SUMMARY
Nuclear pores associate with active protein-coding genes in yeast and have been implicated in transcriptional regulation. Here, we show that in addition to transcriptional regulation, key components of C. elegans nuclear pores are required for processing of a subset of small nucleolar RNAs (snoRNAs) and tRNAs transcribed by RNA Polymerase (Pol) III. Chromatin immunoprecipitation of NPP-13 and NPP-3, two integral nuclear pore components, and importin-β IMB-1, provides strong evidence that this requirement is direct. All three proteins associate specifically with tRNA and snoRNA genes undergoing Pol III transcription. These pore components bind immediately downstream of the Pol III pre-initiation complex, but are not required for Pol III recruitment. Instead, NPP-13 is required for cleavage of tRNA and snoRNA precursors into mature RNAs, whereas Pol II transcript processing occurs normally. Our data suggest that integral nuclear pore proteins act to coordinate transcription and processing of Pol III transcripts in C. elegans.
INTRODUCTION
The nuclear pore is composed of multiple copies of 30 distinct proteins called nucleoporins (Alber et al., 2007). A single pore can contain 500–1,000 individual nucleoporins. Beyond its conventional role in nucleo-cytoplasmic transport, the nuclear pore has been implicated in regulation of RNA Pol II transcription. In S. cerevisiae, the nuclear pore interacts with active genes through transcriptional regulators, and several inducible genes become relocated to the pore upon stimulation (Akhtar and Gasser, 2007; Strambio-De-Castillia et al., 2010). These studies in yeast collectively support the ‘gene gating’ hypothesis, which proposes that steps from transcription to mRNA export are coordinated at the nuclear pore (Blobel, 1985). However, in metazoans the function of the nuclear pore in transcription and RNA processing remain unclear, as does whether active transcription occurs at the nuclear pore.
C. elegans possesses a typical metazoan nuclear pore structure, with highly conserved homologs for 23 of the 30 vertebrate nucleoporins (Galy et al., 2003; Ródenas et al., 2012). Central to our study is NPP-13, the C. elegans ortholog of vertebrate Nup93 (Galy et al., 2003). Nup93 is a remarkably stable integral component of the nuclear pore scaffold. Nup93 has an estimated residence time at the nuclear pore of 70 hours in rat nuclei (Rabut et al., 2004), and a separate study in the rat brain reported an extremely slow cellular turnover rate in the range of months for Nup93 (Savas et al., 2012). Human Nup93 has been shown to associate with the chromatin, but the specific locations of interaction within the genome are not known (Brown et al., 2008). Loss of NPP-13 in C. elegans causes embryonic arrest, but it is not required for nuclear pore assembly nor for normal nuclear import (Galy et al., 2003).
In this short article, we report a surprising and direct requirement of the integral components of the C. elegans nuclear pore for processing of non-coding RNAs. This requirement is specific to Pol III-transcribed small nucleolar RNAs (snoRNAs) and tRNAs, and does not apply to Pol II transcripts. We demonstrate that integral nuclear pore components associate physically and specifically with genes undergoing Pol III transcription. Our results suggest that the C. elegans nuclear pore serves as a physical meeting place for Pol III transcription and the RNA processing machinery.
RESULTS
The integral nucleoporin NPP-13 is required for processing a specific subset of snoRNAs in C. elegans
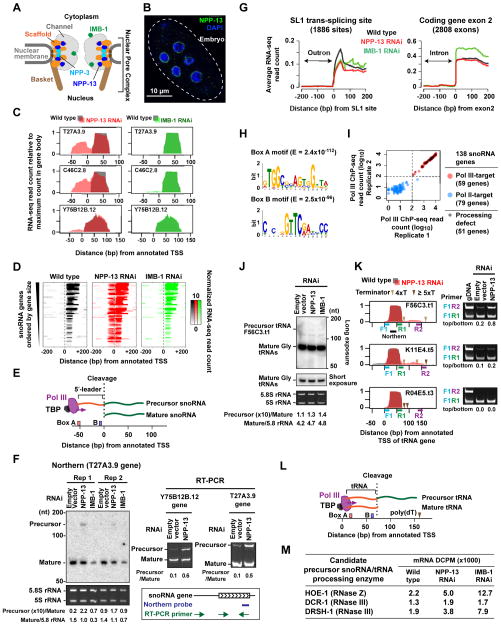
To explore the potential function of C. elegans nuclear pores in gene regulation, we performed total RNA-seq in embryos depleted of NPP-13, the integral nucleoporin localized exclusively at the nuclear pore (Figure 1A, B). The most striking feature of the data was the unexpected appearance of strong RNA signals mapping upstream of the annotated transcription start site (aTSS) of 51 snoRNA genes upon NPP-13 knockdown (Figure 1C, D & Figure S1A). Previous studies had proposed that the C. elegans snoRNAs transcribed by RNA Pol III are generated by cleavage of larger precursor RNAs originating upstream of the aTSS (Li et al., 2008; Xiao et al., 2012) (Figure 1E). We performed Northern Blotting to test if aberrant accumulation of these predicted precursor snoRNAs might account for the upstream RNA-seq signals. Strikingly, a long precursor T27A3.9 snoRNA was detected after NPP-13 knockdown, but not in wild-type embryos (Figure 1F). Consistent with this, RT-PCR yielded predicted precursors for the T27A3.9 and Y75B12B.12 snoRNA loci in NPP-13 knockdown embryos, but only very weak products from wild-type embryos (Figure 1F). The requirement for snoRNA processing was specific to NPP-13. Upon knockdown of IMB-1, a nuclear transport receptor in the importin-β family, only a slight increase in upstream RNA signal was detected (Figure 1C, D & Figure S1B). Thus, at a subset of snoRNA genes, long unprocessed snoRNA precursors accumulate inappropriately upon knockdown of the integral nuclear pore component NPP-13.
Figure 1. NPP-13 is required for snoRNA and tRNA processing.
(A) Schematic of the C. elegans nuclear pore. The locations of nucleoporins studied are indicated, based on the yeast nuclear pore structure (Alber et al., 2007).
(B) Immunofluorescence of NPP-13 in C. elegans embryos.
(C) RNA-seq signal profiles of NPP-13 (red) or IMB-1 (green) knockdown embryos on top of wild-type profile (gray) at selected snoRNA genes. See Figure S1 for all snoRNA genes.
(D) Colorimetric representation of per-base RNA-seq read count at the 51 snoRNA genes exhibiting upstream RNA signals upon NPP-13 knockdown. RNA-seq read counts are normalized by genome-wide average of base coverage (also in G).
(E) Schematic of previously proposed Pol III-transcribed snoRNA processing (Li et al., 2008; Xiao et al., 2012).
(F) Northern blotting and RT-PCR detect uncleaved precursor snoRNAs in NPP-13 knockdown embryos. Densitometry analyses are shown under the images. Ethidium bromide staining of 5.8S rRNA (Pol I target) and 5S rRNA (TFIIIA-dependent Pol III target) shows the loading control.
(G) Average RNA-seq read count profiles at SL1 trans-splice sites and the 5′-end of exon 2 of protein-coding genes.
(H) DNA motifs found in the 51 snoRNA genes showing processing defect upon NPP-13 knockdown. −80 bp to +20 bp from annotated TSS (aTSS) were analyzed. E, Expected value.
(I) Pol III ChIP-seq read counts in 138 annotated snoRNA genes. The maximum read count in the 200 bp region centered on the aTSS were plotted. In order to plot all genes including those with 0 count in the logarithmic space, 1 count was added to all genes. See Supplemental Experimental Procedures for the criterion for dividing snoRNA genes into Pol III and Pol II-targets.
(J) Northern blotting detects precursor tRNA F56C3.t1 around 150 nt. The location of the probe is shown in K. The identity of the band at 125 nt is unknown.
(K) (Left) RNA-seq profiles (same as C) for tRNA genes, F56C3.t1 and K11E4.t5, which show downstream RNA signals, and R04E5.t3, which does not. The location of Pol III terminator poly(dT) tracts is indicated. (Right) RT-PCR detects precursor tRNAs with a long 3′-trailer. The primer locations are shown in the corresponding left panel.
(L) Schematic of tRNA 3′-end processing.
(M) RNA-seq DCPM scores for endonucleases that are predicted to cleave precursor Pol III-snoRNAs.
Processing of Pol II transcripts is normal in embryos depleted of NPP-13
We hypothesized that defect in snoRNA processing might be part of a larger defect in general RNA processing. However, in contrast to the snoRNA transcripts, processing of Pol II transcripts was unaffected by NPP-13 depletion. Both introns and outrons (an intron-like RNA at the 5′-end of pre-mRNA that is trans-spliced) were removed with normal fidelity after NPP-13 knockdown (Figure 1G). Thus, the data suggest that the observed requirement for snoRNA processing was not a secondary consequence of a more general defect in nuclear pore function.
Only snoRNA genes transcribed by Pol III have processing defects upon NPP-13 loss
In an attempt to understand why a specific subset of snoRNA genes were affected by NPP-13 knockdown, we analyzed the DNA sequences around the affected genes. We found that all 51 genes contained the Box A and Box B motifs, which are recognized by the RNA Pol III machinery (Figure 1H, (Schramm and Hernandez, 2002)). We then identified genomic regions bound by Pol III using Chromatin Immunoprecipitation followed by high-throughput sequencing (ChIP-seq) in wild-type embryos. Based on the Pol III ChIP-seq data, we defined 59 out of 138 annotated snoRNA genes to be active targets of Pol III (hereafter ‘Pol III-snoRNA genes’. Supplemental Experimental Procedures, Figure 1I & Table S1). All 51 of the affected snoRNA genes fell within the group of 59 Pol III-snoRNA genes (86%; Figure 1I), indicating that the only snoRNA genes that are transcribed by RNA Pol III exhibit a processing defect upon NPP-13 knockdown.
NPP-13 is also required for efficient processing of tRNA 3′ ends
Because the affected RNAs were all transcribed by Pol III, we asked if the largest class of Pol III-transcribed RNAs, tRNAs, were affected by NPP-13 depletion. Northern Blotting using a probe for glycine tRNAs detected a strong single band of ~70 nt representing the mature glycine tRNAs in wild type, NPP-13 knockdown, and IMB-1 knockdown embryos (Figure 1J). Thus, the majority of glycine tRNAs were successfully processed upon NPP-13 and IMB-1 depletion. However, longer exposure of this blot revealed a faint band at 150 nt in NPP-13 and IMB-1 knockdown embryos, but not in wild-type (Figure 1J). We then more closely interrogated the RNA-seq profile of each of the 609 tRNA genes and identified two tRNA loci, F56C3.t1 (glycine) and K11E4.t5 (lysine), that exhibited unusual RNA-seq signals downstream of the gene bodies upon NPP-13 knockdown (Figure 1K). We found that these two loci naturally produce precursors with an unusually long 3′ trailer, and that removal of this trailer is inhibited by NPP-13 loss (Figure 1L). DNA sequence analysis found that this rare feature is likely caused by a weak Pol III terminator sequence consisting of 4 consecutive Ts immediately downstream of the gene body (Figure 1K). RT-PCR confirmed low levels of the predicted tRNA precursors in wild-type embryos, but much higher levels upon NPP-13 knockdown (Figure 1K). Majority of tRNA genes (78%) have strong terminator sequences of 5 or more Ts immediately downstream of the gene body and do not produce long precursors, as exemplified by R04E5.t3 (Figure 1K). Thus, a rare sequence feature in two transcribed tRNA genes allowed us to determine that NPP-13 is also required for 3′ end processing in at least a subset of tRNA genes.
Endonuclease activities responsible for tRNA and snoRNA processing remain intact in NPP-13 knockdown embryos
We next investigated whether the processing defect of Pol III transcripts might be explained by the absence or inactivity of candidate RNA processing enzymes. In Arabidopsis, a subset of snoRNA genes are transcribed by Pol III, but as a 5′-tRNA-snoRNA-3′ dicistron, a structure similar to the C. elegans Pol III-snoRNAs with the 5′ leader (Kruszka et al., 2003). The resulting plant dicistronic transcripts are cleaved by the endonuclease RNase Z (Barbezier et al., 2009; Kruszka et al., 2003). RNase Z is also the endonuclease that normally cleaves the 3′ trailer from tRNA precursors (Castaño et al., 1985). In yeast, the 5′ end of snoRNA is processed by RNase III (Lee et al., 2003). C. elegans hoe-1 (RNase Z) and dcr-1 (RNase III) and drsh-1 (RNase III) encode the corresponding homologous endonucleases. None of these were down-regulated at the RNA level after NPP-13 knockdown (Figure 1M). Furthermore, 7 of the 59 (12%) of Pol III-snoRNAs exhibited almost complete removal of the 5′ leader RNAs (asterisks in Figure S1A). Thus, the biochemical activity of snoRNA processing machinery is likely to be unaffected by NPP-13 knockdown.
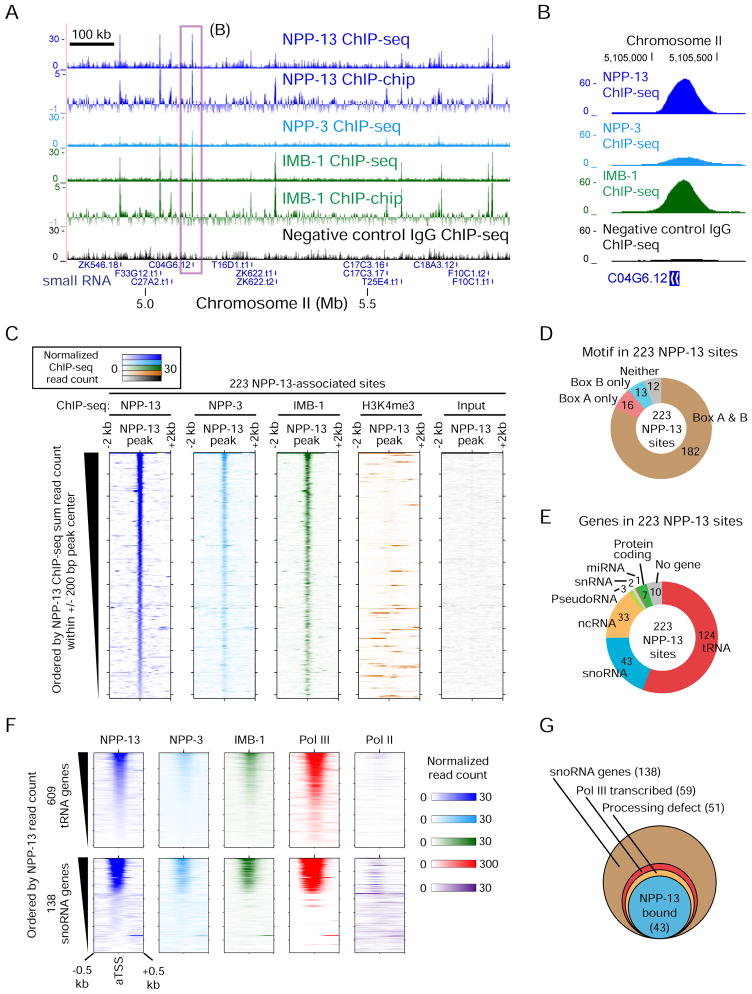
Nuclear pore proteins interact directly with Pol III-snoRNA and tRNA genes
The evidence to this point led us to hypothesize that the very specific requirement for NPP-13 is mediated through a direct interaction between the snoRNA and tRNA loci and the nuclear pore components. To test this, we performed ChIP for NPP-13 in C. elegans embryos (Figure 2A & B). ChIP followed by tiling microarray (ChIP-chip) and ChIP-seq produced sharp signals of enrichment that were concordant across the genome (Figure 2A). We identified 223 statistically significant and highly reproducible NPP-13-associated sites (Figure 2C & Table S2; Supplemental Experimental Procedures). The specificity of the NPP-13 antibodies was confirmed by western blotting (Figure S2A).
Figure 2. Nuclear pore proteins associate with small RNA genes in C. elegans.
(A) ChIP-seq and ChIP-chip profiles of nuclear pore proteins. For ChIP-seq, read counts normalized by average base coverage are shown (also in B, C and F). For ChIP-chip, MA2C scores are shown. The box indicates a region shown in detail in (B). Only small RNA genes are shown under tracks. Signals over the y-axis range are omitted. The specificity of the NPP-13 antibody is validated in Figure S2A.
(B) Nucleoporin ChIP-seq signals at snoRNA gene C04G6.12.
(C) Colorimetric representation of per-base ChIP-seq read count at 223 NPP-13-associated sites. Orientation is according to the genome coordinate.
(D) The number of NPP-13-associated sites harboring Box A or B motifs in the 200 bp region centered on the peak summit.
(E) The class of genes found in the 1 kb region centered on the summit of NPP-13-associated sites. The search was performed first for non-protein coding RNA genes, then for protein-coding genes.
(F) Colorimetric representation of nucleoporin and RNA polymerase ChIP-seq read counts for all annotated tRNA and snoRNA genes.
(G) Summary of 138 snoRNA genes. Of 51 genes showing RNA processing defects upon NPP-13 depletion, 43 are bound by NPP-13 at the level exceeding our peak calling threshold. The remaining 8 genes are still associated with NPP-13 but below the cutoff.
We performed ChIP for two additional nuclear pore-associated proteins, NPP-3 and IMB-1 (Figure 1A). NPP-3 (vertebrate Nup205; yeast Nup192) is also an integral scaffold nucleoporin that interacts directly with NPP-13 and is located exclusively at the nuclear envelope (Alber et al., 2007; Grandi et al., 1997; Hachet et al., 2012). The importin-β protein IMB-1 associates indirectly with NPP-13 through channel nucleoporins (Schrader et al., 2008). ChIP signals of NPP-3 and IMB-1 were nearly identical to the NPP-13 pattern (Figure 2A & B). NPP-3 and IMB-1 were clearly enriched at the 223 NPP-13-associated sites, in contrast to H3 lysine 4 trimethylation (H3K4me3) and input, which were performed in parallel for reference (Figure 2C). These results strongly suggest that 223 sites detected by our ChIP are directly associated with the nuclear pore.
Over 90% of the NPP-13 sites harbored one or both of the Box A and Box B motifs recognized by the Pol III machinery (Figure 2D). Consistent with this, nearly all of the 223 NPP-13 binding sites were located near non-coding RNA genes known to be transcribed by Pol III, including tRNA and snoRNA genes (Figure 2E). NPP-13 interacts with only a subset of all annotated tRNA (124 of 609) and snoRNA genes (43 of 138) (Figure 2F). Strikingly, all 43 NPP-13-associated snoRNA loci were among the 59 Pol III-snoRNA genes. All 43 NPP-13-associated snoRNA loci were also among the 51 snoRNA genes with processing defects upon NPP-13 loss (Figure 2G). Finally, the two tRNA genes with the demonstrated 3′ processing defect upon NPP-13 depletion were also bound by NPP-13 (Figure S2B). Thus, snoRNA and tRNA genes that require NPP-13 for efficient RNA processing are physically associated with the nuclear pore proteins.
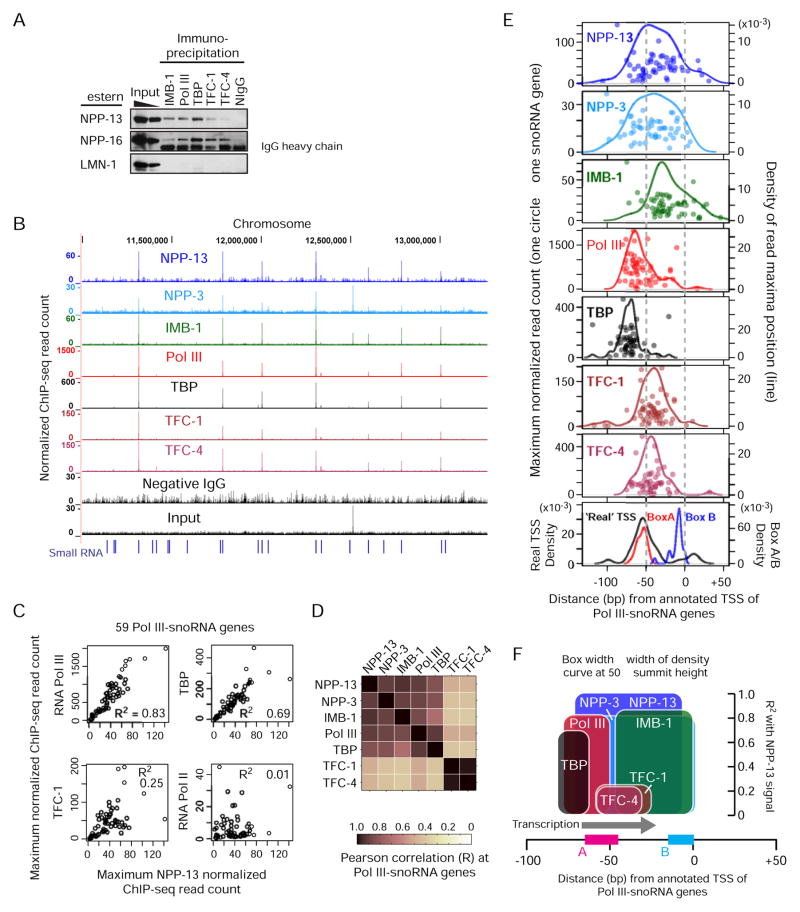
Nuclear pore subunits interact directly with the RNA Polymerase III machinery
We next tested whether nuclear pore proteins and the Pol III machinery components (Pol III, TATA-binding protein TBP, and two TFIIIC proteins TFC-1 and TFC-4) interact directly by co-immunoprecipitation from a cross-linked protein extract. NPP-13 and NPP-16, a component of the basket portion of the nuclear pore co-immunoprecipitated with IMB-1 and each of the Pol III components (Figure 3A). Pol III components, however, did not interact with the nuclear lamina (LMN-1) (Figure 3A). Therefore, Pol III proteins associate specifically with the nuclear pore components, not with the nuclear periphery as a whole.
Figure 3. Chromosomal sites associated with nuclear pore proteins are also bound by the RNA Pol III machinery.
(A) Co-immunoprecipitation of nuclear pore proteins by IMB-1 and Pol III protein antibodies. NIgG, negative control IgG.
(B) ChIP-seq profiles of the Pol III machinery proteins (Pol III, TBP, TFC-1 and TFC-4). ChIP-seq read counts are normalized by average base coverage (also in C and E).
(C) Correlation between ChIP-seq signals at 59 Pol III-snoRNA genes. The maximum signal intensities within 1 kb centered on the aTSS are plotted. R2, the square of Pearson’s correlation coefficient.
(D) Same as C but correlation coefficients (R) for all nuclear pore proteins and Pol III proteins are shown.
(E) Positions of ChIP-seq signal maxima relative to the aTSS. Each circle represents one of 59 Pol III-snoRNA genes. The left y-axis indicates the normalized maximum read count. Lines indicate the Gaussian density estimates for the maximum positions (right y-axis). The bottom box shows density estimates for box A and B motif positions and real TSSs as determined by the 5′-end of the upstream RNA signal detected in NPP-13 knockdown embryos.
(F) Summary of the nuclear pore and Pol III protein binding positions along Pol III-snoRNA genes. The height of boxes indicates the square of the correlation with NPP-13 signals as shown in (C) and (D). See Figure S3 for a hypothetical yet size-scaled model showing how the Pol III transcriptional machinery can be situated in the nuclear pore channel.
We then performed ChIP-seq for each of the Pol III components. The results among all of these Pol III complex members were strikingly consistent, and all were bound strongly to genes that were also associated with the nuclear pore proteins (Figure 3B). Furthermore, at Pol III-snoRNA gene loci across the genome, levels of Pol III and TBP enrichment were strongly correlated with nucleoporin enrichment (R2 = 0.83 for Pol III and 0.69 for TBP with NPP-13; Figure 3C & D). A high degree of correlation between Pol III and nucleoporins was also apparent at tRNA genes (Figure 2F). Unlike the requirement of TFIIIC binding for nuclear peripheral localization of tRNA genes in S. pombe (Noma et al., 2006), TFIIIC proteins showed a weaker correlation with levels of nucleoporin enrichment (R2 = 0.25 for TFC-1; Figure 3C & D). No correlation was observed with Pol II (R2 = 0.01; Figure 3C). Thus, when snoRNA and tRNA genes are associated with the nuclear pore proteins, these genes are also highly occupied by the Pol III machinery, particularly by TBP and Pol III, which are members of the Pol III pre-initiation complex.
Nuclear pore proteins are positioned on genes immediately downstream of the Pol III preinitiation complex
To understand where exactly on the target genes the nuclear pore proteins associate, we determined the position of ChIP-seq signal maxima for each Pol III-snoRNA gene. We focused on snoRNA genes because each gene sequence is unique, unlike the tRNA genes. The actual snoRNA transcript start sites (TSS), inferred by the 5′-end of the upstream RNA-seq signals (Figure 1D), were located around 50 bp upstream of the annotated TSS (aTSS, Figure 3E). TFC-1, which binds to the 3′-end of Box A, and TFC-4, which physically interacts with TFC-1 (Schramm and Hernandez, 2002), exhibited almost identical distributions centered around Box A (Figure 3E). Pol III and TBP bound immediately upstream of the actual TSS as expected, with TBP bound slightly upstream of Pol III (Figure 3E). This inferred organization based on ChIP data is perfectly consistent with the crystal structure and biochemical analyses of the Pol III preinitiation complex (Fernández-Tornero et al., 2010). The binding positions of NPP-13 and NPP-3 were immediately downstream of the Pol III pre-initiation complex (Figure 3E). Interestingly, IMB-1 sites were located about 20 bp downstream of NPP-13/3 sites, perhaps reflecting the fact that IMB-1 does not interact directly with NPP-13 or NPP-3 (Bayliss et al., 2000). The binding configuration suggests that Pol III must pass by the site associated with nucleoporins to transcribe snoRNAs (Figure 3F). We propose a model consistent with these observations in the Discussion
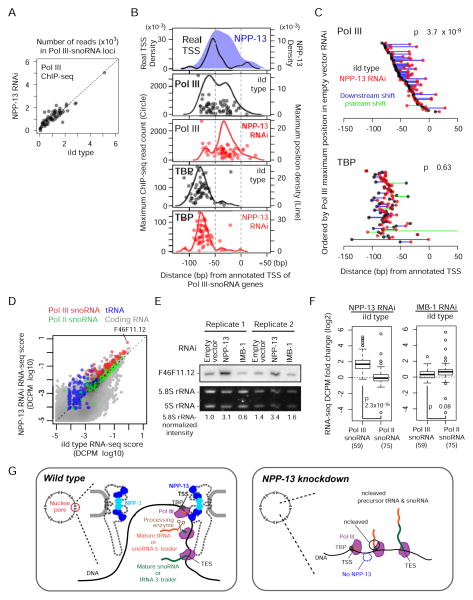
NPP-13 is required for the faithful distribution of Pol III along snoRNA genes
During mRNA transcription, Pol II elongation and mRNA processing are tightly coordinated (Egloff and Murphy, 2008). We next investigated how NPP-13 knockdown, which inhibits efficient Pol III transcript processing, affects Pol III binding and transcription. We found that the degree of Pol III association at snoRNA genes was unchanged after NPP-13 knockdown (Figure 4A). Thus, NPP-13 is not required for Pol III recruitment. However, we observed a striking change in Pol III distribution on genes upon NPP-13 knockdown. After NPP-13 knockdown, Pol III maxima at sites of TBP enrichment were lost at nearly all snoRNA genes (Figure 4B). Instead, most genes now harbored a Pol III maximum 25 bp downstream of the real TSS (-25 bp relative to aTSS). Inspection of individual genes confirmed that Pol III maxima shift downstream upon NPP-13 knockdown (Figure 4C). In contrast, TBP distribution was unchanged after NPP-13 knockdown, indicating that transcription initiation sites remain the same (Figure 4B & C). These results suggest that Pol III elongation kinetics is altered by NPP-13 depletion.
Figure 4. NPP-13 knockdown alters Pol III distribution on snoRNA genes and increases tRNA and snoRNA levels.
(A) Pol III ChIP-seq read counts in NPP-13 knockdown versus wild-type embryos at 59 Pol III-snoRNA genes. Reads overlapping with regions corresponding to -100 bp to the transcription termination site are normalized by average base coverage and plotted.
(B) Positions of Pol III and TBP ChIP-seq signal maxima in wild-type (empty RNAi vector) or NPP-13 knockdown embryos are shown as in Figure 3E. For comparison, the positions of real TSSs and wild-type NPP-13 ChIP-seq maxima used in Figure 3E are shown.
(C) Trajectories of Pol III and TBP maxima shift upon NPP-13 knockdown. Circles represent wild-type and NPP-13-knockdown Pol III maximum positions. Lines connect wild type and NPP-13 knockdown circles of the same gene. P-value is calculated by paired t-test for increased downstream shift.
(D) RNA abundance of NPP-13 knockdown versus wild-type embryos determined by RNA-seq and shown as a DCPM (Depth of Coverage Per base per Million reads) score. Each circle represents a single transcript. Transcripts with DCPM value smaller than 10−5 are not shown.
(E) Northern blotting of snoRNA F46F11.12 (also indicated in D) confirms the increased RNA abundance after NPP-13 knockdown.
(F) Boxplots indicate the log2 fold change of the RNA abundance upon NPP-13 knockdown (left) or IMB-1 knockdown (right). Student’s t-test is used for the statistical analysis. Parentheses indicate the number of transcripts annotated in each category.
(G) A model depicting that the nuclear pore serves as a hub for Pol III transcription and subsequent processing. In wild type C. elegans (left), scaffold nucleoporins associate with snoRNA and tRNA genes that undergo high levels of Pol III transcription. The nascent transcripts are processed by a yet unidentified endonuclease, perhaps in a co-transcriptional manner. Given the stability of NPP-13 and NPP-3 at the nuclear pore, this series of events likely occur in the pore channel although this is still our speculation. In the absence of NPP-13 (right), Pol III can still transcribe the target genes, but the transcripts fail to undergo processing, resulting in producing uncleaved precursor snoRNAs and tRNAs. See also Figure S3 for a size-scaled model.
Pol III transcript abundance increases upon NPP-13 knockdown
We also found that the abundance of most Pol III-snoRNAs, as determined by RNA-seq signals from what would comprise the mature transcript (i.e. excluding the aberrant upstream signals), increased upon NPP-13 knockdown (Figure 4D). Northern blotting for a Pol III-snoRNA gene F46F11.12 confirmed this increase (Figure 4E). The abundance of tRNA also increased upon NPP-13 knockdown (Figure 4D). Knockdown of IMB-1 did not strongly increase the level of Pol III-snoRNAs (Figure 4E & F), indicating that this defect is specific to knockdown of NPP-13. Furthermore, NPP-13 knockdown did not change Pol II-transcribed snoRNA levels (Figure 4D & F), arguing that the defect is further specific to NPP-13-associated Pol III-target genes. These data indicate that NPP-13 antagonizes production of Pol III transcripts, perhaps due to its function in ensuring successful coordination of transcription and RNA processing.
DISCUSSION
Using genomics approaches, we provide evidence for a new role for integral components of the metazoan nuclear pore in RNA processing. We demonstrate that the C. elegans scaffold nucleoporins NPP-13 and NPP-3 associate physically with small RNA genes that are undergoing high levels of Pol III transcription. Given that NPP-13 and NPP-3 are statically localized at the inner surface of the nuclear pore (Krull et al., 2004; Rabut et al., 2004), the most direct interpretation of our data is that scaffold nucleoporins anchor Pol III-transcribing genes inside the nuclear pore (Figure 4G & S3). We show here that NPP-13 is not necessary to recruit Pol III to its target genes, nor for Pol III transcription. Rather, NPP-13 is required for efficient processing of a subset of Pol III transcribed snoRNAs and tRNAs.
While Pol II uses its C-terminal domain (CTD) to coordinate co-transcriptional processing (Egloff and Murphy, 2008), Pol III does not possess a CTD or equivalent structure. Given that NPP-13 and NPP-3 associate just upstream of the cleavage site of snoRNA precursors, we propose a model in which scaffold nucleoporins promote recruitment of RNA processing enzyme(s) to the site of cleavage during transcription (Figure 4G). NPP-13 depletion may disrupt this system by releasing the Pol III-transcribing genes and/or the processing enzymes from the nuclear pore. The accumulation of Pol III very near the cleavage site upon NPP-13 loss may represent a kinetic “pause” caused by the lack of the cleavage event that would normally occur at that location. It is unlikely that the processing defect is caused by overloading the processing machinery with increased Pol III transcripts because there are Pol III-snoRNA genes that are highly upregulated upon NPP-13 knockdown, but do not show any processing defect (e.g. F46F11.12 snoRNA).
Does Pol III transcription occur inside the nuclear pore?
Previous studies in Drosophila showed that ‘mobile’ nucleoporins that constitute the basket filament and channel barrier associate with active Pol II genes, predominantly outside the context of the nuclear pore (Capelson et al., 2010; Kalverda et al., 2010; Vaquerizas et al., 2010). In contrast, our data indicates that the sites of NPP-13 association are bound by Pol III, and that Pol II and H3K4me3 levels are low. These observations could therefore represent two entirely different mechanisms, with mobile nucleoporins involved with Pol II transcription in the nucleoplasm, while static scaffold nucleoporins associate with Pol III transcription for efficient transcript processing, perhaps within the nuclear pore itself. Upon heat shock in C. elegans, NPP-13 also interacts with Pol II-transcribed heat-responsive genes at the pore (Rohner et al., 2013), suggesting an additional function of scaffold nucleoporins in Pol II transcription. It is not known whether the nuclear pore is required for Pol III transcript processing in other organisms, although tRNAs do associate with the nuclear periphery in S. pombe (Noma et al., 2006).
The size of C. elegans nuclear pores has been estimated as intermediate between those of the vertebrates and yeast (the yeast pore channel: ~38 nm diameter, ~37 nm height) (Alber et al., 2007; Cohen et al., 2002; Frenkiel-Krispin et al., 2010; Grossman et al., 2012). Considering that the size of the Pol III pre-initiation complex is 13 × 15 × 17 nm3 (Fernández-Tornero et al., 2010) and the flexible nature of the nuclear pore channel (Grossman et al., 2012), the C. elegans nuclear pore is easily large enough to accommodate the Pol III pre-initiation complex in the channel (see a scaled schematic model in Figure S3). Our data do not strictly exclude the possibility that genomic interactions with NPP-13, NPP-3, and IMB-1 occur in the nucleoplasm, although one would then have to account for the remarkable co-association of all three nuclear pore proteins at the same genomic loci. Further studies will be required to definitively test the hypothesis that Pol III transcribes genes inside the nuclear pore (Figure 4G & S3).
EXPERIMENTAL PROCEDURES
Detailed methods are available in Supplemental Experimental Procedures. Primer and probe sequences are listed in Table S3.
Accession Numbers
ChIP-seq, ChIP-chip, and RNA-seq experiments are listed in Table S4 and submitted to Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession ID GSE42741.
ChIP-seq and ChIP-chip
Chromatin extract was prepared from the N2 strain mixed-stage embryos cross-linked in 2% formaldehyde for 30 min (Ikegami et al., 2010). Five to ten micrograms of antibodies were used for ChIP. Single-end ChIP-seq reads were aligned to the C. elegans reference genome (ce6) accepting only uniquely mapped reads. For ChIP-chip, a C. elegans whole-genome tiling microarray (Ikegami et al., 2010)(GEO ID GPL8647; NimbleGen, Wisconsin) was used.
Total RNA-seq
rRNA-depleted total RNA from mixed-stage embryos were used to generate cDNA libraries and subjected to single-end sequencing. Reads that are uniquely mapped to the ce6/ws190 assembly were used for analyses.
RNAi
RNAi was performed in the liquid S medium by feeding L4-stage N2 worms with bacteria strains expressing double-strand interfering RNA from an inducible vector. In parallel, worms were also fed with bacteria containing the same vector without RNAi inserts (empty vector control).
Supplementary Material
HIGHLIGHTS.
C. elegans nuclear pore components are required for processing of Pol III transcripts
Pol III-transcribed genes are associated with integral components of the nuclear pore
Nucleoporins bind immediately downstream of the Pol III pre-initiation complex
Loss of nuclear pore components alters the distribution of Pol III along genes
Acknowledgments
We thank Virginie Hachet and Pierre Gönczy for antibodies, Piotr Mieczkowski and Hemant Kelkar for high-throughput sequencing, and Brent Chen for technical assistance. This work is supported by modENCODE grant U01 HG004270-03 to JDL, and by Progeria Research Foundation grant #2009-0028 to KI and JDL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Barbezier N, Canino G, Rodor J, Jobet E, Saez-Vasquez J, Marchfelder A, Echeverría M. Processing of a dicistronic tRNA-snoRNA precursor: combined analysis in vitro and in vivo reveals alternate pathways and coupling to assembly of snoRNP. Plant Physiology. 2009;150:1598–1610. doi: 10.1104/pp.109.137968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño JG, Tobian JA, Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3″ terminus of human pre-tRNA-Met(i) (3″ pre-tRNase) J Biol Chem. 1985;260:9002–9008. [PubMed] [Google Scholar]
- Cohen M, Tzur YB, Neufeld E, Feinstein N, Delannoy MR, Wilson KL, Gruenbaum Y. Transmission electron microscope studies of the nuclear envelope in Caenorhabditis elegans embryos. J Struct Biol. 2002;140:232–240. doi: 10.1016/s1047-8477(02)00516-6. [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Fernández-Tornero C, Böttcher B, Rashid UJ, Steuerwald U, Flörchinger B, Devos DP, Lindner D, Müller CW. Conformational flexibility of RNA polymerase III during transcriptional elongation. Embo J. 2010;29:3762–3772. doi: 10.1038/emboj.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol. 2010;395:578–586. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pané N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M. Functional Architecture of the Nuclear Pore Complex. Annual Review of Biophysics. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- Hachet V, Busso C, Toya M, Sugimoto A, Askjaer P, Gönczy P. The nucleoporin Nup205/NPP-3 is lost near centrosomes at mitotic onset and can modulate the timing of this process in C. elegans embryos. Mol Biol Cell. 2012;23:3111–3121. doi: 10.1091/mbc.E12-03-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biology. 2010;11:R120. doi: 10.1186/gb-2010-11-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Krull S, Thyberg J, Björkroth B, Rackwitz HR, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell. 2004;15:4261–4277. doi: 10.1091/mbc.E04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka K, Barneche F, Guyot R, Ailhas J, Meneau I, Schiffer S, Marchfelder A, Echeverría M. Plant dicistronic tRNA-snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNase Z. Embo J. 2003;22:621–632. doi: 10.1093/emboj/cdg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Lee A, Chanfreau G. The roles of endonucleolytic cleavage and exonucleolytic digestion in the 5′-end processing of S. cerevisiae box C/D snoRNAs. Rna. 2003;9:1362–1370. doi: 10.1261/rna.5126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, He H, Wang Y, Zheng H, Skogerbø G, Chen R. In vivo analysis of Caenorhabditis elegans noncoding RNA promoter motifs. BMC Mol Biol. 2008;9:71. doi: 10.1186/1471-2199-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma KI, Cam HP, Maraia RJ, Grewal SIS. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol. 2013;200:589–604. doi: 10.1083/jcb.201207024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ródenas E, González-Aguilera C, Ayuso C, Askjaer P. Dissection of the NUP107 nuclear pore subcomplex reveals a novel interaction with spindle assembly checkpoint protein MAD1 in Caenorhabditis elegans. Mol Biol Cell. 2012;23:930–944. doi: 10.1091/mbc.E11-11-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Structural Basis of the Nic96 Subcomplex Organization in the Nuclear Pore Channel. Mol Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Wang Y, Luo H, Liu L, Wei G, Chen X, Sun Y, Chen X, Skogerbø G, Chen R. A differential sequencing-based analysis of the C. elegans noncoding transcriptome. Rna. 2012;18:626–639. doi: 10.1261/rna.030965.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.