Abstract
Background & objectives:
Mycoplasma pneumoniae is the most important and common cause of community-acquired pneumonia (CAP). The conventional detection methods (culture and serology) lack sensitivity. PCR offers a better approach for rapid detection but is prone to carry over contamination during manipulation of amplification products. Quantitative real-time PCR (qRT-PCR) method offers an attractive alternative detection method. In the present study, qRT-PCR, PCR and serology methods were used to detect M. pneumoniae infection in cases of pneumonias and findings compared.
Methods:
A total of 134 samples consisting of blood (for serology) and respiratory secretions (for PCR and qRT-PCR) from 134 patients were collected. The blood samples were tested for IgG, IgM and IgA using commercially available kits. For standardization of PCR of M. pneumoniae P1 gene was cloned in pGEMTEasy vector. Specific primers and reporter sequence were designed and procured for this fragment. The qRT-PCR assay was performed to prepare the standard curve for M. pneumoniae positive control DNA template and detection in patient samples.
Results:
Of the 134 patients, 26 (19%) were positive for antibodies against M. pneumoniae. IgG was positive in 14.92 per cent (20) cases, IgM in 4.47 per cent (6) and IgA was positive in 5.22 per cent (7) cases. In the qRT-PCR assay 19 per cent (26) samples were positive. Of the 26 qRT-PCR positive samples, nine could be detected by serology. PCR was positive for 25 samples. An extra sample negative by PCR was detected by qRT-PCR. Thus, real-time PCR assay, PCR and serology in combination could detect M. pneumoniae infection in 43 patients.
Interpretation & conclusions:
The study shows that 17 patients were detected by serology alone, 17 were detected by qRT-PCR only and nine patients were positive by both serology and real-time PCR. Of the 134 samples tested, 25 were positive by conventional PCR, but qRT-PCR could detect one more sample that was negative by PCR and serology. These results suggest that a combination of two or three methods may be required for reliable identification of CAP due to M. pneumoniae.
Keywords: Community-acquired pneumonia (CAP), Mycoplasma pneumonia, Quantitative real-time PCR (QRT-PCR), serology
Mycoplasma pneumoniae accounts for as many as 10-30 per cent of all cases of commonly acquired pneumonia (CAP)1 in general population and for 25-71 per cent in closed populations2 such as students and military recruits living in dormitories. M. pneumoniae has been frequently observed in patients suffering with respiratory illness and is also reported to be associated with acute exacerbation of bronchial asthma and chronic obstructive pulmonary disease (COPD)3, acute respiratory distress syndrome (ARDS)4, polyarthritis5, stroke6, Guillain-Barre syndrome7, and coronary artery diseases (CAD)8.
Infections caused by M. pneumoniae have been recognized worldwide. Earlier infections with M. pneumoniae were generally reported to be affecting people between the age of 5 and 25 yr. The incidence of M. pneumoniae pneumonia requiring hospitalization increases with age, highlighting the importance of this pathogen in the elderly hospitalized with pneumonia. Clinically M. pneumoniae pneumonia cannot be differentiated from pneumonia caused by other bacteria and viruses. Mycoplasma infection in India has been reported for 35 per cent patients with community acquired pneumonia in both children as well as in adults9. M. pneumoniae can produce a wide range of clinical symptoms ranging from pneumonia, bronchitis, upper respiratory disease to inapparent infections.
M. pneumoniae grows slowly in culture and can take weeks to grow1,10. Thus, it is difficult to carry out culture analysis for routine diagnosis. Diagnosis of M. pneumoniae infection still relies on conventional serological procedures; however, these are generally non-specific and lack sensitivity. Molecular diagnosis by PCR assays have been described for the detection of M. pneumoniae. Targets reported for PCR are P1 gene11,12, 16SrRNA13 and Tuf gene encoding elongation factor Tu14,15. Real-time PCR16,17,18 based assays targeting some of the above mentioned genes have been described and have an advantage over conventional PCR in terms of sensitivity and specificity19,20. In the present study, a P1 gene based, real-time PCR assay was employed to detect M. pneumoniae infections among Indian patients. Further, conventional PCR and detection of all immunoglobulin classes (IgG, IgM & IgA) were also attempted and results were compared.
Material & Methods
Strain and samples: Standard strain of M. pneumoniae M129 was procured from ATCC, USA. A total of 134 clinical samples consisting of blood and respiratory tract fluids (107 throat swabs, 19 nasopharyngeal aspirates, 6 endotracheal aspirates, and 2 broncoalveolar lavage) were collected between May 2005 and August 2008 from the patients diagnosed to have CAP and admitted at medicine & paediatrics wards of All India Institute of Medical Sciences (AIIMS), New Delhi, India. Serum extracted from blood samples and respiratory tract fluids was stored at -20°C until use. The study protocol was approved by the Institute's ethics committee. Patients included in the study were based on the following criteria:
Inclusion criteria: (i) Community acquired pneumonia (CAP): Presence of at least one of the major clinical criteria (cough, sputum production, fever > 37.8°C) or two of the minor criteria (pleuritic chest pain, dyspnoea, altered mental state, sign of pulmonary consolidation on examination or total leukocyte count of 12000/μl).
(ii) Presence of a new pulmonary infiltrate/ shadow on chest X-ray suggestive of pneumonia at/ within 24 h of hospitalization.
(iii) Patient residing in community.
Exclusion criteria: (i) Hospital acquired pneumonia i.e. pneumonia that developed 72 h after hospitalization or within 7 days of discharge.
(ii) Pulmonary shadow due to a cause other than pneumonia.
All patients fulfilling the inclusion criteria were included.
Culture: The M. pneumoniae standard strain was revived according to ATCC guidelines (www.atcc.org). In brief, the lyophilized culture was resuspended in 6 ml pleropnemonia like organism (PPLO) broth. A single drop was used to inoculate PPLO agar; 3 ml suspension was used to prepare glycerol stocks and stored at -70°C. The remaining 3ml suspension was incubated at 37°C and 5 per cent CO2 incubator till growth was observed with change in colour from red to yellow.
Genomic DNA: PPLO broth culture or samples in PPLO broth after colour change to yellow were used for genomic DNA extraction. Culture (200 μl) was centrifuged at 22,000 g for 15 min to pellet the cells. The pellet was washed with PBS (pH 7.2). After washing twice, pellet was resuspended in 100 μl autoclaved double distilled water. Tubes were placed in boiling water bath for 10 min and then centrifuged for 30 sec to sediment the debris21,22. The supernatant was collected in new micro-centrifuge tubes and stored at -20°C until used.
PCR and positive control preparation: For M. pneumoniae P1 adhesin gene specific primers were used (Table I). The target sequence for amplification was a 543 bp segment of the gene coding for P1 adhesin protein. The 25 μl PCR reaction consisted of 1X PCR buffer (Bangalore Genei, India), 1.5 mM MgCl2 (Bangalore Genei, India), 200 μM dNTPs (MBI Fermentas, USA), 20 pmol of each primer (Sigma-Aldrich, USA), 1 Unit of Taq polymerase (Bangalore Genei, India) and 5 μl of extracted genomic DNA. The reaction was performed in a thermocycler (Perkin Elmer, USA). The target sequence for amplification was a 543bp fragment of the P1 gene and PCR was done as described earlier23. In brief, PCR run consisted of 35 cycles of amplification, each at 94°C for 1 min, 55°C for 1 min and 72°C for 2 min and final elongation step of 10 min at 72°C. A negative control was systematically run in parallel. This method was used for PCR amplification from patient samples. The 543 bp PCR product from M. pneumoniae M129 (ATCC, USA) standard strain was cloned in pGEM-T Easy (Promega, USA) vector according to manufacturer's instructions. The positive clone was confirmed by restriction digestion and sequencing. This clone was used subsequently as a positive control for standardization of M. pneumoniae PCR and real-time PCR assay by serially diluting the plasmid.
Table I.
PCR and real-time PCR primers for P1 adhesin gene of M. pneumoniae

Quantitative real-time PCR assay: The clone once confirmed by sequencing was used to extract DNA in large quantity. For this, a 100 ml Luria Bertani (LB) broth containing ampicillin (100 μg/ml) was inoculated with the glycerol stock. The culture was incubated at 37°C in a shaker. The overnight grown culture was harvested and the plasmid extraction was done using the Qiagen Midi kit (Qiagen, USA). The plasmid DNA extracted was quantified spectrophotometrically. The final concentration of the plasmid DNA was 575 ng/μl. The copy number calculation was done using the following formula:
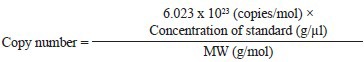
Conc. of std- Obtained spectrophotometrically
MW- Molecular weight of each pGEM-T Easy plasmid with cloned gene.
Using the above formula M. pneumoniae P1 gene cloned in pGEM-T Easy vector was calculated to be 14 × 1010 copies/μl and a set of dilutions were prepared for standard curve preparation. The primers for real time assay were synthesized corresponding to a 73 bp fragment internal to the 543 bp fragment of P1 gene which is detected by Fam-dye labelled reporter sequence. These primers and reporter sequences (Table I) were synthesized by Applied Biosystems, USA. Along with the positive reactions of dilutions of template DNA, a no-template control (NTC) was also included. The reactions were performed in a final volume of 20 μl containing 1 μl 20X assay mix (primers & probe), 10μl 2x TaqMan® Universal Master Mix (enzyme, buffer & dNTPs) and 9 μl DNA diluted in RNase free water. Amplification and product detection were performed with the ABI Prism™ 7700 Sequencing Detection System, USA. Once the reaction for standard curve was finalized, similar reactions were performed for DNA extracted from patients’ throat swab samples.
Serology: Serum was separated from the venous blood samples and stored at -20°C till assayed. Serum specimens collected in acute phase from patients were used to detect IgM, IgA and IgG antibodies against M. pneumoniae by commercial ELISA kits (Verion/Serion, Germany). A titre of IgG >30 U/ml, a titre >1.1RU of IgM and a titre > 14 U/ml of IgA were considered as positive.
Statistical analysis: Fisher exact and Pearson Chi square tests were used to analyse clinical signs and symptoms and demographic data (age & sex).
Results
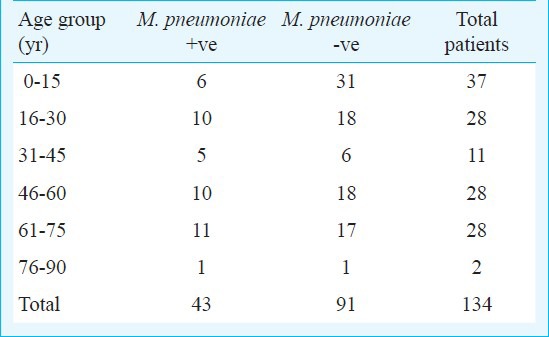
Demographic profile: A total of 134 patients were enrolled in the study. Of these, 92 (69%) were males and 42 (31%) were females. The patients positive for M. pneumoniae by any test (serology, PCR and real-time PCR) consisted of 27 (29%) males and 16 (38%) females. The study group included 37 (28%) paediatric and 97 (72%) adult patients. There were 6 (16%) paediatric and 37 (38%) adult patients positive for M. pneumoniae by any test done in the study. M. pneumoniae positivity using serology and qRT-PCR in different age groups is given in Table II. Overall, real-time PCR assay, PCR and serology in combination could detect M. pneumoniae infection in 43 patients.
Table II.
Age distribution for M. pneumoniae positive (serology & qRT-PCR) and negative patients
PCR and positive control preparation: The M. pneumoniae standard strain maintained in the laboratory was used for genomic DNA extraction. The genomic DNA extracted was used for PCR amplification of 543bp fragment of P1 gene (Fig. a). The PCR amplified product was cloned in pGEM-T Easy vector and confirmed by restriction digestion (Fig. b) and sequencing. This clone was used subsequently as a positive control for standardization of M. pneumoniae PCR and real-time PCR assay. A total of 25 patients were found positive for conventional PCR.
Fig.
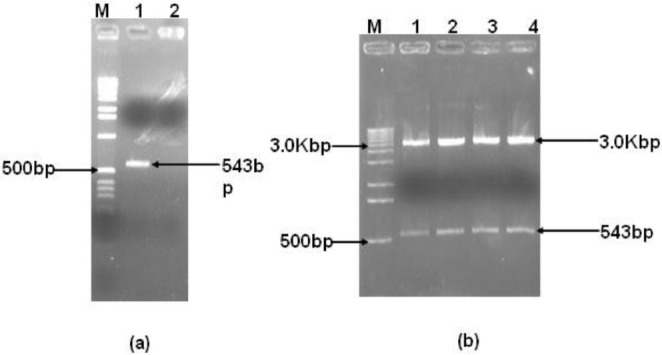
a. PCR amplification of P1 gene fragments for positive control preparation. Lane M: 1 kbp ladder, lane 1: P1 gene 543 bp positive PCR, lane 2: no template control. b. Restriction digestion of positive clones. Lane M: 1 kbp ladder, lanes 1-4: positive clones.
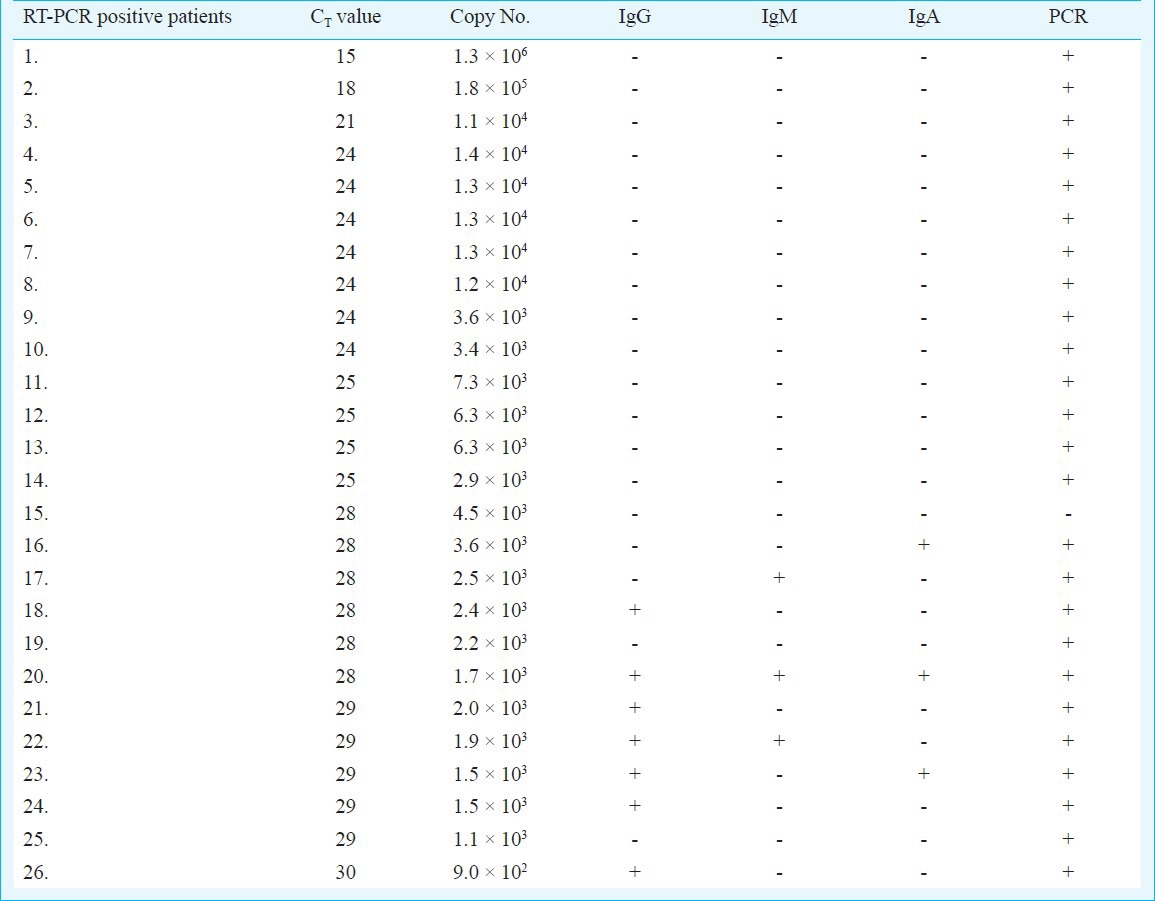
Real-time PCR: In the qRT-PCR assay for M. pneumonia, standard curve was obtained using P1 gene cloned in pGEM-T Easy vector as positive control. The cycle threshold (CT) values obtained for different dilutions of control plasmid were in the range of 10-31 for concentration range of 90 to 9×108 copies/reaction. The minimum detection limit was found to be 90 copies/reaction. The sensitivity and specificity of M. pneumoniae real time PCR assay were 34.6 and 84.2 per cent, respectively. Patient samples using DNA extracted from respiratory secretions (throat swabs, nasopharyngeal aspirates, endotracheal aspirates and broncoalveolar-lavage) were tested with the standardized assays. A total of 26 (19%) samples were found to be positive out of 134 patient samples by real-time PCR assay.
Serology: Of the 134 patients, 26 were positive for any class of immunoglobulins by ELISA; of these, IgG was positive in 20 (14.92%) cases, IgM was positive in 6 (4.47%) cases and IgA was positive in 7 (5.22%) cases. Combination of IgG+IgM+IgA were positive in 2, IgG+IgM in 2 and IgG+IgA in 1 patient.
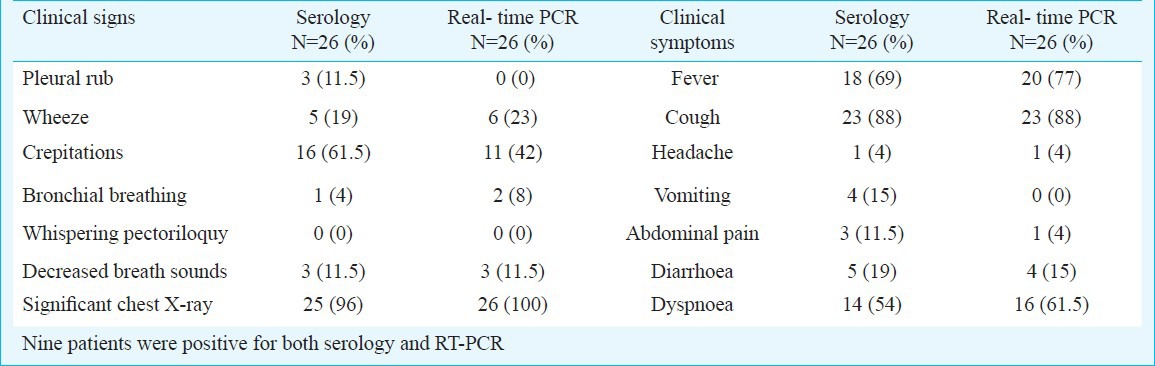
Clinical signs and symptoms: The comparison of clinical data of CAP patients diagnosed for M. pneumoniae based on qRT-PCR assay and serology (Table III), revealed that fever, cough, headache and dyspnoea were equally positive in both the groups. Of the clinical signs crepitations and pleural rub (Table III) was higher in serology positive cases though it was not significant. Wheeze, decreased breadth sounds and significant chest X-ray were equally positive in both the groups. The clinical signs and symptoms for M. pneumoniae positive (serology and qRT-PCR) and M. pneumoniae negative patients did not show any significant difference (Table IV).
Table III.
Clinical signs & symptoms of M. pneumoniae positive [serology (n=17) & qRT-PCR (n=17) and positive for both (n=9)] and negative patients for community acquired pneumonia.
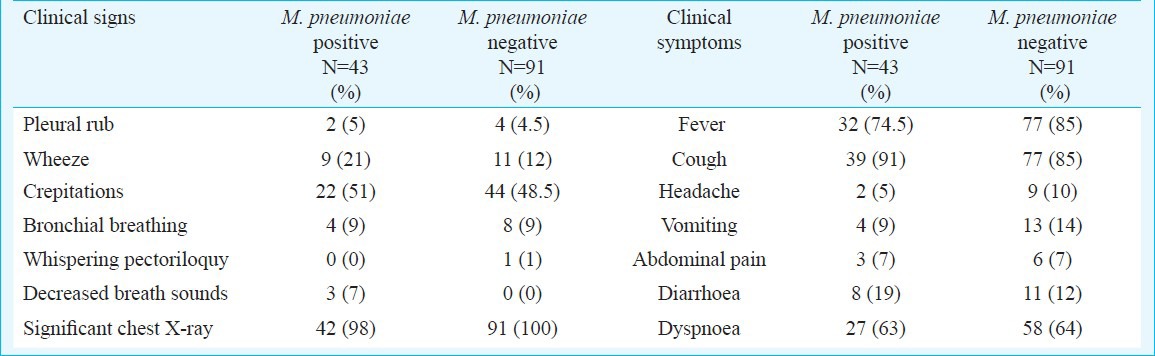
Table IV.
Clinical symptoms and signs for serology and real-time PCR positive patients for community acquired pneumonia
Discussion
Many bacterial and viral infections often share clinical features and symptoms which are difficult to distinguish clinically24,25. A sensitive and effective method of detecting these agents is required so that the correct treatment is offered and unnecessary use of antibiotics can be avoided. The standard laboratory method for the diagnosis of M. pneumoniae as an aetiological agent for CAP has been the culture or serology26. PCR has been shown to be a better diagnostic test than conventional techniques19,27. Real-time PCR significantly reduces time to give results and has an advantage over conventional PCR as detection is performed in a closed system in real time, thus minimizing the risk of contamination. In the present study, a real-time PCR assay that targets the P1 adhesin gene for the diagnosis of M. pneumoniae infections was performed along with serology and conventional PCR. Based on P1 gene detection, qRT-PCR assay was developed to detect M. pneumoniae infections in 134 patients. Using both qRT-PCR and serology, overall 43 (of the 134) patients (32.08%) were found positive. qRT-PCR was positive in 26 of the 134 (19.4%) samples as compared to conventional PCR in 25 of the 134 samples tested. However, 17 cases positive by qRT-PCR were negative by serology (Table V). Serology showed 17 samples positive which were negative by qRT-PCR was negative. Of these 17 cases, only IgG antibody was detected in 11 cases whereas IgA and IgM were absent. In these cases the presence of IgG antibodies alone can be due to past infection and, therefore, real-time PCR missed only six cases of acute infection where IgA/IgM was also detected.
Table V.
Comparison of results of real-time PCR with conventional PCR and serology
Our study showed that clinical signs like pleural rub and crepitations were more in serology positive patients compared to sero negatives though not significantly. Tempelton et al28 reported that none of the patients with M. pneumoniae infections had pulmonary diseases in contrast majority of patients in our study were observed with pulmonary disease. Clinical signs and symptoms among M. pneumoniae positive (serology and qRT-PCR) and negative patients were also compared, but no significant difference was observed. This suggests that M. pneumoniae infection cannot be differentiated from other pulmonary infections on the basis of clinical signs and symptoms. Thus, for better patient management a laboratory diagnosis is important to detect or to rule out M. pneumoniae infection from other respiratory infections.
Tempelton et al28 reported 12 (11%) positive results by qRT-PCR assay for M. pneumoniae infection in a group of 106 patients. Other studies have shown 20-21 per cent positivity by qRT-PCR assay29,30. Our study showed 19.4 per cent (26/134) positive samples by qRT-PCR assay and 18.6 per cent (25/134) by PCR which is close to the reported studies from other geographical locations. One additional case was picked by qRT-PCR which was negative by both PCR as well as serology.
There were 17 samples positive by RT-PCR having high copy number but were not detected by serological test. All these cases were having clinical symptoms for less than one week duration. During early period of infection organism load is usually high in the throat swab sample but detectable amount of antibodies are not generated, and hence such samples are missed by serological methods.
The merit of this study is that three different classes of antibodies, qRT-PCR and PCR have been done in all the cases, whereas none of the earlier studies have looked for IgA, IgM and IgG simultaneously in the same sets of samples. Thurman et al30 used real-time PCR and IgG/IgM antibody detection kit in the investigation of outbreaks of CAP due to M. pneumoniae and showed 81 per cent positive results with IgM antibodies and 54 per cent with IgG/IgM. Morozumi et al31 reported sensitivity and specificity of real-time PCR related to serological assay as 90.2 and 97.9 per cent of real-time PCR positive samples. In our study although both PCR and qRT-PCR showed concordant results, serology was positive in only one third (9/26) of these cases, therefore, sensitivity and specificity of qRT-PCR compared to serological assay (ELISA) was 34.6 and 84.2 per cent, respectively. As reported by Thurman et al30, the sensitivity of the real-time PCR assay reduces with the delay in collection of samples from the onset of the disease.
In conclusion, our study indicated that no single available test was reliable for identification of M. pneumoniae infections. A combination of two or three methods can be the most reliable approach for identification of CAP due to M. pneumoniae, especially in the absence of other suspected respiratory pathogens. However, qRT-PCR may not only detect M. pneumoniae infection early in the course of disease but also quantitates bacterial load and helps to initiate specific antibiotic therapy. The qRT-PCR assay can be used to rule out infection with M. pneumoniae when the samples are tested for new emerging respiratory pathogens.
Acknowledgment
This work was financially supported by the Department of Biotechnology, New Delhi. The authors acknowledge the help provided by Dr Priyanshu, TERI, New Delhi, for conducting real-time PCR experiments, and Dr S.K. Kabra, Department of Pediatrics, All India Institute of Medical Sciences, New Delhi, for providing paediatric patient samples, also acknowledge assistance by Shri Pramod Kumar for basic laboratory work.
References
- 1.Zhang L, Zong Z-Y, Liu Y-B, Ye H, Lv X-J. PCR versus serology for diagnosing Mycoplasma pneumoniae infection: A systematic review & meta-analysis. Indian J Med Res. 2011;134:270–80. [PMC free article] [PubMed] [Google Scholar]
- 2.Waits KB, Talkington DF. Mycoplasma pneumonia and its role as a human pathogen. Clin Microbiol Rev. 2004;17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varshney AK, Chaudhry R, Saharan S, Kabra SK, Dhawan B, Dar L, et al. Association of Mycoplasma pneumoniae and asthma among Indian children. FEMS Immunol Med Microbiol. 2009;56:25–31. doi: 10.1111/j.1574-695X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudhry R, Tabassum I, Kapoor L, Chhabra A, Sharma N, Broor S. A fatal case of acute respiratory distress syndrome (ARDS) due to Mycoplasma pneumoniae. Indian J Pathol Microbiol. 2010;53:557–9. doi: 10.4103/0377-4929.68283. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhry R, Nisar N, Malhotra P, Kumar A, Chauhan VS. Polymerase chain reaction confirmed Mycoplasma pneumoniae arthritis: a case report. Indian J Pathol Microbiol. 2003;46:433–6. [PubMed] [Google Scholar]
- 6.Ngeh J, Goodbourn C. Chlamydia pneumoniae, Mycoplasma pneumoniae, and Legionella pneumophila in elderly patients with stroke (C-PEPS, M-PEPS, L-PEPS): A case-control study on the infectious burden of atypical respiratory pathogens in elderly patients with acute cerebrovascular disease. Stroke. 2005;36:259–65. doi: 10.1161/01.STR.0000152961.11730.d9. [DOI] [PubMed] [Google Scholar]
- 7.Gorthi SP, Kapoor L, Chaudhry R, Sharma N, Perez-Perez GI, Panigrahi P, et al. Guillain-Barre syndrome: association with Campylobacter jejuni and Mycoplasma pneumoniae infections in India. Natl Med J India. 2006;19:137–9. [PubMed] [Google Scholar]
- 8.Goyal P, Kale SC, Chaudhry R, Chauhan S, Shah N. Association of common chronic infections with coronary artery disease in patients without any conventional risk factors. Indian J Med Res. 2007;125:129–36. [PubMed] [Google Scholar]
- 9.Dey AB, Chaudhry R, Kumar P, Nisar N, Nagarkar KM. Mycoplasma pneumoniae and community-acquired pneumonia. Natl Med J India. 2000;13:66–70. [PubMed] [Google Scholar]
- 10.Daxboeck F, Krause R, Wenisch C. Laboratory diagnosis of Mycoplasma pneumoniae infection. Clin Microbiol Infect. 2003;9:263–73. doi: 10.1046/j.1469-0691.2003.00590.x. [DOI] [PubMed] [Google Scholar]
- 11.Tuuminen T, Suni J, Kleemola M, Jacobs E. Improved sensitivity and specificity of enzyme immunoassays with P1-adhesin enriched antigen to detect acute Mycoplasma pneumoniae infection. J Microbiol Methods. 2001;44:27–37. doi: 10.1016/s0167-7012(00)00235-9. [DOI] [PubMed] [Google Scholar]
- 12.Welti M, Jaton K, Altwegg M, Sahli R, Wenger A, Bille J. Development of a multiplex real-time quantitative PCR assay to detect Chlamydia pneumoniae, Legionella pneumophila and Mycoplasma pneumoniae in respiratory tract secretions. Diagn Microbiol Infect Dis. 2003;45:85–95. doi: 10.1016/s0732-8893(02)00484-4. [DOI] [PubMed] [Google Scholar]
- 13.van Kuppeveld FJ, Johansson KE, Galama JM, Kissing J, Bolske G, Hjelm E, et al. 16S rRNA based polymerase chain reaction compared with culture and serological methods for diagnosis of Mycoplasma pneumoniae infection. Eur J Clin Microbiol Infect Dis. 1994;13:401–5. doi: 10.1007/BF01971997. [DOI] [PubMed] [Google Scholar]
- 14.Störmer M, Vollmer T, Henrich B, Kleesiek K, Dreier J. Broad-range real-time PCR assay for the rapid identification of cell-line contaminants and clinically important mollicute species. Int J Med Microbiol. 2009;299:291–300. doi: 10.1016/j.ijmm.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Yogev D, Sela S, Bercovier H, Razin S. Nucleotide sequence and codon usage of the elongation factor Tu (EF-Tu) gene from Mycoplasma pneumoniae. Mol Microbiol. 1990;4:1303–10. doi: 10.1111/j.1365-2958.1990.tb00709.x. [DOI] [PubMed] [Google Scholar]
- 16.Apfalter P, Barousch W, Nehr M, Makristathis A, Willinger B, Rotter M, et al. Comparison of a new quantitative ompA-based real time PCR TaqMan assay for detection of Chlamydia pneumoniae DNA in respiratory specimens with four conventional PCR assays. J Clin Microbiol. 2003;41:592–600. doi: 10.1128/JCM.41.2.592-600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanna M, Fan J, Pehler-Harrington K, Waters C, Douglass P, Stallock J, et al. The Pneumoplex assays, a multiplex PCR-enzyme hybridization assay that allows simultaneous detection of five organisms, Mycoplasma pneumoniae, Chlamydia (Chlamydophila) pneumoniae, Legionella pneumophila, Legionella micdadei, and Bordetella pertussis, and its real-time counterpart. J Clin Microbiol. 2005;43:565–71. doi: 10.1128/JCM.43.2.565-571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raggam RB, Leitner E, Berg J, Mu¨hlbauer G, Marth E, Kessler HH. Single-run, parallel detection of DNA from three pneumonia-producing bacteria by real-time polymerase chain reaction. J Mol Diagn. 2005;7:133–8. doi: 10.1016/S1525-1578(10)60019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardegger D, Nadal D, Bossart W, Altwegg M, Dutly F. Rapid detection of Mycoplasma pneumoniae in clinical samples by real-time PCR. J Microbiol Methods. 2000;41:45–51. doi: 10.1016/s0167-7012(00)00135-4. [DOI] [PubMed] [Google Scholar]
- 20.Niesters HG. Standardization and quality control in molecular diagnostics. Expert Rev Mol Diagn. 2001;1:129–31. doi: 10.1586/14737159.1.2.129. [DOI] [PubMed] [Google Scholar]
- 21.Marques LM, Buzinhani M, Yamaguti M, Oliveira RC, Ferreira JB, Mettifogo E, et al. Use of a polymerase chain reaction for detection of Mycoplasma dispar in the nasal mucus of calves. J Vet Diagn Invest. 2007;19:103–6. doi: 10.1177/104063870701900118. [DOI] [PubMed] [Google Scholar]
- 22.Behbahan NGG, Asasi K, Afsharifar AR, Pourbakhsh SA. Isolation and detection of Mycoplasma gallisepticum by polymerase chain reaction and restriction fragment length polymorphism. Iran J Vet Res. 2005;6:35–41. [Google Scholar]
- 23.Williamson J, Marmion BP, Worswick DA, Kok TW, Tannock G, Hard R, et al. Laboratory. diagnosis of Mycoplasma pneumoniae infection. 4. Antigen capture and PCR-gene amplification for detection of the Mycoplasma: problems of clinical correlation. Epidemiol Infect. 1992;109:519–37. doi: 10.1017/s0950268800050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foy HM. Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin Infect Dis. 1993;17(Suppl 1):S37–46. doi: 10.1093/clinids/17.supplement_1.s37. [DOI] [PubMed] [Google Scholar]
- 25.Saikku P. Atypical respiratory pathogens. Clin Microbiol Infect. 1997;3:599–604. doi: 10.1111/j.1469-0691.1997.tb00464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferwerda A, Moll HA, de Groot R. Respiratory tract infections by Mycoplasma pneumoniae in children: a review of diagnostic and therapeutic measures. Eur J Pediatr. 2001;160:483–91. doi: 10.1007/s004310100775. [DOI] [PubMed] [Google Scholar]
- 27.Tumwasorn S, Nilgate S, Udomsantisuk N. Amplification of P1 gene by polymerase chain reaction for detection of Mycoplasma pneumoniae. J Med Assoc Thai. 2002;85(Suppl 1):S389–98. [PubMed] [Google Scholar]
- 28.Templeton KE, Scheltinga SA, Graffelman AW, Van Schie JM, Crielaard JW, Sillekens P, et al. Comparison and evaluation of real-time PCR, realtime nucleic acid sequence-based amplification, conventional PCR, and serology for diagnosis of Mycoplasma pneumoniae. J Clin Microbiol. 2003;41:4366–71. doi: 10.1128/JCM.41.9.4366-4371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gullsby K, Storm M, Bondeson K. Simultaneous detection of Chlamydophila pneumoniae and Mycoplasma pneumoniae by use of molecular beacons in a duplex real-time PCR. J Clin Microbiol. 2008;46:727–31. doi: 10.1128/JCM.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurman KA, Walter ND, Schwartz SB, Mitchell SL, Dillon MT, Baughman AL, et al. Comparison of laboratory diagnostic procedures for detection of Mycoplasma pneumoniae in community outbreaks. Clin Infect Dis. 2009;48:1244–9. doi: 10.1086/597775. [DOI] [PubMed] [Google Scholar]
- 31.Morozumi M, Nakayama E, Iwata S, Aoki Y, Hasegawa K, Kobayashi R, et al. Simultaneous detection of pathogens in clinical samples from patients with community-acquired pneumonia by real-time PCR with pathogen-specific molecular beacon probes. J Clin Microbiol. 2006;44:1440–6. doi: 10.1128/JCM.44.4.1440-1446.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


