Abstract
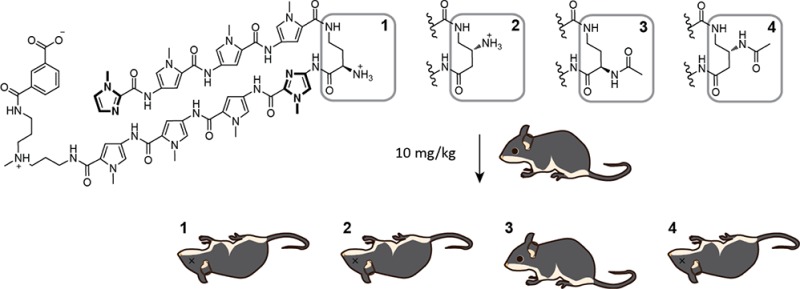
A hairpin pyrrole-imidazole polyamide (1) targeted to the androgen receptor consensus half-site was found to exert antitumor effects against prostate cancer xenografts. A previous animal study showed that 1, which has a chiral amine at the α-position of the γ-aminobutyric acid turn (γ-turn), did not exhibit toxicity at doses less than 10 mg/kg. In the same study, a polyamide with an acetamide at the β-position of the γ-turn resulted in animal morbidity at 2.3 mg/kg. To identify structural motifs that cause animal toxicity, we synthesized polyamides 1–4 with variations at the α- and β-positions in the γ-turn. Weight loss, histopathology, and serum chemistry were analyzed in mice post-treatment. While serum concentration was similar for all four polyamides after injection, dose-limiting liver toxicity was only observed for three polyamides. Polyamide 3, with an α-acetamide, caused no significant evidence of rodent toxicity and retains activity against LNCaP xenografts.
Introduction
Prostate cancer is a major contributor of cancer death in American males.1 The malignant transformation of prostate epithelial tissue is caused by an altered pattern of gene expression driven by the androgen receptor (AR). Clinically, localized prostate cancer is curable by surgery or radiation.2,3 Advanced prostate cancer is treated with systemic therapies that target testosterone signaling (enzalutamide, abiraterone), immunotherapy (sipuleucel T), and taxane-based chemotherapy (docetaxel, cabazitaxel).4 These new agents have shown survival benefits to patients with castration resistant, metastatic disease. However, all patients will eventually progress on these drugs. Resistance to the second-generation antiandrogen enzalutamide and the CYP17 inhibitor abiraterone may be due to the action of splice variants of AR that lack the ligand-binding domain (AR-V).5,6 Therefore, therapy resistant prostate cancer is an unmet clinical need, and novel systemic therapies are needed in patients after these treatments have failed.4
Direct interference of AR-driven transcription at the protein–DNA interface is a strategy that can circumvent resistance conferred by AR-V. Genomic DNA is the predominant target of many chemo- and radiotherapies. The interactions of these therapies with DNA result in the inhibition of DNA-dependent processes that are overactive in cancer cells, such as transcription.7−9 While AR-driven transcription can be inhibited by DNA-targeted agents,10,11 most conventional DNA-targeted therapeutics are genotoxic and can induce secondary malignancies.12 DNA-damaging agents may also contribute to tumor metastasis through effects on noncancerous cells in the tumor microenvironment.13 Small molecules that interact with DNA without genotoxicity could be a significant advance over conventional DNA-targeted therapeutics.
Pyrrole-imidazole (Py-Im) polyamides are minor groove binders that have been shown to affect gene expression in a number of inducible transcription systems.14−20 As noncovalent DNA-binding oligomers, these compounds form specific hydrogen bonds to the minor groove floor with programmable sequence recognition and high affinity.21−23 Py-Im polyamides are toxic to a variety of cancer cell lines, including prostate cancer, and exhibit no apparent genotoxicity.24 A typical hairpin oligomer consists of eight aromatic amino acid rings joined in the middle by a γ-aminobutyric acid (γ-turn).25 While sequence recognition is predominately directed by the antiparallel pairing of N-methylpyrrole and N-methylimidazole carboxamides, structural modifications to the γ-turn, such as substitution at the prochiral α- and β-positions, have been shown to influence the DNA affinity,26 cell uptake, and the biological activity of polyamides in both cell culture and animals.27,28
To date, we have reported the pharmacokinetic (PK) profiles of two eight-ring hairpin-polyamides targeted to the androgen response element half-site 5′-WGWWCW-3′ (W = A or T) in mice. The oligomers 1 and 4 of primary sequence ImPyPyPy-γ-ImPyPyPy∼NHMe∼IPA (Figure 1) differ in substitution at the γ-turn and were found to have distinct PK profiles. Both compounds were bioavailable in serum after intravenous injection for more than 24 h; however, 4 was found to have longer retention in both the serum and tissues. Both compounds were minimally excreted through the feces, but significant renal clearance was exclusive to 4.28 In addition to differences in the PK profiles, hairpins 1 and 4 also exhibited different degrees of toxicity to female C57BL/6J mice. While single subcutaneous administrations of 1 in female mice at 2.5 and 5 mg/kg did not adversely affect the animals, escalated dosing to 10 mg/kg resulted in weight loss greater than 15%. In comparison, 4 caused acute animal toxicity in addition to weight loss at 2.3 and 4.5 mg/kg.28 To dissect the differences in toxicities, there are two variables on the turn that must be sorted out (α- versus β-position and amino versus acetamide substitution).
Figure 1.
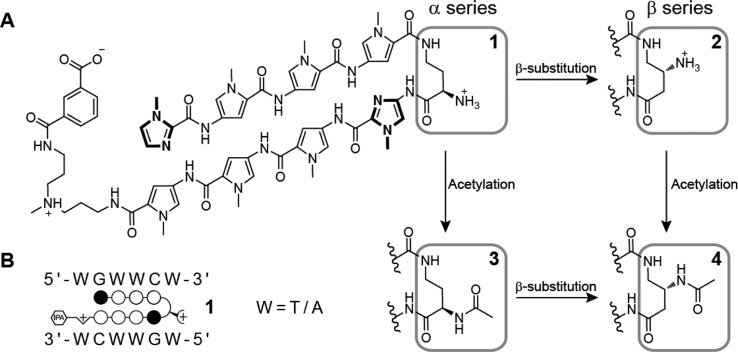
Chemical structures. (A) Structures of polyamides 1–4. The compounds only vary by the amino substitution on the γ-turn unit. (B) The preferred DNA binding sequence of the polyamide core. Polyamide 1 is shown bound to the sequence 5′-WGWWCW-3′. Closed circles represent imidazole units and open circles represent pyrrole units.
More recently, oligomer 1 was found to suppress the growth of LNCaP xenografts in immunocompromised mice after three subcutaneous injections at 1 mg/kg.24 Thus, a systematic toxicity study of 1 and related polyamides with modifications to the γ-turn may yield structures with reduced animal toxicity. In addition, an extensive toxicity study of polyamides in animals to identify target organs of pathology is a necessary step toward translation of this technology into the clinic.
In this paper, we report the animal toxicity of four structurally related polyamides with identical Py-Im sequence but with different substitutions at the γ-turn (Figure 1), a change that does not alter binding sequence preference. We assessed mouse weight, organ histopathology, and serum chemistry in wild-type male mice after single and multiple dosing regimens. Dose-limiting toxicity was observed at the highest dose for three of the four molecules. From this study, we have identified one polyamide that demonstrates no detectable toxicity by histopathology or serum analysis after single or repeated subcutaneous injections.
Results
Selection of Py-Im Polyamides
We synthesized four structurally related polyamides (Figure 1) that have an identical Py-Im sequence ImPyPyPy-γ-ImPyPyPy∼NHMe∼IPA. These polyamides demonstrate thermal stabilization of DNA duplexes containing their target sequence (Figure S1, Supporting Information). Polyamide 1, ImPyPyPy-2-(R)H2Nγ-ImPyPyPy∼NHMe∼IPA, suppressed LNCaP xenografts in mice.24 Polyamide 2, ImPyPyPy-3-(R)H2Nγ-ImPyPyPy∼NHMe∼IPA, differs from 1 in that the γ-turn is substituted at the β-position. Polyamide 3, ImPyPyPy-2-(R)AcHNγ-ImPyPyPy∼NHMe∼IPA, differs from 1 in that the primary amine is acetylated. Polyamide 4, ImPyPyPy-3-(R)AcHNγ-ImPyPyPy∼NHMe∼IPA, incorporates both changes from 2 and 3. Our previous report in female mice showed 1 and 4 both circulated in serum after intravenous injection.28 To determine if 1–4 demonstrated comparable serum levels after subcutaneous injection, male C57BL/6J mice were injected with 10 mg/kg each of 1–4 and blood collected by retroorbital bleed at various time points. All polyamides were bioavailable and detectable up to 24 h after subcutaneous injection (Figure S2, Supporting Information).
Escalating Single Dose Subcutaneous Injections
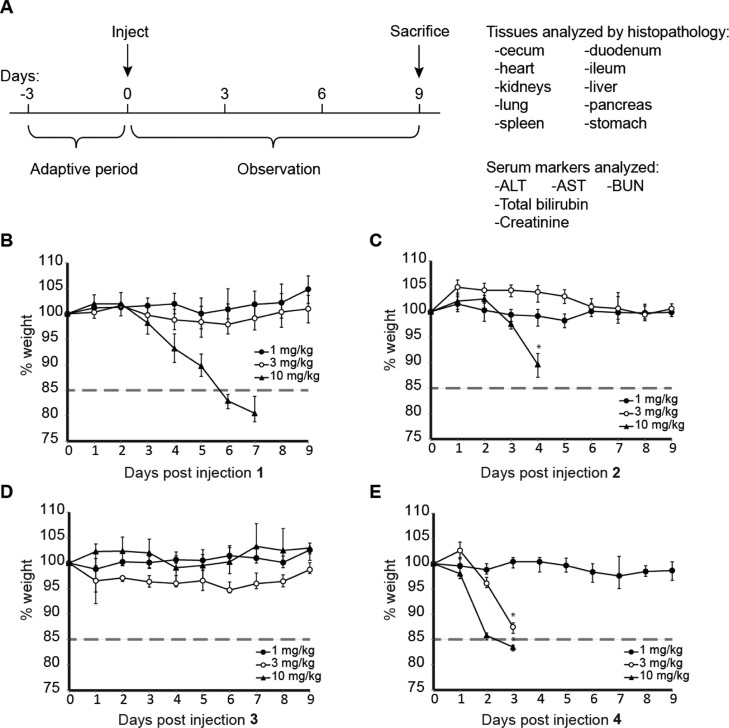
To determine the acute effects of subcutaneous dosing of 1–4 and dose-limiting organ toxicities, 8-week-old male C57BL/6J mice (n = 4 per dosing group) were treated with 1, 3, and 10 mg/kg 1–4 and observed for 9 days and then sacrificed (Figure 2). Representative mice (n = 2 per dosing group unless otherwise noted) were subjected to histopathology analysis by a veterinary pathologist. Blood from all mice was sampled and sent for analysis of serum markers of target organs. Mice treated with 1 and 2 demonstrated significant weight loss only at 10 mg/kg. Polyamide 4 was only tolerated at 1 mg/kg; all mice treated with 4 at 3 or 10 mg/kg exhibited hunched posture, loss of mobility, and acute morbidity. Mice treated with polyamide 2 at 10 mg/kg demonstrated similar morbidity. These mice were euthanized when significant duress was apparent. All other mice, including those treated with 1 at 10 mg/kg and 3 at all concentrations, demonstrated no change in behavior and appearance.
Figure 2.
Experiment setup of single dose weight curve experiments. (A) Male C57BL/6J mice were allowed to adapt to new cage settings for 3 days after arrival and then treated with compound. Animal weights were then monitored for 9 days. Humane end point was defined by visible signs of duress in the animals or weight loss in excess of 15% of original body weight. Weight curves of compounds (B) 1, (C) 2, (D) 3, and (E) 4.
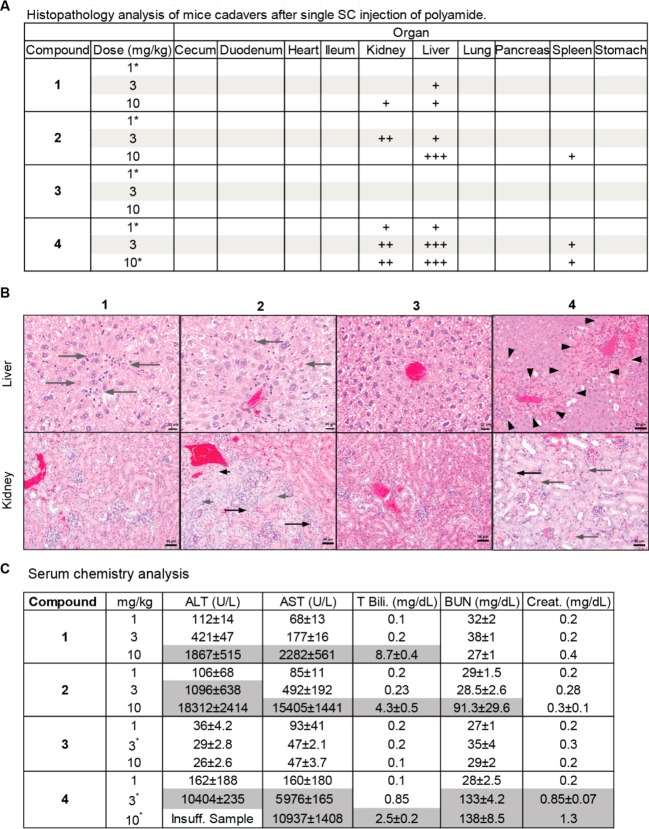
Histopathology revealed lesions consistent with toxicity in the liver, kidney, and spleen in animals receiving a single injection of polyamides 1, 2, and 4. The most severe lesions characterized by diffuse hepatocellular necrosis and apoptosis or multifocal bridging hepatocellular necrosis and apoptosis were identified in animals treated with polyamide 2 at 10 mg/kg and polyamide 4 at both 3 and 10 mg/kg, respectively. Mild hepatocellular necrosis and apoptosis were observed in animals treated with polyamide 1 at doses of 3 and 10 mg/kg, polyamide 2 at 3 mg/kg, and polyamide 4 at 1 mg/kg (Figure 3A). Moderate atypical tubular regeneration (karyomegaly, tubular attenuation, mitotic figures) and/or tubular epithelial necrosis and apoptosis were seen in the kidneys of animals treated with polyamide 2 at 3 mg/kg and polyamide 4 at 3 and 10 mg/kg (Figure 3B). Milder tubular regeneration and karyomegaly was observed in animals treated with polyamide 1 at 10 mg/kg and polyamide 4 at 1 mg/kg. Mild lymphoid apoptosis in the white pulp of the spleen was noted in animals treated with polyamide 2 at 10 mg/kg and polyamide 4 at 3 and 10 mg/kg. Polyamide 3 demonstrated no detectable toxicity at any dose level tested. No lesions consistent with toxicity were observed in the gastrointestinal tract, heart, lung, pancreas, or stomach in any animals.
Figure 3.
(A) Histopathology analysis of sacrificed animals showed primary organ damage in the kidney and liver for compounds 1, 2, and 4. Animals treated with 3 did not exhibit signs of organ damage. * represents n = 1. + = mild damage, ++ = moderate damage, +++ = severe damage. (B) Liver and kidney histopathology of two representative animals treated with compounds 1–4 at 3 mg/kg. Liver: long gray arrow = hepatocellular apoptosis/necrosis, arrowheads = outline area of bridging hepatocellular necrosis/apoptosis. Kidney: short gray arrow = tubular epithelial karyomegaly, long gray arrow = tubular epithelial apoptosis/necrosis, short black arrow = tubular epithelial mitoses, long black arrow = tubular epithelial attenuation. (C) Serum levels of liver damage and kidney damage markers. Significantly elevated markers are shaded in gray. * represents n = 2.
Because the liver and kidneys were identified as the target organs at risk due to toxicity, serum markers for these organ systems were measured (Figure 3C). Mice treated with polyamide 1 demonstrated significant elevation of AST, ALT, and total bilirubin at 10 mg/kg, indicative of acute damage to liver cells, and moderate elevation of ALT at 3 mg/kg. Elevation of creatinine and blood urea nitrogen (BUN) was not observed for any dose level of 1. Polyamide 2 treated mice had severe elevation of AST, ALT, and total bilirubin at 10 mg/kg and to a lesser extent at 3 mg/kg. These mice also had elevated BUN at 10 mg/kg. Mice treated with polyamide 4 demonstrated marked and severe elevations of AST, ALT, and total bilirubin at both 3 and 10 mg/kg. In addition, these mice had significantly elevated creatinine and BUN at 10 mg/kg and elevated BUN at 3 mg/kg. Mice treated with polyamide 3 demonstrated no elevation of these markers at the dose levels tested.
In a previous circulation study, it was found that a cyclic form of a hairpin polyamide targeted to the sequence 5′-WGGWWW-3′ had increased animal toxicity.29 However, in addition to the motif change from hairpin to cycle, the γ-turn of the cyclic compound was also changed from an (R)-2,4-diaminobutyric acid turn to an (R)-3,4-diaminobutyric acid turn. To determine if the toxicity is dependent on the polyamide shape or the γ-turn, we synthesized cyclic polyamide 5 (Figure S3, Supporting Information). The compound was found to be bioavailable after subcutaneous injection at 10 mg/kg and did not cause significant weight loss in animals. However, compound 5 did affect the kidney and liver and caused levels of ALT and AST to increase in a dose-dependent manner.
Multiple-Dose Subcutaneous Injections
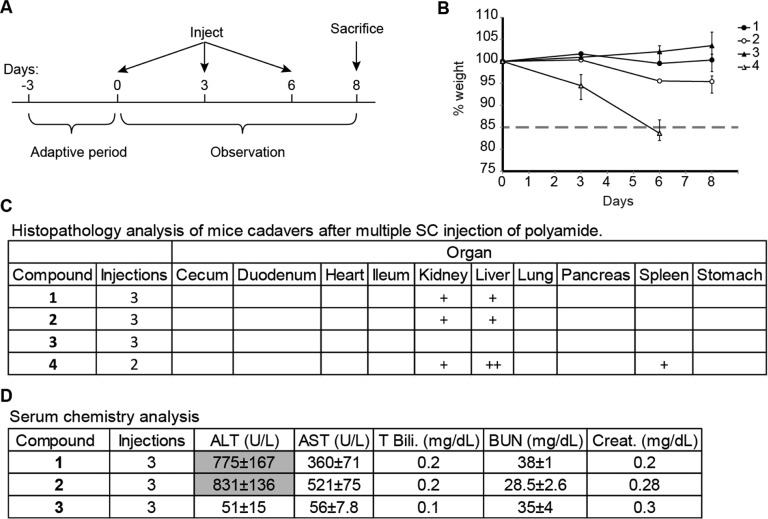
In addition to single dose injections, the effects of repeated dosing of polyamides 1–4 in mice were examined. In this experiment, 8-week-old male C57BL/6J mice (n = 3 per dosing group) were treated with 1 mg/kg of polyamides 1–4 by subcutaneous injection every 3 days, for a cycle of three injections, and then sacrificed 2 days after the final injection (Figure 4A). As in the single dosing experiments, two mice per group were subjected to histopathology analysis and all blood samples were sent for analysis. Mice treated with 1–3 demonstrated no loss in weight or physical morbidities. Two sequential injections of 4 at 1 mg/kg resulted in dramatic weight loss, loss of mobility, and hunched posture within 6 days (Figure 4B). These mice were promptly euthanized.
Figure 4.
Experiment setup of multidose weight curve experiments. (A) Male C57BL/6J mice were treated with compound once every 3 days. Animal weights were then monitored for 9 days. Humane end point was defined by visible signs of duress in the animals or weight loss in excess of 15% of original body weight. (B) Weight curves of compounds 1–4. (C) Histopathology analysis of sacrificed animals after multiple injections of compounds 1–4 at 1 mg/kg. + = mild damage, ++ = moderate damage, +++ = severe damage. (D) Serum levels of liver damage and kidney damage markers after three sc injections of compounds 1–3. Significantly elevated markers are shaded in gray.
Histopathology of the mice treated with polyamide 1 and 2 revealed mild multifocal hepatocellular necrosis and apoptosis in the liver and mild variable tubular attenuation, karyomegaly, and epithelial necrosis and apoptosis in the kidney. There was marked hepatocellular necrosis and apoptosis in the liver and hyaline droplet accumulation in the kidneys of animals treated with polyamide 4 (Figure 4C). Because mice treated with 4 did not tolerate two sequential injections at 1 mg/kg, and single dosing resulted in moderate liver and mild kidney damage at 3 mg/kg, we chose not to test this compound further. Consistent with the findings on histopathology, mice treated with 1 and 2 had elevated AST and ALT (Figure 4D). Mice treated with 3 had no histopathologic lesions consistent with toxicity or alterations in liver and kidney serum markers.
In Vitro Liver Microsomal Stability Assay
Liver pathology was the most striking abnormality and was most severe for 4. To assess if liver pathology was related to the stability of these compounds, we investigated the metabolic stability of these polyamides to liver microsome isolates. Stability to human and mouse liver microsomes with and without NADPH was tested for polyamides 1–4. Polyamide 1–3 all demonstrated high stability (>90% intact) after 1 h incubations (Table S1, Supporting Information). However, less than 5% of polyamide 4 remained intact after 1 h incubation with either human or mouse liver microsomes independent of the presence of NADPH.
Liver Uptake of Fluorescein–Polyamide Conjugates
To determine if the chemical modifications of the γ-turn corresponding to 1–4 could influence liver uptake of polyamides of otherwise identical structure, we synthesized four polyamide analogous to 1–4, but with fluorescein isothiocyanate replacing isophthalic acid at the C-terminus (Figure S4, Supporting Information). Mice treated with FITC–polyamide conjugate 8, which has a γ-turn substitution identical to that of 3, demonstrated less nuclear fluorescence in liver sections than the other FITC–polyamide conjugates (Figure S5, Supporting Information). Mice treated with FITC–polyamide conjugate 9, which has the γ-turn corresponding to 4, demonstrated the most intense nuclear fluorescence in liver sections.
Cellular Uptake and Cytotoxicity
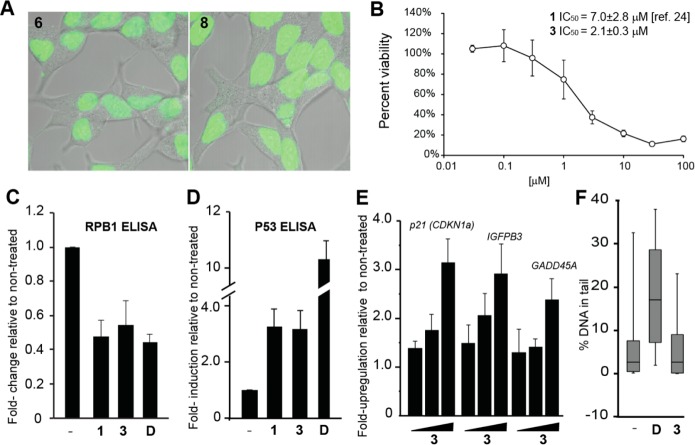
To determine the biological activity of 3 in LNCaP cells we first looked for evidence of nuclear localization using fluorescein analog 8. The fluorescein analog of 1, compound 6, was used as a benchmark. Confocal microscopy of LNCaP cells incubated with 2 μM of 6 or 8 for 24 h showed robust nuclear localization (Figure 5A). The viability of LNCaP cells was also reduced in a dose-dependent manner by 3, with the half-maximal inhibitory concentration at 2.1 ± 0.3 μM (Figure 5B).
Figure 5.
Activity of 3 in LNCaP cells. (A) Nuclear uptake of 6 and 8. (B) Cellular cytotoxicity of 3 toward LNCaP cells after 72 h incubation. (C) RPB1 protein decreases after treatment with 1 or 3 at 10 μM for 72 h or doxorubicin (D) at 1 μM for 24 h. (D) Cellular level of p53 protein increases after treatment 1 or 3 at 10 μM for 72 h or D at 1 μM for 24 h. (E) The p53 responsive genes p21, IGFBP3, and GADD45A are induced by 3 in a dose-dependent fashion (concentrations are 1, 3, 10 μM) after 72 h treatment. (F) Alkaline comet assay shows no increase in genomic fragmentation after prolonged incubation with 3 (48 h, 10 μM). Error bars represents 90% range; boxes represent the upper and lower quartiles and median.
Biological Characterization
Previously we found 1 to affect the RNA polymerase II holoenzyme, leading to the degradation of the large subunit, RPB1, and increase cellular p53 protein.24 Similarly, polyamide 3 reduced RPB1 levels when incubated with LNCaP cells at 10 μM for 72 h (Figure 5C). The level of p53 protein, as well as the transcripts of several p53 target genes, was also increased after treatment with 3 (Figure 5D,E). In addition, treatment of LNCaP cells with 10 μM of 3 for 48 h did not result in increased DNA damage by the comet assay (Figure 5F).
Antitumor Activity
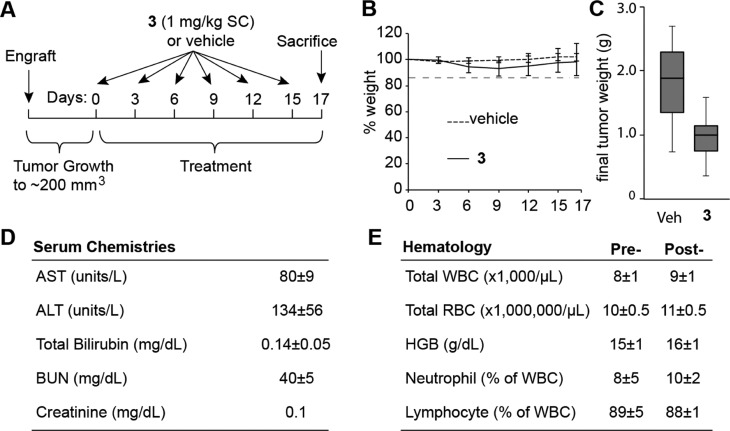
Next, we tested the activity of 3 against LNCaP xenografts in immunocompromised mice. Male NSG immunocompromised mice were engrafted with 2.5 million LNCaP cells. When the tumors reached 200 mm3 (0.5 × L × W2) treatment was initiated. Mice were treated with either 3 (sc, 1 mg/kg in 20% DMSO/normal saline, n = 14) or vehicle (20% DMSO/normal saline, n = 14) once every 3 days for a cycle of six injections. The animals were then sacrificed 2 days after the final injection (Figure 6A). Both groups of animals demonstrated minimal weight loss and no signs of distress during the course of the experiment (Figure 6B). Mice treated with 3 had smaller tumors than those treated with vehicle (T/C = 52.4%) (Figure 6C).
Figure 6.
Activity of 3 against LNCaP xenografts. (A) Timeline of treatment regimen. (B) Mouse weights throughout experiment. (C) Tumor weights at the experimental end point. Error bars represents maximum and minimum; boxes represent the upper and lower quartiles and median. p < 0.01. (D) Serum chemistry analysis of wild-type mice after six injections of 3. Serum levels of AST, ALT, total bilirubin, BUN, and creatinine were found to be within normal limits after compound treatment. (E) Hematological analysis of wild-type mice after six injections of 3. The levels of white blood cells (WBC), red blood cells (RBC), hemoglobin (HGB), neutrophil, and lymphocytes were not significantly affected by polyamide treatment. Values represent average of five animals, and errors are std.
To assess the toxicity of the treatment regimen in healthy animals, male C57BL/6J mice were treated with an identical regimen as the tumor-bearing mice and were sacrificed 2 days after the final injection. Because the liver and kidney were identified as the target organs of toxicity in our previous study, we assessed relevant serum markers for liver and kidney pathology (Figure 6D). Treated mice demonstrated no elevations in AST, ALT, total bilirubin, creatinine, or BUN. To examine if 3 has an effect on circulating blood counts, whole blood was sampled before treatment and at the time of sacrifice. No significant hematologic changes were noted for the total white blood cell, total red blood cell, hemoglobin, neutrophil, or lymphocyte count (Figure 6E).
Discussion
Py-Im polyamides interfere with DNA-dependent processes, including transcription, through noncovalent binding to the minor groove and do not result in significant levels of genotoxicity.24 These characteristics of polyamides may represent an advancement over current DNA-targeted cancer therapies, since development of treatment resistance and secondary diseases has been linked to drug-induced DNA damage.12,13,30 Our recent demonstration of the antitumor efficacy of polyamide 1 against LNCaP xenografts raises the possibility that the Py-Im polyamide technology platform could be developed into a new class of oncologic therapeutics.24 However, a more thorough understanding of the effects of selected polyamides in preclinical animal models is required. Although extensive prior work has demonstrated bioavailability of hairpin polyamides in rodents,28,31−33 the physiological effects of polyamides in an animal model have not been systematically examined. On the basis of our lead polyamide 1, we synthesized three additional polyamides and varied the γ-turn. This chemical change does not alter the DNA target sequence but affects animal toxicity and tissue distribution in mice.28
We find that subtle changes to the substitution on the γ-turn can dramatically impact systemic toxicity of the selected polyamides in rodents. In line with previously published work, compound 1 caused weight reduction in animals treated at 10 mg/kg but caused no other visible side effects.28 Compound 4 led to pronounced deterioration in the animals’ condition at 3 and 10 mg/kg. Initially, the toxicity associated with compound 4 was attributed to the acetylation of the primary amine, since acetylation generally leads to increased toxicity in cell culture.27 However, the unacetylated version of 4, compound 2, also demonstrated marked toxicity toward the animals, while the acetylated version of 1, compound 3, showed no adverse effects, suggesting that the acetylation of the amine is not the sole contributor to differences in toxicity.
Furthermore, a previous study reported that a cyclic polyamide with a (R)-3,4-diaminobutyric acid turn (β-substitution) was more toxic than its hairpin counterpart, which possessed a (R)-2,4-diaminobutyric acid turn (α-substitution). To see if the cyclic version of 1 led to increased animal toxicity, we synthesized 5. This compound was detectable in the serum after sc injection and was found to have less effect on animal weight than 1. This suggests that the increase in polyamide-induced toxicity is associated with the transition of the (R)-2,4-diaminobutyric acid turn to the (R)-3,4-diaminobutyric acid turn. Whether this is a general finding for the entire class of Py-Im polyamides remains to be seen.
To identify the cause of animal morbidity we conducted histopathological analysis on sacrificed animals. We found the liver and kidney to be the main organs of pathology for compounds 1, 2, 4, and 5. Compound 3 caused no detectable organ damage. Liver damage was most pronounced for 2 at 10 mg/kg and 4 at 3 and 10 mg/kg. Compound 5 caused moderate damage to both the liver and kidney at 3 and 10 mg/kg. We further confirmed our histopathology results with serum measurements of ALT, AST, total bilirubin, BUN, and creatinine. The liver damage markers ALT and AST were significantly elevated at higher doses of 1, 2, 4, and 5. Blood urea nitrogen levels were found to be elevated for 2 at 10 mg/kg and 4 at 3 and 10 mg/kg.
In addition to single dose experiments we also examined the effects of 1–4 on animal health after multiple treatments with an injection regimen that was identical to the treatment cycle used in our previous xenograft study.24 We found that compounds 1–3 had minimal effect on animal weight over three injections of 1 mg/kg, while compound 4 caused acute distress in the animals after two injections. Histopathology and serum marker analysis was able to detect liver and kidney damage in animals treated with all compounds except 3.
Since the liver is most affected by polyamides, we speculated that enzymatic degradation of the compounds may contribute to animal toxicity. To test the stability of compounds 1–4 in the liver we conducted microsomal degradation assays with human and mice liver microsomes. Compounds 1–3 were found to be >90% intact after a 60 min incubation with 1 mg/mL of microsomes. Therefore, the reduced liver toxicity of 3 as compared to 1, 2, and 4, may not be explained on the basis of differing stability to liver microsomes. Interestingly, while 4 was previously reported to be stable against rat and human microsomes,34 less than 5% of compound 4 remaining at the end of the assay. This may be explained by the lower amount of enzyme (0.3 mg/mL) used in the previous assay.
The tissue distribution of Py-Im polyamides is affected by structure.28,35 In our previous pharmacokinetic study we showed that 4 had greater localization in the lung, liver, and kidney than 1.28 Thus, differences in liver uptake of compounds 1–4 may contribute to the differences in animal toxicity. To visualize nuclear uptake we synthesized fluorescein analogues of 1–4. Of the four compounds, 8 (the fluorescein analogue of 3) showed the least amount of nuclear localization, which may explain the apparent lack of animal toxicity. A detailed pharmacokinetic analysis of 2 and 3 could provide more information on the structure-dependent bioavailability of these compounds. However, it remains a challenge for the field whether predictable correlation between polyamide structure and tissue distribution can be achieved.
Polyamide 1 was shown to exert cellular toxicity, in part, through the inhibition of transcription.24 In line with previous work, polyamide 3, was also found to affect the cellular level of RPB1 and p53, which suggests that the cytotoxic effects of 3 also stem from transcription inhibition. Furthermore, no increased DNA fragmentation was observed when cells were treated with 3, indicating that the compound interferes with transcription in a nongenotoxic manner.
In addition to exhibiting similar biological activity in cell culture and having reduced animal toxicity, 3 also demonstrated antitumor activity toward LNCaP xenografts. Tumor-bearing animals and wild-type animals were able to sustain six injections of 3 without showing any signs of duress. Further characterization of serum chemistries and hematology markers indicates compound 3 is well-tolerated by the animals.
In conclusion, we have identified a structural motif that affects the animal toxicity of Py-Im polyamides. The transition of the (R)-2,4-diaminobutyric acid turn to a (R)-3,4-diaminobutyric acid turn significantly increases the animal liver and kidney damage caused by polyamides in this study. From the four compounds, we have identified polyamide 3, which contains an acetamide at the α-position of the turn unit and demonstrates no detectable animal toxicity at 10 mg/kg. This compound behaves similarly to 1 in cell culture and retains antitumor activity toward LNCaP xenografts. This second-generation hairpin polyamide provides a promising lead for the development of Py-Im polyamides as anticancer therapeutics.
Experimental Section
Synthesis of Polyamides
Py-Im polyamides 1–4 and 6–9 were synthesized on Kaiser oxime resin (Novabiochem) as previously described.15,36 Complete oligomers were cleaved from resin using 3,3′-diamino-N-methyldipropylamine and purified by reverse-phase HPLC in 0.1% aqueous TFA and acetonitrile.37 Isophthalic acid and fluorescein isothiocyanate conjugates were synthesized as previously described.38 Cyclic polyamide 5 was synthesized on 2-chlorotrityl chloride resin (Bachem) as previously described.39 Deprotection of the γ-turn was performed as described.26 Hairpin polyamides 3, 4, 8, and 9 were acetylated as previously described.26,34 Polyamides 1–9 were purified again by reverse-phase HPLC after final conjugation. All polyamide molecular weight was measured by MALDI-ToF mass spectrometry (Table S2, Supporting Information), and a compound purity of >95% was determined by analytical HPLC.
Chemicals and Animals
Ten percent neutral buffered formalin was purchased from Richard-Allan Scientific, and 6–8-week-old male C57BL/6J mice were purchased from Jackson Laboratories.
Thermal Denaturation Assay
Thermal stabilization of the DNA oligo 5′-TTGCTGTTCTGCAA-3′ by 1–4 (target sequence underlined) was determined as previously described.26
Animal Weight Loss Analysis
All animal experiments were conducted under an approved protocol at the California Institute of Technology. Animals were allowed to adjust for 3 days after arrival before treatment. Compounds were quantified with a UV/vis spectrophotometer using an extinction coefficient of 69 500 M–1·cm–1 at λmax near 315 nm. For single injection weight loss experiments, the animals were separated into three treatment groups receiving 1, 3, or 10 mg/kg of compound in up to 200 μL of a 25% DMSO/saline vehicle, with four animals per group. Animals were monitored daily for weight loss over 9 days and sacrificed. For repeated injection experiments the animals were separated into groups of three and injected with 1 mg/kg of 1–4 once every 3 days and sacrificed 2 days after the last injection. Weight was recorded on days of injection and at the experiment end point.
Animal Histopathology Analysis
Sacrificed animals from weight loss experiments were fixed in 10% formalin and sent for histopathology analysis by IDEXX-RADIL. Histopathologic analysis was performed on the cecum, duodenum, heart, ileum, kidney, liver, lung, pancreas, spleen, and stomach. Tissue analysis was performed as a blind study to the identity of the animals.
Serum Analysis
Serum from treated animals were collected by retroorbital bleeding. Blood samples were centrifuged at 6000 rpm for 5 min to collect the serum. Serum ALT, AST, total bilirubin, BUN, and creatinine levels were sent for analysis at IDEXX-RADIL. Serum analysis was performed as a blind study as to the identity of the animals. For hematology analysis, blood was collected from five male C57BL6/J mice by retroorbital bleed and sent for hematological analysis in K2EDTA-coated BD Microcontainers. The animals were allowed to recover for 1 week before treatment with 3 using the same injection conditions as the NSG mice. At the treatment end point the animals were bled again and euthanized. Blood samples for serum chemistry analysis and hematology analysis were prepared separately. All samples were sent for analysis at IDEXX-RADIL.
Liver Microsomal Stability Analysis
Liver microsomal stability of 1–4 was performed by Apredica. Briefly, each polyamide was incubated with 1 mg/mL human or mouse microsomes at 37 °C. The reaction was incubated in 100 mM KH2PO4, 2 mM NADPH, 3 mM MgCl2 at pH 7.4. Samples were also incubated in the absence of NADPH to detect NADPH-free degradation. After 60 min the samples were mixed with an equal volume of ice-cold methanol stop solution. The mixture was allowed to sit on ice for at least 10 min and mixed with an equal volume of water. The samples were then centrifuged to remove the precipitates and the samples were analyzed by LC/MS/MS. Data represents percent remaining by comparing with time zero concentration. The experiments were performed in duplicate.
Tissue Distribution of Fluorescein Tagged Polyamides
Male C57BL/6J mice (n = 2 per group) were injected with 50 nmol (∼3 mg/kg) of 6–9 and then sacrificed 24 h later. Tissue was excised and processed as previously described.20 The tissues were excised and fixed in 10% neutral buffered formalin for 48 h. They were subsequently cryoprotected in 15% and 30% aqueous sucrose solution (w/v) for 24 h each, embedded in a medium for frozen sections (Sakura) and frozen on dry ice. Fluorescence intensity in liver tissue was assessed by laser confocal microscopy in 10 μm thick sections on slides mounted with a ProLong antifade reagent (Invitrogen).
Cell Viability Assays
LNCaP cells were plated in clear bottom 96-well plates at 5000–7500 cells per well and allowed to adhere for 36–48 h. Compounds were then added in fresh media. Cell metabolic activity was determined by the WST-1 assay (Roche) after 72-h incubation with cells. Quantification was performed on a Perkin-Elmer Victor 3 plate reader. Assays were performed in biological triplicates.
Protein ELISA Assays
Cellular levels of RPB1 and p53 protein in LNCaP cells after treatment with 10 μM 3 for 72 h were determined by ELISA. Cells treated with DMSO vehicle and 1 μM doxorubicin for 24 h were used as control. Cellular RBP1 levels were determined by a RPB1-specific ELISA kit (Cusabio Life Sciences) according to the manufacturer’s instructions. Total cellular p53 protein level was determined with a pan-p53 ELISA kit (Roche) according to manufacturer’s instructions. Assays were performed in biological triplicates.
Quantitative RT-PCR
LNCaP cells were plated in 12-well plates at 50 000 cells per well and allowed to adhere for 36–48 h. The cells were then treated with 1, 3, and 10 μM of 3 for 72 h. Total cellular RNA was extracted using RNEasy columns (Qiagen) following the manufacturer’s protocols. Isolated RNA was reverse transcribed with Transcriptor First Strand cDNA kit (Roche). Quantitative real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7300 instrument. Amplification of p21, IGFBP3, and GADD45A cDNA was measured relative to β-glucuronidase using previously published primers.24 Experiments were performed in biological replicates.
Comet Assay
LNCaP cells were plated in six-well plates at 100 000 cells per well and allowed to adhere for 36–48 h. The cells were then incubated with either 10 μM 3 for 48 h or 5 μM doxorubicin for 4 h. DNA damage was assayed using the Trevigen CometAssay system. Cells were harvested by gentle aspiration with PBS and prepared on slides according to the manufacturer’s protocol. Comet slides were imaged on a confocal microscope (Exciter, Zeiss) at 10× magnification. Images were scored using Comet Assay Lite IV (Perceptive Instruments). More than 100 comets were scored for each condition. DNA damage is reported as percentage of DNA in the tail.
In Vivo Xenograft Experiments
Mice experiments were conducted under an approved protocol by the Institutional Animal Care and Use Committee of the California Institute of Technology. Male C57BL6/J mice and male NSG mice were purchased from The Jackson Laboratory. All animals were maintained on a standard light–dark cycle. LNCaP cells (2.5 million cells) were engrafted in a mixture of 1:1 media and matrigel in the left flank of NSG mice. Tumors were allowed grow to ∼200 mm3 (0.5 × L × W2) before treatment. Py-Im polyamide 3 was administered by sc injection once every 3 d at 1 mg/kg in a 20% (v/v) DMSO:normal saline vehicle solution for six injections. Animals were sacrificed 2 days after the final injection. Animal weight and general health were monitored daily. Fourteen animals were used for each treatment group.
Acknowledgments
This work was supported in part by the National Institutes of Health (Grant GM27681), Sanofi, and the Prostate Cancer Foundation. The authors thank Janet Baer and Gwen E. Williams for helpful discussions. The authors also thank the IDEXX-RADIL team for assistance with serum analysis.
Glossary
Abbreviations
- Py
pyrrole
- Im
imidazole
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BUN
blood urea nitrogen
Supporting Information Available
DNA thermal stabilization of polyamides, serum concentration, characterization of an α-amino polyamide cycle, liver microsomal stability, structure of fluorescein-conjugated polyamides, nuclear localization in the liver, and MALDI-ToF characterization of polyamides. This material is available free of charge via the Internet at http://pubs.acs.org
Author Contributions
F.Y. and N.G.N contributed equally. F.Y., N.G.N., and P.B.D. designed the experiments. F.Y. and N.G.N. conducted animal experiments. B.C.L. and J.W. synthesized the compounds. J.O.S. conducted liver uptake experiments. S.R.H. performed histopathology analysis. F.Y. and N.G.N. performed cytotoxicity, comet, and ELISA experiments. F.Y. conducted confocal experiments. N.G.N. conducted RT-qPCR experimnts. J.L.M. ran thermal denaturation experiments. F.Y., N.G.N., J.O.S., S.R.H., J.L.M., and P.B.D. analyzed data. F.Y., N.G.N., and P.B.D. wrote the paper.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Jemal A.; Center M. M.; DeSantis C.; Ward E. M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 1893–1907. [DOI] [PubMed] [Google Scholar]
- Zelefsky M. J.; Eastham J. A.; Cronin A. M.; Fuks Z.; Zhang Z.; Yamada Y.; Vickers A.; Scardino P. T. Metastasis after radical prostatectomy or external beam radiotherapy for patients with clinically localized prostate cancer: A comparison of clinical cohorts adjusted for case mix. J. Clin. Oncol. 2010, 28, 1508–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tendulkar R. D.; Reddy C. A.; Stephans K. L.; Ciezki J. P.; Klein E. A.; Mahadevan A.; Kupelian P. A. Redefining high-risk prostate cancer based on distant metastases and mortality after high-dose radiotherapy with androgen deprivation therapy. Int. J. Radiat. Oncol., Biol., Phys. 2012, 82, 1397–1404. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Scher H. I. Prostate cancer in 2011: Hitting old targets better and identifying new targets. Nat. Rev. Clin. Oncol. 2012, 9, 70–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Chan S. C.; Brand L. J.; Hwang T. H.; Silverstein K. A.; Dehm S. M. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013, 73, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostaghel E. A.; Marck B. T.; Plymate S. R.; Vessella R. L.; Balk S.; Matsumoto A. M.; Nelson P. S.; Montgomery R. B. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: Induction of steroidogenesis and androgen receptor splice variants. Clin. Cancer Res. 2011, 17, 5913–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C.; Giaccia A. Transformed cells require continuous activity of RNA polymerase II to resist oncogene-induced apoptosis. Mol. Cell. Biol. 1997, 17, 7306–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.; Lippard S. J. RNA polymerase II blockage by cisplatin-damaged DNA—Stability and polyubiquitylation of stalled polymerase. J. Biol. Chem. 2006, 281, 1361–1370. [DOI] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [DOI] [PubMed] [Google Scholar]
- Mantoni T. S.; Reid G.; Garrett M. D. Androgen receptor activity is inhibited in response to genotoxic agents in a p53-independent manner. Oncogene 2006, 25, 3139–3149. [DOI] [PubMed] [Google Scholar]
- Haffner M. C.; Aryee M. J.; Toubaji A.; Esopi D. M.; Albadine R.; Gurel B.; Isaacs W. B.; Bova G. S.; Liu W. N.; Xu J. F.; Meeker A. K.; Netto G.; De Marzo A. M.; Nelson W. G.; Yegnasubramanian S. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 2010, 42, 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arseneau J. C.; Sponzo R. W.; Levin D. L.; Schnipper L. E.; Bonner H.; Young R. C.; Canellos G. P.; Johnson R. E.; DeVita V. T. Nonlymphomatous malignant tumors complicating Hodgkin’s disease. Possible association with intensive therapy. N. Engl. J. Med. 1972, 287, 1119–1122. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Campisi J.; Higano C.; Beer T. M.; Porter P.; Coleman I.; True L.; Nelson P. S. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat. Med. 2012, 18, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan P. B. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001, 9, 2215–2235. [DOI] [PubMed] [Google Scholar]
- Best T. P.; Edelson B. S.; Nickols N. G.; Dervan P. B. Nuclear localization of pyrrole-imidazole polyamide–fluorescein conjugates in cell culture. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 12063–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenyuk B. Z.; Zhang G. J.; Klco J. M.; Nickols N. G.; Kaelin W. G. Jr.; Dervan P. B. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 16768–16773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickols N. G.; Dervan P. B. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickols N. G.; Jacobs C. S.; Farkas M. E.; Dervan P. B. Modulating hypoxia-inducible transcription by disrupting the HIF-1–DNA interface. ACS Chem. Biol. 2007, 2, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskatov J. A.; Meier J. L.; Puckett J. W.; Yang F.; Ramakrishnan P.; Dervan P. B. Modulation of NF-kappa B-dependent gene transcription using programmable DNA minor groove binders. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickols N. G.; Szablowski J. O.; Hargrove A. E.; Li B. C.; Raskatov J. A.; Dervan P. B. Activity of a Py-Im polyamide targeted to the estrogen response element. Mol. Cancer Ther. 2013, 12, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf C. L.; Baird E. E.; Dervan P. B.; Rees D. C. Structural basis for G.C recognition in the DNA minor groove. Nat. Struct. Biol. 1998, 5, 104–109. [DOI] [PubMed] [Google Scholar]
- Kielkopf C. L.; White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B.; Rees D. C. A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science 1998, 282, 111–115. [DOI] [PubMed] [Google Scholar]
- White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B. Recognition of the four Watson–Crick base pairs in the DNA minor groove by synthetic ligands. Nature 1998, 391, 468–471. [DOI] [PubMed] [Google Scholar]
- Yang F.; Nickols N. G.; Li B. C.; Marinov G. K.; Said J. W.; Dervan P. B. Antitumor activity of a pyrrole-imidazole polyamide. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan P. B.; Edelson B. S. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003, 13, 284–299. [DOI] [PubMed] [Google Scholar]
- Dose C.; Farkas M. E.; Chenoweth D. M.; Dervan P. B. Next generation hairpin polyamides with (R)-3,4-diaminobutyric acid turn unit. J. Am. Chem. Soc. 2008, 130, 6859–6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J. L.; Montgomery D. C.; Dervan P. B. Enhancing the cellular uptake of Py-Im polyamides through next-generation aryl turns. Nucleic Acids Res. 2012, 40, 2345–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synold T. W.; Xi B.; Wu J.; Yen Y.; Li B. C.; Yang F.; Phillips J. W.; Nickols N. G.; Dervan P. B. Single-dose pharmacokinetic and toxicity analysis of pyrrole-imidazole polyamides in mice. Cancer Chemother. Pharmacol. 2012, 70, 617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskatov J. A.; Hargrove A. E.; So A. Y.; Dervan P. B. Pharmacokinetics of Py-Im polyamides depend on architecture: Cyclic versus linear. J. Am. Chem. Soc. 2012, 134, 7995–7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehan M. R.; Morse H. R. DNA damage repair and tolerance: A role in chemotherapeutic drug resistance. Br. J. Biomed. Sci. 2013, 70, 31–40. [DOI] [PubMed] [Google Scholar]
- Nagashima T.; Aoyama T.; Yokoe T.; Fukasawa A.; Fukuda N.; Ueno T.; Sugiyama H.; Nagase H.; Matsumoto Y. Pharmacokinetic modeling and prediction of plasma pyrrole-imidazole polyamide concentration in rats using simultaneous urinary and biliary excretion data. Biol. Pharm. Bull. 2009, 32, 921–927. [DOI] [PubMed] [Google Scholar]
- Nagashima T.; Aoyama T.; Fukasawa A.; Watabe S.; Fukuda N.; Ueno T.; Sugiyama H.; Nagase H.; Matsumoto Y. Determination of pyrrole-imidazole polyamide in rat plasma by liquid chromatography–tandem mass spectrometry. J. Chromatogr., B 2009, 877, 1070–1076. [DOI] [PubMed] [Google Scholar]
- Fukasawa A.; Nagashima T.; Aoyama T.; Fukuda N.; Matsuda H.; Ueno T.; Sugiyama H.; Nagase H.; Matsumoto Y. Optimization and validation of a high-performance liquid chromatographic method with UV detection for the determination of pyrrole-imidazole polyamides in rat plasma. J. Chromatogr., B 2007, 859, 272–275. [DOI] [PubMed] [Google Scholar]
- Chenoweth D. M.; Harki D. A.; Phillips J. W.; Dose C.; Dervan P. B. Cyclic Pyrrole-Imidazole Polyamides Targeted to the Androgen Response Element. J. Am. Chem. Soc. 2009, 131, 7182–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harki D. A.; Satyamurthy N.; Stout D. B.; Phelps M. E.; Dervan P. B. In vivo imaging of pyrrole-imidazole polyamides with positron emission tomography. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 13039–13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckett J. W.; Green J. T.; Dervan P. B. Microwave assisted synthesis of Py-Im polyamides. Org. Lett. 2012, 14, 2774–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky J. M.; Nguyen D. H.; Wurtz N. R.; Dervan P. B. Solid-phase synthesis of DNA binding polyamides on oxime resin. Bioorg. Med. Chem. 2002, 10, 2767–2774. [DOI] [PubMed] [Google Scholar]
- Nickols N. G.; Jacobs C. S.; Farkas M. E.; Dervan P. B. Improved nuclear localization of DNA-binding polyamides. Nucleic Acids Res. 2007, 35, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. C.; Montgomery D. C.; Puckett J. W.; Dervan P. B. Synthesis of cyclic Py-Im polyamide libraries. J. Org. Chem. 2013, 78, 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.