RPH (DK045686) and BTR (GM095832).
References
- 1.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zambelli B, Musiani F, Benini S, Ciurli S. Chemistry of Ni2+ in urease: Sensing, trafficking, and catalysis. Acc. Chem. Res. 2011;44:520–530. doi: 10.1021/ar200041k. [DOI] [PubMed] [Google Scholar]
- 3.Mobley HLT, Hausinger RP. Microbial ureases: Significance, regulation, and molecular characterization. Microbiol. Rev. 1989;53:85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witte C-P. Urea metabolism in plants. Plant Sci. 2011;180:431–438. doi: 10.1016/j.plantsci.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Jabri E, Carr MB, Hausinger RP, Karplus PA. The crystal structure of urease from Klebsiella aerogenes. Science. 1995;268:998–1004. [PubMed] [Google Scholar]
- 6.Farrugia MA, Macomber L, Hausinger RP. Biosynthesis of the urease metallocenter. J. Biol. Chem. 2013;288:13178–13185. doi: 10.1074/jbc.R112.446526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MH, Mulrooney SB, Hausinger RP. Purification, characterization, and in vivo reconstitution of Klebsiella aerogenes urease apoenzyme. J. Bacteriol. 1990;172:4427–4431. doi: 10.1128/jb.172.8.4427-4431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park I-S, Carr MB, Hausinger RP. In vitro activation of urease apoprotein and role of UreD as a chaperone required for nickel metallocenter assembly. Proc. Natl. Acad. Sci. USA. 1994;91:3233–3237. doi: 10.1073/pnas.91.8.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moncrief MBC, Hausinger RP. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J. Bacteriol. 1996;178:5417–5421. doi: 10.1128/jb.178.18.5417-5421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park I-S, Hausinger RP. Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J. Bacteriol. 1995;177:1947–1951. doi: 10.1128/jb.177.8.1947-1951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moncrief MBC, Hausinger RP. Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J. Bacteriol. 1997;179:4081–4086. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park I-S, Hausinger RP. Requirement of carbon dioxide for in vitro assembly of the urease nickel metallocenter. Science. 1995;267:1156–1158. doi: 10.1126/science.7855593. [DOI] [PubMed] [Google Scholar]
- 13.Lee MH, Pankratz HS, Wang S, Scott RA, Finnegan MG, Johnson MK, Ippolito JA, Christianson DW, Hausinger RP. Purification and characterization of Klebsiella aerogenes UreE protein: A nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993;2:1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soriano A, Hausinger RP. GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc. Natl. Acad. Sci. USA. 1999;96:11140–11144. doi: 10.1073/pnas.96.20.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang Z, Kuchar J, Hausinger RP. Chemical crosslinking and mass spectrometric identification of sites of interaction for UreD, UreF, and urease. J. Biol. Chem. 2004;279:15305–15313. doi: 10.1074/jbc.M312979200. [DOI] [PubMed] [Google Scholar]
- 16.Quiroz-Valenzuela S, Sukuru SCK, Hausinger RP, Kuhn LA, Heller WT. The structure of urease activation complexes examined by flexibility analysis, mutagenesis, and small-angle X-ray scattering. Arch. Biochem. Biophys. 2008;480:51–57. doi: 10.1016/j.abb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jabri E, Karplus PA. Structures of the Klebsiella aerogenes urease apoprotein and two active-site mutants. Biochemistry. 1996;35:10616–10626. doi: 10.1021/bi960424z. [DOI] [PubMed] [Google Scholar]
- 18.Lam R, Romanov V, Johns K, Battaile K, Wu-Brown J, Guthrie JL, Hausinger RP, Pai E, Chirgadze NY. Crystal structure of a truncated urease accessory protein UreF from Helicobacter pylori. Proteins. 2010;78:2839–2848. doi: 10.1002/prot.22802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong YH, Wong HC, Chuck CP, Chen YW, Sun H, Wong K-B. Assembly of the preactivation complex for urease maturation in Helicobacter pylori: Crystal structure of the UreF/UreH protein complex. J. Biol. Chem. 2011;286:43241–43249. doi: 10.1074/jbc.M111.296830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasper R, Scrima A, Wittinghofer A. Structural insights into HypB, a GTP-binding protein that regulates metal binding. J. Biol. Chem. 2006;281:27492–27502. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- 21.Zambelli B, Stola M, Musiani F, De Vriendt K, Samyn B, Devreese B, Van Beeumen J, Dikiy A, Bryant DA, Ciurli S. UreG, a chaperone in the urease assembly process, is an intrinsically unstructured GTPase that specifically binds Zn2+ J. Biol. Chem. 2005;280:4684–4695. doi: 10.1074/jbc.M408483200. [DOI] [PubMed] [Google Scholar]
- 22.Zambelli B, Musiani F, Savini M, Tucker P, Ciurli S. Biochemical studies on Mycobacterium tuberculosis UreG and comparative modeling reveal structural and functional conservation among the bacterial UreG family. Biochemistry. 2007;46:3171–3182. doi: 10.1021/bi6024676. [DOI] [PubMed] [Google Scholar]
- 23.Boer JL, Quiroz-Valenzuela S, Anderson KL, Hausinger RP. Mutagenesis of Klebsiella aerogenes UreG to probe nickel binding and interactions with other urease-related proteins. Biochemistry. 2010;49:5859–5869. doi: 10.1021/bi1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter EL, Hausinger RP. Characterization of Klebsiella aerogenes urease accessory protein UreD in fusion with the maltose binding protein. J. Bacteriol. 2010;192:2294–2304. doi: 10.1128/JB.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boer JL, Hausinger RP. Klebsiella aerogenes UreF: Identification of the UreG binding site and role in enhancing the fidelity of urease activation. Biochemistry. 2012;51:2298–2308. doi: 10.1021/bi3000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter EL, Boer JL, Farrugia MA, Flugga N, Towns CL, Hausinger RP. Function of UreB in Klebsiella aerogenes urease. Biochemistry. 2011;50:9296–9308. doi: 10.1021/bi2011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Zhong YY, Hyung SJ, Ruotolo BT. Characterizing the resolution and accuracy of a second-generation traveling-wave ion mobility separator for biomolecular ions. Analyst. 2011;136:3534–3541. doi: 10.1039/c0an00987c. [DOI] [PubMed] [Google Scholar]
- 29.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung SJ, Robinson CV. Ion mobility-mass spectrometry analysis of large protein complexes. Nature Prot. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature Prot. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 31.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 32.Hyung SJ, Ruotolo BT. Integrating mass spectrometry of intact protein complexes into structural proteomics. Proteomics. 2012;12:1547–1564. doi: 10.1002/pmic.201100520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benesch JLP, Ruotolo BT. Mass spectrometry: Come of age for structural and dynamical biology. Curr. Opin. Struct. Biol. 2011;21:641–649. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taverner T, Hernandez H, Sharon M, Ruotolo BT, Matak-Vinkovic D, Devos D, Russell RB, Robinson CV. Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc. Chem. Res. 2008;41:617–627. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- 35.Heck AJR. Native mass spectrometry: A bridge between interactomics and structural biology. Nature Meth. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 36.Sharon M, Robinson CV. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 37.Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Protein complexes in the gas phase: Technology for structural genomics and proteomics. Chem. Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 38.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 39.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 40.Benesch JLP. Collisional activation of protein complexes: Picking up the pieces. J. Am. Soc. Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 41.Benesch JLP, Aquilina JA, Ruotolo BT, Sobott F, Robinson CV. Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem. Biol. 13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Sobott F, Robinson CV. Characterising electrosprayed biomolecules using tandem-MS - the noncovalent GroEL chaperonin assembly. Int. J. Mass Spectrom. 2004;236:25–32. [Google Scholar]
- 43.Erba EB, Ruotolo BT, Barsky D, Robinson CV. Ion mobility-mass spectrometry reveals the influence of subunit packing and charge on the dissociation of multiprotein complexes. Anal. Chem. 2010;82:9702–9710. doi: 10.1021/ac101778e. [DOI] [PubMed] [Google Scholar]
- 44.Zhong YY, Hyung SJ, Ruotolo BT. Ion mobility-mass spectrometry for structural proteomics. Exp. Rev. Proteomics. 2012;9:47–58. doi: 10.1586/epr.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 46.Balasubramanian A, Ponnuraj K. Crystal structure of the first plant urease from jack bean: 83 Years of journey from its first crystal to molecular structure. J. Mol. Biol. 2010;400:274–283. doi: 10.1016/j.jmb.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Ligabue-Braun R, Real-Guerra R, Carlini CR, Verli H. Evidence-based docking of the urease activation complex. J. Biomol. Struct. Dyn. 2012 doi: 10.1080/07391102.2012.713782. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.