SUMMARY
MicroRNAs regulate the function of several immune cells but their role in promoting CD8+ T-cell immunity remains unknown. Here we report that miR-155 is required for CD8+ T-cell responses to both virus and cancer. In the absence of miR-155, accumulation of effector CD8+ T cells was severely reduced during acute and chronic viral infections and control of virus replication was impaired. Similarly, Mir155-/- CD8+ T cells were in effective at controlling tumor growth, whereas miR-155 overexpression enhanced the antitumor response. miR-155 deficiency resulted in accumulation of SOCS-1 causing defective cytokine signaling through STAT5. Consistently, enforced expression of SOCS-1 in CD8+ T cells phenocopied the miR-155 deficiency, whereas SOCS-1 silencing augmented tumor destruction. These findings identify miR-155 and its target SOCS-1 as key regulators of effector CD8+ T cells that can be modulated to potentiate immunotherapies for infectious diseases and cancer.
INTRODUCTION
CD8+ T cells are essential effectors in immune responses to intracellular pathogens and cancer (Zhang and Bevan, 2011). Upon stimulation, antigen-specific CD8+ T cells massively expand and differentiate into inflammatory cytokine producing, cytolytic T cells able to eliminate virally infected or transformed cells. As the antigen is cleared, the majority of specific CD8+ effector T cells die (Marrack and Kappler, 2004), whereas only a small number of memory cells survives. The CD8+ T cell response is influenced by a series of costimulatory (and inhibitory) ligands and by multiple soluble mediators such as IL-2 (Boyman and Sprent, 2012). The latter is essential for sustaining an efficient effector response, whereas other cytokines such as IL-7 and IL-15 play crucial roles for the survival of naïve or memory T cells (Cui and Kaech, 2010). Several studies have identified key molecular factors involved in the differentiation from naïve to effector CD8+ T cells, but the contribution of microRNAs (miRs) has just begun to be investigated (Almanza et al., 2010).
miRs are a class of small, non-coding RNAs that impart post-transcriptional gene regulation (Bartel, 2004) through several mechanisms including translational repression and mRNA degradation (Djuranovic et al., 2011). They are important in many physiological processes, in carcinogenesis (Calin and Croce, 2006) and in the immune system (Xiao and Rajewsky, 2009). Early studies in mice deficient for Dicer, an RNAse III enzyme important for mature miR production, revealed that miRs are involved in CD4+ T cell differentiation and strongly influence CD8+ T cell responses (Muljo et al., 2005; Zhang and Bevan, 2010). Specific miRs were shown to regulate both lymphocyte development and function. For instance, miR-181a influences thymocyte selection by modulating the expression of molecules involved in TCR signaling (Li et al., 2007). Moreover, the miR-17~92 cluster regulates B cell development (Ventura et al., 2008), autoimmunity and Th1cell differentiation (Jiang et al., 2011; Xiao et al., 2008).
miR-155 is upregulated upon lymphocyte activation (Haasch et al., 2002) to control cell proliferation and differentiation (O’Connell et al., 2008; Turner and Vigorito, 2008). For instance, miR-155 regulates B cell proliferation, malignancy and antibody production, at least in part through inhibition of activation-induced cytidine deaminase and PU.1 expression (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007). In CD4+ T cells, miR-155 has been shown to suppress differentiation of naïve cells into Th2 by downregulation of c-Maf, to promote Th17 cell mediated inflammation (Kurowska-Stolarska et al., 2011; O’Connell et al., 2010) and to inhibit IFN-γR expression (Banerjee et al., 2010; Martinez-Nunez et al., 2011). In addition to direct modulation of cytokine receptor expression, miR-155 shapes cytokine signaling in several cell subsets via downregulation of SMAD2 (Louafi et al., 2010) and suppressor of cytokine signaling (SOCS-1) (Lu et al., 2009; O’Connell et al., 2010; Wang et al., 2010). Despite the evidence for an important role of miR-155 in a wide spectrum of immune compartments, it is not known if this miRNA, which is highly expressed in antigen-experienced CD8+ T cells (Salaun et al., 2011), influences CD8+ T cells in vivo. In the present study, we have investigated the role of miR-155 during CD8+ T cell responses to viral infection, vaccination and cancer.
RESULTS
CD8+ T cells dynamically regulate miR-155 depending on the magnitude of TCR stimulation or their differentiation state
The strength of TCR signaling has a major impact on the magnitude of CD8+ T cell expansion but not on their differentiation (Zehn et al., 2009). We sought to investigate whether the strength of TCR stimulation affects the expression of miR-155 in CD8+ T cells by transducing human CD8+ T cells with variants of a NY-ESO-1-specific TCR of increasing affinity for its ligand (Derre et al., 2008; Schmid et al., 2010). Comparatively, a mutated low affinity TCR failed to upregulate miR-155 within 48h, whereas TCR variants of higher affinities induced higher levels of miR-155 than the wild type (Figure 1A). Thus, miR-155 expression increased in a TCR affinity-dependent manner in human CD8+ T cells. A similar upregulation was observed for the pri-miR-155 non-coding RNA transcript, BIC (Figure S1). To see if miR-155 was also regulated in an affinity-dependent manner in mouse CD8+ T cells, we activated naïve OT-1 T cells with splenic dendritic cells (DC) loaded with the wild type peptide SIINFEKL (N4) or the weaker altered peptide ligand SIITFEKL (T4) (Daniels et al., 2006). To exclude miR-155 contamination from the DCs, we used Mir155-/- DC which retain normal antigen presenting capabilities (O’Connell et al., 2010). Exposure of OT-1 cells to the WT natural peptide resulted in a strong upregulation of miR-155, while a weaker TCR stimulation by the T4 peptide was less effective (Figure 1B). To assess miR-155 regulation in vivo, we analysed naïve (CD62L+CD44−), effector (CD62L−CD44+) and central memory (CD62L+CD44+) CD8+ T cells following LCMV infection (200 pfu of WE strain). Compared to their naïve counterparts, miR-155 was strongly upregulated in effector cells and to a lower extent in central memory CD8+ T cells 8 days post-infection (Figure 1C). A more detailed kinetics of miR-155 regulation during LCMV infection revealed that numbers of effector cells peaked on day 6, but stayed low in naïve cells (Figure 1D). These results demonstrate that miR-155 is induced in effector CD8+ T cells depending on the strength of stimulation and differentiation.
Figure 1. miR-155 expression is regulated at various stages of CD8+ T cell differentiation in a TCR- affinity dependent manner.
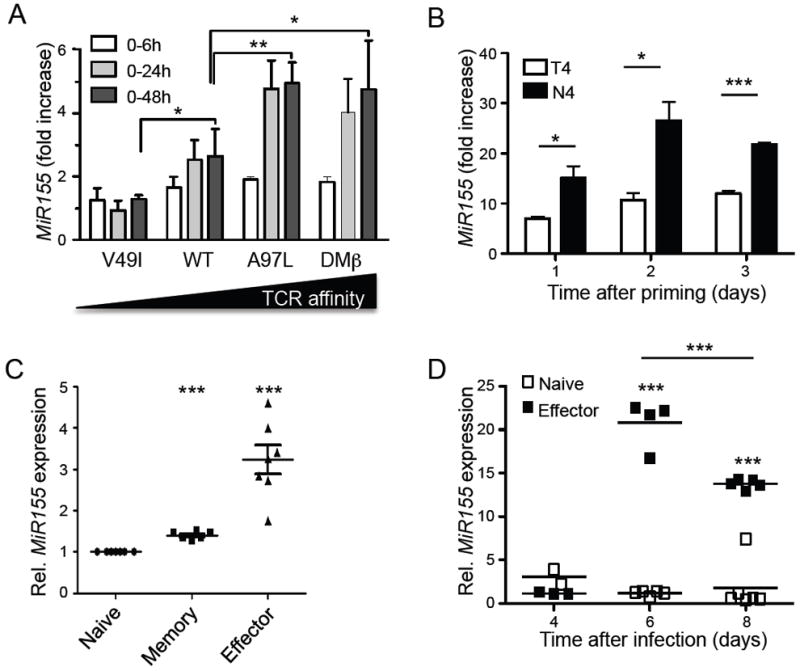
(A) Quantification of miR-155 in human CD8+ T cells transduced with TCRs of increasing affinity following TCR stimulation with multimers (N=3 experiments) as fold increase relative to unstimulated clones. (B) miR-155 expression of naïve mouse OT-1 T cells stimulated with splenic dendritic cells pulsed with the natural SIINFEKL (N4) or weaker SIITFEKL (T4) altered peptide ligand as relative to day 0 unstimulated cells. Data are representative for triplicates in one out of two experiments. (C) miR-155 concentrations in naïve, central memory and effector CD8+ T cells sorted at day 8 after LCMV WE infection as fold change relative to naïve cells. (D) Relative miR-155 expression in splenic naive and effector CD8+ T cells sorted from LCMV infected mice. Symbols represent individual mice and the line is the mean +/- SEM. Data are representative for 2 independent experiments. For human BIC expression please also see Figure S1.
miR-155 promotes the accumulation of anti-viral effector and central memory CD8+ T cells
To determine the role of miR-155 in activated CD8+ T cells, we monitored the expansion of effector cells following acute LCMV WE strain infection in the presence or absence of miR-155. Percentage, number and phenotype of naïve Mir155-/- CD8+ T cells in blood and spleen did not differ from those in wild type mice before infection (Figure S2A and data not shown). In contrast, both percentage and number of total CD8+ T cells as well as virus gp33 tetramer specific CD8+ effector T cells were substantially reduced in spleen and blood of Mir155-/- mice at the peak of the response (Figure 2A, B). Following the expansion of CD44+ effector cells in the blood and spleen from days 6 to 8, we observed impaired effector CD8+ cell accumulation in spleen, liver and blood of Mir155-/- mice (Figure 2C and data not shown). Despite a defect in the magnitude of effector T cell responses, Mir155-/- animals were capable of controlling viral replication and clearing the virus (Figure S2B), as also confirmed by the lack of CD44 upregulation on adoptively transferred naïve LCMV specific P14 T cells (Figure S2C). In line with this result, CD8+ T cells differentiated into phenotypically and functionally cytolytic effector cells similar to wild type cells during LCMV infection (Figure S2A, D, E). Interestingly, circulating T cells in Mir155-/- mice exhibited not only a defect in the expansion at the peak of the immune response but also a more rapid contraction compared to wild type animals (Figure 2D). Moreover, CD127+CD62L+KLRG1− memory cells were strongly reduced in the gp33 and np396 tetramer+ CD8+ T cells in blood, liver and spleen of Mir155-/- mice three months after infection (Figure 2E, S2F and data not shown). Consistent with these findings, IL-2 production, a hallmark of central memory cells, was strongly diminished in Mir155-/- mice after stimulation with gp33 peptide (Figure 2F, S2F). In this immune memory context, it is of interest that we observed a deficient CD4+ effector T cell activation on day 8 of the response in Mir155-/- mice (data not shown). Altogether, these results demonstrate that miR-155 is crucial for a robust T cell expansion but not effector functions as well as for a memory phenotype response upon an acute LCMV infection.
Figure 2. miR-155 is required for optimal effector CD8+ T cell accumulation and memory cell differentiation during acute LCMV infection.
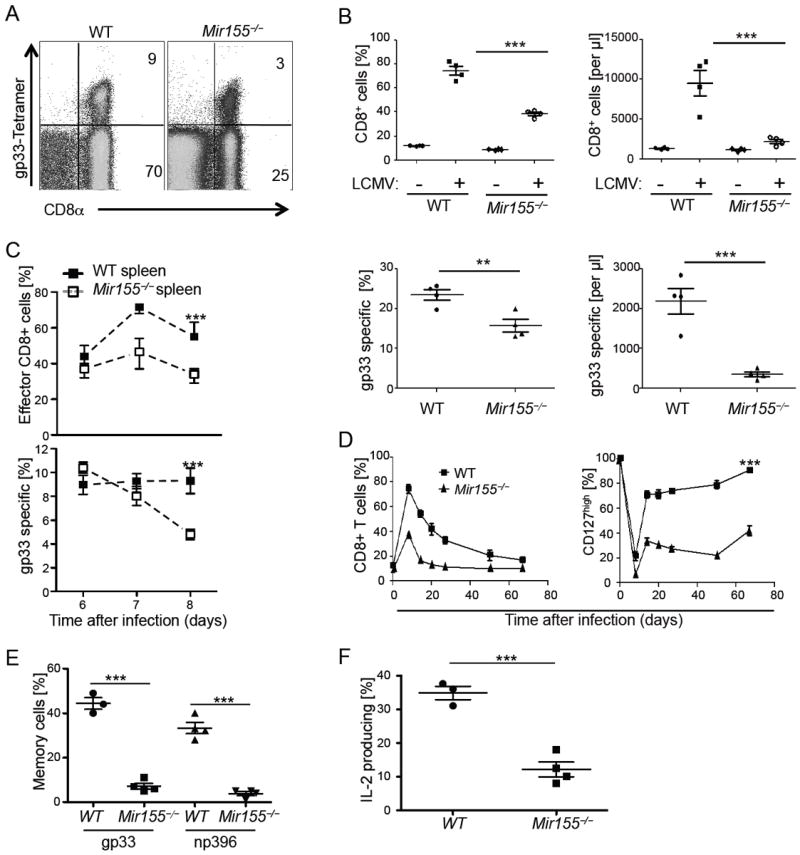
(A) Splenocytes from day 8 infected wild type (WT) and Mir155-/- mice were stained with Kb/LCMV gp33 tetramers and anti-CD8α. (B) Blood percentages and numbers of CD8+ (upper panels) and gp33 tetramer+ cells (lower panels) on day 8 of infection. (C) Percentages of CD44highCD62Llow effector CD8+ T cells gated on lymphocytes in wild type (WT) and Mir155-/- spleen cells (upper) and of gp33 tetramer+ cells within the CD8+ T cells (lower panel) at days 6 to 8. (D) Percentages of total CD8+ T cells (left) and CD127+ cells within tetramer gp33+ CD8+ cells (right panel) in blood at given time points. (E) Percentage of liver CD127highCD62Lhigh tetramer gp33 and np396+ memory cells and (F) IL-2 production upon gp33 peptide restimulation of splenocytes within IFN-γ postitive CD8+ T cells at 3 months past infection. Symbols represent individual mice, and the line is the mean +/- SEM. Representative results of one out of four (A-C) to two (D-F) independent experiments with three to five mice are pictured. Please also see Figure S2.
Intrinsic expression of miR-155 in CD8+ T cells promotes proliferation and limits apoptosis of effector CD8+ T cells
To investigate whether the defective expansion of CD8+ T cells was cell-intrinsic, we cocultured naïve wild type and congenic Mir155-/- OT-1 CD8+ T cells together with peptide pulsed dendritic cells and analysed the OT-1 cell ratio. After 5 days, wild type CD8+ T cells outnumbered Mir155-/- cells and the abundance of dead cells was strongly increased among Mir155-/- T cells (Figure 3A). To assess these parameters in vivo, we co-transferred equal numbers of congenic polyclonal wild type and Mir155-/- CD8+ T cells into either wild type or Mir155-/- hosts, which were then infected with LCMV. Despite the initial low frequency of wild type CD8+ T cells transferred in miR-155 ablated hosts (about 1% of CD8+ T cells in blood before infection), these cells expanded to about 30% of the CD8+ T cells at the peak of the response. In contrast, the frequency of Mir155-/- CD8+ T cells transferred into wild type hosts decreased upon infection (Figure 3B), clearly demonstrating a stronger response of wild type compared to Mir155-/- CD8+ T cells. When Rag2 and common γ chain (γc) deficient hosts were engrafted with a 1:1 mix of wild type and Mir155-/- splenocytes, both populations reached similar frequencies after 2 months, indicating comparable homeostatic expansion (Figure 3C). However, following LCMV infection, wild type T cells again showed an advantage in expansion over their Mir155-/- counterparts. To determine the basis for the impaired accumulation of virus-specific CD8+ T cells in the absence of miR-155, LCMV infected wild type and Mir155-/- mice were pulsed with BrdU and proliferation and apoptosis were measured 4h later. We found that the proliferation of Mir155-/- CD44+ effector CD8+ T cells was decreased compared to wild type cells 6 days after infection (Figure 3D). Additionally, the frequency of proliferating Ki67+ cells within the CD44+CD62L− effector CD8+ T cells was reduced in Mir155-/- mice (Figure 3E). Finally, we observed an increased frequency of AnnexinV+ apoptotic cells in Mir155-/-compared to wild type effector CD8+ T cells 7 days after infection (Figure 3F). Altogether, these data demonstrate a cell-intrinsic role of miR-155 in the proliferation and survival of effector CD8+ T cells in response to LCMV infection but not for homeostatic expansion in lymphopenic hosts.
Figure 3. A cell intrinsic role for miR-155 in promoting effector CD8+T cells.
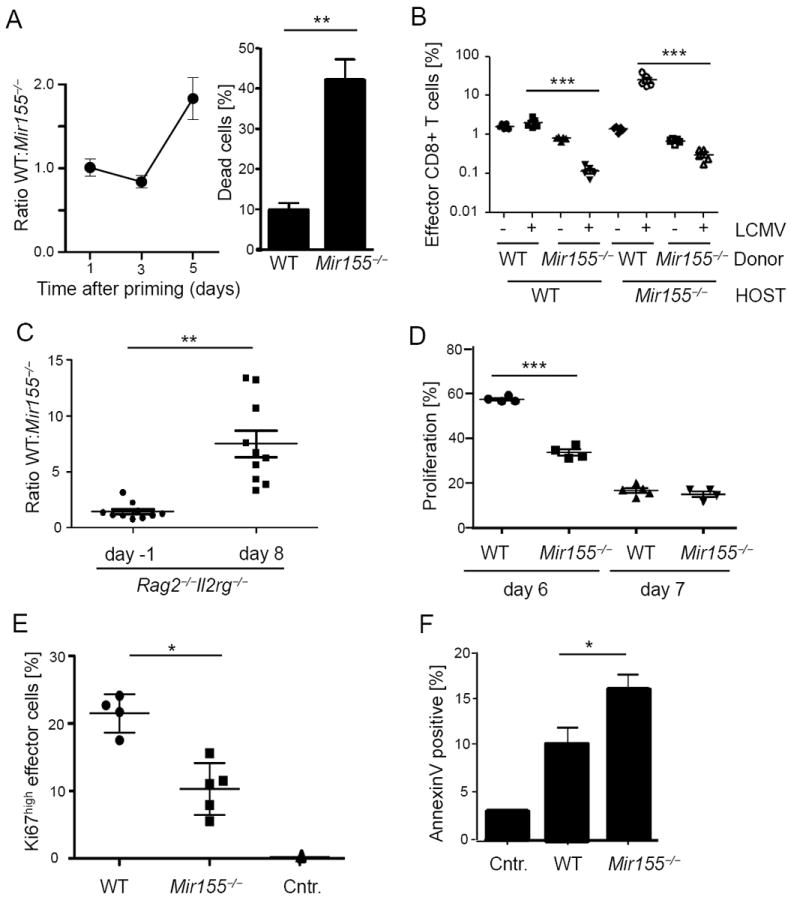
(A) Congenically marked wild type (WT) and Mir155-/- OT-1 cells were competitively cocultured with peptide pulsed dendritic cells and the ratio of populations is pictured at indicated time points (left). On day 5, percentage of trypan blue cells harvested from either WT or Mir155-/- cultures was counted (right graph). Pooled data from three representative experiments are pictured. (B) CD8+ T cells from WT and Mir155-/- mice were cotransferred into WT or deficient hosts before LCMV WE infection and percentages in blood at day 8 were measured. (C) A 1:1 mix of WT and Mir155-/- splenocytes was adoptively transferred into Rag2 and IL2Rγ double deficient mice which were infected with LCMV WE two months after transfer. CD8+ effector T cell ratios at days -1 and 8 post infection are pictured. (D) Proliferating BrdU positive splenic CD44high CD8+ effector T cells at days 6 and 7 post infection. (E) At the same time, cells were stained for the proliferation marker Ki67 (day7) and (F) apoptotic cells were identified by AnnexinV staining. Symbols represent individual mice, and the line is the mean +/- SEM. Representative results from two (B, C) to three (D-F) experiments are pictured.
miR-155 is crucial for effector CD8+ T cell accumulation and virus control in chronic LCMV infection
Based on the strong impairment of effector CD8+ T cell accumulation in low dose LCMV infection, we asked how Mir155-/- mice would respond to high dose and long-lasting antigen exposure, which characterizes chronic infections and cancer. Mice were inoculated with 2×106 pfu of LCMV clone 13, causing a chronic infection for several weeks (Moskophidis et al., 1993; Salvato et al., 1991). While wild type mice mounted a robust effector CD8+ T cell response with high percentages of CD44+CD62L− effector cells that were maintained overtime, Mir155-/- mice progressively lost effector CD8+ T cells (Figure 4A). Interestingly, the remaining CD44+ cells in spleen showed high CD127 and CD62L expression, reminiscent of a memory phenotype (Figure 4B). Percentages and numbers of gp33 tetramer positive cells were also strongly decreased in deficient mice 5 weeks and 3 months post infection (Figure 4B, C and data not shown). At this time, we could not detect cells capable of producing effector cytokines in response to a cocktail of LCMV peptides in miR-155 ablated mice, confirming the loss of most virus specific Mir155-/- CD8+ T cells, and ruling out TCR downregulation that may appear as tetramer negative T cells (Figure 4E and data not shown). Whereas about 50% of wild type cells remained positive for PD-1, associated with T cell exhaustion, PD-1 was barely detectable on Mir155-/- CD8+ T cells 5 weeks upon infection (Figure 4C). Importantly, virus titers were elevated five weeks and two months post infection in miR-155 ablated mice (Figure 4D and data not shown). Finally, wild type but not miR-155 ablated mice showed symptoms of immunopathology such as shivering, hunching and weight loss, suggesting a lower inflammatory response in the absence of miR-155 (Figure 4F and data not shown). These data demonstrate an important role of miR-155 in maintenance and survival of CD8+ effector T cells as well as virus control in chronic virus infections.
Figure 4. miR-155 drives survival of effector cells and sustains the anti-viral response in a chronic LCMV infection.
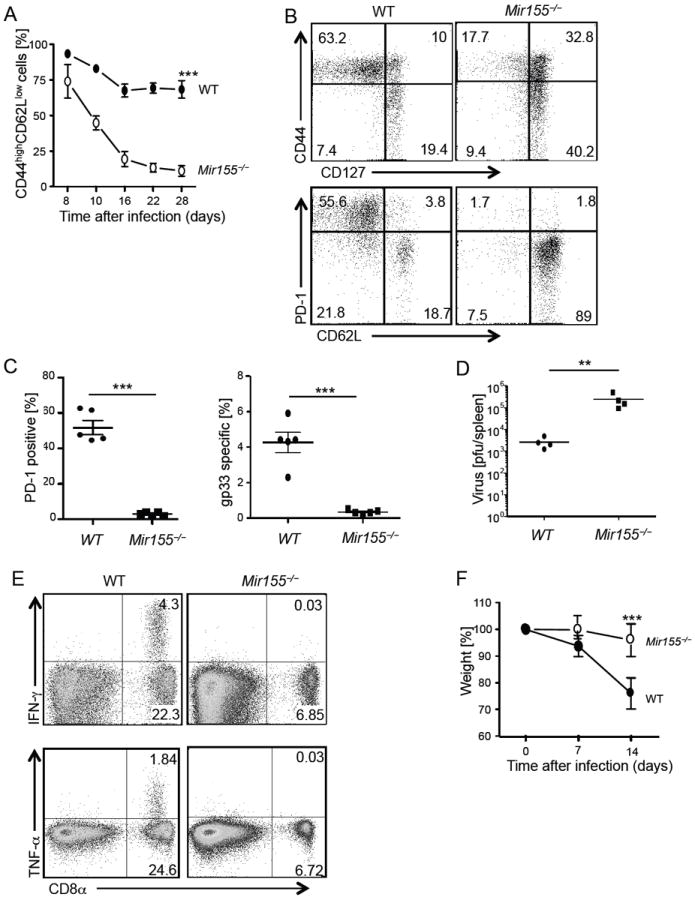
Wild type (WT) and Mir155-/- mice were infected with LCMV clone 13 and phenotype and expansion of effector cells was monitored by flow cytometry. (A) Effector CD44highCD62Llow within blood CD8+ T cells at indicated time points. (B, C) At five weeks, spleenic CD8+ cells were analysed for the indicated activation markers and gp33 tetramer+ cells shown as (B) flow cytometry dot blots from representative mice and (C) graph from one representative out of three experiments. (D) Virus titer and (E) cytokine response upon stimulation with a peptide cocktail was determined in the blood at 2 and 3 months after infection, respectively. (F) Weight of mice was monitored the first 2 weeks post-infection. Symbols represent individual mice, and the line is the mean (C, D). Error bars are given as +/- SEM. Shown are representative results from one out of two experiments with three to five mice per group.
miR-155 expression in CD8+ T cells is crucial for efficient immunization and cancer immunotherapy
Because we observed an important role of miR-155 to sustain CD8+ T cell responses to chronic infection, we looked at the impact of miR-155 on CD8+ T cell dependent anti-tumor immunity, which requires robust CD8+ T cell responses. We examined the role of miR-155 in CD8+ T cells for vaccination, a clinically relevant setting characterized by limited adjuvant-induced inflammation. Polyclonal and OVA-specific OT-1 CD8+ T cells (either wild type or Mir155-/-) were cotransferred into wild type mice before immunization with OVA peptide adjuvanted with IFA and CpG-ODNs. While the ratio of wild type OT-1 to polyclonal cells strongly increased following immunization, there was only a minor increment in the ratio of Mir155-/- OT-1 to polyclonal wild type cells (Figure 5A). We next cotransferred OT-1 cells from both wild type and Mir155-/- backgrounds into wild type mice before immunization. Wild type and Mir155-/- cells were found in similar proportions indicating a comparable survival after adoptive transfer. Following immunization, however, wild type cells accumulated more efficiently than Mir155-/- cells (Figure 5B), whereas upregulation of CD44 and the proportion of cells producing IFN-γ were comparable (Figure S3A, B). Now we were interested, if miR-155 would also be critical for tumor control by CD8+ T cells and engrafted B16 melanoma upon transfer of naïve wild type or Mir155-/- OT-1 T cells and therapeutic vaccination one week later.
Figure 5. miR-155 is crucial for the CD8+ T cell response to peptide vaccination and tumor challenge.
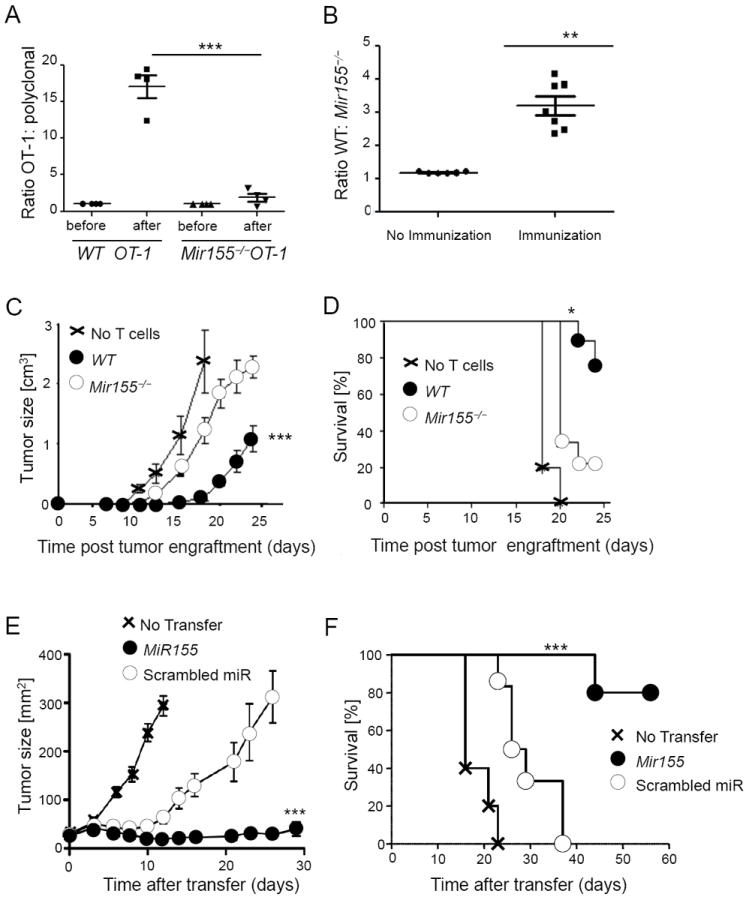
(A) Congenically marked OT-1 and polyclonal CD8+ T cells from wild type (WT) or Mir155-/- backgrounds were transferred into WT hosts, which were immunized with OVA peptide and CpG in IFA. Shown is the ratio of antigen specific versus polyclonal cells in the draining lymph nodes 4 days later. Symbols represent individual mice, and the line is the mean +/- SEM. (B) Ratio of blood WT and Mir155-/- OT-1 CD8+ T cells cotransferred into WT hosts, which were immunized as in (A) on day 7. Symbols represent individual mice, and the line is the mean +/- SEM. Data from 1 out of 2 (A) to 3 (B) independent experiments are pictured. (C, D) Upon adoptive transfer of WT or Mir155-/- OT-1 cells and immunization, mice were engrafted with B16 melanoma cells expressing OVA. Tumor growth (C) and survival (D) are pictured (n=5-9 mice per group, data from 1 out of 2 independent experiments) displayed as mean +/- SEM. (E, F) TCR transgenic pmel CD8+ T overexpressing miR-155 or control scrambled miR were transferred into B16 tumor-bearing mice and (E) tumor growth and (F) survival of mice are shown from one representative out of two experiments with five to eight mice per group displayed as mean +/- SEM. Please also see Figure S3.
Compared to wild type, Mir155-/- CD8+ T cells were less effective in inhibiting tumor growth and ensuring the survival of tumor-challenged mice (Figure 5C, D). Given the inability of T cells to expand and control tumor growth in the absence of miR-155, we hypothesized that enforcing miR-155 expression would augment the anti-tumor activity of CD8+ T cells. Pmel-1 TCR transgenic CD8+ T cells specific for the melanoma antigen gp100 were transduced with a retrovirus encoding miR-155 or scrambled miR and adoptively transferred into tumor-bearing mice in conjunction with gp100 vaccination and IL-2. There were no phenotypic differences between control and miR-155 overexpressing T cells prior to adoptive transfer (Figure S3C). However, overexpression of miR-155 greatly enhanced the anti-tumor responses compared to scrambled miR control (Figure 5E). Notably, 80% of mice receiving miR-155 transduced T cells survived for over 60 days, whereas all mice treated with control cells or left untreated had to be euthanized after less than 40 days due to tumor size (Figure 5F). Taken together, these results show that upon vaccination, Mir155-/- CD8+ T cells differentiated into effector cells but failed to accumulate in normal numbers. Consequently, the anti-tumor response was strongly dependent on miR-155 and could be therapeutically boosted by enforced miR-155 expression.
Targeting of SOCS-1 by miR-155 in effector CD8+ T cells enables cytokine responsiveness and accumulation
miR-155 has been shown to regulate γc-chain cytokine signaling by targeting SOCS-1 expression (D’Souza and Lefrancois, 2003; Lu et al., 2009; Wang et al., 2010). We assessed SOCS-1 regulation in splenic effector CD44+CD62L−CD8+ T cells during the response to acute LCMV infection of wild type and Mir155-/- mice. We found that both wild type and Mir155-/- CD8+T cells downregulated SOCS-1 on days 6 and 8 compared to CD62L+CD44− naïve CD8+ T cells from non-infected mice (Figure 6A). To more directly test if SOCS-1 was regulated by miR-155, we measured SOCS-1 mRNA in wild type and Mir155-/- CD8+ T cells as well as in cells overexpressing miR-155 or scrambled control miR. We found that the amounts of SOCS-1 transcripts were inversely related to the cellular content of miR-155, with the highest concentration of SOCS-1 in Mir155-/- cells and the lowest in miR-155 transduced cells (Figure 6B). These results were further confirmed at the protein level, indicating that miR-155 is a critical regulator of SOCS-1 translation in CD8+ T cells (Figure 6C). To test whether the loss of miR-155 impaired γc chain cytokine signaling in CD8+ T cells by upregulating SOCS-1, we compared STAT5 phosphorylation in response to IL-2, IL-7 or IL-15 in wild type and Mir155-/- cells. Stimulation of naïve and effector CD8+ T cells isolated 8 days after LCMV infection resulted in a limited phosphorylation of STAT5 in miR-155 ablated cells, demonstrating an impaired cytokine signaling (Figure 6D). Diminished STAT5 phosphorylation was not due to differential expression of the cytokine receptor chains CD25, CD122, CD127 or CD132 (Figure S4). To further investigate whether the impaired cytokine signaling was dependent on the higher SOCS-1 concentration in Mir155-/- CD8+ T cells, we transduced wild type and Mir155-/- CD8+ T cells with control or shSOCS-1 lentivirus. Although in vitro activation of T cells diminished the impact of miR-155 on cytokine signaling, we consistently detected a rescue of pSTAT5 generation in shSOCS-1 transfected Mir155-/- cells (Figure 6E). Interestingly, baseline pSTAT5 expression was already higher in wild type than in Mir155-/- cells without additional IL-2 stimulation. The difference between wild type and Mir155-/- cells was still apparent with intermediate, but disappeared with high IL-2 concentrations, demonstrating that saturating amounts of IL-2 overcome the miR-155 and SOCS-1 dependent inhibition of cytokine signaling, as shown for regulatory T cells (Lu et al., 2009). Together, these results demonstrate a dynamic and differentiation-dependent regulation of SOCS-1 during the response to LCMV and suggest that Mir155-/- CD8+ T cells have impaired cytokine signaling due to increased SOCS-1.
Figure 6. SOCS-1 and miR-155 modulate the antiviral CD8+ T cell response and cytokine signaling.
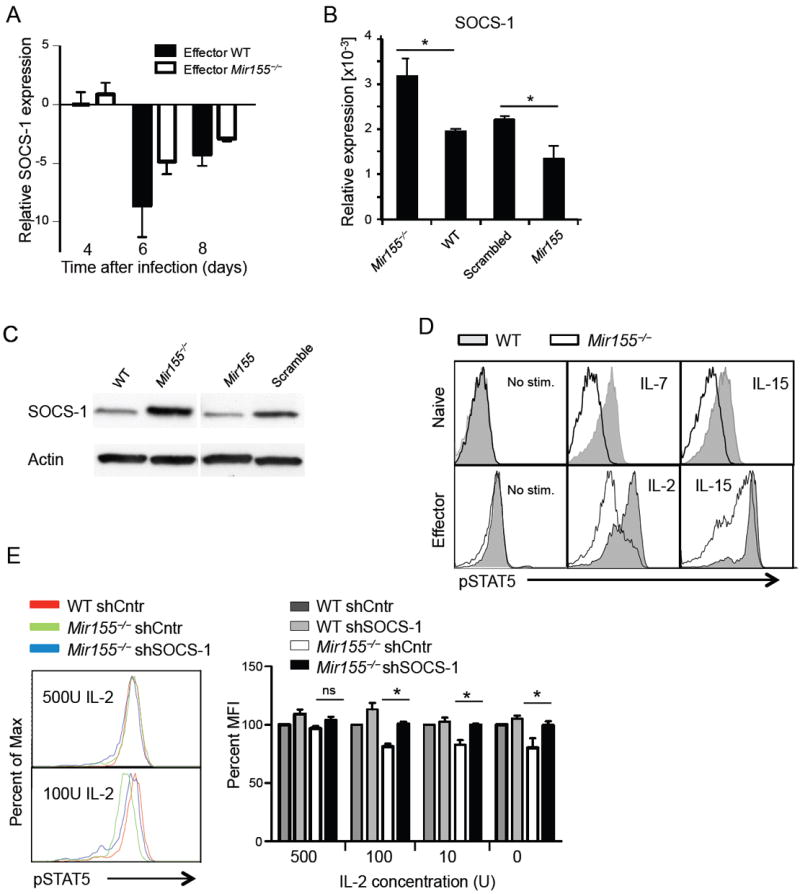
(A) SOCS-1 mRNA concentrations were measured upon LCMV WE infection in purified effector (CD44highCD62Llow) wild type (WT) and Mir155-/- splenic CD8+ T cells by qPCR relative to naïve CD8+ T cells from non-infected mice. (B, C) Regulation of SOCS-1 by miR-155 in naïve CD8+ T cells from WT and miR-155 deleted mice as well as after retroviral transfection with miR-155 overexpressing or control vectors was tested by (B) qPCR (shown as relative to β-actin) and (C) Immunoblot. (D) Naïve or effector T cells from LCMV infected mice were stimulated with indicated cytokines and pSTAT5 was measured by flow cytometry. (E) WT and Mir155-/- T cells were transduced with control or shSOCS-1 lentivirus and pSTAT5 response to IL-2 is shown. The table gives the percentages of MFI normalized to the MFI measured in wild type sh-control cells set to 100%. Representative data from two (A-C) to three (D, E) experiments are pictured as mean +/- SEM. Please also see Figure S4.
SOCS-1 restrains CD8+ T cell responses to virus and cancer
To test whether increased SOCS-1 expression recapitulated the impaired antigen-driven expansion of Mir155-/- CD8+ T cells, SOCS-1 transgenic or wild type P14 CD8+ T cells were adoptively transferred into congenic mice prior to infection with LCMV WE strain. The expansion of SOCS-1 transgenic P14 T cells in blood and spleen was reduced compared to P14 wild type cells (Figure 7A and data not shown). Whereas effector phenotype, granzyme B and cytokine production were not impaired (Figure S5A-C), we detected enhanced apoptosis of SOCS-1 overexpressing cells (Figure 7B), thus phenocopying Mir155-/- CD8+ T cells (Figures 2, 3 and S2). To test if suppression of SOCS-1 could be therapeutically exploited to enhance the CD8+ T cell anti-tumor response Pmel CD8+ T cells transduced with shSOCS-1 were adoptively transferred into tumor-bearing mice. SOCS-1 depletion by the construct was verified by Immunoblot analysis (Figure S5D). An increased expansion of cells expressing shSOCS-1 was detected in the spleen on day 4 compared to control (Figure 7C), associated with profound tumor regression in mice that received shSOCS-1 transduced cells compared to untreated mice or mice treated with control cells (Figure 7D). Together, these results demonstrate that SOCS-1 is negatively regulating the effector CD8+ T cell response to virus and cancer and highlight the importance of SOCS-1 downregulation by miR-155 for efficient CD8+ T cell responses.
Figure 7. SOCS-1 limits the CD8+ T cell response to virus and cancer.
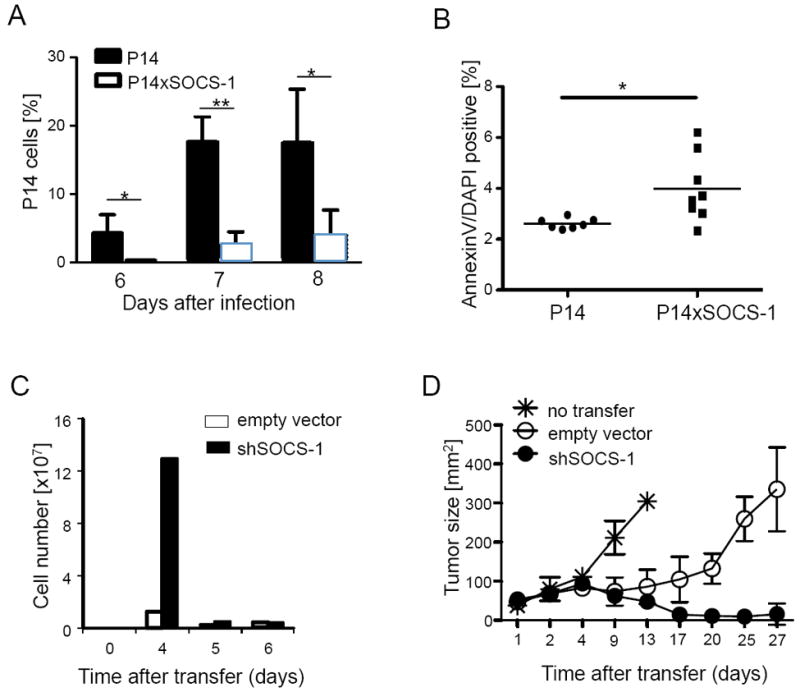
(A) TCR transgenic P14 CD8+ T cells overexpressing SOCS-1 (P14×SOCS-1) or not were adoptively transferred before LCMV WE infection. Data show the percentage of transferred cells in the lymphocyte gate at days 6, 7 and 8 post infection as mean +/- SEM; (B) Apoptotic cells within P14 T cells 7 days upon infection. Symbols represent single mice and the line is the mean. (C, D) TCR transgenic pmel CD8+ T cells were transduced with a retrovirus encoding for a scrambled control or shSOCS-1 mRNA and adoptively transferred into tumor bearing mice. (C) Absolute numbers of donor CD8+ T cells were determined in spleen at days 4 to 6 after adoptive transfer and (D) tumor size of mice was monitored. Data are from one representative out of two independent experiments with two (C) to five mice (D) per group and displayed as mean +/- SEM. Please also see Figure S5.
DISCUSSION
We have shown that miR-155 expression is essential for optimal CD8+ T cell responses towards virus infection, vaccination and cancer. Interestingly, the phenotype of Mir155-/- CD8+ T cells was similar to Dicer deficient CD8+ T cells (Zhang and Bevan, 2010), suggesting that miR-155 is an important, likely non-redundant miRNA for CD8+ effector T cells. Recently, we and others found differential miR-155 expression associated with discrete differentiation stages in CD8+ T cells (Almanza et al., 2010; Salaun et al., 2011; Wu et al., 2007). Here, we showed that miR-155 is highly upregulated in effector CD8+ T cells responding to viral infection but at intermediate concentration in memory cells. This dynamic modulation of miR-155 raises the question of the nature of the regulating factors that are involved. miR-155 is induced by NF-kappaB dependent factors (Kluiver et al., 2007) and AP-1 downstream of B and T cell receptors (Haasch et al., 2002; Yin et al., 2008). Interestingly, we observed that miR-155 expression was proportional to the strength of TCR signaling, suggesting that miR-155 provides competitive fitness to the most avid antigen-specific CD8+ T cells.
Whereas others reported a disadvantage of Mir155-/- naive CD8+ T cells in bone-marrow chimeras (Lu et al., 2009), we found that expansion and long term survival of naïve Mir155-/- CD8+ T cells transferred into lymphopenic hosts were not different from wild type cells. miR-155 was crucial for effector CD8+ T cell proliferation and survival during the peak of LCMV infection, but it did not influence cell survival upon antigen clearance. In contrast to the reported role of miR-155 in other immune cells, CD8+ T cells were not affected in effector functions such as killing or cytokine production and miR-155 deleted mice readily cleared low doses of LCMV. Interestingly, we detected an impaired generation of virus-specific central memory CD8+ T cells as most Mir155-/- cells displayed a terminally differentiated phenotype. Whether this was due to a lack of CD4+ T cell help (Janssen et al., 2003; Shedlock and Shen, 2003), lower cytokine signaling due to higher SOCS-1 expression and/or additional miR-155 targets will be subject of further investigation. In contrast to low-dose LCMV infection, we found that virus-specific Mir155-/- effector cells disappeared during chronic LCMV infection using high doses of clone 13. Under these conditions, wild type CD8+ T cells undergo progressive attrition and display an ‘exhausted’ phenotype with impaired effector functions and high PD-1 expression (Jin et al., 2010; Mueller and Ahmed, 2009; Wherry et al., 2003). We hypothesize that miR-155 is key for the survival of effector CD8+ T cells in conditions of long-term exposure to antigen and inflammation as found in chronic viral infections or cancer. Consistent with the impaired virus control, lack of miR-155 protected from LCMV-induced severe immunopathology, which is in line with reports demonstrating a pivotal role of miR-155 in autoimmune inflammation (Murugaiyan et al., 2011; O’Connell et al., 2010). With regard to T cell survival, miR-155 was shown to inhibit caspase 3 activity in Jurkat T cells (Ovcharenko et al., 2007), and FADD expression in macrophages (Tili et al., 2007). Whether the increased apoptosis of Mir155-/- CD8+ T cells was due to such mechanisms remains to be determined.
In line with our results in virus infection, we demonstrate a central role for miR-155 in tumor specific CD8+ T cells. First, the efficient accumulation of CD8+ T cells by vaccination with adjuvanted peptide required intrinsic miR-155 function, recapitulating our observations in LCMV infection. More importantly, Mir155-/- effector cells were severely impaired in their ability to curb tumor growth. Since these cells acquired full effector functions independently of miR-155, the different tumor control was likely due to defect in the magnitude of tumor-specific T cells, or to higher susceptibility to the suppressive tumor-microenvironment (Klebanoff et al., 2011). Conversely, overexpression of miR-155 greatly increased tumor killing by wild type CD8+ T cells. This indicates that tumor specific CD8+ T cell activity is directly dependent on miR-155 expression.
The accumulation of effector CD8+ T cells is influenced by the cytokine milieu, e.g. their initial expansion is promoted by γc cytokines, which signaling is regulated by SOCS-1 (Cornish et al., 2003). More specifically, CD8+ effector cell accumulation in the LCMV response is reduced if cells lack γc signaling (Decaluwe et al., 2010). Interestingly, IL-2 was found to be critical for the maintenance of effector CD8+ T cells in chronic LCMV infections (Bachmann et al., 2007). Here we show that miR-155 enhances cytokine signaling in CD8+ T cells by targeting SOCS-1. Consequently, Mir155-/- naïve and effector CD8+ T cells failed to mount physiologic levels of pSTAT5 in response to γc cytokines. In line with in vitro experiments using CD4+ T cells (Lu et al., 2009), we observed strong and transient in vivo downregulation of SOCS-1 mRNA in virus-specific CD8+ T cells at day 6 of the response. Interestingly, this occurred in a partially miR-155 independent manner, whereas SOCS-1 protein concentrations of naïve and effector CD8+ T cells were found to be clearly dependent on miR-155. Moreover, we did not detect an impact of miR-155 on pSTAT5 in response to IL-2 in the early effector response in vivo as well as after in vitro priming (unpublished data), whereas the effect was pronounced in naïve and late effector cells. This suggests a miR-155 independent mechanism allowing full cytokine signaling early in the effector response, whereas miR-155 is modulating cytokine signaling in naïve and late effector cells, promoting their accumulation and survival. Interestingly, at the peak of the response, effector CD8+ T cells are reportedly dependent on IL-2 and IL-15 for sustained expansion (D’Souza and Lefrancois, 2003; Sanjabi et al., 2009). We were able to confirm a SOCS-1 dependent inhibition of cytokine signaling in Mir155-/- T cells by transduction with a shSOCS-1 lentivirus in in vitro experiments. Although the formal proof that loss of SOCS-1 in Mir155-/- CD8+ T cells in vivo would restore their function remains to be shown, the observed rescue of STAT5 phosphorylation in Mir155-/- cells back to wild type levels suggests that suppression of SOCS-1 in wild type but not Mir155-/- T cells caused the differences in pSTAT5 signaling. This hypothesis was further supported by our observation that virus specific, SOCS-1 transgenic CD8+ T cells fully differentiated but failed to accumulate to normal numbers at the peak of the LCMV response, which mirrors the phenotype of Mir155-/- T cells and identifies SOCS-1 as an important regulator of CD8+ T cell responses in vivo. Conversely, suppression of SOCS-1 in tumor specific cells increased the accumulation of transferred cells and subsequently was highly therapeutic in limiting growth of established melanoma. Thus, although a negative role for SOCS-1 in CD8+ T cell responses has been suggested before (Chong et al., 2003; Cornish et al., 2003; Davey et al., 2005; Marine et al., 1999; Palmer and Restifo, 2009), we here demonstrate a cell-intrinsic role of SOCS-1 in responses to virus and cancer. Our results of the highly dynamic regulation of SOCS-1 expression in vivo and the strong impact of SOCS-1 alterations in responses to virus and tumor suggests that a major part of the effects caused in CD8+ T cells by miR-155 deletion are due to increased SOCS-1 expression.
In summary, the results presented here identified a crucial cell-intrinsic role of miR-155 and its target SOCS-1 in effector CD8+ T cells, and demonstrated that this miRNA is required for an optimal CTL response to both virus and tumor. Moreover, miR-155 overexpression in tumor specific CD8+ T cells substantially increased their potency, thus providing strong evidence for a clinical potential in the context of therapeutic adoptive T cell transfer.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 mice were from Harlan. Mir155−/− and Rag2−/−Il2rg−/−, TCR transgenic OT-I and P14 mice bearing a transgenic TCR specific for Kb/OVA257-264 or LCMV Db/GP33-41 MHC-I/peptide complexes, respectively, were from Jackson and were bred under specific pathogen free conditions. P14xSOCS-1 transgenic (Seki et al., 2007) and Pmel-1 transgenic mice expressing a TCR specific for an H-2Db-restricted CD8+ T cells epitope from the murine melanoma tumor antigen gp10025-33 or human gp10025-33 have been described (Overwijk et al., 2003). All animal experiments were conducted with the approval of the Lemanic Animal Facility Network (RESAL), NCI (protocol SB-126) or NIAID (protocol LI-10) Animal Use and Care Committees.
Quantitative PCR
Total RNA from subsets was extracted with the miRVana kit (Ambion), and mature microRNAs (miR-155 and controls RNU44 and snoRNA202) were reverse transcribed with TaqMan RT MicroRNA kit (Applied Biosystems) and amplified using Universal Fast Start Rox Probe Master Mix (Roche) and microRNA assay kits in 384 well plates (Applied Biosystems) on an ABI Prism 7900 HT device (Applied Biosystems). RT-PCR for Socs1 was performed with primers from Applied Biosystems. Gene expression was calculated relative to G6PDX.
In vitro priming with dendritic cells
T cells were purified from lymphoid tissues using anti-CD8α beads (Miltenyi) and cocultured with peptide pulsed Mir155-/- DC. In competition experiments, equal numbers of WT and Mir155-/- OT-1 CD8+ T cells were cocultured with WT DC.
Differential stimulation of human CD8+ T cells and microarray
Human T cells transduced with NY-ESO-1 TCR variants (Schmid et al., 2010) were stimulated with 0.01μg/ml (for miR-155 quantification) or 0.002 μg/ml (for BIC microarray) NY-ESO-1157-165 multimer. BIC quantification was done by Miltenyi Biotec.
LCMV infection
Mice were infected intravenously with 200 pfu of the WE strain or 2 × 106 pfu clone 13 of LCMV. Cell counts in blood were performed with TrueCount tubes according to the manufacturer’s instructions (BD). Viral titrations were performed by a standard plaque assay.
Cytokine and cytotoxicity assays
Splenocytes were stimulated with indicated-peptides or a mix of LCMV peptides (gp33, gp70, gp92, gp118, gp276, np166, np205, np235, np396) at 5 μM each or PMA (50 ng/mL) and Ionomycin (500 ng/mL) in the presence of GolgiStop (BD Pharmingen) for analysis in flow cytometry. Cytolytic activity was assessed using the chromium release assay with EL4 pulsed with 1 μM GP33-41 peptide and measured with a TopCount reader (Canberra Packard).
Adoptive cell transfers
Before LCMV infection, CD8+ T cells were purified by magnetic beads sorting (Miltenyi Biotec) and 1×106 purified CD8+ T cells were injected into the tail vein. In the P14xSOCS-1 experiments, 3×104 cells were injected normalized to gp33 tetramer positive populations. For vaccination and tumor challenge, OT-1cells were purified and 1×106 cells were injected intravenously three days before subdermal injection of 1×105 B16 melanoma cells expressing OVA. 7 days later, mice were vaccinated s.c. with 25 μg OVA257-264 peptide and 50 μg CPG-ODN in PBS.
T cell transduction and transfers into tumor bearing mice
Pmel CD8+ T cells were stimulated with anti-CD3 (2μg/ml), anti-CD28 (1μg/ml) for 24h, transduced with retrovirus expressing miR-155 or scrambled miR and expanded for 4 days. To knock down SOCS-1, pmel CD8+ T cells were transduced 2 days after stimulation with lentivirus expressing shSOCS-1 or empty vector and selected in puromycin from days 4-7 prior to transfer into tumor-bearing hosts. 107 cells (miR-155 experiments) or 106 (shSOCS-1 experiments) were injected i.v. into mice bearing B16 tumor in conjunction with 2×107 pfu rvvhgp100 and IL-2 (6X6e4 cu). In shSOCS-1 experiments, mice were sublethally irradiated prior to cell transfer. Tumor volumes were plotted as the product of perpendicular diameters. For rescue of pSTAT5 in miR-155 cells, wild type and Mir155-/- CD8+ T cells were transduced with lentivirus encoding shSOCS-1 or control sh and cultured for one week with IL-2 or IL-15 (R&D systems, 20 ng/ml) before cytokine stimulation (vector sequences upon request).
Flow Cytometry and pSTAT5 measurements
Cells were stained with antibodies of the indicated specificities (eBioscience) or H-2 Db/peptide-loaded MHC. PD-1 PE-Cy7 antibody was from Biolegend. For detection of pSTAT5, cells were stimulated with indicated cytokines (R&D systems) for 18 min and fixed with 0.5% formaldehyde in PBS for 15 min. Upon washing in medium containing 10% FCS, cells were permeabilized in 80% methanol for 20 min, washed and stained with pSTAT5 antibody (BD biosciences) and additional markers (eBiosciences). Labeled cells were analysed on LSR-II (Becton Dickinson).
Immunoblot
Cells were lysed (Cell Signaling Technologies) and immunoblotting was performed using Bio-Rad TGX reagents and protocols on nitrocellulose paper, incubated with antibodies against SOCS-1 (Lifespan Biosciences (Figure 6C) or Imgenex (Figure S6)) and with appropriate HRP-conjugated secondary antibodies (Cell Signal Technologies). Blots were developed using chemiluminescence (Pierce), gel images were captured with Gel Doc XRS (Bio-Rad) and densitometry evaluated using Quantity One software (Bio-Rad).
Proliferation and Apoptosis assay
Mice were injected i.p. with 200 μg BrdU and proliferation was measured in spleen cells 1h (P14xSOCS-1 experiments) or 4h (miR-155 experiments) later by staining with anti-BrdU and anti-ki67+ antibodies. Apoptotic cells were detected by staining splenocytes for AnnexinV upon 1h in vitro incubation at 37 °C (all BD Pharmingen).
Statistics
The two-tailed student’s T test was used to compare two groups, multiple groups’ comparisons were performed with one way analysis of variance (ANOVA) corrected with Bonferroni’s multiple comparison test factor. Statistical survival differences in the tumor experiments were analyzed with the Log-Rank test. Statistical significance is displayed as *: p<0,05, **: p<0,01, ***: p<0,001. Data are displayed as Mean and standard error of mean (SEM) if not indicated otherwise.
Supplementary Material
Acknowledgments
We thank Danny Labes for technical assistance and Immanuel Luescher for providing tetramers. We also thank P. Martiat and B. Badran for critical input. JCD and BS were supported in part by a grant from the MEDIC foundation, RP by NZ FRST, WH and CB by the SNSF. GM, DZ and PR were supported in part by a Prodoc and a Sinergia grants from the SNSF. JY, DCP, NPR and LG were supported by the Intramural Research Programs of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research. TME and SAM were supported by the Intramural Research Program of the US National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Footnotes
The authors have no conflict of interests.
References
- Almanza G, Fernandez A, Volinia S, Cortez-Gonzalez X, Croce CM, Zanetti M. Selected microRNAs define cell fate determination of murine central memory CD8 T cells. PLoS. 2010;5:e11243. doi: 10.1371/journal.pone.0011243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann MF, Wolint P, Walton S, Schwarz K, Oxenius A. Differential role of IL-2R signaling for CD8+ T cell responses in acute and chronic viral infections. European journal of immunology. 2007;37:1502–1512. doi: 10.1002/eji.200637023. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Schambach F, DeJong CS, Hammond SM, Reiner SL. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. European journal of immunology. 2010;40:225–231. doi: 10.1002/eji.200939381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nature reviews. Immunology. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chong MM, Cornish AL, Darwiche R, Stanley EG, Purton JF, Godfrey DI, Hilton DJ, Starr R, Alexander WS, Kay TW. Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation. Immunity. 2003;18:475–487. doi: 10.1016/s1074-7613(03)00078-5. [DOI] [PubMed] [Google Scholar]
- Cornish AL, Chong MM, Davey GM, Darwiche R, Nicola NA, Hilton DJ, Kay TW, Starr R, Alexander WS. Suppressor of cytokine signaling-1 regulates signaling in response to interleukin-2 and other gamma c-dependent cytokines in peripheral T cells. J Biol Chem. 2003;278:22755–22761. doi: 10.1074/jbc.M303021200. [DOI] [PubMed] [Google Scholar]
- Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunological reviews. 2010;236:151–166. doi: 10.1111/j.1600-065X.2010.00926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza WN, Lefrancois L. IL-2 is not required for the initiation of CD8 T cell cycling but sustains expansion. Journal of immunology. 2003;171:5727–5735. doi: 10.4049/jimmunol.171.11.5727. [DOI] [PubMed] [Google Scholar]
- Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- Davey GM, Starr R, Cornish AL, Burghardt JT, Alexander WS, Carbone FR, Surh CD, Heath WR. SOCS-1 regulates IL-15-driven homeostatic proliferation of antigen-naive CD8 T cells, limiting their autoimmune potential. The Journal of experimental medicine. 2005;202:1099–1108. doi: 10.1084/jem.20050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaluwe H, Taillardet M, Corcuff E, Munitic I, Law HK, Rocha B, Riviere Y, Di Santo JP. Gamma(c) deficiency precludes CD8+ T cell memory despite formation of potent T cell effectors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9311–9316. doi: 10.1073/pnas.0913729107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derre L, Bruyninx M, Baumgaertner P, Ferber M, Schmid D, Leimgruber A, Zoete V, Romero P, Michielin O, Speiser DE, Rufer N. Distinct sets of alphabeta TCRs confer similar recognition of tumor antigen NY-ESO-1157-165 by interacting with its central Met/Trp residues. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15010–15015. doi: 10.1073/pnas.0807954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331(6017):550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, Kroeger P, McWeeny K, Halbert DN, Mollison KW, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cellular immunology. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, Wan Y, He L, Li QJ. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HT, Anderson AC, Tan WG, West EE, Ha SJ, Araki K, Freeman GJ, Kuchroo VK, Ahmed R. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14733–14738. doi: 10.1073/pnas.1009731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanoff CA, Gattinoni L, Palmer DC, Muranski P, Ji Y, Hinrichs CS, Borman ZA, Kerkar SP, Scott CD, Finkelstein SE, et al. Determinants of successful CD8+ T-cell adoptive immunotherapy for large established tumors in mice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5343–5352. doi: 10.1158/1078-0432.CCR-11-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, van den Berg A, de Jong D, Blokzijl T, Harms G, Bouwman E, Jacobs S, Poppema S, Kroesen BJ. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta} J Biol Chem. 2010;285:41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Topham DJ, McKay C, Wang D, Parganas E, Stravopodis D, Yoshimura A, Ihle JN. SOCS1 deficiency causes a lymphocyte-dependent perinatal lethality. Cell. 1999;98:609–616. doi: 10.1016/s0092-8674(00)80048-3. [DOI] [PubMed] [Google Scholar]
- Marrack P, Kappler J. Control of T cell viability. Annual review of immunology. 2004;22:765–787. doi: 10.1146/annurev.immunol.22.012703.104554. [DOI] [PubMed] [Google Scholar]
- Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The Interleukin 13 (IL-13) Pathway in Human Macrophages Is Modulated by MicroRNA-155 via Direct Targeting of Interleukin 13 Receptor {alpha}1 (IL13R{alpha}1) J Biol Chem. 2011;286:1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskophidis D, Lechner F, Pircher H, Zinkernagel RM. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 1993;362:758–761. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- Mueller SN, Ahmed R. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8623–8628. doi: 10.1073/pnas.0809818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. The Journal of experimental medicine. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. Journal of immunology. 2011;187:2213–2221. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. The Journal of experimental medicine. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67:10782–10788. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- Overwijk WW, Theoret MR, Finkelstein SE, Surman DR, de Jong LA, Vyth-Dreese FA, Dellemijn TA, Antony PA, Spiess PJ, Palmer DC, et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. The Journal of experimental medicine. 2003;198:569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends in immunology. 2009;30:592–602. doi: 10.1016/j.it.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaun B, Yamamoto T, Badran B, Tsunetsugu-Yokota Y, Roux A, Baitsch L, Rouas R, Fayyad-Kazan H, Baumgaertner P, Devevre E, et al. Differentiation associated regulation of microRNA expression in vivo in human CD8+ T cell subsets. Journal of translational medicine. 2011;9:44. doi: 10.1186/1479-5876-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato M, Borrow P, Shimomaye E, Oldstone MB. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. Journal of virology. 1991;65:1863–1869. doi: 10.1128/jvi.65.4.1863-1869.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S, Mosaheb MM, Flavell RA. Opposing effects of TGF-beta and IL-15 cytokines control the number of short-lived effector CD8+ T cells. Immunity. 2009;31:131–144. doi: 10.1016/j.immuni.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid DA, Irving MB, Posevitz V, Hebeisen M, Posevitz-Fejfar A, Sarria JC, Gomez-Eerland R, Thome M, Schumacher TN, Romero P, et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. Journal of immunology. 2010;184:4936–4946. doi: 10.4049/jimmunol.1000173. [DOI] [PubMed] [Google Scholar]
- Seki Y, Yang J, Okamoto M, Tanaka S, Goitsuka R, Farrar MA, Kubo M. IL-7/STAT5 cytokine signaling pathway is essential but insufficient for maintenance of naive CD4 T cell survival in peripheral lymphoid organs. J Immunol. 2007;178:262–270. doi: 10.4049/jimmunol.178.1.262. [DOI] [PubMed] [Google Scholar]
- Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. Journal of immunology. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- Turner M, Vigorito E. Regulation of B- and T-cell differentiation by a single microRNA. Biochem Soc Trans. 2008;36:531–533. doi: 10.1042/BST0360531. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. Journal of immunology. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. Journal of virology. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Wang X, McBride J, Fewell C, Flemington E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J Biol Chem. 2008;283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458:211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


