Abstract
Tristetraprolin (TTP), a mRNA-binding protein, plays a significant role in regulating the expression of adenylate-uridylate-rich elements (AREs) containing messenger RNAs (mRNAs). Mice deficient of TTP (TTP−/−) develop a systemic autoimmune inflammatory syndrome characterized by cachexia, conjunctivitis and dermatitis. Interleukin-12 (IL-12) plays a crucial role in immune defense against infectious and malignant diseases. In the present study, we found increased production of IL-12 during endotoxic shock and enhanced Th1 cells in TTP knockout mice. The levels of IL-12 p70 and p40 protein as well as p40 and p35 mRNA were also increased in activated macrophages deficient of TTP. In line with these findings, overexpression of TTP suppressed IL-12 p35 and p40 expression at the mRNA and promoter level, while surprisingly had little effects on their mRNA stability. Our data showed that the inhibitory effects of TTP on p35 gene transcription were completely rescued by overexpression of NF-κB p65 and c-Rel but not the p50 in activated macrophages. Our data further indicated that TTP acquired its inhibition on IL-12 expression through blocking nuclear translocation of NF-κB p65 and c-Rel while enhancing p50 upon stimulation. In summary, our study reveals a novel pathway through which TTP suppresses IL-12 production in macrophages, resulting in suppression of Th1 cell differentiation. This study may provide us with therapeutic targets for treatment of inflammatory and autoimmune disorders.
Introduction
IL-12 is the first heterodimeric cytokine in the IL-12 family composed of two heterologous chains, a 35-kDa light chain (known as p35 or IL-12α) and a 40-kDa heavy chain (known as p40 or IL-12β) (1). It is mainly produced by activated monocytes, macrophages and dendritic cells (DCs) during innate and adaptive immune responses (2). IL-12 plays an essential role in initiation of antigen-specific Th1 cell responses, in activation of NK cells, and in induction of opsonic as well as complement-fixing antibodies (3). Similar to other pro-inflammatory cytokines, the production of IL-12 is regulated strictly by positive and negative regulatory mechanisms. Products from microorganisms, including bacteria, parasites, fungi, double-stranded RNA, bacterial DNA and CpG-containing oligonucleotides, are strong inducers of IL-12 by macrophages, monocytes and DCs (2). Our previous work demonstrate that the expression of IL-12 p35 and p40 subunit is differentially regulated by IFN-γ signaling pathway through interferon regulatory factor 1 (IRF-1) and IRF-8 at the level of transcription (4, 5). T cells can also enhance the production of IL-12 by a cell-cell contact mechanism through ligands of the tumor necrosis factors (TNF) family (6). Meanwhile, anti-inflammatory cytokines such as IL-10, transforming growth factor-β and prostaglandin E2 suppress IL-12 production at different levels (2). Up to date, the molecular mechanisms of transcriptional IL-12 gene expression have been studied extensively, and some important mediators and effectors have been well identified, including NF-κB (7–11). Our previous study also demonstrate that NF-κB, especially the p65 and c-Rel subunit, activates IL-12 p35 gene transcription (5). NF-κB plays a variety of roles in cell survival, differentiation and proliferation, and is known to be involved in innate immune responses (12). Dysregulation of NF-κB activity has been linked to cancer, inflammatory and autoimmune diseases, viral infection, and improper immune development (13–15). Whether IL-12 is regulated at the level of post-transcription and what is the role of NF-κB in the posttranscriptional regulation of IL-12 is currently unknown.
Messenger RNA (mRNA) decay is a critical posttranscriptional regulation step to control the expression of many inflammation and cancer-associated genes (16). These transcripts are targeted for rapid mRNA degradation by adenylate-uridylate-rich elements (AREs) motifs present in the mRNA 3’ untranslated region (3’UTR). Tristetraprolin (TTP), one of the best-characterized ARE-binding proteins, functions to mediate mRNA rapid decay (17, 18). It is involved in the regulation of inflammatory responses at the posttranscriptional level. TTP binds to AREs within the 3’UTR causing destabilization of mRNAs encoding tumor necrosis factor-α (TNF-α) (19), granulocyte-macrophage colony-stimulating factor (GM-CSF) (20), cyclooxygenase 2 (21), interleukin-2 (22), interleukin-10 (23) and the chemokine CXCL1 (24). The mRNAs encoding TNF-α and GM-CSF are stabilized in TTP-deficient mice and in cells derived from these deficient mice (17, 20). Overproduction of these cytokines in TTP knockout mice results in a severe systemic inflammatory response including arthritis, autoimmunity and myeloid hyperplasia (25, 26). Collectively, all evidence indicates that TTP is a critical protein involved in the control of inflammation and maintenance of homeostasis. Though TTP functions as RNA-binding protein promoting mRNA degradation, it has been reported that TTP also regulates gene expression at the level of transcription. Schichl et al reported that independent of its mRNA destabilizing effect TTP inhibited several NF-κB-dependent promoter activities including IL-8 promoter and tissue factor promoter (27). Ling et al also reported that TTP repressed NF-κB activity by directly binding to the p65 subunit and by recruiting histone deacetylases to promoters of the NF-κB target genes (28). Several NF-κB target genes such as IL-1β and TNF-α were also suppressed by TTP through inhibition of NF-κB activity.
It has been reported that silencing TTP expression in mouse macrophage cell line J774A.1 cells led to an increase in IL-12 expression (29). However, it is unknown whether this is true in primary macrophages and how TTP regulates IL-12 expression. In this study, using TTP deficient mice, we demonstrate that the levels of IL-12 in serum are enhanced in TTP knockout mice during endotoxic shock. There are also increased Th1 cells in mice deficient of TTP. TTP suppresses IL-12 expression through blocking nuclear translocation of NF-κB p65 and c-Rel while enhancing p50 in activated macrophages. Our study reveals a novel pathway through which TTP suppresses IL-12 production in macrophages, resulting in suppression of Th1 cell differentiation. This study may provide us with therapeutic targets for treatment of inflammatory and autoimmune disorders.
Materials and Methods
Mice
Conventional TTP knockout mice (26) were backcrossed into C57Bl6 mice more than 26 generations. Female TTP−/− mice and their wild type littermates at 4–6 week old were used in the experiments. All mice were housed in cages with filter tops in a laminar flow hood and fed food and water ad libitum at Saint Louis University Animal Facilities in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication no. 85-23, 1985).
Cells
Both murine macrophage cell line RAW264.7 (RAW cells) and human macrophage cell line THP-1 cells were obtained from ATCC (Manassas, VA) and were maintained in RPMI 1640 supplemented with 2 mM glutamine, 100 U/ml penicillin and streptomycin, and 10% FBS (Sigma-Aldrich, St. Louis, MO; endotoxin, N-myristoyltransferase 10.0 EU/ml) (complete medium). Mouse peritoneal macrophages were obtained by lavage 3 days after injection of sterile 3% thioglycolate broth (1 ml i.p. per mouse) and plated in 24-well tissue culture plates (1 × 106 cells/well) with RPMI 1640 complete medium. Mouse bone marrow-derived macrophages (BMDMs) were generated from bone marrow cells and cultured with RPMI 1640 complete medium containing 10 ng/mL M-CSF for one week. The spleens were sampled and single cell suspensions were prepared by mechanical mincing, as reported previously (30).
Plasmids
Human IL-12 p35 and p40 promoters were described previously (5). The expression vectors for TTP, CMV hTTP.HA (19) and CMV mTTP.HA (31), were originally cloned in Perry Blackshear’s laboratory. The expression vectors for NF-κB p50, p65 and c-Rel were originally provided by K. Murphy (Washington University, St. Louis, MO) (32). All plasmid DNAs were prepared with Qiagen EndoFree Maxiprep kits.
Antibodies and reagents
Rabbit anti-TTP antibody (N-terminal) was purchased from Sigma-Aldrich. Mouse anti-NF-κB p65, c-Rel and p50 monoclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rat anti-HA monoclonal antibody (clone 3F10) was obtained from Roche Applied Science (Indianapolis, IN). Secondary antibodies conjugated to Alexa Fluor 488 and Alexa Fluor 568 were obtained from Invitrogen (Eugene, OR). Recombinant mouse IFN-γ was purchased from Genzyme (Boston, MA). M-CSF was purchased from R&D Systems. All other chemicals if not otherwise stated were obtained from Sigma-Aldrich (St. Louis, MO).
Quantitative real-time PCR
RNA extraction and reverse-transcription reactions were carried out as previously described (33). Quantitative real-time PCR (qRT-PCR) was performed by a modified protocol (34). The following primers were used for PCR amplification of the mouse p35 cDNA sense: acctgctgaagaccacagatgaca, antisense: tagccaggcaactctcgttcttgt; mouse p40 cDNA sense: acctgtgacacgcctgaagaagat, antisense: tcttgtggagcagcagatgtgagt; mouse TTP cDNA sense: aatccctcggaggactttggaaca, antisense: agttgcagtaggcgaagtaggtga; mouse TNF-α cDNA sense: agccgatgggttgtaccttgtcta, antisense: tgagatagcaaatcggctgacggt; mouse CCL5 cDNA sense: gatggacatagaggacacaact, antisense: tgggacggcagatctgaggg; mouse IL-27 p28 cDNA sense: ctctgcttcctcgctaccac, antisense: ggggcagcttcttttcttct, and mouse GAPDH cDNA sense: aactttggcattgtggaagg, antisense: acacattgggggtaggaaca. All measurements were performed in triplicate. The average cycle threshold for untreated cells was around 38 and for no RNA control condition above 40 cycles.
ELISAs
Supernatants from murine peritoneal macrophage and BMDMs cultures were harvested at 24 h after IFN-γ and LPS stimulation and stored at −70°C. Mouse IL-12 p70, IL-12 p40, IL-10 and TNF-α were detected using BD OptEIA ELISA kits according to the manufacturer’s instructions. Concentrations were calculated by regression analysis of a standard curve.
Primary transcript measurement
To determine the primary transcript rates of IL-12 p35 and p40 gene as well as CCL5 and IL-27 p28 gene, cDNAs were synthesized with random primers with 1 µg DNase-treated RNA extracted from BMDMs treated with IFN-γ and LPS for different times. The primers used for primary transcript were: mouse p35 sense (intron3): tcacctcagcttctccttatgt, antisense (exon4): gttccagtggtaaacaggtcttc; Mouse p40 sense (intron2): tgtgtcaagacactgagtgaaa, antisense (exon3): ccaggtgatgtcatcttcttca; Mouse CCL5 sense (intron1): taaagagcccagcatagctggcaa, antisense (exon2): acgactgcaagattggagcacttg; Mouse IL-27 p28 sense (exon3): atctcgattgccaggagtgaacct, antisense (intron3): aaatcccagctccctctcctttgt. qRT-PCR was performed by a modified protocol as described previously (34).
Transfection assay
Transient transfections were performed as described previously (34). Briefly, for each condition a 1×107 RAW cell or THP-1 cell suspension were mixed with 16 µg total DNA (including reporter, effector, internal control, and carrier DNA), and electroporated at 975 microfarad and 300 V in RPMI 1640 medium without serum. The transfected cells were resuspended in RPMI 1640 complete medium containing 10 mM chloroquine and incubated for 48 h prior to harvesting. To measure luciferase activity, cells were pelleted and resuspended in lysis buffer. Luciferase activity was measured in cell lysates. We routinely observe about 50% cell death after electroporation and about 30% transfection efficiency by electroporation. 2∼3 × 106 transfected cells were used to extract RNA for measurement of mRNA expression.
Immunofluorescence and confocal microscopy
For immunofluorescence analyses, transfected RAW cells were grown on glass coverslips for 48 hrs and then treated with LPS plus IFN-γ. One hour after treatment, cells were fixed for 20 min at room temperature with 1% paraformaldehyde and permeabilized in 0.1% Triton X-100 for 1min. After blocking in PBS containing 5% FBS, cells were co-labeled with anti-HA and anti-NF-κB primary antibodies at 4 °C overnight. Cells were washed three times in block buffer and subsequently incubated with the indicated fluorophore-conjugated secondary antibodies for 1 hour. Next, cells were labeled with 4′,6′-diamidino-2-phenylindole (DAPI) for nuclear staining, and coverslips were mounted on glass slides using ProLong Anti-Fade mounting medium (Invitrogen). Cells were examined with a Confocal Laser-Scanning Microscope (Zeiss LSM 510 META, Thornwood, NY), which was mounted on an inverted microscope equipped with a 63 × Plan-pochromat oil-immersion objective lens (1.5 NA). Images were acquired by sequential excitation at 488 nm and 568 nm laser lines. Instrument settings were kept constant for each replicate.
Cell extracts and Western blotting
For NF-κB p65, c-Rel and p50 nuclear translocations, cells were grown and transfected in 10-cm dishes. Forty-eight hours after transfection, cells were stimulated with LPS plus IFN-γ for 1 h. Cells were harvested, and both cytoplasmic and nuclear extracts were prepared as described before (34). 100 µg proteins were separated by 10% SDS-PAGE, transferred electrophoretically to PVDF membranes and blocked in 5% nonfat milk in Tris buffer (pH 8.0). Primary antibody was added at a concentration of 1 µg/ml in blocking buffer containing 5% milk and left overnight at 4°C. After extensive washing, secondary antibody conjugated to horseradish peroxidase (HRP) was added at a 1:5000 dilution in 5% nonfat milk in Tris buffer. After extensive washing, blots were subjected to ECL detection (PerkinElmer Life Sciences, Boston, MA).
Statistical analysis
Student’s test was performed wherever applicable. Standard deviation of the mean is shown unless otherwise indicated. *: p<0.05, **: p<0.01, ***: p<0.001 between two groups.
Results
Mice deficient of TTP are susceptible to endotoxic shock and produce more IL-12
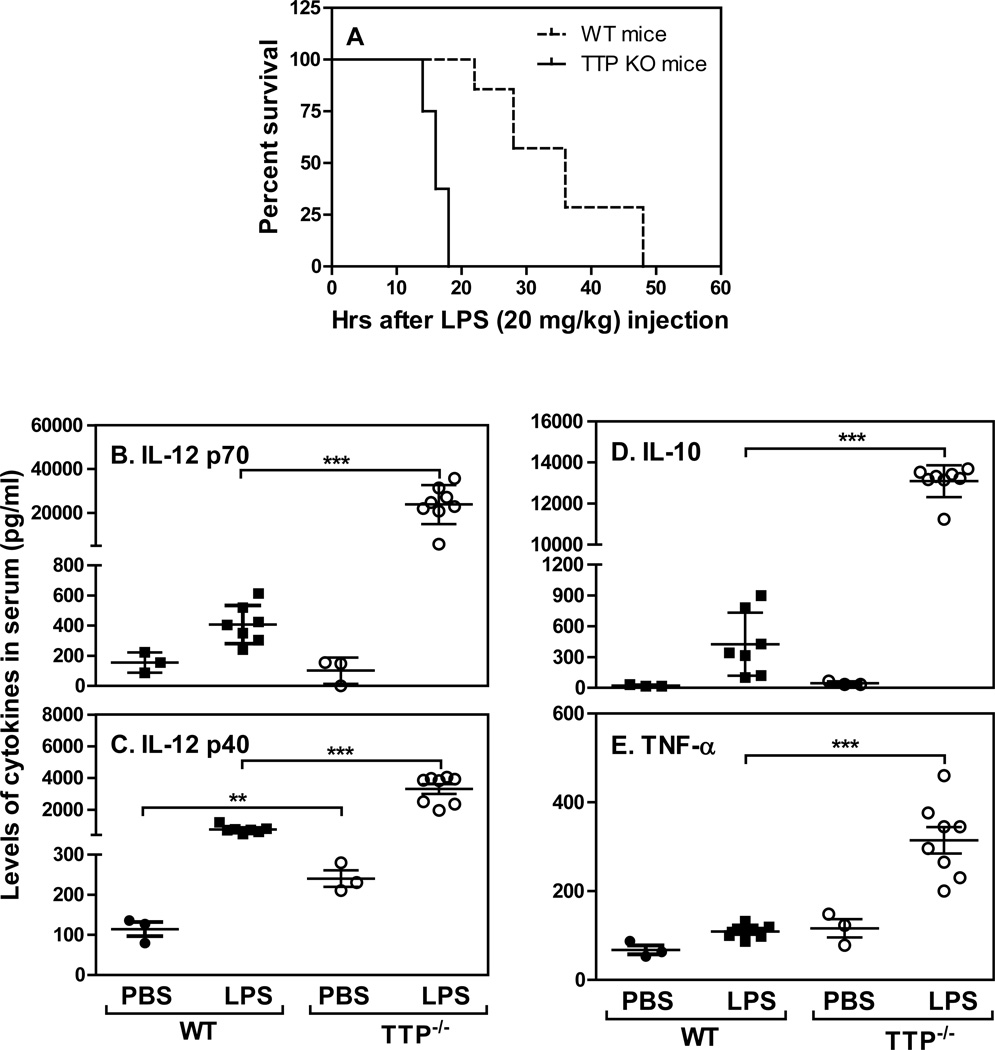
It has been shown that TTP promotes the degradation of many proinflammatory cytokines such as TNF-α, IL-6 and IL-1 (26) which play important roles in the pathogenesis of endotoxic shock. Mice specifically deficient of TTP in macrophages were highly susceptible to low dose of LPS challenge and displayed an increased in TNF-α production from macrophages (35). However, in contrast to conventional TTP KO mice (35), macrophage-specific TTP conditional knockout mice did not exhibit an early-onset, severe inflammatory phenotype. To determine if the conventional TTP KO mice are also susceptible to LPS challenge, we injected intraperitoneally LPS into WT and TTP KO mice and observed survival. All TTP−/− mice died within 20 hours whereas WT mice survived close to 50 hours after LPS injection (Fig. 1A), indicating that similar to macrophages-specific TTP KO mice the conventional TTP KO mice are also susceptible to LPS challenge. We next measured the levels of IL-12 p70 and p40 as well as TNF-α and IL-10 in sera of both WT and TTP KO mice 18 hours after LPS injection. The levels of IL-12 p70 and p40 were significantly increased in TTP KO mice compared to WT mice during endotoxic shock (Figs. 1B & C), indicating that the loss of TTP results in an increase in IL-12. As expected, the levels of IL-10 (Fig. 1D) and TNF-α (Fig. 1E) were enhanced in TTP knockout mice after LPS injection (23, 36).
Figure 1. Mice deficient of TTP are susceptible to endotoxic shock and produce more cytokines.
Ten age and gender-matched conventional TTP deficient and WT littermates were injected with 20 mg/kg LPS in equal amount of PBS. Survival was closely monitored in every 2 h up to 48 h (A). The sera was collected at 18 h after LPS injection and used to measure the levels of IL-12 p70 (B), IL-12 p40 (C), IL-10 (D), and TNF-α (E) by ELISA. Each dot represents one mouse in each group.
TTP suppresses IL-12 production and Th1 cell differentiation
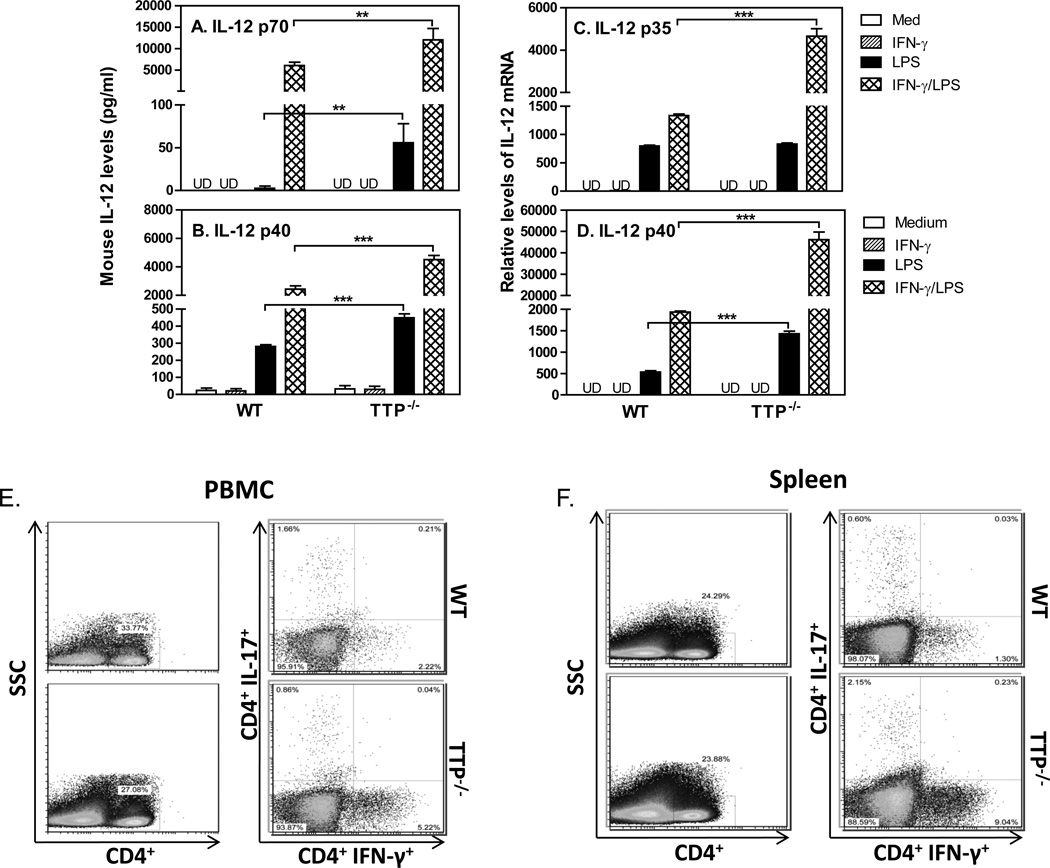
To determine if TTP inhibited IL-12 production we isolated peritoneal macrophages from WT and TTP KO mice and treated the cells with IFN-γ and LPS, followed by measuring IL-12 protein and mRNA expression. In agreement with our previous studies (4, 5), IFN-γ and LPS synergistically enhanced IL-12 p70 (Fig. 2A) and p40 (Fig. 2B) production by WT macrophages, whereas IL-12 production was further enhanced in macrophages deficient of TTP. Since biological IL-12 consists of two different subunits, p35 and p40, next we measured the expression of p35 (Fig. 2C) and p40 (Fig. 2D) mRNA in peritoneal macrophages. Consistent with the enhanced IL-12 protein production in TTP KO cells, both p35 and p40 mRNA expression were induced by IFN-γ and LPS treatment in WT cells and further enhanced in TTP KO cells, suggesting that TTP suppresses IL-12 p35 and p40 expression at the level of transcription. The suppression of IL-12 by TTP compelled us to investigate if Th1 cell development was affected since IL-12 is an essential cytokine for Th1 cell differentiation and proliferation. We purified PBMCs and collected single splenic cells from peripheral blood and spleen, respectively, and then detected the IL-17-producing and IFN-γ-producing CD4+ T cells by flow cytometry. The percentages of IFN-γ+Th1 cells were increased about two-fold in PBMCs of TTP KO mice compared to WT mice (Fig. 2E), which even further increased in splenic cells of TTP KO mice (1.3% Th1 cells in WT spleen and over 9% Th1 cells in spleens of TTP KO mice). However, the patterns of Th17 cells were different between PBMCs and spleens, with reduced Th17 cells in PBMCs and increased Th17 cells in spleens of TTP KO mice. Nevertheless, these data suggest that the increased Th1 cells in TTP KO mice may be due to an increase in IL-12 production.
Figure 2. TTP inhibits IL-12 production and Th1 cells.
1 × 106 mouse peritoneal macrophages isolated from WT and TTP−/− mice were incubated in 24-well plate with 1 ml RPMI 1640 complete medium. The cells were stimulated with IFN-γ (10 ng/ml), LPS (1 µg/ml) or IFN-γ plus LPS for 24 hours, followed by collection of supernatant to measure the levels of IL-12 p70 (A) and IL-12 p40 (B) with ELISA, or by RNA extraction to quantify IL-12 p35 (C) and IL-12 p40 (D) mRNA expression with qRT-PCR. qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each sample and further normalized to the results from the untreated group (Medium). Data shown are mean plus SD from three independent experiments. CD4+ T cells were purified by negative selection from PBMCs and spleens of untreated WT and TTP−/− mice. The CD4+ T cells were stimulated with PMA and Ionomycin for 3–4 hours before staining. IFN-γ-producing CD4+ T cell populations were analyzed by FACS in PBMCs (E) and spleens (F) from WT and TTP−/− mice by gating on CD4+ cells. Data represent one of three experiments with similar results.
TTP inhibits IL-12 p35 and p40 mRNA expression at the level of transcription
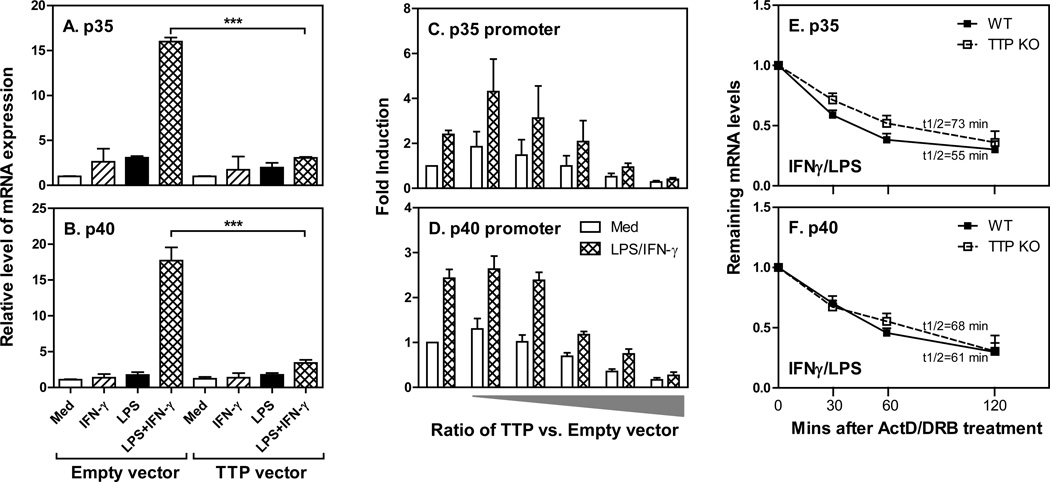
To further confirm the inhibitory effects of TTP on IL-12 expression we overexpressed TTP in THP-1 cells and measured p35 and p40 mRNA expression after treatment with IFN-γ and LPS. LPS and IFN-γ synergistically induced both p35 (Fig. 3A) and p40 (Fig. 3B) mRNA expression in human monocytes, and overexpression of TTP significantly suppressed their expression, indicating that TTP indeed inhibits IL-12 expression in both human and mouse monocytes/macrophages. To explore the molecular mechanisms of TTP-mediated suppression of IL-12 expression we co-transfected different amounts of TTP with either human IL-12 p35 promoter- or p40 promoter-driven luciferase vector into THP-1 cells and measured luciferase activity after treatment. Both p35 (Fig. 3C) and p40 (Fig. 4D) promoters were activated by IFN-γ and LPS. Furthermore, overexpression of TTP dose-dependently suppressed p35 and p40 promoter activity (Figs. 3C&D), suggesting that TTP inhibits IL-12 expression at the level of transcription. Since TTP is one of the best characterized RNA-binding proteins important for promoting mRNA degradation, to rule out whether the posttranscriptional mechanism was involved, we measured the half-life of p35 (Fig. 3E) and p40 (Fig. 3F) mRNA in TTP KO macrophages after blocking de novo RNA synthesis with Actinomycin D (ActD) and 5,6-dichlorobenzimidazole riboside (DRB), and compared it to WT cells. The half-life of p35 and p40 mRNA was similar between WT and TTP KO cells, further confirming that the suppression of IL-12 by TTP is regulated at the transcriptional level.
Figure 3. Transcriptional regulation of IL-12 p35 and p40 mRNA by TTP.
Five micrograms of CMV empty or hTTP vectors were transfected into THP-1 cells by electroporation. The transfected cells were treated with IFN-γ (10 ng/mL), LPS (1 µg/mL), or IFN-γ plus LPS for 4 hours, followed by extraction of total RNA for measurement of human IL-12 p35 (A) or p40 (B) mRNA expression. For promoter analysis, 10 × 106 THP-1 cells were transiently co-transfected by electroporation with 5 µg of IL-12 p35 or p40 full length promoter-luciferase construct in together with increased ratio of TTP expressing vector (from 1 µg to 8 µg) vs. empty vector (from 10 µg to 2 µg) as illustrated. Forty hours later, the transfected cells were treated with IFN-γ (10 ng/mL), LPS (1 µg/mL) or IFN-γ plus LPS for 7 h, followed by lysis of the cells and measurement of p35 (C) and p40 (D) luciferase activity by luminometer. The average raw luciferase expression values for the empty vector without any treatment are 2890 for p35 promoter and 1226 for p40 promoter, respectively. Results shown are mean plus SD of three independent experiments. For measuring mRNA half-life, 3 × 106 BMDMs differentiated from bone marrow cells of WT and TTP−/− mice were treated with IFN-γ and LPS for 2 hrs, and then added ActD (10 µg/ml) and DRB (50 µM), followed by collecting total RNA at 0, 30, 60, and 120 min to measure the remaining levels of p35 (E) and p40 (F) mRNAs by qRT-PCR. Data were normalized relative to GAPDH mRNA expression levels and further normalized to the results obtained at 0 min after ActD/DRB treatment in each condition, which was set as 1. Data shown are mean plus SD from three independent experiments.
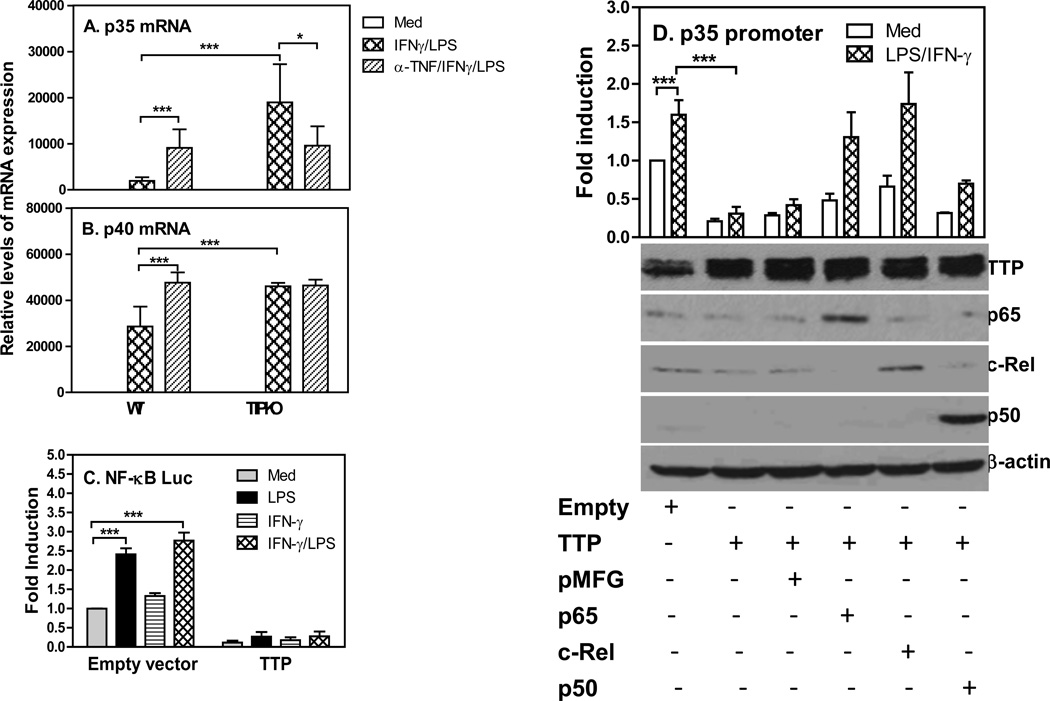
Figure 4. TTP-mediated inhibition of IL-12 is rescued by NF-κB overexpression.
2 × 106 BMDMs differentiated from bone marrow cells of WT and TTP−/− mice were pretreated with 20 µg TNF-α neutralizing antibody for 1 h, followed by treatment with IFN-γ (10 ng/ml) and LPS (1 µg/ml) for 4 h. Total RNA was extracted for measuring p35 (A) and p40 (B) mRNA expression by qRT-PCR. Data were normalized relative to GAPDH mRNA expression levels and further normalized to the results obtained under untreated condition (med) in WT BMDMs. Data shown are mean plus SD from three independent experiments. (C) 10 × 106 RAW cells were transiently transfected by electroporation with NF-κB–luciferase vector and TTP expression vector. Forty hours later, the transfected cells were treated with IFN-γ (10 ng/mL), LPS (1 µg/mL) or IFN-γ plus LPS for 7 h, followed by measurement of luciferase activity by luminometer. The raw luciferase values were normalized to the results obtained from NF-κB-Luc and CMV empty co-transfection group. Results shown are mean plus SD of four independent experiments. (D) 10 × 106 RAW cells were transiently transfected with IL-12 p35 full length promoter-luciferase construct and TTP vector along with NF-κB p65, c-Rel or p50 construct, as indicated. Forty hours later, the transfected cells were treated with IFN-γ (10 ng/mL) plus LPS (1 µg/mL) for 7 h, followed by lysis of the cells and measurement of luciferase activity. The raw luciferase values were normalized to the results obtained from p35 promoter and CMV empty co-transfection group (upper panel in D). Results shown are mean plus SD of three independent experiments. The transfected RAW cells from each condition were lysised for Western blot to determine the levels of TTP, NF-κB p65, c-Rel and p50 protein. β-Actin was used as loading control (lower panel in D).
NF-κB mediates the inhibitory effects of TTP on IL-12
Our previous study demonstrates that the expression of IL-12 is mediated through NF-κB signaling pathway in activated macrophages (5). Since TNF-α can activate NF-κB signaling pathway and its levels was increased in TTP knockout mice (Fig. 1E), to determine the role of TNF-α in TTP-related IL-12 reduction, we first neutralized TNF-α and then measured the levels of IL-12 expression in macrophages stimulated with IFN-γ and LPS. As shown in Figures 4A&B, neutralizing TNF-α enhanced the expression of p35 and p40 mRNA expression in WT macrophages, which is consistent with previous reports that TNF-α inhibits IL-12 expression in macrophages and dendritic cells (37–39). Interestingly, neutralizing TNF-α partially inhibited the expression of p35 mRNA but not the p40 mRNA in TTP−/− macrophages, indicating that in difference of WT macrophages TNF-α contributes partially to the overproduction of IL-12 in TTP knockout cells. To confirm the effects of TTP on NF-κB activation, we co-transfected TTP expression vector and NF-κB-consensus tandem repeat linked luciferase vector into RAW cells and measured luciferase activity after treatment with LPS and INF-γ. NF-κB-driven luciferase activity was significantly enhanced by LPS or IFN-γ and LPS in CMV empty vector transfected cells, whereas overexpression of TTP almost completely abolished NF-κB luciferase activities (Fig. 4C). Next, we wanted to know if the suppression of IL-12 production by TTP was mediated through altering NF-κB signaling. Since IL-12 p40 subunit is always produced in large quantity compared to the p35 subunit, the levels of p35 expressed in macrophages actually determine the amount of biologically active form of IL-12. Therefore, we focused on the effects of NF-κB on TTP-mediated inhibition of p35 transcription by co-transfecting human p35 promoter and TTP expression vector with different members of NF-κB family into macrophages, followed by measurement of luciferase activity after treatment with IFN-γ and LPS. Overexpression of TTP inhibited p35 promoter activation (Fig. 4D). Interestingly, this inhibition was completely reversed when NF-κB p65 and c-Rel were introduced into the cells but not the NF-κB p50 (Fig. 4D), suggesting that the inhibitory effects of TTP on IL-12 expression is mediated through NF-κB p65 and c-Rel. To confirm successful overexpression of the TTP and NF-κB family members after transfection, we measured the protein levels of TTP and NF-κB p65, p50 and c-Rel by western blot from the above transfected cells. As shown in the lower panel in Figure 4D, the protein expression of TTP, NF-κB p65, c-Rel and p50 were markedly increased compared to their respective controls. Collectively, these results suggest that TTP acts through NF-κB p65 and c-Rel to exert its inhibitory effects on IL-12 production.
TTP blocks NF-κB nuclear translocation
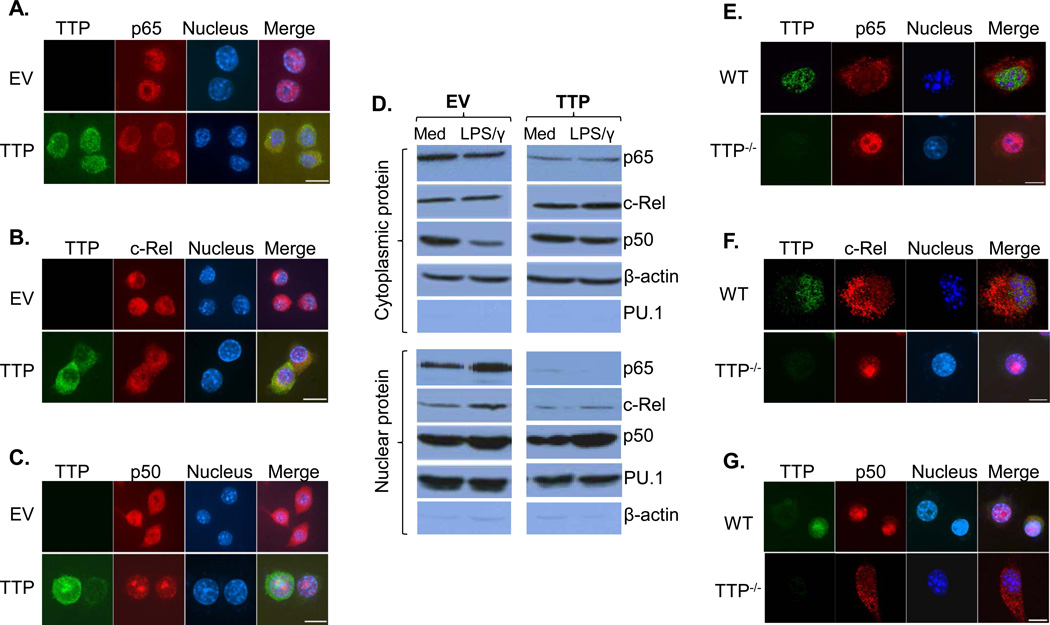
TTP is known to be able to shuttle between cytoplasm and nucleus (28). We reasoned that TTP may inhibit IL-12 production via affecting NF-κB nuclear translocation as reported previously (27). We tracked the subcellular localizations of distinct NF-κB members in cells with or without TTP overexpression. As shown in Fig. 5A, confocal microscopy revealed a major nucleus distribution of NF-κB p65 in empty vector transfected cells treated with LPS plus IFN-γ, whereas p65 nuclear translocation was almost completely blocked after overexpression of TTP. In line with this finding, western blot results showed that IFN-γ and LPS treatment induced nuclear p65 levels in cells transfected with empty vector but not in cells overexpressing TTP (Fig. 5D). In addition, nuclear translocation of the NF-κB c-Rel displayed a similar pattern as the p65 though at a less extent, in that overexpression of TTP partially blocked c-Rel nuclear expression (Fig. 5B). Western blot also indicated that the induction of c-Rel in nucleus was abolished in TTP-overexpressing cells upon treatment (Fig. 5D, lower panel). Interestingly, p50 showed a different pattern compared to p65 and c-Rel, in that the p50 was distributed in both cytoplasm and nucleus in activated cells transfected with empty vector but appeared to be translocated into nucleus after overexpressing TTP (Fig. 5C). To further confirm the effects of TTP on NF-κB nuclear translocation, we examined the nuclear localization of NF-κB members in WT and TTP−/− macrophages treated with IFN-γ and LPS. NF-κB p65 (Fig. 5E) and c-Rel (Fig. 5F) were increased in nucleus in TTP−/− macrophages compared to WT macrophages stimulated with IFN-γ and LPS, whereas nuclear p50 (Fig. 5G) was decreased in TTP−/− cells compared to WT cells. Taken together, these results indicate that the inhibitory effects of TTP on IL-12 production are mediated through blocking NF-κB nuclear translocation, especially the p65 and c-Rel, in activated macrophages.
Figure 5. TTP blocks NF-κB nuclear translocation.
10 × 106 RAW cells were transiently transfected with HA-tagged TTP or control vector. 40 hrs later, the transfected cells were treated with IFN-γ (10 ng/mL) plus LPS (1 µg/mL) for 1 h, followed by immunostaining with anti-HA antibody (green) and anti-NF-κB p65 antibody (red) (A), or anti-NF-κB c-Rel antibody (red) (B), or anti-NF-κB p50 (red) antibody (C). DAPI labeling was used to identify the cell nucleus (blue). Scale bars = 10 µm. (D) Western blot analysis of the endogenous p65, c-Rel and p50 protein in cytoplasmic and nuclear extracts prepared from TTP overexpressing RAW cells stimulated with IFN-γ (10 ng/mL) plus LPS (1 µg/mL) for 1 h. β-Actin was used for the cytoplasmic protein loading control and PU.1 was used for the nuclear protein loading control. 1 × 106 BMDMs differentiated from bone marrow cells of WT and TTP−/− mice were treated with IFN-γ (10 ng/mL) plus LPS (1 µg/mL) for 1 h, followed by immunostaining with anti-TTP antibody (green), and anti-NF-κB p65 antibody (red) (E), anti-NF-κB c-Rel antibody (red) (F), or anti-NF-κB p50 antibody (red) (G). DAPI labeling was used to identify the cell nucleus (blue). Scale bars=10 µm. Data shown represent one of three independent experiments with similar results.
TTP suppresses NF-κB dependent gene expression
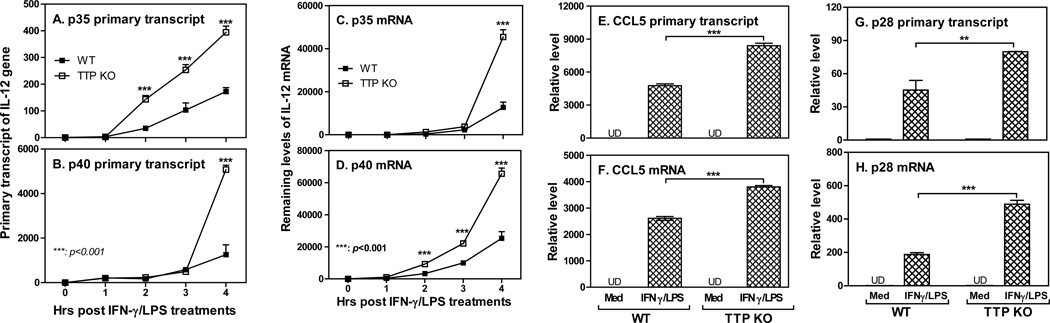
Ours and others studies demonstrate that TTP can inhibit NF-κB activity, indicating a broader effect of TTP on NF-κB dependent gene expression. We first wanted to confirm whether IL-12 p35 and p40 gene transcription was affected by TTP using TTP−/− macrophages. Both p35 (Fig. 6A) and p40 (Fig. 6B) primary transcript rates were significantly increased in cells deficient of TTP compared to WT cells. The expression of p35 (Fig. 6C) and p40 (Fig. 6D) mRNA were also markedly enhanced in macrophages lacking TTP. These results confirmed the overexpression data (Figs. 3A–D) that the inhibitory effect of TTP on IL-12 was mediated at the level of transcription. To determine if other NF-κB dependent genes were also affected by TTP deficiency, we respectively measured the primary transcript rates (Figs. 6E&G) and mRNA expression (Figs. 6F&H) for CCL5 and IL-27 p28 in macrophages stimulated with IFN-γ and LPS. The primary transcript and mRNA expression of CCL5 and p28 were increased in cells deficient of TTP, suggesting that TTP indeed has a broader effect on NF-κB dependent gene expression.
Figure 6. TTP suppresses NF-κB dependent gene expression.
2 × 106 mouse peritoneal macrophages isolated from WT and TTP−/− mice were incubated in each of 12-well plates with 2 ml RPMI 1640 complete medium. The cells were stimulated with IFN-γ (10 ng/ml) and LPS (1 µg/ml) for different times as indicated, followed by collection of total RNA to measure p35 (A) and p40 (B) primary transcript as well as their mRNA expression (C&D) by qRT-PCR. qPCR data were normalized relative to GAPDH mRNA expression levels in each sample and further normalized to the results from the untreated group (0). CCL5 primary transcript (E) and mRNA expression (F) as well as IL-27 p28 primary transcript (G) and mRNA expression (H) were measured from the above cDNA generated from cells treated with IFN-γ and LPS for 4 h. Data shown are mean plus SD from three independent experiments.
Discussion
The IL-12 family cytokines, including IL-12, IL-23, IL-27 and IL-35, are important mediators of inflammatory diseases (2, 40). As first member of the family, IL-12 is essential for the differentiation and proliferation of Th1 cells important for cellular immune responses against intracellular infection (2). Our previous studies demonstrate that IL-12 expression is induced during endotoxic shock (41) and the induction of IL-12 from macrophages depends on two signals, LPS/TLR4 and IFN-γ/IRFs (5, 8). In this study, we found that both IL-12 p70 and p40 proteins were enhanced in TTP KO mice during endotoxic shock (Figs. 1B&C) and in TTP KO macrophages stimulated with LPS or LPS plus IFN-γ (Figs. 2A–D). As described previously (42), TNF-α levels were also enhanced in TTP KO mice (Fig. 1E). We also measured an increase of IL-10 production in TTP KO mice during endotoxic shock (Fig. 1D). It seems that this increased production of anti-inflammatory IL-10 could not sufficiently override the detrimental effects of the proinflammatory cytokines, such as TNF-α and IL-12, resulting in death of the TTP knockout mice. The reduced survival of TTP KO mice during endotoxic shock is also consistent to a recent report using conditional TTP KO mice with specific deletion of TTP expression in macrophages, though at a less extent (35). The suppression of IL-12 expression by TTP has also been observed previously in mouse macrophage cell line J774 cells after knocking down TTP expression with shRNA (29). Nevertheless, the increased IL-12 production in TTP knockout mice could lead to an increase in differentiation and proliferation of Th1 cells (Figs. 2E & F), which may involve in the development of autoinflammation seen in TTP knockout mice (42).
Regulation of cytokine expression can be roughly grouped into transcriptional and posttranscriptional regulations. Posttranscriptional regulation of cytokine production is mediated by multiple steps, including nuclear export, cytoplasmic localization, translation initiation and mRNA decay (43). These regulatory pathways are coordinated to control the expression of proinflammatory and anti-inflammatory cytokines to turn on or off inflammatory responses (43). Roughly, about 5–8% of human gene transcripts are regulated by control of mRNA stability (44). One of mRNA decay mechanisms is ARE-binding proteins binding to the 3’UTR of target genes causing mRNA degradation. We previously found that TTP inhibits IL-23 expression by promoting IL-23 p19 mRNA degradation (33). Since IL-12 and IL-23 share the p40 subunit, we wondered whether TTP inhibited IL-12 expression through a similar mechanism. Surprisingly, the half-life of p35 and p40 mRNA was similar between WT and TTP KO macrophages (Figs. 3E&F), suggesting that TTP-mediated inhibition of IL-12 expression is not through affecting mRNA stability. Moreover, we found that TTP inhibits both p35 and p40 gene transcription as shown by promoter assay (Figs. 3C&D) and primary transcript measurement (Figs. 6A&B), further indicating that the inhibition of IL-12 expression by TTP is regulated at the level of transcription, which is different from IL-23. It is worth noting that there was a discrepancy of p40 protein (two fold) (Fig. 2B) and mRNA expression (twenty fold) (Fig. 2D) between WT and TTP−/− macrophages treated with IFN-γ and LPS, suggesting that translational regulation of p40 may also play a role in TTP-mediated inhibition of IL-12 production under this condition.
Transcription factor NF-κB has been shown to be able to control IL-12 expression and numerous genes involved in inflammatory reactions and immune regulatory functions (5, 45). Since TNF-α has been known to activate NF-κB signaling, we wanted to determine if the increased IL-12 is due to a second effect of enhanced TNF-α in TTP knockout mice. Neutralizing TNF-α in activated macrophages confirmed previous studies that TNF-α inhibits IL-12 expression (Figs. 4A&B). Interestingly, neutralizing TNF-α only partially inhibited the expression of p35 mRNA but not the p40 mRNA in TTP−/− macrophages treated with IFN-γ and LPS, indicating that in difference of WT macrophages TNF-α may contribute partially to the overproduction of IL-12 in TTP knockout cells. It also seems that p40 and p35 are regulated differently by TNF-α. More study is needed to further investigate the differential regulation of p35 and p40 mRNA by TNF-α in TTP−/− macrophages. To confirm whether NF-κB is involved in suppression of IL-12 expression by TTP, we overexpressed NF-κB molecules along with TTP and found that overexpression of c-Rel and p65 but not p50 can completely recover p35 gene transcription (Fig. 4D, upper panel). Furthermore, overexpression of TTP directly suppressed NF-κB consensus sequence-driven luciferase activity (Fig. 4C). Our results indicate that TTP-mediated suppression of IL-12 expression acts through affecting NF-κB activity, especially the c-Rel and p65. It is not suppressing that p50 overexpression could not rescue the inhibition of IL-12 by TTP since p50-p50 homodimer has been shown to be able to suppress target gene transcription as a transcription suppressor. We focused on the p35 promoter is because that the p40 subunit is a shared subunit with IL-23 and always produced in large quantity compared to the p35 subunit. Our data further indicate that suppression of NF-κB signaling by TTP is achieved by blocking p65 and c-Rel nuclear translocation as evidenced by confocal (Figs. 5A–C & 5E–G) and western blot (Fig. 5D) analyses, which is also in consistence with recent studies indicating that TTP is involved in the p65 trafficking between nuclear and cytoplasm in MEF and HEK293 cells (27, 28). To determine the specificity of TTP in inhibition of IL-12 expression, we measured two NF-κB targeted genes, chemokine CCL5 and IL-27 p28 with both genes being previously demonstrated to be regulated by NF-κB pathway (46, 47). As expected, both CCL5 (Fig. 6E) and p28 (Fig. 6G) primary gene transcript and mRNA (Figs. 6F&H) were induced by IFN-γ and LPS. More interestingly, their primary gene transcript and mRNA expression were further enhanced in TTP−/− macrophages, indicating that the inhibitory effects of TTP on NF-κB activity could have broad effects on NF-κB dependent gene expression, which is also in consistence with the concept postulated by Martin’s group (27).
In summary, we demonstrate, for the first time, that TTP inhibits IL-12 production through suppression of p35 and p40 transcription via selectively blocking nuclear translocations of NF-κB p65 and c-Rel. Suppression of IL-12 by TTP leads to increased Th1 cells which may contribute to the chronic inflammation seen in TTP knockout mice. Our findings reveal an important role for TTP in regulation of immune responses as a resolution molecule, which may help us design better strategies to control chronic inflammation.
Acknowledgment
We would like to thank Deborah J. Stumpo for her technique support in performing this project and Daniel Hoft for discussion in preparation of the manuscript.
Research reported in this publication was supported by the National Institutes of Health under Award Number CA163808 to J.L. and HL098794 to M.F. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used in this article
- ActD
Actinomycin D
- DRB
5,6-dichlorobenzimidazole riboside
- BMDMs
bone marrow-derived macrophages
- DC
dendritic cell
- qRT-PCR
quantitative real-time PCR
- t1/2
half life
- UD
undetectable
- EV
empty vector
- TNF-α
tumor necrosis factor-α
- IFN-γ
interferon-γ
- α-TNF
TNF-α neutralizing antibody
- WT
wild type
- KO
knockout
- 3’UTR
3’ untranslated region
- PBMC
peripheral blood mononuclear cell.
Footnotes
Disclosures
The authors have no financial conflict of interest.
References
- 1.Kobayashi M, Fitz L, Ryan M, Hewick RM, Clark SC, Chan S, Loudon R, Sherman F, Perussia B, Trinchieri G. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. The Journal of experimental medicine. 1989;170:827–845. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 3.Trinchieri G. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu Rev Immunol. 1995;13:251–276. doi: 10.1146/annurev.iy.13.040195.001343. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Guan X, Tamura T, Ozato K, Ma X. Synergistic activation of interleukin-12 p35 gene transcription by interferon regulatory factor-1 and interferon consensus sequence-binding protein. The Journal of biological chemistry. 2004;279:55609–55617. doi: 10.1074/jbc.M406565200. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Cao S, Herman LM, Ma X. Differential regulation of interleukin (IL)-12 p35 and p40 gene expression and interferon (IFN)-gamma-primed IL-12 production by IFN regulatory factor 1. The Journal of experimental medicine. 2003;198:1265–1276. doi: 10.1084/jem.20030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz O, Edwards AD, Schito M, Aliberti J, Manickasingham S, Sher A, Reis e Sousa C. CD40 triggering of heterodimeric IL-12 p70 production by dendritic cells in vivo requires a microbial priming signal. Immunity. 2000;13:453–462. doi: 10.1016/s1074-7613(00)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Murphy TL, Cleveland MG, Kulesza P, Magram J, Murphy KM. Regulation of interleukin 12 p40 expression through an NF-kappa B half- site. Mol Cell Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma X, Chow JM, Gri G, Carra G, Gerosa F, Wolf SF, Dzialo R, Trinchieri G. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. The Journal of experimental medicine. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plevy SE, Gemberling JH, Hsu S, Dorner AJ, Smale ST. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol Cell Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeire K, Heremans H, Vandeputte M, Huang S, Billiau A, Matthys P. Accelerated collagen-induced arthritis in IFN-gamma receptor-deficient mice. J Immunol. 1997;158:5507–5513. [PubMed] [Google Scholar]
- 11.Grazia Cappiello M, Sutterwala FS, Trinchieri G, Mosser DM, Ma X. Suppression of Il-12 transcription in macrophages following Fc gamma receptor ligation. J Immunol. 2001;166:4498–4506. doi: 10.4049/jimmunol.166.7.4498. [DOI] [PubMed] [Google Scholar]
- 12.Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 13.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 14.Brasier AR. The NF-kappaB regulatory network. Cardiovasc Toxicol. 2006;6:111–130. doi: 10.1385/ct:6:2:111. [DOI] [PubMed] [Google Scholar]
- 15.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 16.Sanduja S, Blanco FF, Young LE, Kaza V, Dixon DA. The role of tristetraprolin in cancer and inflammation. Front Biosci. 2012;17:174–188. doi: 10.2741/3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 18.Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. The Journal of biological chemistry. 2001;276:47958–47965. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]
- 19.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 21.Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetraprolin binds to the 3'-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J Biol Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 22.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 23.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. The Journal of biological chemistry. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datta S, Biswas R, Novotny M, Pavicic PG, Jr., Herjan T, Mandal P, Hamilton TA. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J Immunol. 2008;180:2545–2552. doi: 10.4049/jimmunol.180.4.2545. [DOI] [PubMed] [Google Scholar]
- 25.Phillips K, Kedersha N, Shen L, Blackshear PJ, Anderson P. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc Natl Acad Sci U S A. 2004;101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 27.Schichl YM, Resch U, Hofer-Warbinek R, de Martin R. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. The Journal of biological chemistry. 2009;284:29571–29581. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang J, Lei T, Song Y, Yanes N, Qi Y, Fu M. RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. The Journal of biological chemistry. 2009;284:29383–29390. doi: 10.1074/jbc.M109.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jalonen U, Nieminen R, Vuolteenaho K, Kankaanranta H, Moilanen E. Down-regulation of tristetraprolin expression results in enhanced IL-12 and MIP-2 production and reduced MIP-3alpha synthesis in activated macrophages. Mediators Inflamm. 2006:40691. doi: 10.1155/MI/2006/40691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian X, Zhang J, Liu J. Tumor-secreted PGE2 inhibits CCL5 production in activated macrophages through cAMP/PKA signaling pathway. The Journal of biological chemistry. 2011;286:2111–2120. doi: 10.1074/jbc.M110.154971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. The Journal of biological chemistry. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 32.Murphy TL, Cleveland MG, Kulesza P, Magram J, Murphy KM. Regulation of interleukin 12 p40 expression through an NF-kappa B half-site. Mol Cell Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian X, Ning H, Zhang J, Hoft DF, Stumpo DJ, Blackshear PJ, Liu J. Posttranscriptional regulation of IL-23 expression by IFN-gamma through tristetraprolin. Journal of immunology. 2011;186:6454–6464. doi: 10.4049/jimmunol.1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Qian X, Ning H, Yang J, Xiong H, Liu J. Activation of IL-27 p28 gene transcription by interferon regulatory factor 8 in cooperation with interferon regulatory factor 1. J Biol Chem. 2010;285:21269–21281. doi: 10.1074/jbc.M110.100818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu LQ, Stumpo DJ, Blackshear PJ. Myeloid-specific tristetraprolin deficiency in mice results in extreme lipopolysaccharide sensitivity in an otherwise minimal phenotype. Journal of immunology. 2012;188:5150–5159. doi: 10.4049/jimmunol.1103700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaljo B, Kratochvill F, Gratz N, Sadzak I, Sauer I, Hammer M, Vogl C, Strobl B, Muller M, Blackshear PJ, Poli V, Lang R, Murray PJ, Kovarik P. Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. Journal of immunology. 2009;183:1197–1206. doi: 10.4049/jimmunol.0803883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodge-Dufour J, Marino MW, Horton MR, Jungbluth A, Burdick MD, Strieter RM, Noble PW, Hunter CA, Pure E. Inhibition of interferon gamma induced interleukin 12 production: a potential mechanism for the anti-inflammatory activities of tumor necrosis factor. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13806–13811. doi: 10.1073/pnas.95.23.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma X, Sun J, Papasavvas E, Riemann H, Robertson S, Marshall J, Bailer RT, Moore A, Donnelly RP, Trinchieri G, Montaner LJ. Inhibition of IL-12 production in human monocyte-derived macrophages by TNF. Journal of immunology. 2000;164:1722–1729. doi: 10.4049/jimmunol.164.4.1722. [DOI] [PubMed] [Google Scholar]
- 39.Zakharova M, Ziegler HK. Paradoxical anti-inflammatory actions of TNF-alpha: inhibition of IL-12 and IL-23 via TNF receptor 1 in macrophages and dendritic cells. Journal of immunology. 2005;175:5024–5033. doi: 10.4049/jimmunol.175.8.5024. [DOI] [PubMed] [Google Scholar]
- 40.Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu Rev Immunol. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Malkani G, Shi X, Meyer M, Cunningham-Runddles S, Ma X, Sun ZS. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. 2006;74:4750–4756. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki Y, Orellana MA, Schreiber RD, Remington JS. Interferon-gamma: the major mediator of resistance against Toxoplasma gondii. Science. 1988;240:516–518. doi: 10.1126/science.3128869. [DOI] [PubMed] [Google Scholar]
- 43.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 44.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Ma X. Interferon regulatory factor 8 regulates RANTES gene transcription in cooperation with interferon regulatory factor-1, NF-kappaB, and PU.1. The Journal of biological chemistry. 2006;281:19188–19195. doi: 10.1074/jbc.M602059200. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Guan X, Ma X. Regulation of IL-27 p28 gene expression in macrophages through MyD88- and interferon-gamma-mediated pathways. The Journal of experimental medicine. 2007;204:141–152. doi: 10.1084/jem.20061440. [DOI] [PMC free article] [PubMed] [Google Scholar]