Abstract
Amelogenesis imperfecta describes a group of inherited disorders that results in defective tooth enamel. Two disorders associated with human amelogenesis imperfecta are the point mutations T21→I or P40→T in amelogenin, the dominant protein present in ameloblasts during the early stages of enamel biomineralization. The biophysical properties of wildtype murine amelogenin (M180) and two proteins containing the equivalent mutations in murine amelogenin, T21→I (M180-I) and P41→T (M180-T), were probed by NMR spectroscopy. At low protein concentration (0.1 mM) M180, M180-I, and M180-T are predominately monomeric at pH 3.0 in 2% acetic acid and neither mutation produces a major structural change. Chemical shift perturbation studies as a function of protein (0.1 to 1.8 mM) and NaCl (0 to 400 mM) concentration show that the mutations affect the self-association properties by causing self-assembly at lower protein or salt concentrations, relative to wildtype amelogenin, with the largest effect observed for M180-I. Under both conditions, the premature self-assembly is initiated near the N-terminus, providing further evidence for the importance of this region in the self-assembly process. The self-association of M180-I and M180-T at lower protein concentrations and lower ionic strengths than wildtype M180 may account for the clinical phenotypes of these mutations, defective enamel formation.
Keywords: amelogenesis imperfecta, intrinsic disorder, amelogenin, enamel, biomineralization, nanospheres
Introduction
Dental enamel, the 1–2 mm layer of elongated, nanorod-like hydroxyapatite crystals on the outer surface of the tooth, is repeatedly exposed to masticatory, parafunctional, and occasional impact loading in the bacteria-filled environment of the mouth. Exposed to such stresses and unable to self-repair or remodel itself, it is not surprising that enamel is the hardest tissue in the vertebrate body [1,2]. The strength and robust mechanical properties of enamel are due to a unique combination of high mineral content and high degree of architectural organization [3]. Ninety-seven percent of mature enamel, by weight, consists of carbonated hydroxyapatite crystals elongated in the c-axial direction and tightly packed into parallel arrays, called enamel rods or prisms, that are intricately interwoven into an unique lattice [4,5]. The central protein guiding the nucleation, growth, and organization of these enamel rods into enamel is amelogenin [5–9]. This process, amelogenesis, occurs over three main stages (secretory, transition, and maturation) [10] starting with an amelogenin-filled (~90%) extracellular matrix in the secretory-stage [5–7,11,12]. During the transition-stage proteases, such as enamelysin (MMP-20) and kallikrein (KLK-4), cleave the amelogenin and at the end of the maturation-stage hard and tough enamel, largely devoid of organic material, remains.
Amelogenin is a low-molecular-weight protein (~ 20 kDa depending on the species) with amino and carboxyl terminal regions that are highly conserved among species [13]. Associated with the termini sequence conservation are charged amino acid residues, suggesting that these regions serve important functional roles for mediating enamel biomineralization [14]. Aside from the C-terminal region (the charged residues in the N-terminal region are dispersed), the protein is highly hydrophobic in nature and contains a central region enriched in the amino acids P, L, H, and Q (HQP-rich region). Indeed, variations in the length of the HQP-rich region largely accounts for the variations in size of amelogenin among different species. This hydrophobic region is responsible for one of the unique properties of amelogenin, its ability, under the correct environmental conditions in the secretory-stage, to self-assemble into a unique quaternary structure, 40 – 100 nm in diameter, called a nanosphere [12].
Nanosphere formation alters the physical properties of amelogenin (hydrophobicity and electrostatics) [5,15,16] and has been observed both in vivo [12] and in vitro [11]. These structures are believed to be biologically significant because mutations in amelogenin disrupt the properties and size of nanospheres in vitro [17–19] with some of the same mutations observed to cause malformed enamel in knock-in mice in vivo [20]. Nanosphere assembly is hypothesized to occur via a progressive accretion of amelogenin molecules into larger units [21,22], a process that depends on the interplay between protein concentration and the properties of the solution (ionic strength, pH, solutes, and temperature) [22–26]. Solution NMR studies tracking the self-assembly process in murine amelogenin suggests that assembly is initiated near the N-terminus (Y12-I51) followed by a region near the C-terminus (L141-T171) [26]. Regions near the N- and C-termini, M1-M42 and S157-K173, respectively, have been identified by yeast two-hybrid assays [27] as essential for nanosphere self-assembly. These same regions are highly conserved among species [13] and it has been postulated that upon nanosphere formation these termini are surface exposed [11,20,24,27], increasing the nanosphere’s solubility and enhancing its interactions with calcium phosphate [14]. Indeed, removing the C-terminus of amelogenin results in a protein with a reduced affinity for hydroxyapatite and a weaker ability to inhibit crystal growth [28,29].
Mutations to six genes (AMELX, ENAM, KLK4, MMP20, WDR72, and FAM83H) have been linked to diverse clinical phenotypes (hypomaturation, hypoplasia, and hypocalcification) associated with amelogenesis imperfecta [30], a group of hereditary conditions that affects the quantity and quality of enamel [31,32]. Out of the six amelogenesis imperfecta associated genes, the majority of genetic mutations identified to date are in AMELX [18] and include two separate missense mutations that result in single amino acid substitutions, T21→I or P40→T, in the primary sequence of human amelogenin. The phenotype of the T21→I mutation is hypomineralized/hypomatured enamel with brown discoloration while the phenotype of the P40→T is hypomatured enamel with discoloration. Because these two mutations are in the N-terminal region of amelogenin, a region that may play a direct role in the early stages of amelogenin self-assembly [26] and enamel formation [33], examining the biophysical properties of amelogenin containing these point substitutions may provide information on the molecular mechanism of defective enamel formation [18,34].
Previously, using two proteins containing the equivalent mutations in murine amelogenin, T21→I (M180-I) and P41→T (M180-T), optical spectroscopy (CD, fluorescence), transmission light spectroscopy, and dynamic light scattering were used to show that these mutations affected the self-assembly properties of amelogenin relative to the wild type (M180) [18,34]. To probe the finer molecular details of how these two point mutations affect self-assembly, M180, M180-I, and M180-T were characterized in solution using NMR spectroscopy under conditions that promote self-assembly, increasing protein concentration and ionic strength [22,26,35]. Such an analysis was assisted by the previous assignment of the backbone 1HN and 15N amide resonances for M180 [36]. These earlier assignments made it possible to identify the specific residues involved in the early stages of nanosphere assembly by following perturbations to their amide chemical shifts in the 1H-15 N HSQC spectra of M180, M180-I, and M180-T as a function of protein or NaCl concentration. Such perturbations, typically chemical shift and/or intensity reductions to the backbone 1HN and 15N resonances [26,37], reflect changes in the chemical environment of nuclei at interaction interfaces. To determine if the altered self-assembly properties of M180-I and M180-T may be due to dynamic changes affected by the point mutations, backbone amide relaxation measurements were also made for M180, M180-I, and M180-T under conditions where all three proteins were monomeric in solution (pH 3.0, 0.1 mM protein concentration). To assess the size of the major species in solution under conditions where 1H-15N HSQC spectra were no longer tractable (2000:1 molar ratio of NaCl:protein), dynamic light scattering (DLS) experiments were performed.
Materials and methods
All chemicals were purchased from Research Products International Corporation (Mount Prospect, IL) except trifluroacetic acid (TFA) and HPLC grade acetonitrile (Sigma Chemical Company, St. Louis, MI).
Protein expression and purification
The gene for full-length murine amelogenin [38], codon-optimized for Escherichia coli expression, was synthesized (Genescript, Piscataway, NJ) between NdeI and BamHI restriction endonuclease sites to allow insertion into the expression vector pET 11d (Novagen, Madison, WI). The gene was designed such that the gene product contained a 12-residue, uncleavable, N-terminal histidine tag (MRGSHHHHHHGS) to facilitate purification by metal-affinity chromatography. The parent amelogenin gene was used to generate two variant genes (Genescript, Piscataway, NJ) that translated into protein products containing a single amino acid substitution, T21→I or P41→T. The recombinant plasmids were transformed into E. coli BL21(DE3)pLysS cells (Novagen, Madison, WI) following a heat shock method. Using the antibiotics chloramphenicol (34 µg/mL) and ampicillin (100 µg/mL), uniformly 15N-labeled protein was prepared using an autoinduction protocol [39] and uniformly 13C- and 15N-labeled protein was prepared using a M9 minimal medium protocol [36]. The protein was purified using modifications to a previously reported protocol [40] beginning with the resuspension of the frozen cell pellet in 32 mL of NTA-wash buffer (8 M urea, 100 mM NaPO4, pH 8.2). The cells were then lysed by three passes through a French Press (SLM Instrument, Urbana, IL) followed by sonication for 1 min. Insoluble cell debris was removed by centrifugation at 25000g for 1 h and the supernatant loaded onto a 20 mL Ni-NTA affinity column (Qiagen, Valencia, CA). The column was washed sequentially with 40 mL of NTA-wash buffer followed by 40 mL of buffer (6 M urea, 100 mM NaPO4, pH 8.2) containing increasing concentrations of imidazole (10, 20, 50, 500 mM). The amelogenin eluted primarily in the fraction containing 500 mM imidazole. This fraction was applied onto a Varian Dynamax Microsorb 300-5 C-18 reverse phase column (250 × 21.4 mm) (Agilent Technologies, Santa Clara, CA) with the following purification gradient applied after loading the material using 100% Buffer A (Buffer A = water in 0.1% TFA, Buffer B = 70% aq. CH3CN in 0.1% TFA, flow rate 5 mL/min: Step 1 – 0.5 CV, 100% Buffer A; Step 2 – 0.2 CV, linear gradient 0 to 20% Buffer B; Step 3 – 2.5 CV, linear gradient 20 to 100% Buffer B). The fractions containing amelogenin (~ 80% Buffer B) were pooled and frozen (80 °C). After lyophilization, the material was resuspended in the final NMR buffer (2% CD3CO2D, 7% D2O/91% H2O, pH 3.0) [26] and the protein concentrations measured using the Bradford assay (BioRad, Hercules, CA) [41]. Yields of purified protein were ~40 mg/L using the M9 minimal medium protocol and ~70 mg/L using the autoinduction protocol.
NMR data collection and processing
All NMR data was collected at 20 °C using a Varian Inova-750 spectrometers and Biopack pulse programs. Three-dimensional HNCACB and HNN data were collected on the M180-I and M180-T samples to verify some amide chemical shift assignments that were otherwise based primarily on comparison to the 1H-15 N HSQC spectrum of previously assigned M180 [36]. For the dilution studies, initial 1.8 mM samples of amelogenin were prepared in the final NMR buffer using the Bradford assay [41] (BioRad, Hercules, CA) to estimate the protein concentration. The initial 1H-15 N HSQC was collected on a 250 µL sample in a Shigemi NMR tube (Shigemi Inc., Allison Park, PA) with subsequent spectra collected following serial dilutions to 0.9, 0.45, and 0.1 mM. For the NaCl titration studies, a 2 M NaCl stock solution was prepared in NMR buffer. Seven and one half microliter aliquots of the sodium chloride stock solution were added directly to the amelogenin sample (0.25 mM (300 µL)) and, following gentle agitation, a high resolution 1H-15N HSQC spectrum (20 °C) was immediately acquired. Spectra were recorded in the absence of salt and at NaCl:protein molar ratios of 400, 800, 1200, 1600, and 2000 to one. At the end of the titration, due to dilution, the final concentration of amelogenin and NaCl was 0.20 mM and 400 mM, respectively, in a final volume of 375 µL. Nitrogen-15 R1 and R2 values were measured using previously described experiments [42,43] with seven different time delays (s): 0.0, 0.1, 0.2, 0.3. 0.4, 0.6, 0.8 (R1); 0.01, 0.03, 0.05, 0.07, 0.09, 0.11, and 0.13 (R2).Values for R1 and R2 and an estimate of their associated errors were obtained using the rate analysis function in Sparky (v3.115) [44] by fitting the measured peak heights to a two-parameter exponential function, I(t) = I0exp(-t*R1,2). Steady-state {1H}-15N heteronuclear NOE values (NOE = Isat/Iunsat) were measured in triplicate from the ratios of 1H-15N HSQC cross peak volumes in spectra recorded in the presence (Isat ) and absence (Iunsat) of 2.5 seconds of proton presaturation prior to the 15N excitation pulse. All NMR data was processed using Felix2007 (Felix NMR, Inc, San Diego, CA) and Sparky [44] software with the 1H, 13C, and 15N chemical shifts referenced to DSS (DSS = 0 ppm) using indirect methods [45].
Dynamic light scattering (DLS)
The DLS measurements were obtained at 20 °C on M180, M180-I, and M180-T at concentrations of 0.2 mM in 2% acetic acid, 400 mM NaCl, pH 3.0, using a Brookhaven Instruments 90 Plus (Brookhaven Instruments Corporation, Holtsville, NY) equipped with a 657 nm 35 mW laser using a 90° scattering angle with acquisition times of 1 to 3 minutes. Protein solutions were mildly centrifuged prior to analysis.
Results
Structural effects of the point mutations
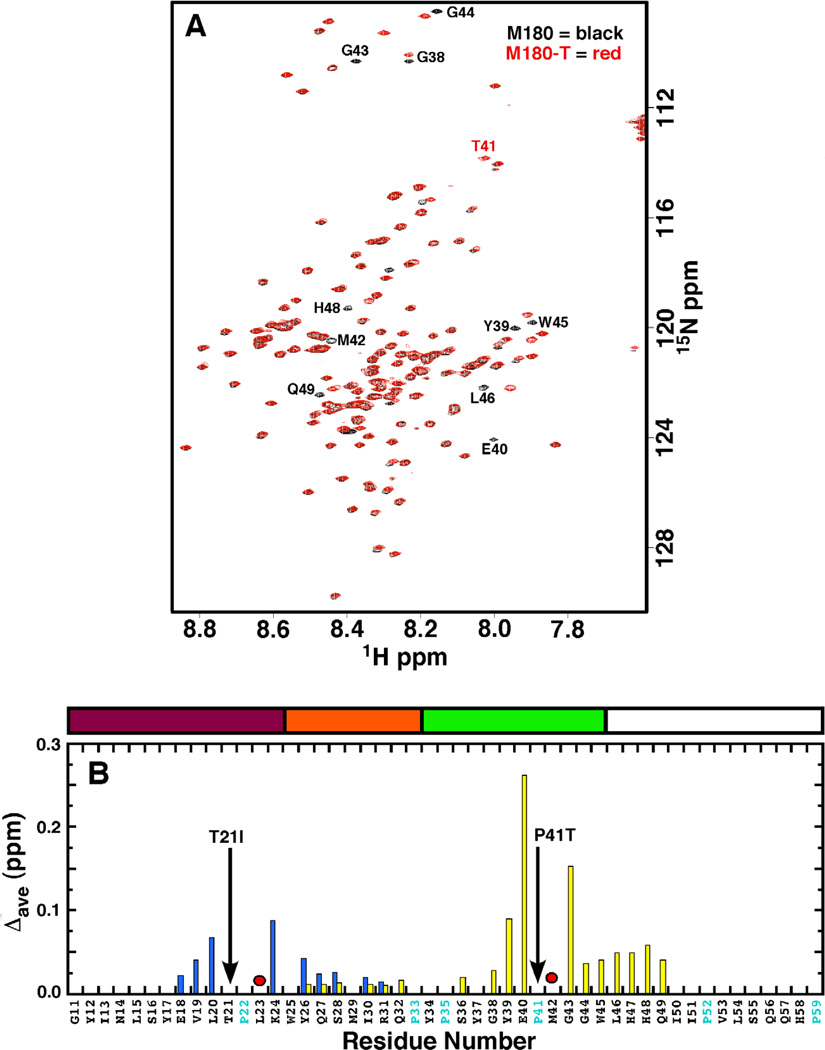
Figure 1A is an overlay of the 1H-15 N HSQC spectrum of M180 with M180-T under conditions where M180 was previously shown to be monomeric in solution (0.1 mM protein concentration, 2% acetic acid, pH 3, 20 °C) [26]. Aside from a few resonances, labeled in Fig. 1A, the spectra for M180 and M180-T overlay well, as did the 1H- N15 HSQC spectra for M180 and M180-I (data not shown), suggesting that the amino acid substitutions had little effect on the global structure of amelogenin. The similar amide line widths and chemical shifts, along with good magnetization transfer in the three-dimensional backbone assignment experiments, suggest that M180-I and M180-T are also monomeric in solution under these same conditions. The chemical shift differences are so slight it is possible to readily assign the 1H-15 N HSQC spectra of M180-I and M180-T primarily using the previous amide assignments for M180 [36]. Using these assignments it is possible to map the location of the resonances that do not overlay well onto the primary amino acid sequence of amelogenin by plotting the average combined chemical shift perturbation in the 1H-15N HSQC spectra of M180-1 and M180-T relative to M180 as shown in Fig. 1B. For both point mutations the chemical shift perturbations are small, less than 0.3 ppm, with only two residues perturbed greater than 0.1 ppm. All of the perturbed residues are clustered around the mutated residue with the perturbations slightly greater for M180-T then M180-I. Such a highly localized chemical shift effect as a result of a mutation is consistent with an instrinsically disordered protein, as reported for porcine [46] and murine [26] amelogenin. It is likely that local electronic differences due to the amino acid substitutions in the mutated proteins are responsible for the observed minor chemical shift perturbations.
Fig. 1.
(A) Overlay of the 1H-15N HSQC spectrum of M180 (black) and M180-T (red). Data collected under similar experimental conditions (0.1 mM protein concentration, 2% acetic acid, pH 3.0, 20 °C) at a 1H resonance frequency of 750 MHz. Residues with significantly perturbed chemical shifts are labeled. (B) Plot of the average combined chemical shift perturbation in the 1H and 15N resonances of M180- (blue) and M180-T (yellow) relative to M180 at a protein concentration of 0.1 mM. Significant chemical shift perturbations were only observed in the N-terminal region. The positions of the point mutations are shown with a solid black arrow. Amide resonances that could not be unambiguously tracked in the mutant proteins are identified with a red circle. The average chemical shift change = Δave = [((Δ1HN)2 + (Δ15N/5)2)/2)1/2 Above the plot is a schematic illustration of identified regions in the protein [16] (see Fig. 2 caption for description). Part of the murine amelogenin sequence is shown with the proline residues highlighted in cyan and the residues numbered sequentially starting at P2 (P2 -D180) after the 12-residue, N-terminal poly-histidine tag.
Protein concentration effects on self-assembly
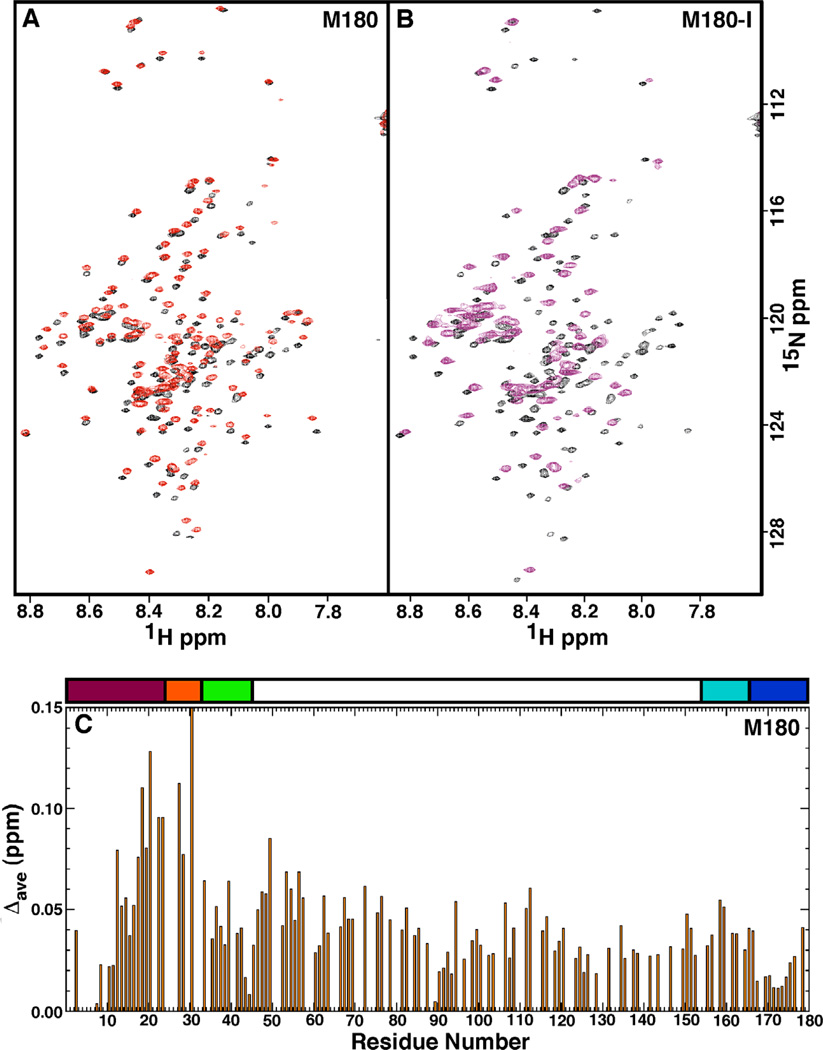
The acquisition of 1H-15 N HSQC spectra as a function of protein concentration is routinely used to assess concentration dependent self-association or transient aggregation of NMR samples. Here, it was clearly observed that the 1H-15 N HSQC spectra of M180, M180-I, and M180-T changed as a function of protein concentration indicating that aggregation occurred as the protein concentration increased. Figure 2A shows an overlay of the 1H-15 N HSQC spectra of M180 collected at 0.1 (black) and 1.8 (red) mM protein concentration. While the general pattern of the two 1H-15N HSQC spectra are similar, they do not overlay well indicating self-association is occurring at higher M180 concentrations. Furthermore, the line widths of the cross peaks in the 1.8 mM 1H-15N HSQC spectrum are slightly greater than for the 0.1 mM 1H-15 N HSQC spectrum, another feature characteristic of the formation of a larger molecular weight complex. The chemical shift perturbations were tracked, allowing the assignment of the 1H-15N HSQC spectrum collected at 1.8 mM M180 concentration. A plot of the average combined chemical shift perturbation in the 1HN and 15N resonances of M180 at 0.1 and 1.8 mM protein concentration is shown in Fig. 2C. The plot indicates that all the chemical shift perturbations are small, with the majority less than ~ 0.06 ppm and all the perturbations less than 0.15 ppm. The chemical shift perturbations that are greater than 0.06 ppm are clustered in the N-terminal region of the protein, primarily between I13 – E40.
Fig. 2.
(A) Overlay of the 1H-15N HSQC spectrum of M180 at a protein concentration of 0.1 mM (black) and 1.8 mM (red). (B) Overlay of the 1H-15N HSQC spectrum of M180-I at a protein concentration of 0.1 mM (black) and 1.8 mM (purple). Data collected under similar experimental conditions (0.1 mM protein concentration, 2% acetic acid, pH 3.0, 20 °C) at a 1H resonance frequency of 750 MHz. (C) Plot of the average combined chemical shift perturbation in the 1H and 15N resonances of M180 between a protein concentration of 0.1 mM and 1.8 mM. The average chemical shift change = Δave = [((Δ1HN)2 + (Δ15N/5)2)/2)]1/2. Above the plot is a schematic illustration of the various regions of the protein [16]. The majority of the protein, colored white, is composed of the HQP-rich region due to the many histidine, glutamine, and proline residues. The first N-terminal 45-residues consists of the tyrosine-rich amelogenin polypeptide (TRAP), a proteolytic fragment of the hydrolysis of amelogenin by metalloproteinase 20 (enamelysin). The TRAP region has been further sub-divided into the protein-protein interaction region (magenta), linker region (orange), and lectin-like binding tri-tyrosine region (green). The C-terminal contains a hydrophobic section that is also cleaved by enamelysin (cyan) and a hydrophilic mineral-binding region (blue).
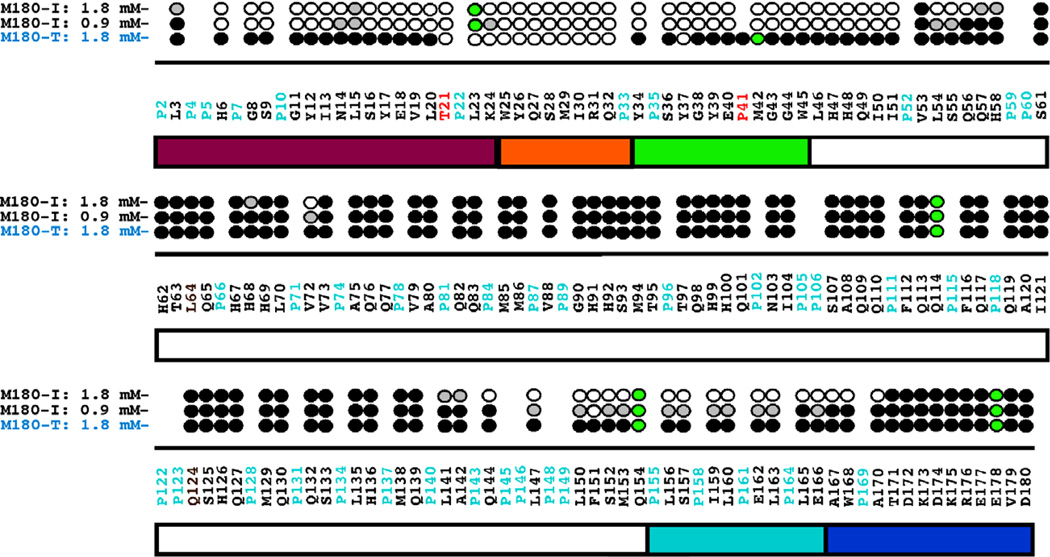
Figure 2B shows an overlay of the 1H-15N HSQC spectra of M180-I collected at 0.1 (black) and 1.8 (purple) mM protein concentration. In contrast to the features in the 1H-15N HSQC spectra of M180 over the same concentration range, many of the amide cross peaks have disappeared (59) and the line widths of those that remain are significantly broader for M180-I at the higher protein concentration. These latter two features indicate that M180-I has self-associated to form a larger molecular weight complex. The disappearance of amide cross peaks in the 1H-15N HSQC spectrum at high protein concentrations is likely due to restricted motion at a protein-protein interface as interpreted previously for M180 as a function of increasing NaCl and CaCl2 concentrations [26,47]. For M180-T over the same concentration range, the line widths of the amide cross peaks were also broader at the higher protein concentration, however, significantly fewer cross peaks disappeared completely (12, 1H-15N HSQC spectra not shown) suggesting that M180-T self-associated to a degree intermediate between M180 and M180-I. It was possible to track the disappearance of most of the amide cross peaks in the 1H-15N HSQC spectra over the concentration range and this data is summarized for M180-I and M180-T in Fig. 3. At 1.8 mM concentration, the amide resonances that disappear in the 1H-15N HSQC spectrum of M180-T are all localized near the N-terminus between T21 – Y37. In contrast, M180-I was much more sensitive to concentration. At only 0.9 mM most of the amide resonances near the N-terminus of M180-I (H6 – I51) have already disappeared or are significantly reduced in intensity and the intensity of a small swath of amide resonances have been reduced in a region near the C-terminus (L147 – E166). At 1.8 mM concentration, the C-terminal swath for M180-I has expanded to include amide resonances between L142 – A170 and the effected N-terminal region has grown to include amide resonances between L3 – H58. These results suggest that the mutated proteins self-assemble at a lower protein concentration than the wildtype protein with self-assembly initiated at lower protein concentration for M180-I than M180-T.
Fig. 3.
Summary of the amide resonances that partially disappear (grey-filled circles) or completely disappear (open circles) in the 1H-15N HSQC spectra of 15N-labelled M180-I and M180-T as a function of increasing protein concentration. Amide cross peaks whose intensity change little over the concentration range are indicated by solid circles and cross peaks that could not be tracked unambiguously are indicated by green-filled circles. The full murine amelogenin sequence is shown with the proline residues highlighted in cyan and the site of the point mutations highlighted in red.
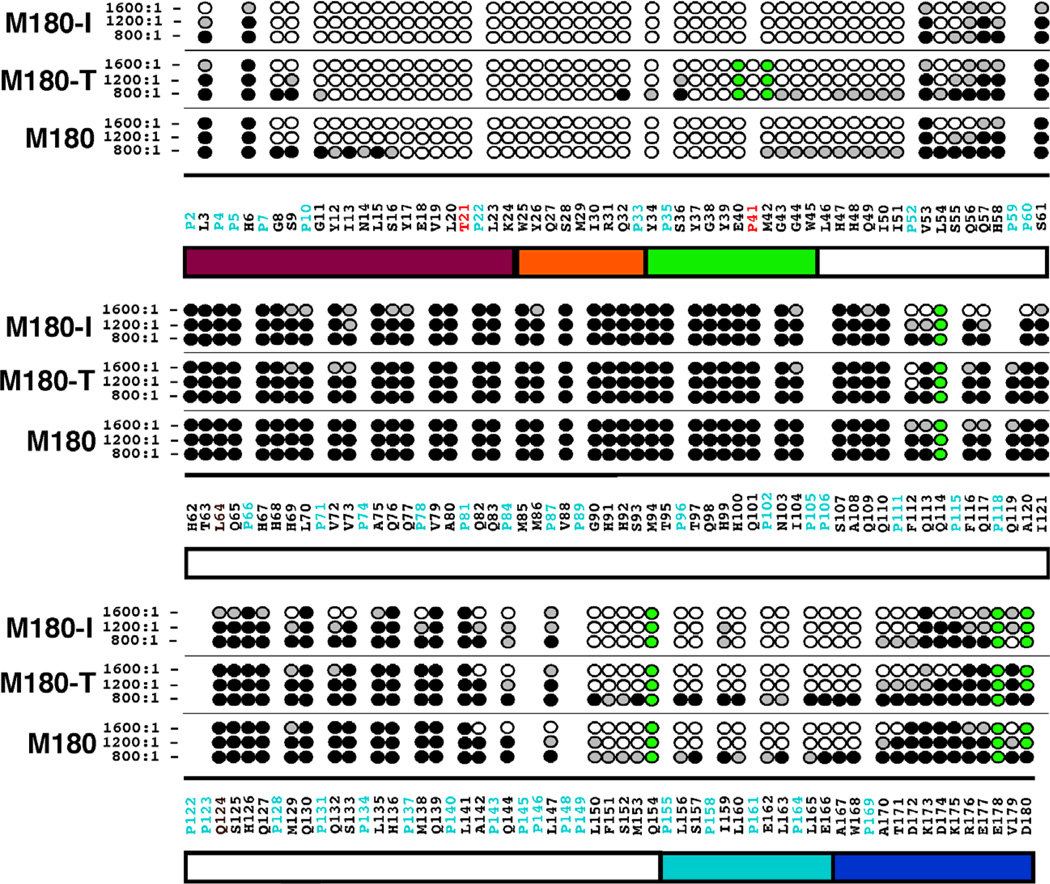
Sodium chloride effects on self-assembly
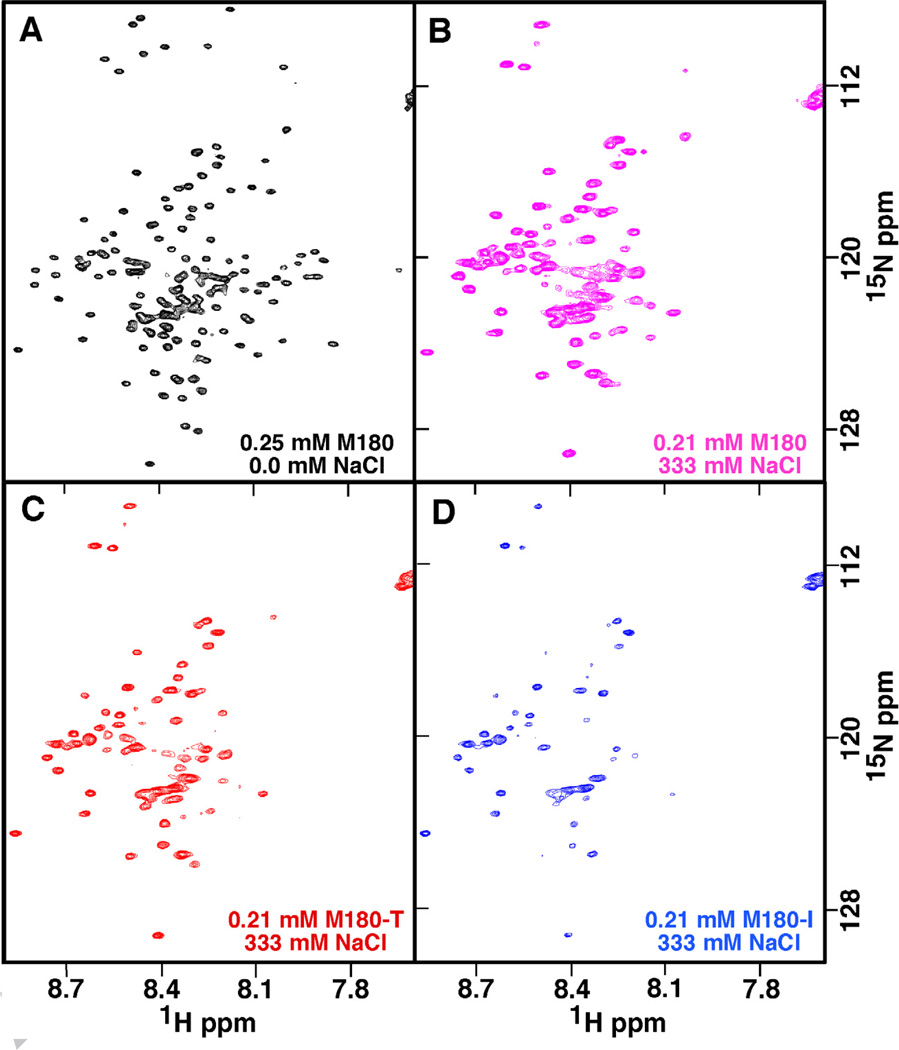
By monitoring the disappearance of amide cross peaks in the 1H-15N HSQC of M180 as a function of increasing NaCl or CaCl2 concentration, it was previously shown that increasing ionic strength affected a stepwise self-assembly of amelogenin into larger complexes [26]. This is illustrated for M180 in Figs. 4A and 4B. At a NaCl:M180 molar ratio of 1600:1, a significant number of the M180 amide cross peaks have disappeared and the line widths of the remaining resonances are broader. To determine if the point mutations M180-I and M180-T affected this salt-induced self-assembly process, similar NaCl NMR titration experiments were performed with M180-I and M180-T. For M180-T the substitution only marginally accelerated the self-assembly process. This is illustrated in the 1H-15N HSQC spectrum for M180-T at a 1600:1 molar ratio in Fig. 4C where a few additional amide cross peaks disappear or are weaker relative to the equivalent spectrum for M180 (Fig. 4B). On the other hand, for M180-I, NaCl significantly accelerated the self-assembly process. This is illustrated for M180-I at a 1600:1 molar ratio in Fig. 4D where significantly fewer amide cross peak are present relative to the equivalent spectrum for M180 (Fig. 4B).
Fig. 4.
The 1H-15N HSQC spectra of M180, M180-I, and M180-T at various NaCl concentrations illustrating the different effects NaCl has on the self-association properties of each protein. (A) Spectrum of M180 (0.25 mM) with no NaCl. (B – D) Spectra of 0.21 mM M180 (B), M180-T (C), and M180-I (D) collected at a salt:protein molar ratio of ~1600:1. All the data were collected under identical conditions (1H resonance frequency of 750 MHz, 20 °C, 2% acetic acid (pH 3.0)) and similarly processed. The spectra in B, C, and D are shown at identical contour levels.
The sequential disappearance of amide cross peaks were tracked as a function of the molar ratio of NaCl present, allowing the identification of regions of the protein responsible for initiating self-assembly. Figure 5 summarizes the NaCl titration data at three different NaCl:protein molar ratios, 800:1, 1200:1, and 1600:1. At the first titration point, 400:1, there is little change in the 1H-15N HSQC spectra for all three proteins aside from some mild line broadening (hence, this data is not shown). The 1H-15N HSQC spectra for M180 and M180-T are similarly affected by NaCl between 800:1 and 1600:1 with amide cross peaks disappearing primarily from a region near the N- and C-termini. At a 1600:1 molar ratio, the region affected at the N-terminus is the same (G8 – H58) while the affected C-terminal region is slightly larger in M180-T (A142 – K175) than in M180 (A142 – T171). On the other hand, over the same titration range, wider regions of the N- (L3 – S61) and C-termini (A142 – D180) of M180-I are affected. Furthermore, additional residues in the central portion of M180-I disappear, primarily those in a region from F112 – K138. At the final titration point (data not shown), a 2000:1 molar ratio, the 1H-15N HSQC spectrum for M180 changes little relative to the spectrum at 1600:1, and indeed, was observed to change little up to a molar ratio of 3000:1 where the species was previously interpreted to be a dimer or at most a trimer [26]. On the other hand, essentially all the amide resonances for M180-I and M180-T disappear into the “noise” at a 2000:1 molar ratio, observations that could be explained by the formation of complexes larger than dimers/trimers. In an attempt to verify this hypothesis, dynamic light scattering data was collected for all three proteins at the highest salt concentration (2000:1 molar ratio of NaCl:protein) to estimate the size of the major species in solution. These DLS data show no correlation between the diameter of the major species formed by self-assembly (M180-T (14.8 ± 0.4 nm) > M180 (12.9 ± 0.6 nm) > M180-I (11.9 ± 0.4 nm)) and the disappearance of amide cross peaks at a 2000:1 molar ratio (M180-I = M180-T > M180). The relatively narrow range of measured diameters for all three proteins, 12 – 15 nm, suggests that M180-I and M180-T also form dimer or trimer complexes at this salt concentration with the diameter differences possibly attributable to shape differences (eg: compact versus elongated) or equilibrium differences (eg: dimer versus trimer population). Consequently, the complete disappearance of all the amide resonances in the 1H-15N HSQC spectra of M180-I and M180-T is likely not a result of increased molecular weight but due to other factors such as a more rigid interface encompassing the central region of the protein or transient association of dimers and trimers.
Fig. 5.
Summary of the amide resonances that partially disappear (grey-filled circles) or completely disappear (open circles) in the 1H-15N HSQC spectrum of 15N-labelled M180, M180-I, and M180-T as a function of increasing NaCl concentration. Amide cross peaks whose intensity change little over the concentration range are indicated by solid circles and cross peaks that could not be tracked unambiguously are indicated by green-filled circles. The full murine amelogenin sequence is shown with the proline residues highlighted in cyan and the site of the point mutations highlighted in red. Underneath the primary sequence is a schematic illustration of the various regions of the protein (see Fig. 2 caption for description).
Backbone amide relaxation studies
To determine if the impaired self-assembly properties of M180-I and M180-T are related to dynamic changes affected by the amino acid substitutions, R1, R2, and heteronuclear NOE measurements were made for M180, M180-I, and M180-T under conditions where all three proteins were monomeric. Figures S1 and S2 illustrate the measured R1 and R2 values, respectively, for each tractable residue of M180, M180-I, and M180-T.Relative to the rest of protein, there are no significant differences in the R1 and R2 values at or near the site of the single T21→I or P41→T substitution. Table 1 provides a summary of the average R1 and R2 values for each of the three proteins. The average R1 values are all within the standard deviation for each protein with the average value for M180-I, 754 msec−1, smaller than the average for M180 and M180-T.Likewise, the average R2 values are all within the standard deviation for each protein with the average for M180-I, 214 msec−1 , smaller than the averages for M180 and M180-T. For each of the three proteins, the heteronuclear NOE values were all less than 0.6 with most of the values negative (data not shown). In general, heteronuclear NOE values less than ~0.6 are indicative of significant motion in the ps - ns timescale, a characteristic of a flexible, intrinsically disordered protein.
Table 1.
Average R1 and R2 values for M180, M180-I, and M180-T.
| Protein | R1 (ms−1) | R2 (ms−1) |
|---|---|---|
| M180 | 794 ± 98 | 290 ± 67 |
| M180-I | 754 ± 101 | 214 ± 82 |
| M180-T | 801 ± 104 | 275 ± 81 |
Discussion
At 0.1 mM protein concentration, the NMR data presented here show that M180-I and M180-T are monomeric, intrinsically disordered proteins and that the single T21→I or P41→T substitution has little, if any, effect on the protein’s structure. However, at higher protein and NaCl concentrations, the 1H-15N HSQC spectra of M180-I and M180-T were significantly different when compared to the spectrum for wildtype M180. These spectral differences at equivalent protein and NaCl concentrations indicated that the mutant proteins began to self-assemble at lower concentrations than M180 with the self-assembly occurring at earlier stages for M180-I than M180-T as the concentrations were increased. Measuring the degree of self-association simply on the basis of the number of amide cross peaks that disappear in the 1H-15N HSQC spectra in going from 0.1 mM to 1.8 mM yields an order of self-association of M180-I (59) > M180-T (12) > M180 (0) as a function of increasing protein concentration. A similar measurement in going from no salt to a 800:1 molar ratio of NaCl:protein would be M180-I (54) > M180-T (24) > M180 (21) as a function of increasing salt concentration. Another major spectral difference between the mutant proteins and M180 is at a NaCl:protein molar ratio of 2000:1 where essentially all the amide cross peaks in the 1H-15N HSQC spectra of M180-I and M180-T disappear while the 1H-15N HSQC spectrum of M180 at 1600:1 looks essentially the same at 2000:1 and at 3000:1 [26]. Given that the DLS data suggests that the size of all three protein complexes are roughly similar at a 2000:1 molar ratio, the NMR data shows the mutations alter the biophysical interactions of the assembled complexes in environments of high ionic strength.
An advantage of using NMR spectroscopy to study protein self-assembly is the ability to map the sequence location of the specific amino acid residues driving the process via chemical shift/intensity perturbation experiments [26,37]. As a function of increasing protein concentration, M180 displayed only chemical shift perturbations localized near the N-terminus (I13 – E40) while M180-I and M180-T both displayed significant intensity perturbations (disappearing cross peaks). For M180-T these changes were localized to a similar area near the N-terminus (T21 – Y37). For M180-I, intensity losses were observed near both the N-terminus (L3 – H58) and C-terminus (L142 – A180) with the N-terminus effected first at a lower protein concentration. Note that the effected residues are similar in all cases, but, in comparison to M180, onset occurs earlier for M180-I and somewhat earlier for M180-T as a function of protein concentration. The N-terminal region of M180 was also the region first effected in the process of self-assembly induced by increasing salt concentrations [26], a region identified in playing a role in porcine amelogenin self-assembly induced by 2,2,2-trifluoroethanol [48], and a region implicated in self-assembly at a more physiological pH (5.5) by NMR experiments using a series of truncated murine amelogenin sequences [49]. Hence, the concentration dependent data presented here adds to the growing body of evidence implicating the importance of the N-terminal region of amelogenin in the self-assembly process.
Previously, on the basis of fluorescence spectroscopy, CD, and DLS studies, both M180 and M180-T were reported to exist as monomers while M180-I formed small oligomers at ~ 0.02 mM protein concentrations [35]. The latter observations were made at pH 5.8 in 25 mM sodium acetate buffer at 25 °C, environments known to promote amelogenin self-assembly [26,35], and illustrate that M180-I has a greater predisposition to self-assembly than M180 and M180-T, consistent with the data presented here. While the NMR R1 and R2 relaxation data presented here show no unusual dynamic perturbations around the point mutations, it is interesting to note that lower average R1 and R2 values were observed for M180-I over M180 and M180-T that perhaps may be related to M180-I’s greater predisposition to self-assemble.
Hydrophobic intermolecular interactions are believed to be a major driving force responsible for protein association/aggregation [50]. Consequently, increasing the hydrophobicity of the N-terminus by replacing T21 and P41 with a more hydrophobic amino acid residue might be expected to facilitate self-association [35]. A greater tendency to self-assemble has also been observed with human amelogenin containing a single P70→T substitution [19]. Using the hydrophobicity index of Kyte and Doolittle [51], the T21→I substitution represents a 5.2 unit increase in hydrophobicity while the P41→T substitution is a smaller increase of 0.9 units. The approximately five-fold difference in hydrophobicity increase in M180-I over M180-T may account for the stronger tendencies of M180-I to self-associate over M180-T. Regardless of the physical explanation, the observation that M180-I and M180-T self-associate at lower protein concentrations and lower ionic strengths than M180 may account for the defective enamel associated with amelogenesis imperfecta. An increased tendency to self-associate could impair amelogenesis in a number of ways. For example, premature intracellular self-association may hinder amelogenin secretion, a mechanism recently reported for the Y34→H amelogenin mutation associated with amelogenesis imperfecta [52]. Alternatively, proteolytic processing and protein-mineral interactions, both hypothesized to involve regions near the N- and C-termini of amelogenin, may be impeded in a more tightly associated complex [18,34]. Indeed, a more tightly associated complex may hinder the disassembly of amelogenin nanospheres, a process that may be necessary for proper enamel formation [53,54].
Supplementary Material
Highlights.
The T21→I or P40→T point mutations alter amelogenin’s self-association properties.
Relative to wildtype amelogenin, self-assembly in the point mutations begin at lower ionic strength and protein concentrations.
Premature self-assembly initiated near the N-terminus.
Acknowledgements
This research was supported by NIH-NIDCH Grant number DE-015347. It was performed at the Pacific Northwest National Laboratory, a facility operated by Battelle for the U.S. Department of Energy, and included access to the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. DOE Biological and Environmental Research program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://
References
- 1.Ten Cate AR. Oral histology: developement, structure, and function. Mosby: St. Louis; 1994. [Google Scholar]
- 2.Shore RC, Robinson J, Kirkham J, Brookes SJ. In: Dental Enamel from Formation to Destruction. Robinson C, Kirkham J, Shore RC, editors. Boca Raton: CRC Press; 1995. p. 151. [Google Scholar]
- 3.White SN, Luo W, Paine ML, Fong H, Sarikaya M, Snead ML. J. Dent. Res. 2001;80:321–326. doi: 10.1177/00220345010800010501. [DOI] [PubMed] [Google Scholar]
- 4.Hunter G. Curr. Opin. Mat. Sci. 1996;1:430–435. [Google Scholar]
- 5.Margolis HC, Beniash E, Fowler CE. Crit. Rev. Oral Biol. Med. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 6.Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. J. Biol. Chem. 1990;255:9760–9768. [PubMed] [Google Scholar]
- 7.Uchida T, Tanabe T, Fukae M, Shimizu M, Yamada M, Miake K, Kobayashi S. Histochemistry. 1991;96:129–138. doi: 10.1007/BF00315983. [DOI] [PubMed] [Google Scholar]
- 8.Beniash E, Simmer JP, Margolis HC. J. Struct. Biol. 2005;149:182–190. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine ML, Shaw WJ. J. Cryst. Growth. 2007;304:407–415. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmer JP, Fincham AG. Crit. Rev. Oral Biol. Med. 1995;6:84–108. doi: 10.1177/10454411950060020701. [DOI] [PubMed] [Google Scholar]
- 11.Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC. J. Struct. Biol. 1994;112:103–109. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- 12.Fincham AG, Moradian-Oldak J, Diekwisch TGH, Lyaruu DM, Wright JT, Slavkin PBHC., Jr. J. Struct. Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 13.Toyosawa S, O'hUigin F, Figueroa F, Tichy H, Klein J. Proc Natl Acad Sci USA. 1998;95:13056–13061. doi: 10.1073/pnas.95.22.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moradian-Oldak J, Tan J, Fincham AG. Biopolymers. 1998;46:225–238. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 15.Dempsey CE. Biochim. Biophys. Acta. 1990;1031:143–161. doi: 10.1016/0304-4157(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 16.Moradian-Oldak J. Matrix Biol. 2001;20:293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- 17.Moradian-Oldak J, Paine ML, Lei YP, Fincham AG, Snead ML. J. Struct. Biol. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 18.Lakshminarayanan R, Bromley KM, Lei YP, Snead ML, Moradian-Oldak J. J. Biol. Chem. 2010;285:40593–40603. doi: 10.1074/jbc.M110.131136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu L, Uskokovic V, Le T, DenBensten P, Huang Y, Habelitz S, Li W. Arch. Oral Biol. 2011;56:331–336. doi: 10.1016/j.archoralbio.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Matrix Biol. 2001;20:273–292. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- 21.Wen HB, Fincham AG, Moradian-Oldak J. Matrix Biol. 2001;20:387–395. doi: 10.1016/s0945-053x(01)00144-5. [DOI] [PubMed] [Google Scholar]
- 22.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 23.Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. Biopolymers. 1994;34:1339–1347. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- 24.Moradian-Oldak J, Leung W, Fincham AG. J. Struct. Biol. 1998;122:320–327. doi: 10.1006/jsbi.1998.4008. [DOI] [PubMed] [Google Scholar]
- 25.Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. J. Struct. Biol. 2007;160:57–69. doi: 10.1016/j.jsb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchko GW, Tarasevich BJ, Bekhazi J, Snead ML, Shaw WJ. Biochemistry. 2008;47:6571–6582. doi: 10.1021/bi8018288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paine ML, Snead ML. J. Bone Min. Res. 1997;12:221–227. doi: 10.1359/jbmr.1997.12.2.221. [DOI] [PubMed] [Google Scholar]
- 28.Aoba T, Moreno EC. Calcif. Tissue Int. 1987;41:86–94. doi: 10.1007/BF02555250. [DOI] [PubMed] [Google Scholar]
- 29.Moradian-Oldak J, Bouropoulos N, Wang L, Gharakhanian N. Matrix Biol. 2002;21:197–205. doi: 10.1016/s0945-053x(01)00190-1. [DOI] [PubMed] [Google Scholar]
- 30.Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, Hart PS, Hart TC. Cells Tissues Organs. 2011;194:279–283. doi: 10.1159/000324339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witkop CJ, Jr., Kuhlmann W, Sauk J. Oral Surg. Oral Med. Oral Pathol. 1973;36:367–382. doi: 10.1016/0030-4220(73)90215-6. [DOI] [PubMed] [Google Scholar]
- 32.Ravassipour D, Hart PS, Hart TC, Ritter AV, Yamauchi M, Gibson C, Wright JT. J. Dent. Res. 2000;79:1476–1481. doi: 10.1177/00220345000790070801. [DOI] [PubMed] [Google Scholar]
- 33.Le Norcy E, Kwak S-Y, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. Cells Tissues Organs. 2011;194:188–193. doi: 10.1159/000324827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bromley KM, Lakshminarayanan R, Lei YP, Snead ML, Moradian-Oldak J. Cells Tissues Organs. 2011;194:284–290. doi: 10.1159/000324342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bromley KM, Kiss AS, Lokappa SB, Lakshminarayanan R, Fan D, Ndao M, Evans JS, Moradian-Oldak J. J. Biol. Chem. 2011;286:34643–34653. doi: 10.1074/jbc.M111.250928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchko GW, Bekhazi J, Cort JR, Valentine NB, Snead ML, Shaw WJ. Biomol. NMR Assign. 2008;2:89–91. doi: 10.1007/s12104-008-9092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuiderweg ERP. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 38.Snead ML, Lau EC, Zeichner-David M, Fincham AG, Woo SL, Slavkin HC. Biochem. Biophys. Res. Commum. 1986;129:812–818. doi: 10.1016/0006-291x(85)91964-3. [DOI] [PubMed] [Google Scholar]
- 39.Studier WF. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Simmer JP, Lau EC, Hu CC, Bringas P, Santos V, Aoba T, Lacey M, Nelson D, Zeichner-David M, Snead ML, Slavkin HC, Fincham AG. Calcif. Tissue Int. 1994;54:312–319. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- 41.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 42.Farrow NA, Muhandiram DR, Singer AU, Pascal SM, Kay LE, Gish G, Shoelson SE, Pawson T, Forman-Kay JD. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 43.Buchko GW, Daughdrill GW, de Lorimier R, Rao BK, Isern NG, Lingbeck JM, Taylor JS, Wold MS, Gochin M, Spicer LD, Lowry DF, Kennedy MA. Biochemistry. 1999;38:15116–15128. doi: 10.1021/bi991755p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; 1999. [Google Scholar]
- 45.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 46.Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, Evans JS. Biochemistry. 2009;48:2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buchko GW, Tarasevich BJ, Roberts J, Snead ML, Shaw WJ. Biochim. Biophys. Acta. 2010;1804:1768–1774. doi: 10.1016/j.bbapap.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ndao M, Dutta K, Bromley KM, Lakshminarayanan R, Sun Z, Rewari G, Moradian-Oldak J, Evans JS. Protein Sci. 2011;20:724–734. doi: 10.1002/pro.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Ramirez BE, Liao X, Diekwisch TGH. PLos ONE. 2011;6:e24952. doi: 10.1371/journal.pone.0024952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller S, Janin J, Lesk AM, Chothia C. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 51.Kyte J, Doolittle RF. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 52.Barron MJ, Brookes SJ, Kirkham J, Shore RC, Hunt C, Mironov A, Kingswell NJ, Maycock J, Shuttleworth CA, Dixon MJ. Hum. Mol. Genet. 2010;19:1230–1247. doi: 10.1093/hmg/ddq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tarasevich BJ, Lea S, Shaw WJ. J. Struct. Biol. 2010;169:266–276. doi: 10.1016/j.jsb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarasevich BJ, Perez-Salas U, Masica DL, Philo J, Kienzle P, Krueger S, Majkrzak CF, Gray JL, Shaw WJ. J. Phys. Chem. B. 2013;117:3098–3109. doi: 10.1021/jp311936j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.