Abstract
Endocytic trafficking plays an important role in signal transduction. Signal transducer and activator of transcription 3 (STAT3) has been localized to endosomal structures and is dependent upon endocytosis for downstream function, but the nature of the interaction between STAT3 and endosomes is poorly defined. Interleukin-6 (IL-6) treatment time courses combined with pharmacologic and temperature inhibition, pulse-chase assays, and in vitro kinase assays allow us to evaluate the role of endosomes in the initiation, modulation, amplification and persistence of STAT3 signal transduction and transcription. We demonstrate that IL-6-induced STAT3 activation is initiated by direct interaction with internal structures upstream of the lateendosome and that persistent STAT3 signaling depends upon recurrent activation from endocytic structures. Further, we show that STAT3 subcellular localization is not dependent upon endocytic trafficking. Instead, STAT3 interacts with endosomes transiently and relocates to the nucleus by an endosome-independent mechanism. Finally, we establish STAT3 serine 727 phosphorylation as dependent upon endocytic trafficking and crosstalk with the mitogen-activated protein kinase (MAPK) signaling system. These data reveal endosomes as central to the genesis, course and outcome of STAT3 signal transduction and transcription.
Keywords: STAT3, MAPK, Erk1/2, IL-6, Signaling Endosome
Introduction
Signal transduction over large distances, as may exist between the distal axon and cell body in neurons, cannot occur by diffusion alone within physiologically relevant time frames [1, 2]. Instead, signaling systems demonstrably employ endocytic structures to properly target and transport signals across vast intracellular expanses [3]. These endosomes are additionally involved in regulating the spatial and temporal dynamics of signal initiation, propagation and cessation, which has led to the formation of the signaling endosome hypothesis [4]. Most simply, the signaling endosome hypothesis posits that ligand-bound receptor complexes at the plasma membrane are internalized into endosomal structures to which the proteins necessary for signal transduction, sorting, and transport have been recruited [1, 5, 6]. These signaling endosomes then serve as intracellular platforms from which activated signals are moved to distinct subcellular locations, propagated and then terminated through lysosomal degradation [7].
Importantly, the spatial challenges of signaling within neurons also exist within smaller cell populations. According to random walk modeling data, the predicted spatial range for free diffusion of signal transduction proteins, such as STAT3, before dephosphorylation and inactivation is 200 nm [2]. Distances greater than 200 nm readily exist between the plasma membrane and the nucleus within nearly all cells [1, 2]. Consequently, a facilitated mechanism for the trafficking and targeting of signaling proteins during transduction is necessary and endosomes likely fill this role.
Endosomes have been experimentally implicated in nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), epidermal growth factor (EGF) and transforming growth factor beta (TGF-β) signal transduction [6, 8–10]. As the importance of endocytic trafficking in signal transduction has become clearer, the need to understand the relationship between endosomes and the signaling proteins interacting with these structures has become essential. Within these systems, endosomes have been shown to facilitate intracellular signal transport to discrete subcellular location across a broad range of distances, to maintain signal fidelity through the preservation of ligand-receptor binding within the endosomal lumen, and to provide a physical substrate for the accumulation and interaction of proteins involved in signaling, sorting and transport, and to terminate signal propagation through the separation of ligand from receptor and lysosomal degradation [3, 6, 8, 11]. While the necessity of endosomes in signal transduction has been clearly demonstrated, the nature of interactions between signaling proteins and endosomal structures is largely undescribed.
We have employed IL-6 signaling to gain insight into the endosomal regulation of STAT3 and MAPK signal initiation, crosstalk and regulation. The IL-6 receptor has no inherent signaling capacity and initiates signal transduction through dimerization of the integral signaling protein gp130 upon ligand binding [12]. Following gp130 dimerization, constitutively associated Janus kinase (JAK) proteins phosphorylate the gp130 cytoplasmic tail to recruit and then activate the STAT and MAPK signaling systems [12].
Members of the STAT protein family are unique in their capacity to rapidly transmit signals from plasma membrane to nucleus without the involvement of a second-messenger signaling cascade [13]. Preformed STAT3 dimers recruited to the gp130 cytoplasmic tail are phosphorylated upon tyrosine residue 705 (Y705) by JAK, resulting in a structural change that permits DNA binding [14]. In the unstimulated state, STAT3 constitutively shuttles between the cytoplasm and nuclear compartments with the aid of alpha importins and exportin-1 [15–17]. STAT3 Y705 phosphorylation enables transcription and reduces the rate of nuclear export, increasing STAT3 concentration within the nuclear compartment [16]. STAT3 phosphorylation upon serine residue 727 (S727) facilitates an interaction with the transcriptional co -activators CBP and P300, maximizing transcriptional activity [18, 19].
MAPK signaling is initiated by the recruitment of Shc and SHP-2 to the cytoplasmic tail of gp130 following IL-6 treatment [12, 20, 21]. Signaling then continues through the adaptor protein Grb2, the GTPases Sos and Ras, and the kinases Raf, MEK1/2, and Erk1/2. Activated Erk1/2, a serine/threonine kinase, then phosphorylates and initiates transcription through a wide range of transcription factors, of which CREB and Elk-1 are the most widely recognized.
MAPK signal transduction has been closely associated with endosome-based signaling whereas STAT3 signal transduction has not [6]. Recently, STAT3 was localized to endosomal structures and endocytosis was demonstrated to be necessary for STAT3 transcriptional activity within the IL-6 signaling system [22]. In addition, STAT3 nuclear localization following receptor tyrosine kinase activation is demonstrably dependent upon endocytosis and peripheral cytoplasmic activation [23, 24]. However, substantial questions regarding the nature of STAT3 interaction with endosomal structures remain.
We hypothesize that the internalization and endocytic trafficking of activated IL-6 receptor complexes regulates STAT3 signal initiation, crosstalk, transcription and subcellular translocation. To evaluate this hypothesis, we performed the following experiments. First, we identified the subcellular loc ale from which STAT3 is activated. Second, we determined the extent to which endocytic trafficking regulates STAT3 signal propagation, signal persistence, transcriptional activity, crosstalk with the MAPK signaling system, and subcellular localization. Third, we examined whether or not endosomes directly amplify STAT3 signals.
Experimental Procedures
Cell Culture, Inhibitors, Antibodies and Reagents
The HepG2 human hepatocytoma cell line was acquired from ATCC and grown in Dulbecco’s Modified Eagle’s Media (DMEM) with 10% fetal bovine serum (FBS) and 40 μg/ml penicillin-streptomycin at 37 °C in a 5% CO2 environment. All cell culture reagents were purchased from Mediatech (Monassas, VA). Human recombinant IL-6 was purchased from R&D Systems (Minneapolis, MN). The inhibitors 2′-Amino-3′-methoxyflavone(PD 98059); 6-Nitrobenzo[b]thiophene-1,1-dioxide (STATTIC); and 2-(1,1-Dimethylethyl)-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinolin-7-one (Pyridone 6; P6); were purchased from Calbiochem (San Diego, CA), 2-Chloro-10-[3′-(dimethylamino)propyl]phenothiazine, HCl (Chlorpromazine, CPZ) and (Monensin) were purchased from Sigma Aldrich (St. Louis, MO). Antibodies against STAT3 (9132, 9139), phospho-STAT3 Y705 (9131), phospho-STAT3 S727 (9136), Erk1/2 (9102) and phospho-Erk1/2 (9101, 9106) were purchased from Cell Signaling Technologies (Beverly, MA). Antibodies against rab5, rab9, and dynein were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody against clathrin heavy chain was purchased from Affinity Bioreagents (Rockford, IL). The E-cadherin antibody was a kind gift from Dr. Bruce Horazdovsky (Mayo Clinic) and the histone H3 antibody was a kind gift from Dr. Zhiguo Zhang (Mayo Clinic). Rabbit and mouse HRP conjugated secondary antibodies were purchased from Jackson Immunoresearch (West Grove, PA).
Western Blotting
Samples were mixed 1:1 in Laemmli Sample Buffer (2% SDS (w/v), 10% glycerol, 60 mM Tris-Cl pH 6.8, 0.01% bromophenol blue (w/v)) containing 20% DTT (v/v), and boiled at 100° C for 5 minutes. Samples and a protein standard for molecular weight calibration were loaded onto either 7.5% or 10% polyacrylamide Tris-HCl gels with 5% stacking gels and run for 15 hours at 90V under constant voltage at 4 °C. Gels were then transferred to 0.45 μm nitrocellulose (Whatman Protran BA85; Maidstone, Kent) for 3 amp-hours under constant current at 4 °C, washed in ddH2O and blocked in 4% bovine serum albumin in tris-buffered saline (TBS; 20 mM Tris, 137 mM NaCl, pH 7.5 in ddH2O) for one hour at room temperature while rocking. Antibodies diluted 1:1000 in TBS were incubated with membranes while rocking at 4 °C overnight. The following morning, membranes were washed three times quickly with TBS-T (1x TBS with 0.002% Tween-20) and then once for 30 minutes at room temperature while shaking. Membranes were then incubated with HRP-conjugated secondary antibodies diluted 1:15000 in TBS for 1 hour at room temperature while shaking. Following secondary incubation, membranes were washed as before with TBS-T, developed using West Pico Chemiluminescent Substrate per manufacturer’s instruction (Pierce; Rockford, IL) and detected by film exposure (Denville HyBlot CL; Metuchen, NJ).
Whole Cell Lysate Activity Profile
HepG2 cells plated at a density of 2.5 × 105 cells per well in 6 -well plates were starved in serum-free DMEM for 3 hours at 37 °C. Inhibitors – 25 μM Mek1/2 inhibitor PD98059, 80 μM STAT3 tyrosine 705 inhibitor STATTIC, 100 μM JAK inhibitor P6, 50 mM NH4Cl, 2 μM Na+/H+ ionophore monensin, and 40 μM clathrin-mediated endocytosis inhibitor chlorpromazine – were applied to cells for 15 minutes at 37 °C. For temperature restriction, 20 mM HEPES was added to the starvation media and cells were incubated at either 4 °C or 15 °C for 15 minutes prior to IL-6 treatment. 20 ng/mL IL-6 was added directly to the starvation media and cells were incubated for 5, 10, 15, 30, or 45 minutes at 37 °C. Cells were then placed on ice, media was aspirated and cells were lysed in 100 μL RIPA buffer (1% Nonidet P-40 (v/v), 1% deoxycholic acid sodium salt (w/v), 0.1% sodium dodecyl sulfate (w/v) and 0.002 M EDTA in 1x TBS pH 8.0 with 1 mM PMSF, 10 μg/mL aprotinin, 1 μg/mL leupeptin, 1 mM NaVO3 and 1 mM NaF added prior to use). Cells were scraped into 1.5 mL microfuge tubes, bath sonicated for one minute at 4 °C and protein concentration was determined using the bicinchoninic acid protein assay kit from Pierce (Rockford, IL). 20 μg protein from each sample was loaded onto 7.5% Tris-HCl gels, separated by electrophoresis, transferred and blotted as described.
Pulse-Chase
HepG2 cells in 6-well plates were serum-starved as described above. Cells were pulsed with 20 ng/mL IL-6 for 5 minutes at 37 °C, quickly washed twice with warm DPBS, and then chase media (warm starvation media without IL-6) was applied for 5, 10, 15, 30 or 45 minutes. To prevent cell-surface initiated STAT3 activity, cells were exposed to a low pH environment that releases receptor-bound IL-6 by adding two pH 4.0 Dulbecco’s phosphate buffered saline (DPBS) washes following the pulse period. To inhibit prolonged JAK1 activity, 100 μM P6 was included in the chase media. Following the chase time course, cells were placed on ice, media was aspirated, and the cells were lysed in RIPA buffer. Lysate was prepared and proteins characterized as described.
Luciferase Assay
Luciferase constructs containing the STAT3 responsive M67 c-Fos sis-inducible element (M67 SIE) promoter (a kind gift from Dr. Keith Bible, Mayo Clinic), AP-1 promoter (a kind gift from Dr. Dan Billadeau, Mayo Clinic) and Elk-1 promoter (Panomics; Fremont, CA) were used to assess the transcriptional response of HepG2 cells following IL-6 treatment. Prior to elecroporation, 19.5 μg of reporter construct was mixed with 500 ng of Renilla luciferase control vector (a kind gift of Dr. Richard Bram, Mayo Clinic) and precipitated by mixing with 1/10 volume 3M sodium acetate pH 5.2 and 2 volumes of ice-cold 100% ethanol followed by a 5 minute incubation on dry ice and centrifugation at 16,000 x g for 10 minutes at 4° C. DNA pellets were air dried and resuspended in 50 μL DMEM with 10 mM HEPES pH 7.4. HepG2 cells plated at a density of 4.0 × 106 in 10 cm plates 18 hours prior to electroporation were scraped into 15 mL conical tubes, spun down, and resuspended in 350 μL electroporation media (RPMI with 5% (v/v) fetal calf serum, 5% (v/v) calf serum, 0.1 mM 2-mercaptoethanol, 25 mM HEPES pH 7.4 and 2 mM glutamine). DNA and cell mixtures were combined, incubated at room temperature for 10 minutes, and then transferred to 4.0 mm gap electroporation cuvettes (BTX 640; Holliston, MA). Samples were electroporated in a BTX T820 ElectroSquarePorator in LV mode with 1 70 ms pulse at 150 V. Samples were then flicked ten times, incubated at room temperature for ten minutes, and plated into 3 cm dishes containing DMEM with 10% FBS and incubated at 37 °C. The next day, cells were split into 24-well plates with 50,000 cells per well. Two days after electroporation cells were starved in serum-free DMEM for three hours at 37 °C. Inhibitors were applied for 15 minutes prior to IL-6 treatment as previously described. Cells treated with IL-6 received 20 ng/mL for 3 hours or 6 hours. Following treatment, cells were placed on ice, aspirated, washed in DPBS and lysed in 40 μL Passive Lysis Buffer included with the Dual Luciferase Reporter Assay kit (Promega; Madison, WI). Firefly (reporter) and Renilla (control) luciferase activity was then detected using a GloMax Luminometer (Promega) according to manufacturer’s instruction. All treatments and experiments were run in triplicate. Samples were normalized by dividing reporter activity by control activity and fold change was calculated by dividing this ratio by the ratio of untreated controls.
Cellular Fractionation
Cells plated in 15 cm plates were starved in serum-free DMEM for 3 hours at 37 °C and then treated with 20 ng/mL IL-6 for the indicated time period. Following treatment, cells were placed on ice, aspirated, washed with DPBS, and scraped into 1.5 mL microfuge tubes with 1 mL MES buffer (150 mM NaCl, and 25 nM 2-(N-Morpholino) ethanesulfonic acid sodium salt (Sigma M5057) pH to 6.5 in ddH2O with 1 mM PMSF, 10 μg/mL aprotinin, 1 μg/mL leupeptin, 1 mM NaVO3 and 1 mM NaF added fresh prior to use) and placed on ice. Subcellular fractions were isolated as described and resuspended in either RIPA lysis buffer or MES buffer depending upon use [25]. All fractions were used immediately for activity based assays or stored at −20 °C for western blot characterization.
Immunofluorescence
HepG2 cells plated on poly-d-lysine coated glass coverslips in 6-well plates at a density of 150,000 cells per well were serum starved, treated with inhibitors, and 20 ng/mL IL-6 was applied as described above. Cells were then placed on ice, immediately fixed with ice-cold 2:1 methanol:acetone for 15 minutes at room temperature, and then washed three times with PBS pH 7.5 for 5 minutes each at room temperature. Blocking solution (10% horse serum, 1% BSA, and 0.1% saponin in PBS pH 7.5) was then added to cells for 1 hour at room temperature. Primary antibody against STAT3 was diluted 1:400 in PBS pH 7.5 with 0.1% saponin and 75 μL was applied to coverslips overnight at 4 °C. The following day, coverslips were washed three times with PBS containing 0.1% saponin for 5 minutes at room temperature. FITC-conjugated secondary antibody was diluted 1:200 in PBS with 0.1% saponin and 75 μL was applied to coverslips for 45 minutes at room temperature in the dark. Coverslips were then washed twice with PBS containing 0.1% saponin for 5 minutes, twice with PBS for 5 minutes at room temperature, and then mounted onto slides using DAPI hardmount (Vector Labs; Burlingame, CA). Images were collected with a Zeiss LSM 510 confocal microscope and 63x/1.2 NA C-Apochromat lens. STAT3 nuclear staining was quantified by imaging 25 nuclei (identified by DAPI stain) from 5 fields of cells under each experimental condition with identical microscope settings and measuring the mean fluorescence intensity in the appropriate channel.
Endosome In Vitro Kinase Assay
HepG2 cells plated at 4.0 × 106 cells per plate in 6 15 -cm plates were starved in serum-free DMEM for 3 hours at 37° C. Two plates of cells were left untreated while two plates of cells were used for each 5 minute or 10 minute 20 ng/mL IL-6 treatment at 37 °C. Following treatment, cells were placed on ice, washed once with DPBS, scraped into microfuge tubes with MES buffer and Balch homogenized as described. Samples were then spun at 1000 x g for 10 minutes at 4 °C and supernatants were transferred to new microfuge tubes. Centrifugation was repeated to remove contaminants and supernatants were transferred to ultracentrifuge tubes and spun at 115,000 x g for 60 minutes at 4 °C. Supernatant was discarded and the remaining ‘Endosomes’ pellet, a combination of the large and small vesicle fractions highly enriched in endosomes, was resuspended in 100 μL MES buffer and protein concentration was determined by BCA. For the kinase assay, 20 μg endosomes were mixed with 500 ng His-tagged human recombinant STAT3 (Abcam; Cambridge, MA) and 25 μM ATP in kinase assay buffer (50 mM NaCl, 25 mM HEPES pH 7.5, 25 mM MgCl2, 10 mM MnCl2) for 30 minutes at 30 °C. 80 μM STATTIC was added directly to the kinase assay when used. To stop the reaction, equal volume NDG lysis buffer was added to the samples and they were immediately placed on ice. Ni+-NTA beads washed with MES were then added to the samples and rotated overnight at 4 °C to isolate protein targets. For M67 SIE pulldown experiments, 50 mM EDTA was used to stop the reaction. Protein was pulled down by the addition of 500 ng biotinylated M67 SIE oligo to each sample and neutravidin beads as described. Following overnight incubation, samples were spun down at 1000 x g and supernatants were removed as the “no pulldown” fraction and mixed with 35 μL sample buffer containing DTT. Beads were then washed three times with MES buffer before adding 35 μL elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0) and 35 μL sample buffer with DTT. All samples were boiled at 100 °C for 5 minutes, spun down at 1000 x g for one minute and then run on 7.5% Tris-HCl SDS-PAGE gels to separate proteins. Gels were transferred and blotted as described to detect changes in target phosphorylation.
Oligonucleotide Binding Assay
Oligonucleotides containing a known STAT3 DNA binding sequence were synthesized with the forward oligonucleotide biotinylated on the 3′ end [26].
Forward Oligo
5′ – TTTGGTATTCCCGGAAATGTTTTTGGTATTCCCGGAAATGTTTTTTTTTTTTTTT – 3′
Reverse Oligo
5′ – AAACCATAAGGGCCTTTACAAAAACCATAAGGGCCTTTACAAAAAAAAAAAA AAA – 3′
A 100 ng/μL oligo stock was made by mixing the two oligos in annealing buffer, heating the mixture to 72 °C for 10 minutes, and allowing the stock to cool to room temperature over a four hour period. Following 20 ng/mL IL-6 treatment, the nuclear fraction and the ‘endosomes’ fraction, consisting of the combined 16,000 x g and 115,000 x g fractions, were isolated as described. 20 μg protein from each sample was mixed with 500 ng biotinylated oligonucleotide and rotated at 4 °C overnight. The following day, samples were added to 50 μL neutravidin beads (Pierce) and rotated for 2 hours at room temperature. Samples were then spun down at 1000 x g. Supernatants were removed as the ‘no pulldown’ fraction and mixed with 35 μL sample buffer with DTT. Beads were washed three times with MES buffer and eluted in 75 μL sample buffer containing DTT. All samples were boiled at 100 °C, spun down and run on 7.5% Tris-HCl SDS-PAGE gels to separate proteins. Gels were transferred to nitrocellulose and probed as described.
Statistics
Statistical analyses were performed on data from the luciferase and immunofluorescence experiments. Two-way ANOVA tests were run in Sigma Stat 3.0 using the Student-Neuman-Keuls method for pairwise comparison with significance set at p < 0.05. All errors are reported as 95% confidence intervals.
Results
IL-6 Initiates STAT3 and Erk1/2 Signaling and Crosstalk
Hepatocytes are highly sensitive to IL-6 given their role in the acute phase response, making them a good model in which IL-6 induced signaling may be studied [12]. Serum-starved HepG2 cells were treated with 20 ng/mL IL-6 by direct addition of cytokine to the media for 5, 10, 15, 30 or 45 minutes. Whole cell lysates were then analyzed by western blot to define the phosphorylation profiles of STAT3 and Erk1/2 activation. STAT3 is phosphorylated at two residues, tyrosine 705 (Y705) and serine 727 (S727), upon activation. Phosphorylation of Y705 is obligatory for STAT3 transcriptional activity and is the most widely recognized marker for activation [13]. Additional phosphorylation of S727 is required for maximal STAT3 trancriptional output, which does not occur through Y705 phosphorylation alone [13]. Erk1/2 activitation is characterized by phosphorylation of threonine 202 and tyrosine 204 in Erk1 and threonine 185 and tyrosine 187 in Erk2.
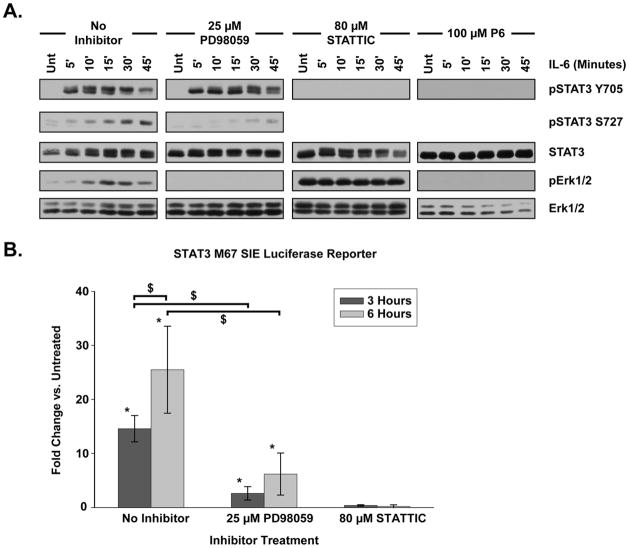
Within 5 minutes of IL-6 treatment, STAT3 was extensively phosphorylated upon Y705 with activation peaking between 10 and 15 minutes post-treatment and gradually decaying by 45 minutes (Fig. 1A). Phosphorylation of S727 began 5 minutes post-treatment, progressed comparatively slowly, and existed for the duration of the time course (Fig. 1A). A STAT3 molecular weight shift observed 5 minutes after IL-6 treatment was increasingly prevalent as the time course progressed, consistent with post-translational modification. Unlike STAT3, a basal level of Erk1/2 activation was present in the untreated state. Erk1/2 phosphorylation peaked 15 minutes after IL-6 treatment followed by a gradual decay in activation over the next 30 minutes (Fig. 1A).
Figure 1. IL-6 Activates STAT3 and Erk1/2.
(A) HepG2 cells were treated with IL-6 (20 ng/mL) for the indicated period of time. STAT3 and Erk1/2 activation was assessed by phosphorylation profile. The MEK1/2 inhibitor PD 98059, the STAT3 inhibitor STATTIC, and the broad spectrum JAK inhibitor P6 were all applied for 15 minutes prior to IL-6 treatment. Blots shown are representative of at least three separate, replicate experiments. (B) M67 SIE luciferase reporter activity in HepG2 cells treated with IL-6 for 3 or 6 hours. Cells were incubated with inhibitors for 15 minutes prior to IL-6 treatment. Fold change is compared against untreated samples. PD98059 treated cells exhibited a significant reduction in luciferase activity as compared to cells without inhibitor at both 3 and 6 hours post-IL-6. No significant luciferase activity was detected in STATTIC treated cells at any time period. Data were analyzed by two-way ANOVA with Student-Neuman-Keuls (SNK) pairwise comparison. Error bars indicate 95% confidence intervals. * indicates significant difference from untreated, p < 0.05. $ indicates significant difference between specified treatments, p < 0.05.
Transcription is a hallmark of both STAT3 and MAPK signal transduction. Many target genes of these pathways contain multiple promoter sequences to which both STAT3 and MAPK-associated transcription factors, such as Elk-1, may bind [27]. To avoid ambiguity in measuring the transcriptional response of these pathways, luciferase reporter assays with promoters responsive to only either STAT3 or MAPK transcription factors were utilized. Luciferase reporter constructs containing promoters to which AP-1, Elk-1 and STAT3(M67 SIE) bind were electroporated into HepG2 cells. Cells were serum-starved, treated with IL-6 for 3 or 6 hours and luciferase activity within the cellular lysates was compared to untreated controls. No change in AP-1 or Elk-1 luciferase reporter activity was detected upon IL-6 treatment (data not shown) despite robust Erk1/2 phosphorylation (Fig 1). Further, cells with 10% serum returned to the media or cells treated with 100 ng/mL EGF for 6 hours had no detectable AP-1 or Elk-1 luciferase response (data not shown). In contrast, IL-6 induced a significant increase in STAT3 transcription, as M67 SIE luciferase activity demonstrated a 14.59 ± 2.44 fold change 3 hours and a 25.51 ± 8.05 fold change 6 hours after application (Fig. 1B).
To delineate the IL-6-induced STAT3 and Erk1/2 signal transduction and transcription pathways, we employed the pharmacologic inhibitors P6, STATTIC, and PD 98059 [28–30]. P6 (100 μM), a broad spectrum JAK inhibitor [30], prevented both STAT3 and Erk1/2 activation by IL-6, indicating that both signaling pathways are downstream of JAK activity (Fig. 1A). STATTIC (80 μM), a small molecule inhibitor that binds the STAT3 SH2 domain to inhibit phosphorylation [29], abolished IL-6 induced STAT3 Y705 phosphorylation following IL-6 treatment (Fig. 1A). In addition, no significant M67 SIE luciferase activity was detected 3 or 6 hours after IL-6 application in cells treated with STATTIC, indicating that M67 SIE output was entirely a product of STAT3 transcriptional activity(Fig. 1B). Treatment with the MEK1/2 inhibitor PD 98059 (25 μM) eliminated IL-6 induced Erk1/2 activation [28](Fig. 1A).
Noticeable changes occurred in STAT3 and Erk1/2 signal transduction when the opposite pathway was inhibited, suggesting crosstalk between the STAT3 and MAPK signaling pathways. STATTIC inhibition of STAT3 resulted in persistent Erk1/2 activation in the absence of IL-6, which was unaltered by IL-6 treatment (Fig. 1A). PD 98059 inhibition of Erk1/2 did not alter the overall phosphorylation state of STAT3 Y705, but instead slowed the rate of STAT3 molecular weight shift (Fig. 1A). Importantly, PD 98059 markedly reduced IL-6-induced STAT3 S727 phosphorylation and significantly inhibited STAT3 transcription as measured by M67 SIE luciferase activity 3 hours or 6 hours following IL-6 treatment (Fig. 1A, 1B). Within the EGF and NGF receptor signaling systems, activated MAPK members Erk1/2 and MEK1 have been shown to interact with STAT3 and phosphorylate S727, yielding an increase in STAT3 transcriptional output [31, 32]. Consequently, the reduction of STAT3 phosphorylation and transcription we observe following MEK1/2 inhibition by PD98059 leads us to conclude that crosstalk between the MAPK and STAT3 signaling pathways is involved in regulating STAT3 transcriptional activity within the IL-6 signaling system.
STAT3 and Erk1/2 Associate with Endosomes
Subcellular fractions were isolated from IL-6 treated cells to evaluate the localization of STAT3 and Erk1/2. Balch homogenization and differential centrifugation were employed to yield five subcellular fractions – a nuclear fraction, a ghost fraction, a cytoplasmic fraction, a 16,000 x g fraction and a 115,000 x g fraction(25). Early and late endosomal structures as well as clathrin-coated vesicles were highly enriched within the 16,000 x g fraction as indicated by the presence of rab5, rab9, and clathrin heavy chain, respectively (Fig. 2A). The 115,000 x g fraction predominantly contained clathrin -coated vesicles and some early endosomes (Fig. 2A). Dynein, a retrograde transport protein, was also highly enriched within this fraction, suggesting the presence of mobile, vesicular structures (Fig. 2A). Further, negative staining and transmission electron microscopy of the pooled 16,000 x g and 115,000 x g fractions, referred to as the endosome fraction, revealed numerous membrane-bound organelles, consistent with an endosome-enriched population (Fig. 2B).
Figure 2. STAT3 Associates with Vesicular Structures.
Blots shown are characteristic of results seen within at least three separate and replicate experiments. (A) Subcellular fractions isolated from IL-6 (20 ng/mL) treated HepG2 cells using Balch homogenization and differential centrifugation as described within the materials and methods section. (B) Transmission electron microscopy of the combined 16,000 x g and 115,000 x g “endosomes” fraction after phosphotungstic acid negative staining, showing and enriched population of membrane-bound organelles. (C) HepG2 cells were treated with IL-6 (20 ng/mL) for the indicated period of time and subcellular fractions were isolated. The endosomes and nuclear fractions were prepared as described and incubated with a biotinylated M67 SIE oligonucleotide to test the DNA binding capacity of STAT3 within these fractions. Protein bound to DNA was separated by pull down with neutravidin beads and identified as ‘SIE Pulldown’. Protein that did not bind DNA and precipitate with neutravidin beads is identified as ‘No Pulldown’.
Prior to IL-6 activation, STAT3 localized to all of the subcellular fractions, but was particularly concentrated within the 16,000 x g and 115,000 x g vesicular fractions (Fig. 2A). Except for the cytoplasmic fraction, Erk1/2 was present within each of the fractions. Following IL-6 treatment, phosphorylated STAT3 localized to the vesicular and ghost fractions and persisted through the 45 minute time point (Fig. 2A). Total STAT3 protein and Y705 activated STAT3 rapidly localized to the nucleus (Fig. 2A). These data indicate that following IL-6 treatment, STAT3 is activated throughout the subcellular space and rapidly moves to the nucleus. Notably, activated STAT3 was present in and moved through the vesicular fractions as the IL-6 time course progressed. Contrary to expectation, activated Erk1/2 did not relocate to the nucleus but instead remained within the 16,000 x g vesicular fraction for the duration of the time course (Fig. 2A). This accounts for the lack of MAPK transcriptional activity observed following IL-6 treatment.
To demonstrate the transcriptional relevance of STAT3 isolated from the endosome-enriched fractions, a biotinylated oligo containing the M67 SIE promoter sequence was used to pull down STAT3 competent to bind DNA. Following IL-6 treatment, the nuclear and endosomes fractions were isolated and incubated with the oligo. As compared to untreated controls, greater amounts of STAT3 protein was pulled down with the oligo from the endosomes fraction of IL-6 treated cells, indicating that STAT3 localized to these fractions can bind DNA (Fig. 2C). From these data we conclude that transcriptionally competent, activated STAT3 interacts with endosomal structures.
Endocytosis is Necessary for STAT3 and Erk1/2 Signal Transduction
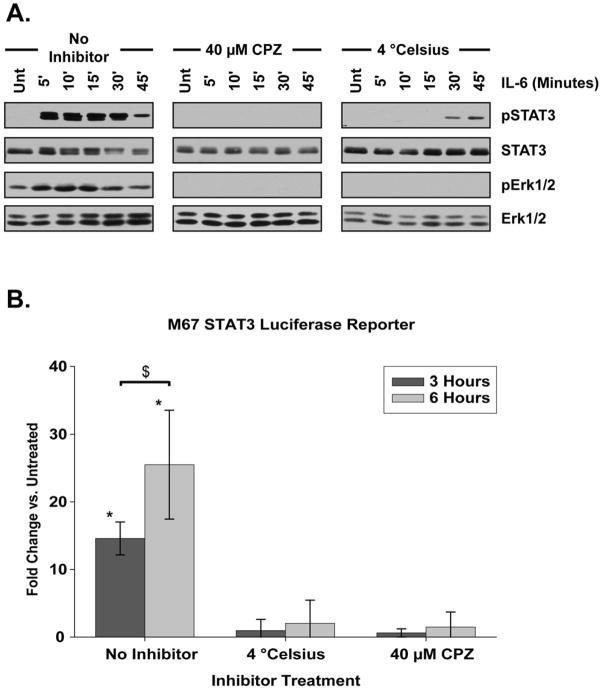
In canonical IL-6 signaling models, both STAT3 and Erk1/2 are activated at the plasma membrane [12, 13]. Accordingly, blocking endocytosis, the first stage of cargo maturation along the endocytic pathway, should not affect the phosphorylation of either STAT3 or Erk1/2. The cationic amphiphilic drug chlorpromazine (CPZ) and reduction of cellular temperature to 4 °C both inhibit internalization [33–35]. Treatment with 40 μM CPZ abolished both STAT3 and Erk1/2 phosphorylation upon IL-6 treatment(Fig. 3A). Temperature restriction to 4 °C prior to and during IL-6 treatment also abolished Erk1/2 activation and diminished and delayed STAT3 activation (Fig. 3A). Importantly, under both conditions cellular viability was not decreased (data not shown) and overall protein levels of both STAT3 and Erk1/2 remained unaltered (Fig. 3A). No significant STAT3 transcription following IL-6 treatment as assessed by M67 SIE luciferase reporter activity could be detected in samples either held at 4 °C or treated with CPZ (Fig. 3B). Together, these data demonstrate that STAT3 and Erk1/2 are not activated at the plasma membrane, contrary to canonical IL-6 signaling models.
Figure 3. STAT3 Requires Endocytosis for Activation.
(A) Phosphorylation profile of STAT3 and Erk1/2 following IL-6 treatment (20 ng/mL) for the indicated period of time. Cells were treated with clathrin-mediated internalization inhibitor chlorpromazine (CPZ) or held at 4° C for 15 minutes prior to IL-6 application. Three separate replicate experiments are represented by the blots shown. (B) M67 SIE luciferase reporter activity in IL-6 treated cells that were held at 4° C, inhibited with CPZ, or treated without inhibitors for the period of time indicated. Inhibition and temperature manipulation was applied for 15 minutes prior to IL-6 treatment. Fold change is compared against cells not treated with IL-6. Neither 4 °C restricted or CPZ treated cells exhibited a significant increase in luciferase activity as compared to untreated cells either 3 or 6 hours following IL-6 treatment. Data were analyzed by two-way ANOVA and pairwise comparisons were made using the SNK method. Error bars indicate 95% confidence intervals. * indicates significantly different from untreated, p < 0.05. $ indicates significant difference between specified treatments, p < 0.05.
STAT3 is Activated from Endocytic Structures
Following internalization, material is sorted within early endosomes and typically sent along a recycling or degradation pathway [36, 37]. Material sorted into the recycling pathway moves into recycling endosomes and is then reintegrated into the plasma membrane [36, 37]. Material moving through the degradation pathway is trafficked from early endosomes into late endosomes/multivesicular bodies [36, 37]. Material within the late endosome intravesicular lumen is then transferred into the lysosome and degraded [36, 37].
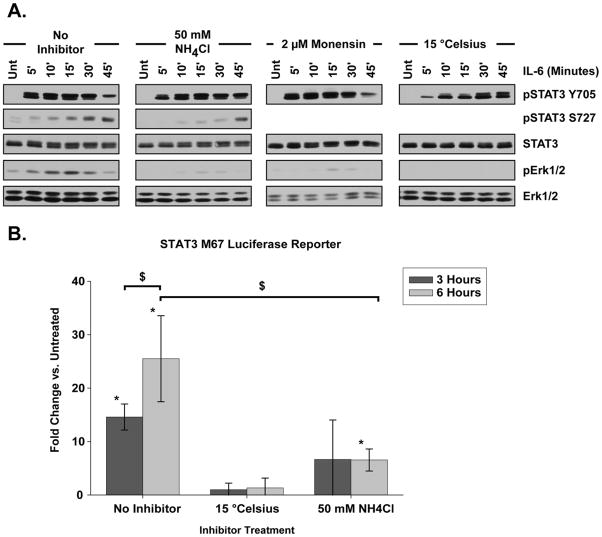
Since STAT3 and Erk1/2 are not activated from the plasma membrane, pharmacologic inhibition and temperature regulation were used to manipulate endocytic trafficking and assess the impact upon signal transduction. Reduction of cellular temperature to 15 °C allows internalization but prevents endocytic trafficking beyond the early endosome stage [38]. When HepG2 cells were held at 15 °C, the rate of IL-6 induced STAT3 Y705 phosphorylation was noticeably delayed (Fig. 4A). Further, the STAT3 molecular weight shift seen within 5 minutes of IL-6 treatment in cells at 37 °C did not occur in cells held at 15 °C until 45 minutes following IL-6 application (Fig. 4A). No significant STAT3 transcriptional activity was detected within cells held at 15 °C and treated with IL -6 (Fig. 4B).
Figure 4. STAT3 is Activated from Endocytic Structures.
(A) STAT3 and Erk1/2 phosphorylation profile after IL-6 treatment (20 ng/mL) for the indicated time period. Disruption of endocytic trafficking was achieved by altering vesicular pH with ammonium chloride (NH4Cl) or monensin treatment, or holding cells at 15° C for 15 minutes prior to IL-6 application. Blots shown are representative of at least three separate replicate experiments. (B) M67 SIE luciferase reporter activity in cells treated with IL-6 for the amount of time indicated. Inhibition or temperature manipulation was done for 15 minutes prior to IL-6 application. Fold change is compared to cells untreated with IL-6. No significant increase in luciferase activity over untreated controls was detected in 15 °C restricted cells at either 3 or 6 hours following IL-6 treatment or in NH4Cl treated cells 3 hours after IL-6 treatment. NH4Cl treated cells exhibited a significant increase in luciferase activity over untreated cells 6 hours post-IL-6, but this level of activity was significantly when compared to cells without inhibitor. Error bars indicate 95% confidence intervals and data were analyzed by two-way ANOVA with pairwise comparisons made using the SNK method. * indicates significantly different from untreated, p < 0.05. $ indicates significant difference between specified treatments, p < 0.05.
As vesicles traffic from the early endosome to late endosome and the lysosome, their pH is progressively reduced through the facilitated movement of hydrogen ions into the lumen by H+-ATPase pumps [39–41]. Ammonium chloride (NH4Cl) and monensin prevent vesicle acidification by directly increasing vesicular pH or reducing the capacity of vesicle associated H+-ATPase pumps, respectively [40, 42]. Under these conditions trafficking of vesicles between the early endosomal, late endosomal and lysosomal compartments is effectively halted, while endocytosis and vesicular movement into early endosomes still takes place. Previous work has demonstrated 50 mM NH4Cl and 2 μM monensin effective concentrations for the disruption of endocytic trafficking in cell culture applications [43–45]. When applied to HepG2 cells, we found that both NH4Cl and monensin altered the phosphorylation profile of STAT3 Y705 and substantially reduced Erk1/2 activation upon IL-6 treatment (Fig. 4A). NH4Cl also delayed and reduced the extent of STAT3 S727 phosphorylation, in correspondence with the decline in Erk1/2 activity (Fig. 4A). The extent of STAT3 Y705 phosphorylation was unchanged between inhibitor treated and untreated cells, but NH4Cl and monensin delayed both the molecular weight shift and decay in Y705 activation as compared to cells treated with IL -6 in the absence of inhibition. A significant increase in STAT3 transcriptional activity was only detected in NH4Cl treated cells 6 hours following IL-6 application (Fig. 4B). However, the level of IL-6 induced STAT3 transcriptional activity within NH4Cl treated cells as compared to cells without inhibitor was significantly reduced (Fig. 4B). Together, these data demonstrate STAT3 is activated from a vesicular structure upstream of the late endosomes whereas Erk1/2 is activated from a vesicular structure downstream of the early endosome. When taken with the observation that neither STAT3 nor Erk1/2 are activated when trafficking from the plasma membrane is restricted, these data lead us to conclude that IL-6 induced signal transduction must be initiated from an internal structure.
STAT3 Subcellular Localization is Not Dependent Upon Endocytic Trafficking
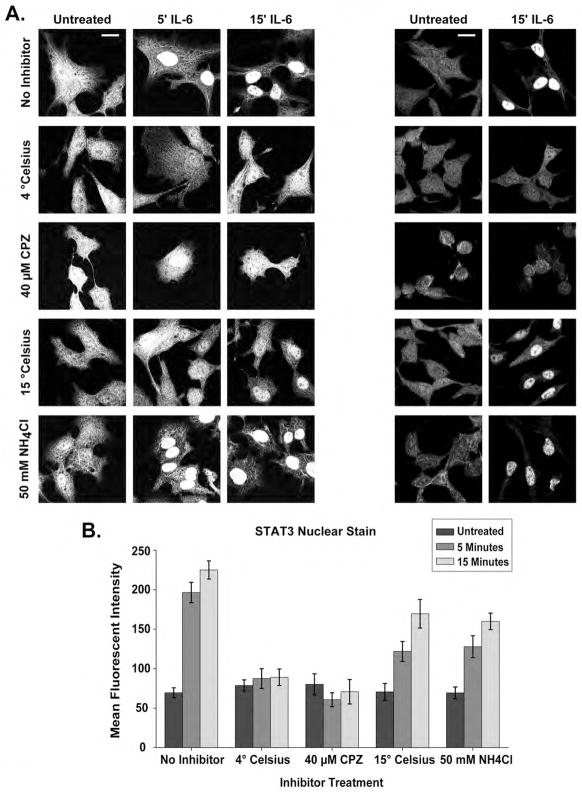
STAT3 rapidly translocates to the nucleus upon IL-6-induced activation. To determine the roles internalization and endocytic trafficking play in STAT3 subcellular localization, we evaluated the mean fluorescent intensity (MFI) of STAT3 nuclear staining following IL-6 treatment and inhibition of endocytosis and trafficking. At rest, STAT3 staining was punctate but homogeneous throughout the cytoplasm (Fig 5A). In the absence of inhibitors, STAT3 nuclear MFI at 5 minutes and 15 minutes following IL-6 was significantly (ANOVA 5′ p < 0.0001, 15′ p < 0.0001) greater than cells at rest, but the 5 minute and 15 minute treatments were not significantly different than one another (Figure 5A, 5B). STAT3 was observed predominantly within the nucleus 5 minutes and 15 minutes after IL-6, but substantial amounts could be observed within the cytoplasm in punctuate and fibrillary staining patterns (Figure 5A). When endocytosis was inhibited with CPZ or 4 °C temperature restriction, IL-6 stimulation did not result in significant STAT3 nuclear accumulation at any time point as measured by MFI (Figure 5A, 5B). This observation was expected given that STAT3 is not activated when internalization is inhibited (Fig. 3). Surprisingly, STAT3 translocated to the nucleus upon IL-6 treatment despite disruption of endocytic trafficking by 15 °C temperature restriction or 50 mM NH4Cl demonstrating an MFI significantly greater than untreated cells at both 5 and 15 minute time points following IL-6 (ANOVA 5′ P < 0.001, 15′ P < 0.001) (Figure 5A, 5B). Cells treated with IL-6 at 37 °C, however, had a significantly greater increase in STAT3 nuclear accumulation when compared to 15 °C temperature restriction or NH4Cl inhibition at all time points after stimulation (Two-Way ANOVA, Unt vs. 15 °C P < 0.001 at 5′ and 15′, Unt vs. NH4Cl P < 0.001 at 5′ and 15′) (Fig. 5B). Again, the amount of STAT3 reaching the nucleus in IL-6 treated cells without inhibition was significantly higher at both the 5 and 15 minute time points (Fig 5A, 5B). These data further support the assertion that STAT3 is activated from an internal structure following endocytosis as no significant subcellular relocalization is seen when internalization is inhibited. Surprisingly, IL-6 still initiates significant STAT3 movement into the nucleus when trafficking along the endocytic pathway is inhibited by 15 °C temperature restriction and NH4Cl treatment. From these data we conclude that endosomes function predominantly as a platform for STAT3 activation following IL-6 stimulation rather than a vector of transport.
Figure 5. STAT3 Rapidly Localizes to the Nucleus Following IL-6 Treatment.
(A) STAT3 staining within HepG2 cells under various inhibitor treatments and temperature restriction following IL-6 treatment. Panels on the left demonstrate STAT3 staining in both the nuclear and cytoplasmic compartments. Panels on the right demonstrate fields from which mean fluorescent intensity of STAT3 nuclear staining was calculated. (B) Mean fluorescent intensity of STAT3 nuclear staining under the treatment conditions in (A) calculated as described in the Experimental Procedues. Error bars represent 95% confidence intervals and significance is discussed within the Results section. Scale bars are equivalent to 20 um.
Temporally Persistent STAT3 Activation Arises from Endocytic Structures
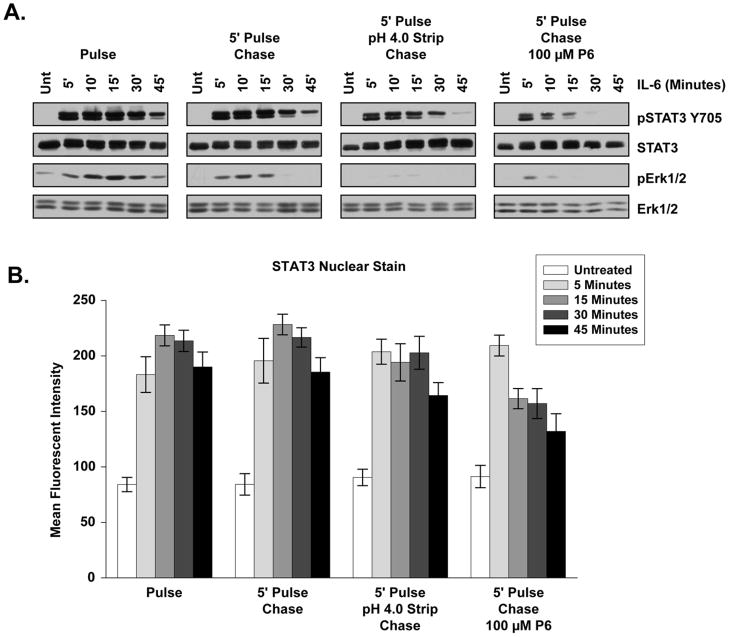
STAT3 activation and nuclear relocation was rapid and long-lasting following IL-6 treatment (Figs. 1, 2, 5). This persistent STAT3 phosphorylation may be the consequence of a single activation event or persistent, recurrent activation following IL-6 stimulus. To examine the temporal dynamics of STAT3 activation in the context of receptor complex internalization, various iterations of pulse-chase experiments were performed.
The pulse-chase condition tested the necessity of continuous IL-6 receptor complex activation at the plasma membrane for persistent STAT3 signaling. Serum-starved cells were treated with IL-6 added to the media for 5 minutes (Pulse) followed by two quick DPBS washes and reapplication of starvation media without IL-6 (Chase) for the indicated period of time. Little difference in the extent or persistence of the STAT3 Y705 phosphorylation profile was seen between pulse-chase treated cells (Fig. 6A, 5′ Pulse-Chase) and those with constant IL-6 exposure for the duration of the time course (Fig 6A, Pulse). As determined by STAT3 nuclear MFI, no significant difference was detected in STAT3 nuclear aggregation between the pulse or pulse-chase treatments at any time point (Fig. 6B). From this result we conclude that the entirety of IL-6 receptor binding required for STAT3 activation and nuclear aggregation occurred within 5 minutes of treatment and continuous activation of IL-6 receptor complexes at the plasma membrane was unnecessary for persistent STAT3 activation and nuclear translocation over the 45 minute time course. The extent of Erk1/2 signaling was reduced in the pulse-chase condition (Fig. 6A). Erk1/2 activation may arise from a subset of receptor complexes internalized at a slower rate than those which activate STAT3, require continuous ligand binding and internalization of receptor complexes from the plasma membrane, or require greater receptor occupancy than STAT3 for maximal activation.
Figure 6. Persistent STAT3 Activation Arises from Endocytic Structures.
(A) STAT3 and Erk1/2 phosphorylation profile following pulse or pulse-chase 20 ng/mL IL-6 treatment as described in the Experimental Procedures and the Results sections. Blots shown are representative of results seen in at least three independent experiments. (B) Mean fluorescent intensity of STAT3 nuclear staining across a time course of 20 ng/mL IL-6 pulse and pulse-chase conditions. Error bars indicate 95% confidence intervals and significance is discussed within the Results section.
To evaluate the necessity of endocytosis for signal propagation, low pH washes were used to strip IL-6 from the plasma membrane following the 5 minute pulse. The IL-6 receptor does not bind IL-6 at pH less than 5.0 [46], so pH 4.0 DPBS was used during the wash stage of pulse-chase experiments to strip away any receptor-bound IL-6 residing at the plasma membrane. When compared to cells treated under the pulse or pulse-chase conditions, low pH-stripped cells showed diminished persistence of STAT3 signal activation at 45 minutes but did not exhibit noticeable changes in the overall extent of activation (Fig. 6A). Low pH-stripped cells demonstrated a small but significant decrease in STAT3 nuclear staining when compared to pulse and pulse-chase conditions 45 minutes after the chase (Two Way ANOVA, Pulse vs. Low pH P < 0.05, Pulse-Chase vs. Low pH p < 0.05) (Fig. 6B). At the same time, Erk1/2 activation was almost completely eliminated in surface-stripped cells, further indicating that MAPK signal activation requires greater receptor occupancy or arises from a population of receptor-complexes internalized later than those that initiate STAT3 signaling. Since STAT3 Y705 phosphorylation and nuclear translocation was not substantially altered across the time course by low pH washes, the IL-6 receptor-complexes responsible for STAT3 activation must be protected from plasma membrane manipulation within 5 minutes (Fig. 6A, 6B). From these data we conclude that persistent STAT3 activation arises from an internal structure (Fig. 6A). These observations also suggest that Y705 phosphorylated STAT3 and S727 phosphorylated STAT3 are temporally and spatially distinct protein species. STAT3 protein that is Y705 phosphorylated rapidly translocates to the nucleus, which would prevent MAPK interaction and S727 phosphorylation on the same protein. The reduction in STAT3 activation and nuclear aggregation after low pH washes and a 45 minute chase also suggests some level of delayed receptor internalization is involved in the persistence of STAT3 Y705 phosphorylation that may be tied to MAPK signaling.
To test whether STAT3 signal persistence and nuclear inclusion is the product of a single activation event or continual reactivation, activated receptor complexes were inhibited by the inclusion of the JAK inhibitor P6 within the media during the chase stage. The addition of P6 to the chase media dramatically reduced the extent and persistence of STAT3 Y705 activation, which was completely absent at chase time points beyond 15 minutes(Fig. 6A). Addition of P6 to the chase media resulted in a very significant reduction in STAT3 nuclear inclusion at all chase time points after 5 minutes when compared to all other treatments (Two-Way ANOVA, P < 0.001 comparing P6 to all other treatments) (Fig. 6B). These data indicate that persistent STAT3 signal transduction and nuclear inclusion requires continuous cytoplasmic stimulation from activated receptor complexes within an endosomal structure and is not dependent upon continuous activation from the plasma membrane.
Transcriptionally Competent STAT3 is Directly Activated from Endosomal Structures
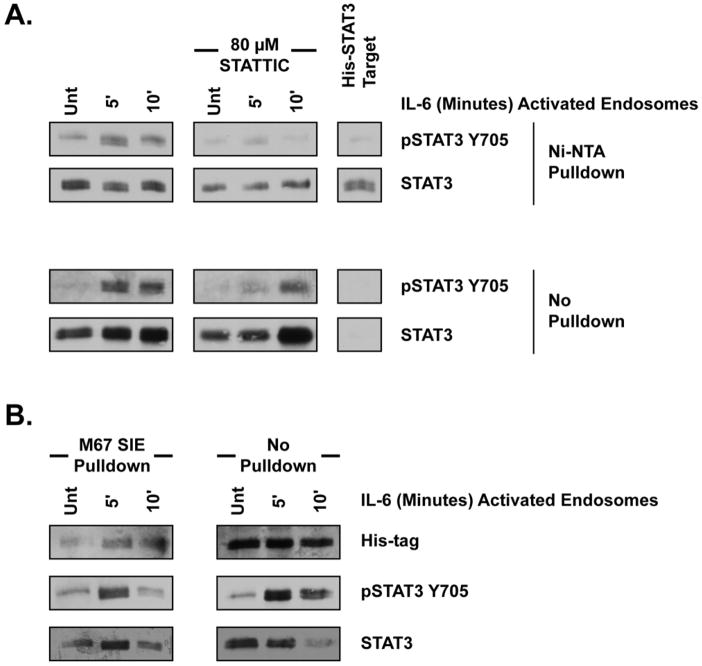
The inhibition of endocytosis, disruption of endocytic trafficking, STAT3 nuclear relocalization and pulse-chase experiments all suggest STAT3 is activated from vesicular structures within the cytoplasm (Figs. 3, 4, 5, 6). To directly test the capacity of endosomes to activate STAT3, in vitro kinase assays were performed utilizing exogenous STAT3 target and IL-6 activated endosomes. Endosomes were isolated as pooled 16,000 x g and 115,000 x g fractions from HepG2 cells at restor following 5 or 10 minute treatment with 20 ng/mL IL-6 at 37 °C. These endosomes were then incubated with recombinant His-tagged STAT3 target for 30 minutes at 30 °C and the target isolated using Ni-NTA beads. STAT3 target incubated with IL-6-activated endosomes experienced substantially greater Y705 phosphorylation than target incubated with endosomes from untreated cells (Fig. 7A). Endosomes from cells treated with IL-6 for 5 minutes demonstrated the greatest kinase activity, corresponding with the time period in which an initial STAT3 activating event occurs, as demonstrated by the pulse-chase experiments(Fig 6A).. To rule out the potential false positive created by dimerization between His-STAT3 and phosphorylated, endogenous STAT3 from the endosomes fraction, 80 μM STATTIC was added directly to the kinase assay. STATTIC prevented phosphorylation of the STAT3 target, ensuring target phosphorylation was the product of the in vitro assay itself (Fig. 7A). Untreated STAT3 target was only detected within the Ni-NTA pulldown fraction, indicating the entire amount of target used was isolated by the Ni-NTA beads (Fig. 7A). Substantial STAT3 in the resting and phosphorylated state was also detected within the ‘no pulldown’ fraction, indicating a clear separation of His-STAT3 from endogenous STAT3 within the endosomes fraction (Fig. 7A). To determine the transcriptional relevance and capacity of endosome activated STAT3, biotinylated oligonucleotide containing the M67 SIE promoter was used to isolate STAT3 from the kinase reactions instead of Ni-NTA beads. The presence of the His-tag within the pull-down fraction indicates that the STAT3 target was capable to bind DNA following phosphorylation by endosome-associated receptor complexes(Fig. 7B). These experiments demonstrate that STAT3 is directly activated by endosomes and endowed with the capacity to bind DNA, suggesting that transcriptionally-relevant STAT3 signals arise from endosomal structures.
Figure 7. Endosomes Directly Activate STAT3.
(A) Phosphorylation profile of His-STAT3 by in vitro kinase assay. Endosomes isolated from HepG2 cells treated with 20 ng/mL IL-6 as indicated were mixed with His-tagged recombinant human STAT3 target for 30 minutes at 30 C. Ni+-NTA pulldown indicates protein isolated with Ni+-NTA agarose beads. No pulldown indicates protein that did not bind Ni+-NTA beads. Blots shown are representeative of at least three, replicate experiments. (B) M67 SIE oligo binding following an endosome in vitro kinase assay as in (A). At the conclusion of the kinase assay, 500 ng biotinylated M67 SIE oligo was used to pull down protein with DNA binding capacity as described within experimental procedures. ‘Pulldown’ indicates protein bound to the biotinylated oligo and isolated with neutravidin beads. ‘No Pulldown’ indicates protein that did not bind the biotinylated oligo.
Discussion
Three important aspects of STAT3 endosome-based signaling are demonstrated in this study. First, STAT3 signal activation, signal persistence, and transcriptional activity arise from endocytic structures. Second, crosstalk between the MAPK and STAT3 signaling systems is requisite for maximal STAT3 transcriptional activity, and these events depend upon Erk1/2 activation from late endosomal structures. Third, the subcellular localization of activated STAT3 is not strictly dependent upon endocytic trafficking.
Previous work has localized STAT3 to vesicular, endosomal structures and shown the relevance of internalization upon STAT3 transcriptional activity within the IL-6 signaling system [22]. In support of these observations, our data confirm STAT3 localization to endosomal structures and the necessity of internalization for transcriptional activity (Figs. 2, 3). Localization alone does not demonstrate the necessity of endosomes for the initiation or regulation of STAT3 signal transduction. If STAT3 is activated at the plasma membrane, then any transcriptional reduction upon inhibition of endocytosis is a consequence of trafficking deficits. Under these circumstances, endosomes would function as a mechanism for transport of activated STAT3 and would not play a direct role in STAT3 activation. Indeed, internalization is not necessary for STAT3 activation by receptor tyrosine kinases but is required for STAT3 nuclear localization within these systems [23, 24]. This is consistent with the signaling endosome hypothesis and its supporting work, which claims endosomes responsible for signal trafficking and fidelity [1, 5, 6, 11].
Our observations taken together, however, indicate that endosomes function primarily as a mechanism for local STAT3 signal initiation, amplification and modification rather than a vector for intracellular transport and distribution within the IL-6 system. We clearly demonstrate internalization itself as crucial for IL-6-induced STAT3 activation since STAT3 signal transduction, localization and transcription are all abrogated when endocytosis is inhibited (Figs. 3, 5). Transcriptionally relevant STAT3 also interacts with endosomes (Fig. 2) and these structures demonstrate the capacity to directly phosphorylate STAT3 and induce DNA binding (Fig. 7). That STAT3 activation follows internalization fundamentally contradicts canonical signaling models in which STAT3 is activated at the plasma membrane [12, 13]. While STAT3 activation is rapid, characteristic and prolonged STAT3 signaling requires recurrent IL-6 activation from an internal structure, not the plasma membrane, as shown in the pulse-chase studies (Fig. 6). In addition, endosomes containing activated IL-6 receptor complexes augment STAT3 transcriptional activity through MAPK activation and subsequent S727 phosphorylation. The STAT3 S727 residue is flanked by an Erk1/2 kinase phosphorylation sequence [47], -P-X-S-P-, and has been shown to be phosphorylated by both MEK1 and Erk1/2 following receptor tyrosine kinase activation [31, 32]. We demonstrate that PD98059 MAPK inhibition and endocytic traffic restriction substantially reduce both STAT3 S727 and Erk1/2 activation following IL-6 treatment (Fig 1A, 1B, 4A, 4B). These observations indicate that STAT3 S727 phosphorylation and transcriptional modulation following IL-6 treatment is a consequence of interaction with the MAPK signaling system and that this interaction takes place upon late endosomal structures (Figs. 1, 4). Finally, we found STAT3 nuclear localization reduced but not eliminated when endocytic trafficking was halted(Fig. 5). As a consequence, endosomes cannot be the sole or primary means of STAT3 subcellular distribution in response to IL-6. This finding also suggests that interaction between STAT3 and endosomes is brief and that STAT3 moves away from endosomal structures following signal initiation.
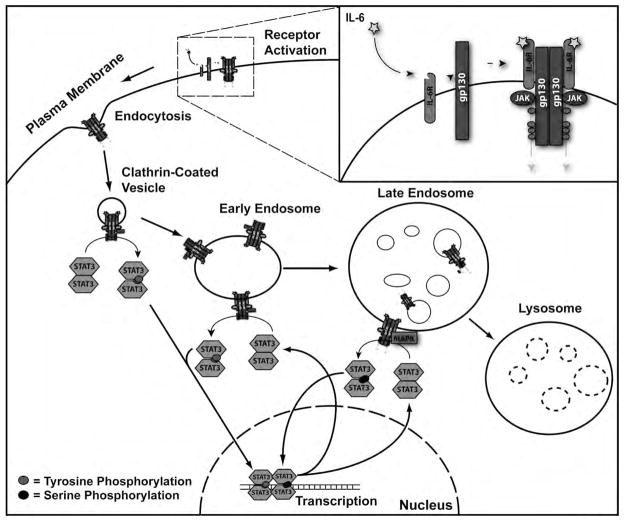
Our data support a model in which IL-6 ligand binding at the plasma membrane induces receptor complex internalization into an endosomal structure (Fig. 8). These endosomes then function as beacons of activation as they move through the endocytic pathway, directly and catalytically phosphorylating local cytoplasmic populations of STAT3 through brief, transient interactions. Activated STAT3 then utilizes a means of subcellular transportation separate from the endocytic pathway to reach the nucleus and initiate transcription. The rate of STAT3 diffusion is rapid, but short-lived [2]. Provided the point of STAT3 phosphorylation is within 200 nm of the final target, we propose local activation and diffusion away from IL-6 signaling endosomes as a probable mechanism for STAT3 nuclear localization. Previous work has shown STAT3 to constitutively shuttle between the cytoplasm and nucleus, and nuclear accumulation the consequence of a phosphorylation-induced conformational change within STAT3 that prevents nuclear export [15–17]. Recurrent, perinuclear activation of cytoplasmic STAT3 from an endosome would shift the balance between STAT3 nuclear import and export towards a higher rate of import, resulting in STAT3 nuclear aggregation. Upon reaching the late endosome, IL-6-receptor complexes induce MAPK signaling to further modify STAT3 signaling and transcriptional activity through S727 phosphorylation. Following transcription, STAT3 returns to the cytoplasm whereupon it is reactivated by JAK1 or MAPK upon IL-6 signaling endosomes. As activated STAT3 receptor complexes are degraded within the lysosome and protein inhibitors of STAT3 signaling are produced, STAT3 signal propagation is terminated. The disparate manner in which receptor tyrosine kinase and cytokine receptor systems utilize endosomes warrants substantially greater investigation into the role these structures play in signal transduction.
Figure 8.
Model of Endocytic Regulation of IL-6-Induced STAT3 Signaling.
Acknowledgments
We thank Dr. Bruce Horazdovsky, Dr. Zhiguo Zhang, Dr. Keith Bible, and Dr. Dan Billadeau for providing materials, Dr. Larry Pease and Dr. Jennifer Westendorf for the use of equipment, Mike Bell for his assistance with electroporation. Mary Pendergast, Reghann Corey, Kim Cook, Rachel Bergstrom and Dr. William Schmalstieg have been invaluable in their critical reading of this manuscript. This work was supported by Donald and Frances Herdrich (CLH) and an early career development award from Mayo Clinic (CLH). CLG was supported by the Mayo Graduate School and the Kern Predoctoral Neuroscience Fellowship.
References
- 1.Howe CL, Mobley WC. Long-distance retrograde neurotrophic signaling. Curr Opin Neurobiol. 2005;15:40–48. doi: 10.1016/j.conb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Howe CL. Modeling the signaling endosome hypothesis: why a drive to the nucleus is better than a (random) walk. Theor Biol Med Model. 2005;2:43. doi: 10.1186/1742-4682-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron. 2003;39:69–84. doi: 10.1016/s0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- 4.Beattie EC, Zhou J, Grimes ML, Bunnett NW, Howe CL, Mobley WC. A signaling endosome hypothesis to explain NGF actions: potential implications for neurodegeneration. Cold Spring Harb Symp Quant Biol. 1996;61:389–406. [PubMed] [Google Scholar]
- 5.Howe CL, Mobley WC. Signaling endosome hypothesis: A cellular mechanism for long distance communication. J Neurobiol. 2004;58:207–216. doi: 10.1002/neu.10323. [DOI] [PubMed] [Google Scholar]
- 6.Howe CL, Valletta JS, Rusnak AS, Mobley WC. NGF signaling from clathrin-coated vesicles: evidence that signaling endosomes serve as a platform for the Ras-MAPK pathway. Neuron. 2001;32:801–814. doi: 10.1016/s0896-6273(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 7.Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
- 8.Heerssen HM, Pazyra MF, Segal RA. Dynein motors transport activated Trks to promote survival of target-dependent neurons. Nat Neurosci. 2004;7:596–604. doi: 10.1038/nn1242. [DOI] [PubMed] [Google Scholar]
- 9.Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 10.Sorkin A. Internalization of the epidermal growth factor receptor: role in signalling. Biochem Soc Trans. 2001;29:480–484. doi: 10.1042/bst0290480. [DOI] [PubMed] [Google Scholar]
- 11.Ye H, Kuruvilla R, Zweifel LS, Ginty DD. Evidence in support of signaling endosome-based retrograde survival of sympathetic neurons. Neuron. 2003;39:57–68. doi: 10.1016/s0896-6273(03)00266-6. [DOI] [PubMed] [Google Scholar]
- 12.Kamimura D, Ishihara K, Hirano T. IL-6 signal transduction and its physiological roles: the signal orchestration model. Rev Physiol Biochem Pharmacol. 2003;149:1–38. doi: 10.1007/s10254-003-0012-2. [DOI] [PubMed] [Google Scholar]
- 13.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 14.Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol Biosyst. 2006;2:536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 15.Herrmann A, Vogt M, Monnigmann M, Clahsen T, Sommer U, Haan S, Poli V, Heinrich PC, Muller-Newen G. Nucleocytoplasmic shuttling of persistently activated STAT3. J Cell Sci. 2007;120:3249–3261. doi: 10.1242/jcs.03482. [DOI] [PubMed] [Google Scholar]
- 16.Pranada AL, Metz S, Herrmann A, Heinrich PC, Muller-Newen G. Real time analysis of STAT3 nucleocytoplasmic shuttling. J Biol Chem. 2004;279:15114–15123. doi: 10.1074/jbc.M312530200. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci U S A. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun W, Snyder M, Levy DE, Zhang JJ. Regulation of Stat3 transcriptional activity by the conserved LPMSP motif for OSM and IL-6 signaling. FEBS Lett. 2006;580:5880–5884. doi: 10.1016/j.febslet.2006.09.054. [DOI] [PubMed] [Google Scholar]
- 19.Schuringa JJ, Schepers H, Vellenga E, Kruijer W. Ser727-dependent transcriptional activation by association of p300 with STAT3 upon IL-6 stimulation. FEBS Lett. 2001;495:71–76. doi: 10.1016/s0014-5793(01)02354-7. [DOI] [PubMed] [Google Scholar]
- 20.Giordano V, De Falco G, Chiari R, Quinto I, Pelicci PG, Bartholomew L, Delmastro P, Gadina M, Scala G. Shc mediates IL-6 signaling by interacting with gp130 and Jak2 kinase. J Immunol. 1997;158:4097–4103. [PubMed] [Google Scholar]
- 21.Kim H, Baumann H. Dual signaling role of the protein tyrosine phosphatase SHP-2 in regulating expression of acute-phase plasma proteins by interleukin-6 cytokine receptors in hepatic cells. Mol Cell Biol. 1999;19:5326–5338. doi: 10.1128/mcb.19.8.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah M, Patel K, Mukhopadhyay S, Xu F, Guo G, Sehgal PB. Membrane-associated STAT3 and PY-STAT3 in the cytoplasm. J Biol Chem. 2006;281:7302–7308. doi: 10.1074/jbc.M508527200. [DOI] [PubMed] [Google Scholar]
- 23.Bild AH, Turkson J, Jove R. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. Embo J. 2002;21:3255–3263. doi: 10.1093/emboj/cdf351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kermorgant S, Parker PJ. Receptor trafficking controls weak signal delivery: a strategy used by c-Met for STAT3 nuclear accumulation. J Cell Biol. 2008;182:855–863. doi: 10.1083/jcb.200806076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.German CL, Howe CL. Preparation of biologically active subcellular fractions using the Balch homogenizer. Anal Biochem. 2009 doi: 10.1016/j.ab.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath CM, Wen Z, Darnell JE., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 27.Ramos JW. The regulation of extracellular signal-regulated kinase (ERK) in mammalian cells. Int J Biochem Cell Biol. 2008;40:2707–2719. doi: 10.1016/j.biocel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 29.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 30.Pedranzini L, Dechow T, Berishaj M, Comenzo R, Zhou P, Azare J, Bornmann W, Bromberg J. Pyridone 6, a pan-Janus-activated kinase inhibitor, induces growth inhibition of multiple myeloma cells. Cancer Res. 2006;66:9714–9721. doi: 10.1158/0008-5472.CAN-05-4280. [DOI] [PubMed] [Google Scholar]
- 31.Lim CP, Cao X. Regulation of Stat3 activation by MEK kinase 1. J Biol Chem. 2001;276:21004–21011. doi: 10.1074/jbc.M007592200. [DOI] [PubMed] [Google Scholar]
- 32.Ng YP, Cheung ZH, Ip NY. STAT3 as a downstream mediator of Trk signaling and functions. J Biol Chem. 2006;281:15636–15644. doi: 10.1074/jbc.M601863200. [DOI] [PubMed] [Google Scholar]
- 33.Haigler HT, Willingham MC, Pastan I. Inhibitors of 125I-epidermal growth factor internalization. Biochem Biophys Res Commun. 1980;94:630–637. doi: 10.1016/0006-291x(80)91279-6. [DOI] [PubMed] [Google Scholar]
- 34.Wang LH, Rothberg KG, Anderson RG. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weigel PH, Oka JA. Temperature dependence of endocytosis mediated by the asialoglycoprotein receptor in isolated rat hepatocytes. Evidence for two potentially rate-limiting steps. J Biol Chem. 1981;256:2615–2617. [PubMed] [Google Scholar]
- 36.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stenmark H, Olkkonen VM. The Rab GTPase family. Genome biology. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Punnonen EL, Ryhanen K, Marjomaki VS. At reduced temperature, endocytic membrane traffic is blocked in multivesicular carrier endosomes in rat cardiac myocytes. Eur J Cell Biol. 1998;75:344–352. doi: 10.1016/s0171-9335(98)80067-8. [DOI] [PubMed] [Google Scholar]
- 39.Yamashiro DJ, Maxfield FR. Acidification of endocytic compartments and the intracellular pathways of ligands and receptors. J Cell Biochem. 1984;26:231–246. doi: 10.1002/jcb.240260404. [DOI] [PubMed] [Google Scholar]
- 40.Saermark T, Flint N, Evans WH. Hepatic endosome fractions contain an ATP-driven proton pump. Biochem J. 1985;225:51–58. doi: 10.1042/bj2250051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshansky V, Futai M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol. 2008;20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galloway CJ, Dean GE, Marsh M, Rudnick G, Mellman I. Acidification of macrophage and fibroblast endocytic vesicles in vitro. Proc Natl Acad Sci U S A. 1983;80:3334–3338. doi: 10.1073/pnas.80.11.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Layer PG, Shooter EM. Binding and degradation of nerve growth factor by PC12 pheochromocytoma cells. J Biol Chem. 1983;258:3012–3018. [PubMed] [Google Scholar]
- 44.Saxena S, Howe CL, Cosgaya JM, Steiner P, Hirling H, Chan JR, Weis J, Kruttgen A. Differential endocytic sorting of p75NTR and TrkA in response to NGF: a role for late endosomes in TrkA trafficking. Molecular and cellular neurosciences. 2005;28:571–587. doi: 10.1016/j.mcn.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 45.Mollenhauer HH, Morre DJ, Rowe LD. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochimica et biophysica acta. 1990;1031:225–246. doi: 10.1016/0304-4157(90)90008-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nesbitt JE, Fuller GM. Dynamics of interleukin-6 internalization and degradation in rat hepatocytes. J Biol Chem. 1992;267:5739–5742. [PubMed] [Google Scholar]
- 47.Gonzalez FA, Raden DL, Davis RJ. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- 48.Chung J, Uchida E, Grammer TC, Blenis J. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol. 1997;17:6508–6516. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuringa JJ, Dekker LV, Vellenga E, Kruijer W. Sequential activation of Rac-1, SEK-1/MKK-4, and protein kinase Cdelta is required for interleukin-6-induced STAT3 Ser-727 phosphorylation and transactivation. J Biol Chem. 2001;276:27709–27715. doi: 10.1074/jbc.M009821200. [DOI] [PubMed] [Google Scholar]