DNA polymerase (pol) β is a member of the X-family of DNA polymerases and plays an important role in base excision repair that cleanses the genome of simple base lesions.[1] It has been the subject of extensive studies examining its key roles in repair[2] and cancer.[3]
In an effort to identify features of the pol β active site that modulate nucleotide binding, we have made modifications in the triphosphate moiety and studied the effect on enzyme binding, specificity, and chemistry.[4] Replacement of the Pα-O-Pβ bridging oxygen with a carbon atom prevents catalysis,[4b] whereas modification of the Pβ-O-Pγ bridging oxygen alters leaving group properties.[4a] The introduction of these substituents may also enable entirely new bonding (or repulsive) active site interactions, not present with the natural nucleoside triphosphate, and thus could inform mechanistic insight as well as inhibitor design seeking to exploit pol β as a drug target.[4a]
Previously we studied several α,β-methylene-dNTP analogues as probes for the capture of pre-insertion substrate complexes with pol β and found that α,β-CF2-analogues have an apparent equilibrium dissociation constant (Kd) that is two orders of magnitude greater than the corresponding α,β-CH2-analogues and the β,γ-dGTP-CF2-analogue.[4a, 4b] To further explore the molecular basis of pol β nucleotide binding, the dimethylated (α,β-C(Me)2-, (5)) and the monofluorinated (R/S)-(α,β-CHF-, (6a/b)) dATP analogues have been synthesized and their Kd values determined. In addition, the X-ray crystallographic structures of ternary substrate complexes of pol β and DNA primer template formed from α,β-CF2-dATP[4b] (7) and 6a/b have been solved. Interestingly, only one diastereomer of 6 is found in the active site (Figure 1).
Figure 1.
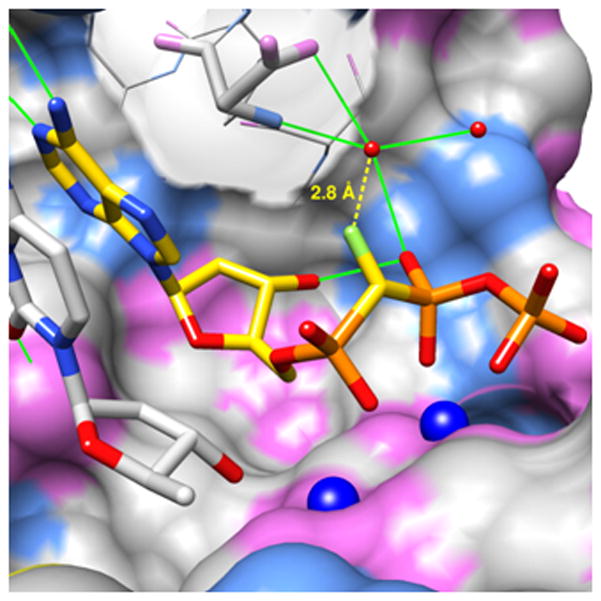
X-ray crystal structure of the DNA pol β active site containing (S)-α,β-CHF-dATP. The solvent-excluded pol β active site surface is colored by atom (carbon, gray; nitrogen, light blue; oxygen, pink; magnesium, dark blue spheres). The surface of Asp276 has been made transparent. The fluorine atom (light green) is 2.8 Å from the oxygen of a conserved water molecule (small red sphere) that is within hydrogen bonding distance (green solid lines) of the oxygen of an adjacent water (2.6 Å), the carboxyamide nitrogen (2.8 Å) and a carboxylate anion oxygen of Asp276 (2.9 Å), as well as the phosphoryl oxygen of Pβ (2.8 Å) which accepts a hydrogen bond from the 3′-OH (2.8 Å). The 3′-primer terminal nucleoside is shown (gray carbons), but its backbone is omitted for clarity.
5 and 6a/b were prepared by reaction of 2′-deoxyadenosine 5′-tosylate with the tris(tetrabutylammonium) salt of the corresponding bisphosphonic acid followed by enzymatic phosphorylation of the resulting dA 5′-bisphosphonate[4b] (Scheme 1). In a previously reported synthesis of 6a/b in which (R/S)-α,β-CHF-dADP (4a/b) was phosphorylated with p-nitrobenzyl phosphormorpholidate,[5] an approximately 1:1 mixture of α,β-CHF-dATP diastereomers was observed in the 19F NMR spectrum. Similarly, phosphorylation of 4a/b using nucleoside diphosphate kinase and a catalytic amount of ATP generates both diastereomers as observed in the 19F and 31P (Pα) NMR spectra. Although precise quantification is not possible due to signal overlap, by 19F NMR analysis the isomers are in roughly similar abundance, indicating that the enzymatic phosphorylation did not significantly enrich a particular isomer.
Scheme 1.
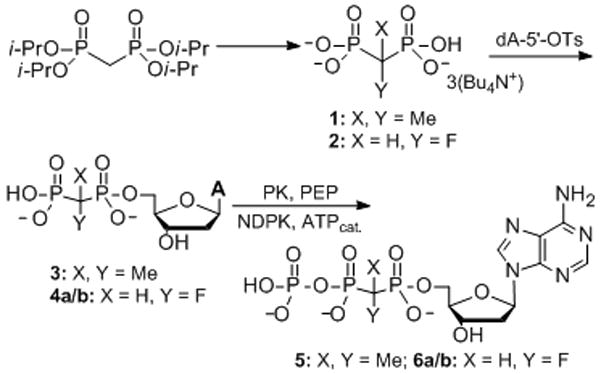
Synthesis of α,β-CXY-dATP analogues.
As expected, 5 and 6a/b are not pol β substrates (SI, Figure S25) and inhibit gap-filling DNA synthesis by pol β in a concentration dependent manner (SI, Figures S26 and S28). The apparent equilibrium binding constant for 6a/b is approximately 10 μM, similar to that of the natural substrate and significantly lower than that of the α,β-CF2-dATP analogue (Figure 2).[4b] The α,β-CH2-dATP analogue binds with about a 10-fold higher affinity[4b, 6] than 6a/b consistent with a correlation between the basicity of the phosphonate moiety and the strength of inhibitor binding.[4b] However, α,β-C(Me)2-dATP (5) which has two electron-donating substituents on the α,β-methylene, has a Kd of approximately 1100 μM and does not conform to this trend.
Figure 2.
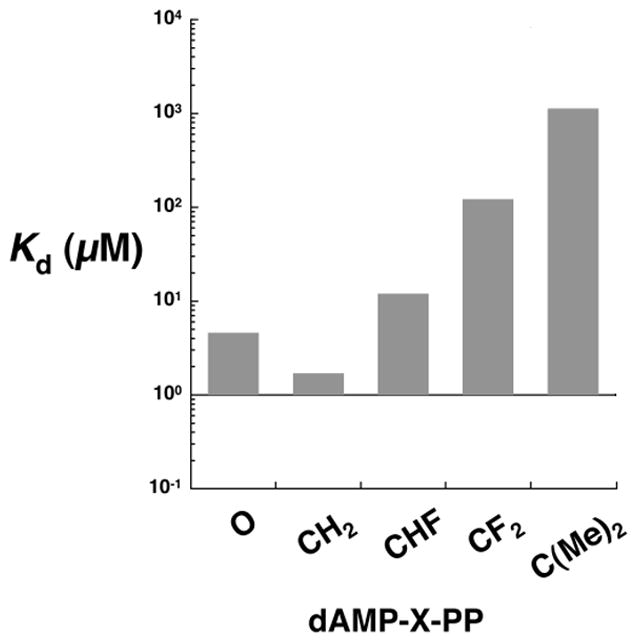
DNA pol β dissociation constants of dATP and α,β-methylene analogues. The data for dATP, α,β-CH2-dATP, and α,β-CF2-dATP were previously published.[4b, 9] The Ki (i.e., Kd) values were determined as described previously.[8]
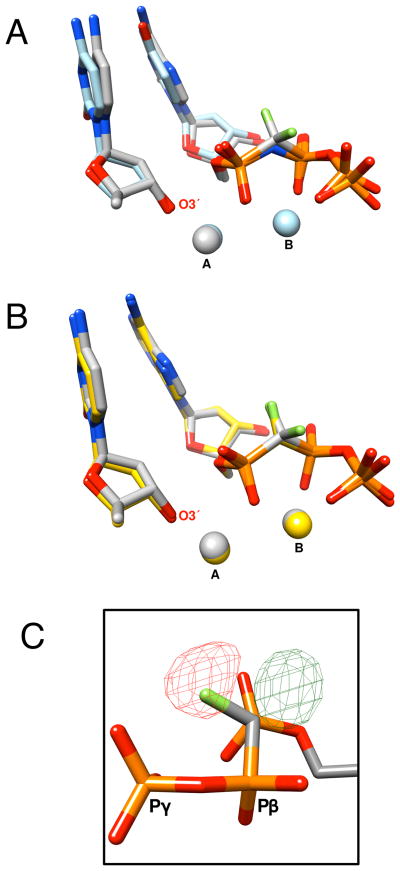
The crystallographic structures of ternary complexes of pol β with incoming α,β-CF2-dATP (7) (PDB ID 3TFR) or α,β-CHF-dATP (6) (PDB ID 3TFS) opposite a templating thymidine, resolved at 2.00 Å (SI Table S1) were next determined. All corresponding atoms in the active site of these structures superimpose well with previously determined ternary complex structures of pol β where the reaction was trapped by deletion of the nucleophilic 3′-OH on the primer terminus or by using a nitrogen in place of the α,β-bridging oxygen (Figure 3A).[8] Thus, α,β-CHF-dATP and 7 are well accommodated in the polymerase active site as has been shown previously for α,β-CF2-dTTP.[4b]
Figure 3.
A) The ternary complex crystallographic structures of pol β with an incoming α,β-CF2-dATP (7) (gray carbons) and α,β-NH-dUTP (light blue carbons; PDB ID 2FMS)[8] were superimposed using all 326 Cα (rmsd = 0.29 Å). The incoming nucleoside triphosphates are shown as well as the primer terminal nucleoside (O3′ is labeled). The position of the active site Mg2+ (spheres) are colored according to the respective carbons. A and B refer to the catalytic and nucleotide binding metal, respectively. B) The ternary complex crystallographic structures of pol β with an incoming 7 (gray carbons) and α,β-CHF-dATP (6b) (yellow carbons) were superimposed using all 326 Cα (rmsd = 0.15 Å). C) A difference density map generated using the R-CHF isomer shows both positive (green, contoured at 3.2σ) and negative density (red, contoured at 4.0σ) in the vicinity of the position of the proposed fluorine atom indicating that the orientation for the R-isomer is not correct.
Molecular docking of 5 and 6 with pol β-DNA binary complex using Autodock Vina[7] suggests that the unexpectedly low binding affinity of 5 can be attributed to an unfavorable steric interaction, specifically a clash of the methyl groups of 5 with a structural water bound to Asp276 (PDB ID 3TFS water 3). When docked in the absence of this structural water, the lowest energy conformations for both 5 and 6 overlap well with the coordinates of the 6 found in the crystal structure, whose conformation is congruent with those of DNA pol β-bound α,β-CF2-dTTP[4b] and α,β-NH-dUTP.[8] However, when docking using a macromolecule file prepared to include the water bound to Asp276, 5 is unable to achieve the experimentally observed conformation while the geometry of 6 is unperturbed (SI, Figure S27).
Incubation of crystals of a binary DNA complex of pol β with the α,β-CHF-dATP diastereomer mixture (6a/b) resulted in a complex in which only the (S)-CHF-stereoisomer (6b) could be found by X-ray analysis (Figures 1 and 3B). Modeling (R)-α,β-CFH-dATP (6a) into the electron density failed to account for the observed density (Figure 3C). The close overlap of the CF2- and the (S)-CHF- conformations indicates that exclusion of 6a analogue is not the result of an unattainable binding conformation or destabilizing steric interaction (Figure 3B). As we reasoned previously,[4d, 4e] stabilization of as little as ~1 kcal/mol could be sufficient to generate the selectivity observed in the crystal structures.
In an earlier study, we reported a series of stereospecifically formed pol β-DNA-dGTP ternary complexes that provided evidence for a stabilizing polar interaction between the active site Arg183 and the chiral phosphonate C-F in CHF, CMeF, and CClF dGTP-β,γ analogues.[4d, 4e] Unlike the β,γ-CXF-dGTP-DNA-pol β ternary complexes, in this structure no direct stabilizing interactions with active site residues can be identified. The structure does, however, reveal that the (S)-isomer fluorine is within 2.8 Å (B-factor: 15.4 Å2) of an oxygen in a structural water molecule, present in both structures (for CF2 distance = 2.9 Å, B-factor: 15.1 Å2), that is within hydrogen bonding distance of several key atoms (a carboxylate anion oxygen and carboxyamide nitrogen of Asp276, phosphoryl oxygen of Pβ; Figure 1). Observing no similar interactions with the pro-(R)-fluorine in the CF2-structure, one might consider a weak interaction of the (S)-fluorine in 6b with structural water as the source of its stabilization in the complex relative to 6a, however other factors might be invoked[10] and further work will be necessary to clarify the origin of the observed stereospecificity.
In summary, our results show the influence of a structural water bound to Asp276 on the nucleotide binding properties of DNA pol β. Kd determinations for a series of α,β-bridging substituted dATP analogues display a correlation with the basicity of the bisphosphonate moiety and the binding affinity to pol β. This trend, however, does not extend to the α,β-C(Me)2-dATP analogue which displays a larger than expected Kd attributed to an unfavorable steric interaction of the methyl substituents with the structural water bound to Asp276. The X-ray crystal structure of α,β-CHF-dATP in complex with pol β and DNA contains only one diastereomer despite the presence of both diastereomers in the crystallization mixture. Comparison of this structure with that of α,β-CF2-dATP raises the possibility of a non-covalent weak interaction between the ligand and the active site structural water proximal to Asp276. Our structural observations may be worth taking into account in PDB studies[11] searching for fluorine/protein interactions.
Supplementary Material
Acknowledgments
This research was supported by NIH grant 5U19CA105010. We thank Ms. Inah Kang for assistance in preparing the manuscript.
Contributor Information
Dr. Charles E. McKenna, Email: mckenna@usc.edu.
Dr. Samuel H. Wilson, Email: wilson5@niehs.nih.gov.
References
- 1.Beard WA, Wilson SH. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 2.Sobol RW, Horton JK, Kühn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 3.Starcevic D, Dalal S, Sweasy JB. Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 4.a) Sucato CA, Upton TG, Kashemirov BA, Osuna J, Oertell K, Beard WA, Wilson SH, Florian J, Warshel A, McKenna CE, Goodman MF. Biochemistry. 2008;47:870–879. doi: 10.1021/bi7014162. [DOI] [PubMed] [Google Scholar]; b) Upton TG, Kashemirov BA, McKenna CE, Goodman MF, Prakash GKS, Kultyshev R, Batra VK, Shock DD, Pedersen LC, Beard WA, Wilson SH. Org Lett. 2009;11:1883–1886. doi: 10.1021/ol701755k. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chamberlain BT, Upton TG, Kashemirov BA, McKenna CE. J Org Chem. 2011;76:5132–5136. doi: 10.1021/jo200045a. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Batra VK, Pedersen LC, Beard WA, Wilson SH, Kashemirov BA, Upton TG, Goodman MF, McKenna CE. J Am Chem Soc. 2010;132:7617–7625. doi: 10.1021/ja909370k. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) McKenna CE, Kashemirov BA, Upton TG, Batra VK, Goodman MF, Pedersen LC, Beard WA, Wilson SH. J Am Chem Soc. 2007;129:15412–15413. doi: 10.1021/ja072127v. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Prakash GKS, Zibinsky M, Upton TG, Kashemirov BA, McKenna CE, Oertell K, Goodman MF, Batra VK, Pedersen LC, Beard WA, Shock DD, Wilson SH, Olah GA. Proc Natl Acad Sci U S A. 2010;107:15693–15698. doi: 10.1073/pnas.1007430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn GM, Langston SP. Tetrahedron Lett. 1991;32:6425–6428. [Google Scholar]
- 6.Liang F, Jain N, Hutchens T, Shock DD, Beard WA, Wilson SH, Chiarelli MP, Cho BP. J Med Chem. 2008;51:6460–6470. doi: 10.1021/jm800692a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trott O, Olson AJ. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH. Structure (Camb) 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radhakrishnan R, Arora K, Wang Y, Beard WA, Wilson SH, Schlick T. Biochemistry. 2006;45:15142–15156. doi: 10.1021/bi061353z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunitz JD, Schweizer WB. Chem--Eur J. 2006;12:6804–6815. doi: 10.1002/chem.200600092. [DOI] [PubMed] [Google Scholar]
- 11.a) Dalvit C, Vulpetti A. ChemMedChem. 2011;6:104–114. doi: 10.1002/cmdc.201000412. [DOI] [PubMed] [Google Scholar]; b) Carosati E, Sciabola S, Cruciani G. J Med Chem. 2004;47:5114–5125. doi: 10.1021/jm0498349. [DOI] [PubMed] [Google Scholar]; c) Dunitz JD, Taylor R. Chem--Eur J. 1997;3:89–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.