Abstract
Background & objectives:
Pipistrellus ceylonicus bat species is widely distributed in South Asia, with additional populations recorded in China and Southeast Asia. Bats are the natural reservoir hosts for a number of emerging zoonotic diseases. Attempts to isolate bat-borne viruses in various terrestrial mammalian cell lines have sometimes been unsuccessful. The bat cell lines are useful in isolation and propagation of many of the viruses harboured by bats. New stable bat cell lines are needed to help such investigations and to assist in the study of bat immunology and virus-host interactions. In this study we made an attempt to develop a cell line from P. ceylonicus bats.
Methods:
An effort was made to establish cell line from embryo of P. ceylonicus species of bat after seeding to Dulbecco's modified eagle medium (DMEM) supplemented with 10 per cent foetal bovine serum; a primary cell line was established and designated as NIV-BtEPC. Mitochondrial DNA profile analysis was done using cyt-b and ND-1 gene sequences from the cell line. Phylogenetic tree was constructed using neighbour-joining algorithm for cyt-b and ND-1 genes with 1000-bootstrap replicates.
Results:
NIV-BtEPC cell line was susceptible to Chandipura (CHPV) and novel adenovirus (BtAdv-RLM) isolated from Rousettus leschenaulti from India but did not support multiplication of a number of Bunyaviruses, Alphaviruses and Flavivirus. This might be useful for isolation of a range of viruses and investigation of unknown aetiological agents.
Interpretation & conclusions:
In this study, a new bat cell line was developed from P. ceylonicus. This cell line was successfully tested for the susceptibility to Chandipura and BtAdv-RLM virus isolated from bats. The approach developed and optimised in this study may be applicable to the other species of bats and this established cell line can be used to facilitate virus isolation and basic research into virus-host interaction.
Keywords: Arboviruses, cell lines, cytochrome-b gene, nicotinamide adenine dinucleotide dehydrogenase subunit-1, Pipistrellus ceylonicus
Bats are reservoir of many highly infectious pathogens like Ebola, Marburg, severe acute respiratory syndrome (SARS), coronavirus (CoV) and Nipah viruses1. During 2001, Nipah virus outbreak was reported in India2. Adjoining countries Bangladesh and China had recorded bat species harbouring Nipah virus and isolated the virus3. West Nile virus and Parainfluenza virus type-2 were isolated from frugivorous bat Rousettus leschenaulti, while kyasanur forest disease (KFD) virus was isolated from Cynopterus sphinx bat species from India4,5.
Pipistrellus ceylonicus bat species is largely distributed in South Asia. It is presently found in Bangladesh, India (Andhra Pradesh, Bihar, Goa, Gujarat, Jharkhand, Karnataka, Kerala, Madhya Pradesh, Maharashtra, Rajasthan, Tamil Nadu and West Bengal), Pakistan (Punjab and Sind) and Sri Lanka (Central, Eastern, Uva and Western provinces)6,7. This species can be found in varied habitats from arid regions to humid Montane forests. It roosts in human habitations in both rural and urban areas, in old dilapidated buildings, crevices and cracks in walls, tree hollows, trees holes, caves, wells, old temples and under overhanging ledges.
Recently, bat derived adenoviruses have been isolated in China, Japan and Germany8,9,10, with remarkable genetic diversity. This points out the requirement of bat originated cell line for virus isolation. Although DNA viruses have been discovered in bats, but the majority of viruses identified in bats are RNA viruses11. In addition, PCR screening has revealed the presence of seven genetically different gamma herpes viruses and one novel beta herpes virus in seven different European bat species in the family Vespertilionidae12. Another study using PCR analysis has identified a novel gamma herpes virus in a serotine bat (Eptesicus serotinus) in Hungary during an investigation of a sick bat13. Looking at the requirement of better cell line for the virus isolations, an effort was made to develop a cell line from bat P. ceylonicus for virus isolation and other studies.
Material & Methods
This study was conducted during March 2010 at National Institute of Virology (NIV), Pune which is responsible for investigation of viral disease outbreaks including zoonosis in India. The study protocol was approved by the Institutional Animal Ethical Committee (IAEC) permitting the use of bat for development of cell line. Capturing of bats was done after getting permission of Chief Conservator of Forests, Maharashtra State, India.
Development of cell line: A female P. ceylonicus bat was collected from the institute's building floor, that was probably dehydrated and moribund. On dissection, she was found in gravid status. Embryo and body organs were removed aseptically. These were washed with sterile phosphate buffer saline (PBS) and chopped finely and seeded in 25cm2 bottles with Dulbecco's modified eagle medium (DMEM) (Gibco BRL, USA). Cells were supplemented with DMEM and 10 per cent foetal bovine serum (FBS) (Gibco BRL, USA), 100 U penicillin/ml, and 0.1 mg streptomycin/ml at 37°C in an incubator supplemented with 5 per cent CO2 to initiate the cell line growth. The medium was prepared in the laboratory. The chemicals required were procured from Sigma, USA. The cells from other organs except embryo showed bacterial contamination and were discarded. Live cells from embryo were attached to the surface of wall of culture bottle in a few hours and dead cells were discarded next day and medium was changed. After 3rd day, trypsinization was done and cells were split into new culture bottles. The embryo cells of P. ceylonicus bat were screened for Nipah, Ebola and Marburg viruses (procured from NIV, Pune virus repository) by real-time RT-PCR and after establishment of cell line, therefore again tested for these pathogens14,15,16. Mycoplasma infection was screened using Universal Mycoplasma Detection Kit (ATCC, USA) as per manufacturer's instruction. The developed cell line was named NIV-BtEPC.
Susceptibility of cell line to viruses: The viruses used in the study to determine the susceptibility of the bat cell line NIV-BtEPC are mentioned in the Table. Virus susceptibility experiments for NIV-BtEPC cells were done in comparison with Vero E6 cells (ATCC, USA); after infection with different groups of viruses, these were observed for cytopathic effects (CPE). The virus stocks were prepared in the Vero E6 cell line and the dosages used to infect the Vero E6 and bat cells are mentioned in the Table. The viruses used in the study were confirmed by RT-PCR before the experiments (data not shown).
Table.
Comparative cytopathic profile of various viruses in NIV-BtEPC P. ceylonicus and Vero E6 cells lines
DNA extraction from bat tissues and bat cell line: At the seventh passage level, trypsinized cells were harvested from the bottles and pellet was washed twice in PBS. Total genomic DNA was extracted using QlAmp DNA mini kit (Qiagen, Germany), according to the manufacturer's instructions. Resulting DNA was re-suspended in 50 μl diethylpyrocarbonate (DEPC) treated nucleases free water. Similarly, DNA was also extracted from the liver tissues of other bat species viz. R. leschenaulti (Mahabaleshwar), C. sphinx (Siliguri, West Bengal), Pteropus giganteus (Vageshwar, Maharashtra), Megaderma lyra (Siliguri, West Bengal), which were collected earlier with the permission of Principle Chief Conservator of Forests from various parts of India during the survey of emerging viruses from bats. DNA was also extracted from other cell lines, which are used in this laboratory for various purposes viz. Vero E6 and C6/36 cell lines for comparison.
Polymerase chain reaction (PCR) for amplification of mitochondrial gene and sequencing: Each DNA sample of individual bats and cell lines was amplified and sequenced for two mitochondrial genes, Cytochrome b gene (cyt-b) and Nicotinamide adenine dinucleotide dehydrogenase subunit 1 (NADH1/ND1). The partial Cyt-b gene was amplified using two primers (L-14724, 5’-CGA AGC TTG ATA TGA AAA ACC ATC GTT G3’) and (H-15149, AAA CTG CAG CCC CTC AGA ATG ATA TTT GTC CTC A-3’), which were earlier used for species determination17,18. The letters L and H refer to the light and heavy strands and the number refers to the position of the 3’ base of the primer in the complete mitochondrial DNA sequence. PCR was performed using Platinum Taq DNA polymerase kit (Invitrogen, USA). Amplification included 3’ initial denaturation at 95°C, followed by 35 cycles at 95°C (45 sec), 50°C (45 sec), 72°C (1 min 30 sec), with a final extension at 72°C (30 min).
Subsequent PCR amplifications of partial ND-1 gene were carried out using ER65 (L-2985, 5’CCT CGA TGT TGG ATC AGG 3’) and ER66 (H-4419, 5’GTA TGG GCC CGA TAG CTT-3’) primers. PCR conditions used were as follows; 95°C- 5 min, 40 cycles of 94°C- 30 sec, 50°C for 30 sec and 72°C for 90 sec; and a final 10 min extension at 72°C.
The amplified products were analyzed on 2 per cent agarose gels in Tris-acetate EDTA buffer (0.04 M Tris-acetate, 1 mM EDTA) and stained with ethidium bromide and visualized on ultraviolet transilluminator at 302 nm. The bands of appropriate sizes (486 bp product of Cyt-b gene and 800 bp of ND-1 gene) were recovered from the gel using QIAgen DNA Gel Extraction Kit (Qiagen), as per the manufacturer's protocol. Direct sequencing of the amplified product was performed with the primers L-14724 and H-15149 in case of Cyt-b, while in ND-1 along with ER-65 and ER-66, two internal primers ER-89 (5’-CTC TAT CAA AGT AAC TCT TTT ATC AGA C-3’) for the H-strand and ER-175 (5’-GGC TGG GCC TCA AAC TCN AAA TA3’) for the L-strand were also used, by using Big Dye terminator kit in an ABI 3100 automated DNA sequencer (Applied Biosystems, Inc., USA.
Phylogenetic data analysis: The sequences obtained for all the bat species and cell lines were analyzed by BLAST (http://blast.nci.nlm.nih.gov) and curetted using the software KODON (Applied Maths, USA)19. Curetted sequences were aligned using Clustal W program from MEGA v4.0 software20. Phylogenetic tree was constructed using neighbour-joining algorithm for Cyt-b and ND-1 genes with 1000-bootstrap replicates. Other representative sequences of Rousettus, Cynopterus, Pteropus and Megaderma bats were included from GenBank (http://blast.nci.nlm.nih.gov) for phylogenetic tree and distance analysis.
Results
The culture originated from embryo divided very fast during first 8-10 passages, the cell doubling time was of approximately 12 h. During this period, formation of small button like mass due to aggregation of cells was observed. Therefore, initially the culture was subcultured at an interval of 2-3 days. This was continued until the 10th passage level.
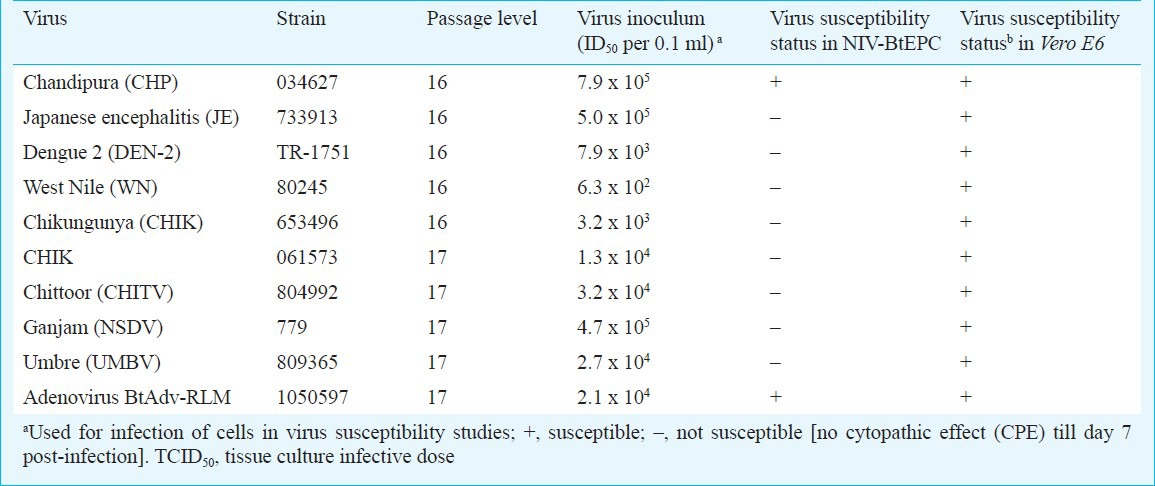
After 5th passage, the cell line was cryopreserved and after three days could be revived again and established properly. The aggregation of cells and small mass formations was not seen in the cell line after cryopreservation. This phenomenon gradually disappeared as the passage levels increased. At early passages between 1 and 5 the morphology study showed presence of more than one type of cells (Fig. 1), however, the later passages showed monomorphic type of cells (Fig. 2A).
Fig. 1.
NIV-BtEPC P. ceylonicus monolayer culture, showing heterogeneous cell population (arrow), at P-8, stained with Giemsa stain [100X].
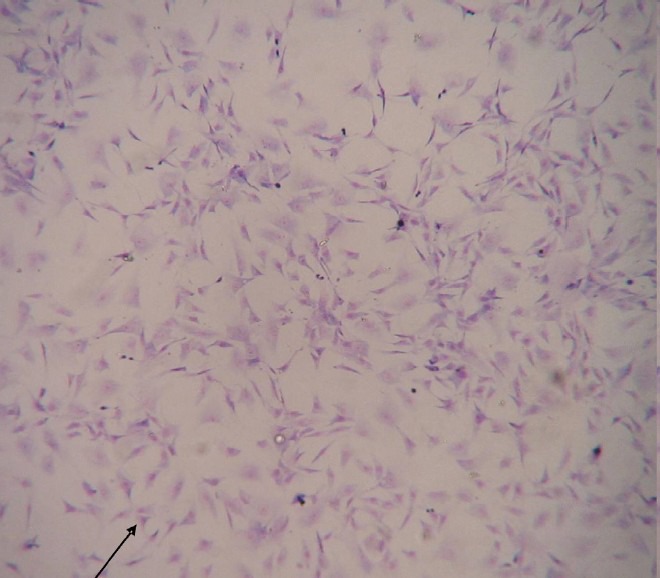
Fig. 2.
Comparison of susceptibility of Vero E6 and NIV-BtEPC cells after 1 days post infection with Chandipura virus (CHPV). (A) NIV-BtEPC control uninfected cells showing monolayer (arrow) at 10X amplification. (B) NIV-BtEPC cells infected with Chandipura virus showing cell rounding and detachments, foci formation and disintegration of cells monolayer (arrow) at 20X amplification. (C) Vero E6 control uninfected cells showing monolayer (arrow) 10X amplification. (D) Vero E6 cells infected with Chandipura virus showing cell rounding, detachments and disintegration of cells monolayer (arrow) 10X amplification.
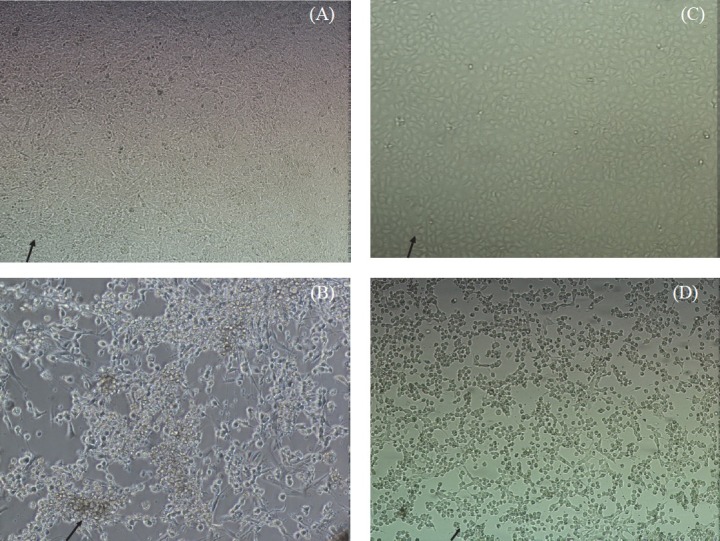
After 25th passage level, the susceptibility of this cell line to various viruses was determined. The cell line was found refractory to support the growth of alphavirus (Chikungunya), flaviviruses (dengue virus- DEN-2, West Nile, Japanese encephalitis) and bunyaviruses (Chittoor, Umbre, Ingwavuma and Ganjam virus). It showed cytopathic effects only with Chandipura (rhabdoviruses) and a novel adenovirus isolated from R. leschenaulti bats from India21 (Table). CPE was observed after eight hours of virus infection in the case of CHPV and progressed to cell rounding and detachments within 24 h for NIV-BtEPC cells. At 3rd post-infection day (PID) the total cell sheet was detached (Fig. 2A, B, C, D). Similarly, in the case of CHPV infected Vero E6 cells CPE started at 8th hour and totally detached at 24th hour as reported earlier22. CPE was characterized by cell rounding, foci formation and disintegration of cells and finally cell death. In case of NIV-BtEPC cells infected with Adenovirus, CPE was obvious after 2nd PID. Cells became rounded and clumped after 48 h and detached slowly but not 100 per cent. In Vero E6 cells adenovirus infected cells showed CPE, starting on the 3rd PID (Fig. 3A, B, C, D).
Fig. 3.
Comparison of susceptibility of Vero E6 and NIV-BtEPC cells after post infection of 3 days with BatAdV. (A) NIV-BtEPC control cells at passage 25 showing monomorphic (arrow) cell sheet at 20X amplification at 20X amplification. (B) NIV-BtEPC cells infected with Bat AdV showing cell rounding and cell clumping and detachment at 3rd PID (arrow) at 20X amplification. (C) Vero E6 control uninfected cells showing monolayer (arrow) at 20X amplification. (D) Vero E6 cells infected with Bat AdV showing cell rounding and cell clumping at 3rd PID (arrow).
DNA extraction and PCR amplification of partial Cyt-b and ND-1 genes products from bat tissues and bat cell line: The use of L-14724 and H-15149 primers resulted in successful PCR amplification of a 486 bp product of Cyt-b gene and 800 bp of ND-1 gene. The PCR products were sequenced to verify the authenticity of the PCR products and to obtain the fragment nucleotide sequences of these samples.
Phylogenetic analysis of partial Cyt-b and ND-1 genes of bat cell line and other representative bat species: Phylogenetic tree construction revealed formation of two clear lineages viz. 1 of Vespertilionidae family and 2 of Pteropodidae, Rhinolophidae, and Megadermatidae. R. leschenaultii, P. giganteus, C. sphinx and M. lyra bat species were placed in their respective family and genus. Cell line sequence was placed in family Vespertilionidae, genus Pipistrellus with other Pipistrellus bat sequences. This finding was consistent for both ND-1 and Cyt-b genes (Figs 4 and 5). Bat cell line nucleotide sequences showed highest homology with Pipistrellus species of bats with 86 to 87 per cent and 80 to 84 per cent for Cyt-b and ND-1 genes, respectively. Next close sequence was Pteropus with 75 per cent and Pipistrellus cell line sequences with 68 per cent homology for Cyt-b and ND-1 gene, respectively.
Fig. 4.
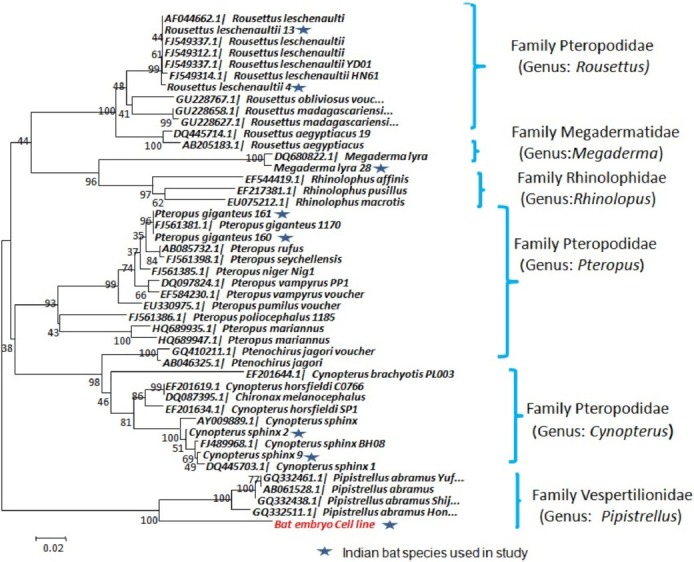
Phylogenetic tree of Cyt-b gene of P. ceylonicus cell line and other bat species collected from India and sequences available from GenBank showing formation of clear grouping of bat cell line sequence among Pipistrellus genus.
Fig. 5.
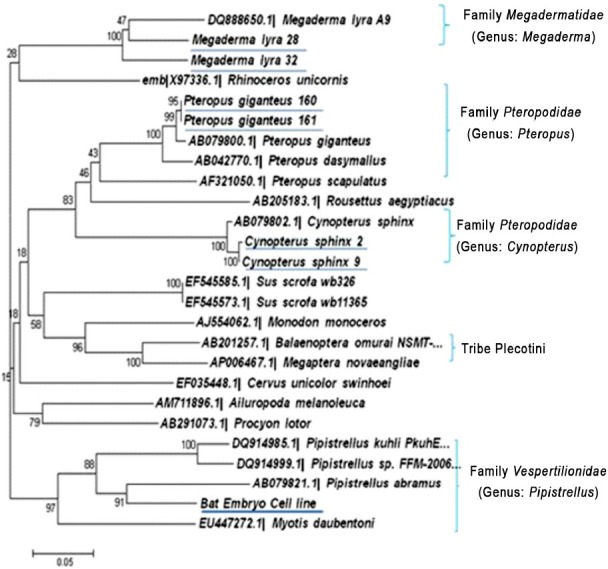
Phylogenetic tree of ND-1 gene of P. ceylonicus cell line and other bat species collected from India showing the formation of separate lineages of genus Pipistrellus sequences.
Discussion
Recent evidence of bat-borne viruses crossing the species barrier and causing severe diseases in humans and other animals, suggests the urgent need to establish cell lines from various bat species to facilitate virus isolation and understand virus-host interaction. Many bat-borne viruses like Ebola and Nipah cause fatal infections in human but reservoirs remain healthy. Therefore, understanding the bat immune responses and their role in maintaining the symbiotic presence of a large number of viruses, apparently without causing clinical disease is important23. Bats harbour many infectious agents ranging from risk group-2 to -41. Cell lines have been established from Egyptian fruit bat (R. aegyptiacus), which is fully permissive for a highly attenuated poxvirus, modified vaccinia Ankara (MVA)24. Myotis bat primary kidney cell line was reported to be susceptible for bat derived adenovirus isolation25. The Pteropus bat derived cell line was found susceptible for Hendra and Nipah viruses26. The present cell line NIV-BtEPC was selectively susceptible to rhabdo and adenoviruses. Therefore, it would be worthwhile to examine the susceptibility of this cell line to Nipah, SARS, CoV and Ebola viruses. At present, isolates of these highly pathogenic viruses are not available in India but a novel adenovirus was isolated from R. leschenaulti bat21 and Nipah viral RNA was detected from P. giganteus species of bats from India (NIV unpublished data). The multiplication of Chandipura virus (family: Rhabdoviridae) in NIV-BtEPC cell line also suggests that this species of bats perhaps may support multiplication of rabies virus, which is a member of the same family and has been isolated and detected in many bat species27. Highly divergent mitochondrial lineages provide strong evidence for cryptic species diversity in Indian bats. Close relatedness of cell line mitochondrial homology with Pipistrellus group of bats proved the identity of this cell line. Looking at the infectious and unknown disease spectrum of viruses, there is a need of developing new cell lines from the bats.
In conclusion, a new cell line NIV-BtEPC was establised from P. ceylonicus bat species which could be used for research on bat harbouring viruses and their isolation.
Acknowledgment
Authors thank the Principal Chief Conservator of Forests, Maharashtra and West Bengal for kind permission to collect samples from bats in Mahabaleshwar, Maharashtra, and Siliguri, West Bengal, India, and express gratitude to Dr A.C. Mishra, Former Director, National Institute of Virology for critical review of the manuscript. Authors also thank Shri U.K. Shende for technical help and to Shriyut M. Karanjawane, S.V. Gopale, G.K. Chopade and M. Holeppanavar, the bat capture team for their assistance in the field.
References
- 1.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–45. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ, et al. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis. 2006;12:235–40. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong HT, Abdullah S, Tan CT. Nipah virus and bats. Neurol Asia. 2009;14:73–6. [Google Scholar]
- 4.Pavri KM, Singh KR. Kyasanur Forest disease virus infection in the frugivorous bat, Cynopterus sphinx. Indian J Med Res. 1968;56:1202–4. [PubMed] [Google Scholar]
- 5.Pavri KM, Singh KR, Hollinger FB. Isolation of a new parainfluenza virus from a frugivorous bat, Rousettus leschenaulti, collected at Poona, India. Am J Trop Med Hyg. 1971;20:125–30. doi: 10.4269/ajtmh.1971.20.125. [DOI] [PubMed] [Google Scholar]
- 6.Khan MA. Status and distribution of bats in Bangladesh with notes on their ecology. Zoos’ Print J. 2001;16:479–83. [Google Scholar]
- 7.Korad V, Yardi K, Raut R. Diversity and distribution of bats in the Western Ghats of India. Zoos’ Print J. 2007;22:2752–8. [Google Scholar]
- 8.Li Y, Ge X, Zhang H, Zhou P, Zhu Y, Zhang Y, et al. Host range, prevalence and genetic diversity of adenoviruses in bats. J Virol. 2010;84:3889–97. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda K, Hondo E, Terakawa J, Kiso Y, Nakaichi N, Endoh D, et al. Isolation of novel adenovirus from fruit bat (Pteropus dasymallus yayeyamae) Emerg Infect Dis. 2008;14:347–9. doi: 10.3201/eid1402.070932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonntag M, Muhldorfer K, Speck S, Wibbelt G, Kurth A. New adenovirus in bats, Germany. Emerg Infect Dis. 2009;15:2052–5. doi: 10.3201/eid1512.090646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong S, Lau S, Woo P, Yuen KY. Bats as a continuing source of emerging infections in humans. Rev Med Virol. 2007;17:67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wibbelt G, Kurth A, Yasmum N, Bannert M, Nagel S, Nitsche A, et al. Discovery of herpesviruses in bats. J Gen Virol. 2007;88:2651–5. doi: 10.1099/vir.0.83045-0. [DOI] [PubMed] [Google Scholar]
- 13.Molnar V, Janoska M, Harrach B, Glavits R, Palmai N, Rigo D, et al. Detection of a novel bat gamma herpes virus in Hungary. Acta Vet Hung. 2008;56:529–38. doi: 10.1556/AVet.56.2008.4.10. [DOI] [PubMed] [Google Scholar]
- 14.Guillaume V, Lefeuvre A, Faure C, Marianneau P, Buckland R, Lam S, et al. Specific detection of Nipah virus using real-time RT-PCR (TaqMan) J Virol Methods. 2004;120:229–37. doi: 10.1016/j.jviromet.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, et al. Discovery of Swine as a host for the Reston ebolavirus. Science. 2009;325:204–6. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- 16.Towner JS, Amman BR, Tara K, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linacre A, Lee JC. Species determination: the role and use of the cytochrome B gene. Methods Mol Biol. 2005;297:45–52. [PubMed] [Google Scholar]
- 18.Mayer F, von Helversen O. Cryptic diversity in European bats. Proc Biol Sci. 2001;268:1825–32. doi: 10.1098/rspb.2001.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yadav PD, Vincent MJ, Khristova M, Kale C, Nichol ST, Mishra AC, et al. Genomic analysis reveals Nairobi sheep disease virus to be highly diverse and present in both Africa, and in India in the form of the Ganjam virus variant. Infect Genet Evol. 2011;11:1111–20. doi: 10.1016/j.meegid.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 21.Raut CG, Yadav PD, Towner JS, Amman BR, Erickson BR, Cannon DL, et al. Isolation of a novel adenovirus from Rousettus leschenaultii bats from India. Intervirology. 2012;55:488–90. doi: 10.1159/000337026. [DOI] [PubMed] [Google Scholar]
- 22.Jadi RS, Sudeep AB, Kumar S, Arankalle VA, Mishra AC. Chandipura virus growth kinetics in vertebrate cell lines, insect cell lines & embryonated eggs. Indian J Med Res. 2010;132:155–9. [PubMed] [Google Scholar]
- 23.Crameri G, Todd S, Grimley S, McEachern JA, Marsh GA, Smith C, et al. Establishment, immortalisation and characterisation of Pteropid bat cell lines. PLoS One. 2009;4:e8266. doi: 10.1371/journal.pone.0008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan I, Horn D, Oehmke S, Leendertz FH, Sandig V. Cell lines from the Egyptian fruit bat are permissive for modified vaccinia Ankara. Virus Res. 2009;145:54–62. doi: 10.1016/j.virusres.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Ge X, Zhang H, Zhou P, Zhu Y, Zhang Y, et al. Host range, prevalence and genetic diversity of adenoviruses in bats. J Virol. 2010;84:3889–97. doi: 10.1128/JVI.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banyard AC, Hayman D, Johnson N, McElhinney L, Fooks AR. Bats and lyssaviruses. Adv Virus Res. 2011;79:239–89. doi: 10.1016/B978-0-12-387040-7.00012-3. [DOI] [PubMed] [Google Scholar]
- 27.King A, Davies A, Lawrie A. The rabies viruses of bats. Vet Microbiol. 1990;23:165–74. doi: 10.1016/0378-1135(90)90146-m. [DOI] [PubMed] [Google Scholar]


