Abstract
Melanocortin receptor accessory proteins (MRAPs) modulate signaling of melanocortin receptors in vitro. To investigate the physiological role of brain-expressed Melanocortin 2 Receptor Accessory Protein 2 (MRAP2), we characterized mice with whole body and brain-specific targeted deletion of Mrap2, both of which develop severe obesity at a young age. Mrap2 interacts directly with Melanocortin 4 Receptor (Mc4r), a protein previously implicated in mammalian obesity, and it enhances Mc4r-mediated generation of the second messenger cyclic AMP, suggesting that alterations in Mc4r signaling may be one mechanism underlying the association between Mrap2 disruption and obesity. In a study of humans with severe, early-onset obesity, we found four rare, potentially pathogenic genetic variants in MRAP2, suggesting that the gene may also contribute to body weight regulation in humans.
Membrane-expressed G protein-coupled receptors (GPCRs) modulate cellular responses to numerous physiological stimuli. The melanocortin receptors (MCRs) are a subfamily of GPCRs that mediate signaling in response to the pro-opiomelanocortin-derived peptides, adrenocorticotropic hormone (ACTH) and α-melanocyte–stimulating hormone (αMSH) and their competitive antagonists, agouti and agouti-related protein. The MCRs mediate a diverse range of physiological functions: MC1R is involved in skin pigmentation, MC2R plays a critical role in the hypothalamic-pituitary-adrenal axis, MC3R and MC4R are involved in energy homeostasis and MC5R is implicated in exocrine function (1).
There is increasing recognition that accessory proteins can modulate GPCR trafficking, as well as ligand binding and signaling (2). An accessory protein for MC2R, MC2R accessory protein (MRAP), is required for the trafficking of MC2R to the surface of adrenal cells and for signaling in response to ACTH (3, 4). Loss of either MC2R or MRAP in humans causes severe resistance to ACTH, with resulting glucocorticoid deficiency (5, 6).
All mammals have a paralogous gene, MRAP2, which, like MC3R and MC4R, is predominantly expressed in the brain (7), most prominently in the pons and cerebellum, but including in regions involved in energy homeostasis such as the hypothalamus and brainstem (Fig. S1, A–C). Within the paraventricular nucleus of the hypothalamus (PVN), Mrap2 and Mc4r mRNAs are co-expressed in many cells (Fig. S1D). We hypothesized that Mrap2 might modulate signaling through a melanocortin receptor and potentially affect energy homeostasis. We therefore performed targeted deletion of Mrap2 in mice, using Cre-lox-mediated excision of the 100 bp exon 3 (which encodes the highly conserved transmembrane domain (7)) to create mice with normal levels of an mRNA predicted to encode a truncated protein that includes the first 55 amino acids of Mrap2, with the transmembrane domain replaced by 11 aberrant amino acids specified by the out-of-frame exon 4, followed by a stop codon (Fig. S1, E–H). Normal levels of the mutant mRNA indicate preservation of Mrap2-containing neurons in null mice, although these neurons likely do not express the predicted mutant protein, since mutant Mrap2 mRNA, but not protein, is present in cells transfected with the same Mrap2 mutant construct used to create the null mice (Fig. S1I).
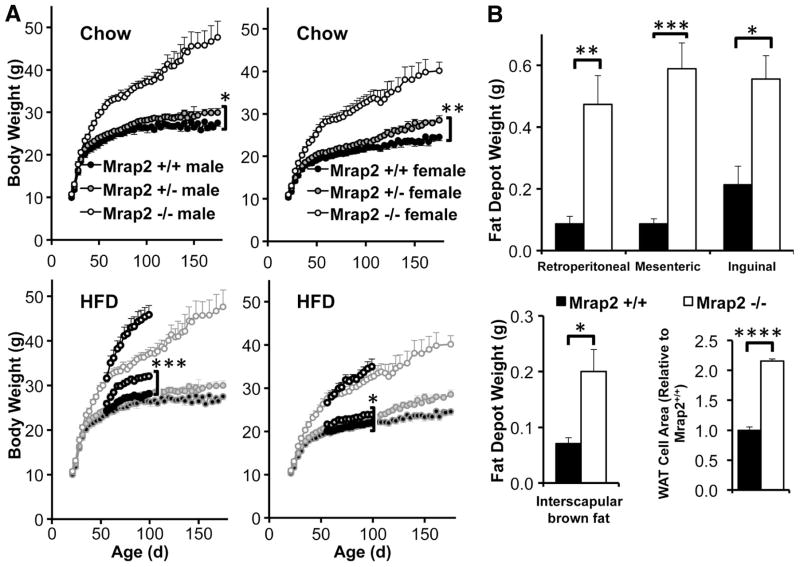
Mrap2 null mice appeared normal at birth, with normal weight gain and post-weaning food intake during early life (0–32 days and 23–32 days, respectively), although young Mrap2−/− male mice trended toward greater weight and food intake with advancing age (Fig. S1J). However, null mice of both genders gradually became extremely obese on a diet of regular chow ad libitum (Fig. 1A, S2A). Heterozygous mice were significantly heavier than wild-type animals on standard chow (160–175 days; males, Mrap2+/+ 26.0±0.4 g, Mrap2+/− 29.9±0.9 g; females Mrap2+/+ 24.5±0.9 g, Mrap2+/− 28.1±0.7 g), and at younger ages (56–95 days) on a high fat diet (Fig. 1A). In addition, Mrap2−/− mice had increased length (Fig. S1K) and per cent of weight due to fat, and decreased per cent of weight due to lean mass (Fig. S1L). Both genders of Mrap2−/− mice had increased visceral adiposity, over twice the normal white adipose tissue cell size, enlarged brown adipose tissue depots, normal liver histology on a regular chow diet, but much greater hepatic steatosis compared with wild-type mice on a high fat diet (Fig. 1B, Fig. S2, A–B). Adult Mrap2 null mice had, as expected, elevated leptin concentrations corresponding to their increased fat mass, which normalized with diet-induced weight normalization (Fig. S2C). Obese adult mice had normal fasting insulin (Fig. S2D) and normal tolerance to intraperitoneal glucose injection (Fig. S2E). Mrap2 has been postulated to play a role in the adrenal response to ACTH (8). We therefore measured diurnal rhythmicity and stress responsiveness of the adrenal axis in Mrap2 null mice, which were normal (Fig. S2F). Thyroid hormone levels were also normal (Table S1). Epinephrine and norepinephrine excretion were reduced in male Mrap2−/− mice only (Fig. S2G), but Ucp1 mRNA concentrations increased appropriately in both genders of null mice following exposure to 4°C for 18h (Fig. S2H). Hypothalamic Agrp mRNA concentration was reduced in Mrap2 null mice, whereas Pomc mRNA was normal (Fig. S2I).
Fig. 1.
Phenotype of Mrap2−/− mice. (A) Weight curves for Mrap+/+ vs. Mrap+/− vs. Mrap2−/− mice on standard chow (Chow, upper panels: male n= 9 vs. 28 vs. 15, female n=12 vs. 18 vs. 10) or high fat diets (HFD, ages 56–95 days, lower panels, superimposed on standard chow curves: male n= 10 vs. 8 vs. 10, female n=7 vs. 12 vs. 7). For both genders the weight curves of Mrap+/+ and Mrap+/− mice on standard chow differ significantly at older (161–175 days) ages, and at younger ages (56–95 days) on a high fat diet. ). *p=.02, **p=.001, ***p=.0003. (B) Fat depots on standard chow diet. Upper panel: White adipose tissue (WAT) weights in Mrap+/+ vs. Mrap2−/− (males and females, ages 117–122 days, n=5 vs. 4, respectively). Lower left panel: BAT weight in Mrap+/+ vs. Mrap2−/− mice (males and females, ages 117–122 days, n=5 vs. 4). Lower right panel: WAT cell size in in Mrap+/+ vs. Mrap2−/− mice (females, 50 cells counted from each mouse). *p=.009, **p=.003, ***p=.0003, ****p<.00001.
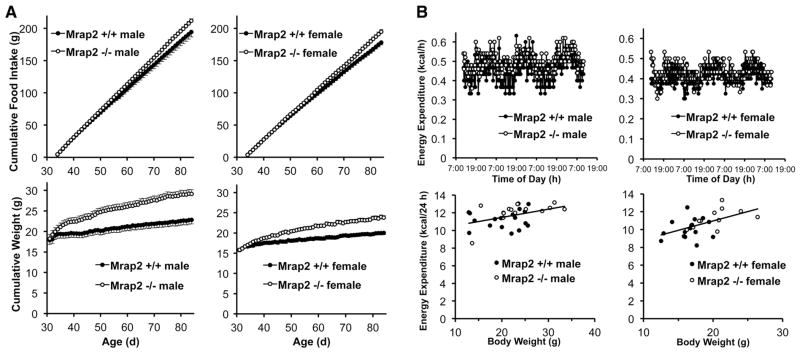
To characterize the mechanisms underlying the obesity in these mice, we measured food intake under a variety of conditions. At 42 (Fig. S2J) and 84 (Fig. S2K) days of age, when Mrap2−/− mice were clearly overweight, no difference in food intake was detected between the two genotypes when analyzed over a 4-day interval. Obesity was not caused by more efficient absorption of calories in null mice (Fig. S2L). Only when monitored daily over 50 days (ages 34–84 days) was a subtle increase in cumulative food intake discernable in the null animals (Fig. 2A), with the onset of obesity preceding hyperphagia (Fig. 2A and Fig. S2M). To further understand the contribution of hyperphagia to obesity in Mrap2−/− mice, we limited their food intake to that amount consumed by their normal siblings (pair feeding). Even when fed the same amount of chow, null mice gained more weight than did wild-type mice (Fig. S2N and Fig. S2O). Only when the amount of food intake in null mice was further restricted to 10% (females) and 13% (males) less than that of wild-type mice was there equivalent weight gain (Fig. S2P) in the two genotypes. To determine whether the late onset hyperphagia in Mrap2−/− mice (Fig. 2A) could simply be the consequence of an increased body mass at this older age caused by a separate metabolic defect, we switched null mice to ad libitum access to chow after 40 days of restricted feeding (upward arrow, Fig. S2P). During the first 24 h of ad libitum feeding, food intake almost doubled in null mice (from 2.9±0.1 to 5.6±0.5 g/d in males, and from 2.8±0.1 to 5.3±0.2 g/d in females), with a corresponding marked increase in body weight. Thus, hyperphagia develops in an age-dependent manner in older mice, independent of body weight. Consistent with this, young (age 38–45 days) Mrap2−/− mice had an intact anorectic response to the melanocortin receptor (Mc4r and Mc3r) agonist, MTII (Fig. S2Q), corresponding to their normal ad libitum food intake at this age.
Fig. 2.
Energy balance in Mrap2−/− mice. (A) Cumulative food intake (upper panels) and weight (lower panels) in ad libitum fed Mrap+/+ vs. Mrap2−/− males (n=10 vs. 11) and females (n=11 vs. 8). (B) Energy expenditure in ad libitum-fed Mrap+/+ vs. Mrap2−/− mice. Upper panels, continuous measurement over 3 days, males (n=3 vs. 4), females (n=4 vs. 3), ages 30–34 days. Lower panels: body weight vs. energy expenditure, integrated over 24 h, males (n=18 vs. 14, ages 30–45 days), females (n=16 vs. 11, ages 30–42 days). Analysis by ANCOVA showed no differences between genotypes (males, p=.38, females, p=.67).
We hypothesized that young Mrap2−/− mice might display abnormal energy expenditure because obesity develops early during ad libitum feeding prior to the onset of hyperphagia, persists in mutant mice pair-fed to a normal dietary intake, and is abolished only by underfeeding. To explore this, we measured energy expenditure and respiratory exchange ratio (RER) by indirect calorimetry, as well as locomotor activity and core body temperature, in young (30–45 days of age) wild-type and Mrap2 null mice, just as their weights began to diverge (Fig. 2A). Surprisingly, the wild-type and mutant mice had indistinguishable 24h total energy expenditure, as analyzed by ANCOVA (9) (Fig. 2B). There were also no differences between Mrap2+/+ and Mrap2−/− mice in RER (Fig. S2R), locomotor activity (Fig. S2S), or core body temperature at 22°C (Fig. S2T), with both genotypes exhibiting the expected increase in all three parameters during the active night period. Following exposure to 4°C for 18h, null and wild-type mice became significantly hypothermic to the same extent (Fig. S2T).
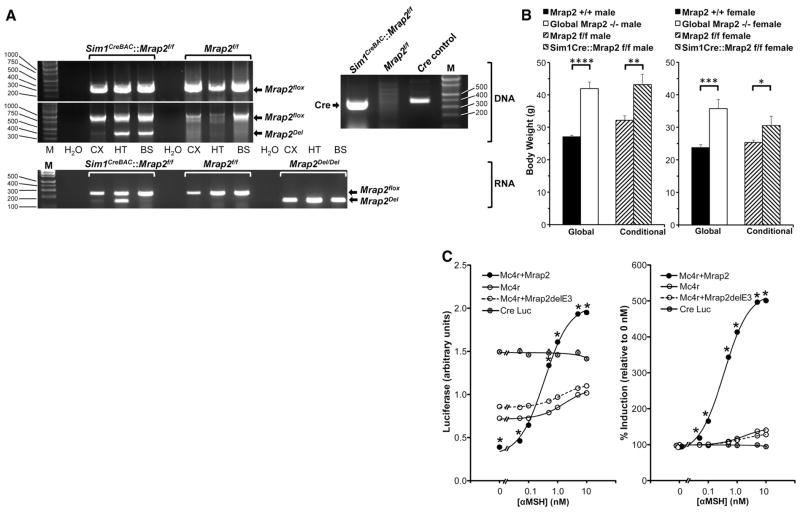
Since 1) MRAP is essential for signalling through MC2R (3, 4), 2) MRAP’s paralog, Mrap2, is expressed principally in the brain, and 3) Mc2r’s paralog, Mc4r, has a key role in energy balance in Sim1-containing neurons (10), we asked whether deletion of Mrap2 causes obesity in part by altering signaling through centrally-expressed Mc4r. We created a Sim1Cre::Mrap2flox/flox mouse with conditional deletion of Mrap2 exclusively in these neurons, and expression of Mrap2Del mRNA only in hypothalamus and not cerebral cortex or brainstem (Figs. S3A, 3A). Like global null mice, conditional mutants were similarly obese (Fig. 3B), and pair-feeding to a normal dietary intake only partially reversed their obesity (Fig. S3B).
Fig. 3.
Interaction between Mrap2 and Mc4r. (A) Conditional deletion of Mrap2 in Sim1neurons. Top right, Cre DNA analysis by PCR. HT DNA from Sim1CreBAC::Mrap2f/f mice contains Cre (374 bp), but from Mrap2f/f mice does not. Molecular weight marker (M) on right (bp). Top left, Mrap2 DNA analysis in Sim1CreBAC::Mrap2f/f and Mrap2f/f mice by PCR. Both genotypes contain floxed, intact Mrap2 DNA in CX, HT, and BS (314 bp in upper electropherogram, and 1013 bp in lower electropherogram, molecular weight markers on left). Only Sim1CreBAC::Mrap2f/f mice contain Mrap2Del (400 bp, lower electropherogram), and only in HT and BS, but not in CX, consistent with fluorescent reporter data (Fig. S3A). No PCR products are present without added DNA (H2O). Bottom, Mrap2 mRNA expression in Sim1CreBAC::Mrap2f/f and Mrap2f/f mice by RT-PCR. Both genotypes express floxed, intact Mrap2 mRNA in CX, HT, and BS (247 bp). Only Sim1CreBAC::Mrap2f/f mice express Mrap2Del mRNA (147 bp), and only in HT. Global Mrap2Del/Del mice express Mrap2Del mRNA in all 3 sites. (B) Body weights of Mrap2+/+ (male n=6, female n=11), Mrap2−/− (male n=11, female n=7), Mrap2f/f (male n=8, female n=12) and conditional Sim1CreBAC::Mrap2f/f (male n=8, female n=7) mice, all age 133 days. *p=.04, **p=.007, ***p=.0002, ****p<.0001. (C) Effect of Mrap2 on Mc4r signaling. Left panel: Level of cAMP reporter activity (CRELuc) in CHO cells alone, or co-transfected with Mc4r, with or without Mrap2 or the Mrap2 knockout construct, Mrap2delE3, 5 h following exposure to 0–10 nM αMSH (n=3/group). Right panel: cAMP activity of these same constructs, expressed as percent induction following 0–10 nM αMSH, relative to 0 nM αMSH. *p<.0001, Mc4r+Mrap2 vs. Mc4r at same [αMSH], by ANOVA. For most data points, error bars are obscured by symbols.
If Mrap2 facilitates the action of Mc4r, then Mc4r deficiency should create an equivalent or more severe obesity phenotype than does Mrap2 deficiency, depending on the degree to which Mrap2 interferes with Mc4r function. Supporting this, Mrap2+/− mice of both genders were less obese than either Mc4r+/− or doubly heterozygous mice (Fig. S3C). The differences between Mc4r+/− and doubly heterozygous mice were not statistically significant, although the latter trended toward being heavier. Among homozygous knockouts, those with Mc4r deficiency alone were more obese than those with Mrap2 deficiency alone (Fig. S3C). The Mc4r knockout mice were more obese than mice with deletion of both Mc4r and Mrap2 (in males, with a trend in females), suggesting that Mrap2 may promote weight gain through both Mc4r-dependent and Mc4r-independent actions.
To determine whether mouse Mrap2 and Mc4r can interact directly, we co-immunoprecipitated transiently expressed, N-terminally Myc-tagged Mrap2 and N-terminally GFP-tagged Mc4r in CHO cells (devoid of endogenous Mrap, Mrap2 and melanocortin receptors). We found that mouse Mrap2 and Mc4r interact (Fig. S3D), consistent with previous data (7). We next investigated the impact of Mrap2 on Mc4r (Fig. 3C) and Mc3r (Fig. S3E) signaling. The combined expression of Mc4r and Mrap2 in CHO cells suppressed basal PKA signaling compared with Mc4r alone (Fig. 3C, left panel), as previously reported with the human orthologs (7). But in contrast to that report (which used NDP-MSH), we found that αMSH caused a 5-fold increase above basal PKA activity (Fig. 3C, right panel) compared with less than a 2-fold increase with Mc4r alone or Mc4r plus the Mrap2 null construct, Mrap2delE3 (our in vitro model for in vivo disruption of Mrap2). The presence of Mrap2 increased signaling through Mc3r at the two highest αMSH doses (Fig. S3D). These findings suggest Mrap2 may alter signalling through Mc4r and perhaps other receptors.
To investigate whether alterations in MRAP2 are associated with human obesity, we sequenced the coding region and intron/exon boundaries of MRAP2 in obese and control individuals from the Genetics of Obesity Study (GOOS) cohort (11) and the Swedish obese children’s cohort (12). Four rare heterozygous variants were found in unrelated, nonsyndromic, severely obese individuals, that were absent from cohort-specific controls and 1000 genomes (Table), with all but one in the C-terminal region of the protein (Fig. S4). In three of these subjects, no pathogenic variants were found in the coding region or intron/exon boundaries of all known nonsyndromic human obesity genes (Table S2). Only one of the variants (E24X) is clearly disruptive, and overall few rare variants were found in the obese cohorts, indicating that MRAP2 mutations, if they contribute to severe human obesity, do so rarely.
Table .
MRAP2 variants detected in obese subjects and controls
| MRAP2 variant | Subjects with variant | * Subject Sex/Age/BMI/BMI SDS | Controls with variant | ** MAF: European American | ** MAF: African American | *** Polyphen prediction |
|---|---|---|---|---|---|---|
| E24X | 1/488 | M/19/63/4.7 | 0/488 | 0.000% (0/8600) | 0.000% (0/4406) | damaging |
| N88Y | 1/376 | M/11/29.6/3.3 | 0/376 | 0.000% (0/8600) | 0.000% (0/4406) | possibly damaging |
| L115V | 1/488 | M/5/24/4.2 | 0/488 | 0.012% (1/8600) | 0.000% (0/4406) | benign |
| R125C | 1/488 | F/8/29/3.5 | 0/488 | 0.047% (4/8600) | 0.045% (2/4406) | possibly damaging |
Subject Sex (Male[M], Female[F])/Age (y)/BMI (kg/m2)/SDS (standard deviation score)
MAF (minor allele frequency, NHLBI exome variant server http://evs.gs.washington.edu/EVS/)
Polyphen (http://genetics.bwh.harvard.edu/pph2/)
In summary, we have found that global or brain-specific inactivation of Mrap2 causes obesity in mice and that rare heterozygous variants in MRAP2 are associated with early-onset, severe obesity in humans. The mechanism(s) by which Mrap2 exerts its effects on body weight regulation remain to be firmly established but likely involve altered signaling through Mc4r and perhaps other melanocortin receptors. Under conditions comparable to those we describe whereby Mrap2 greatly enhances cAMP signaling through Mc4r, Sebag et al. (new reference) have found that the zebrafish ortholog of Mrap2 (zMRAP2b) similarly affects zMC4R signaling. This evolutionary conservation, plus the extreme disease phenotype caused by loss of Mrap2 function, supports the importance of Mrap2 in vertebrate biology.
Supplementary Material
Acknowledgments
We thank T. Nguyen for DNA analysis, H. Feldman and A. Fleisch for statistical advice; M. Mulcahey for thyroid assays, H. Turkova for catecholamine assays, S. Cabi for creating the software program used to analyze calorimetry data, M. Geibel for bioinformatics analyses, and D. Margulies, B. Lowell, J. Flier, and Boston Children’s Hospital Endocrinology Division scientists for helpful discussions. We are indebted to the patients and their families for their participation and to the physicians involved in the Genetics of Obesity Study (GOOS) and the Swedish obese children’s cohort study. This work was supported by grants from the National Institutes of Health (J.A.M.), the Timothy Murphy Fund (J.A.M.), NARSAD (M.A.), the Wellcome Trust (I.S.F., S.O’R.), Medical Research Council (I.S.F., L.F.C. [grant number G0802796]), NIHR Cambridge Biomedical Research Centre (I.S.F., S.O’R.), and R01DK075787 (J.N.H.). S.O’R. is a paid Scientific Advisor for Pfizer, Inc. in the area of cardiometabolic disease. Until 2010, J.A.M. was on the Board of, and was a paid Scientific Advisor for, Correlagen Diagnostics, Inc., a company whose projects included molecular diagnostic tests related to obesity.
Footnotes
Supplementary Online Materials
Materials and Methods
References (13–18) [Note: The numbers refer to additional references cited only within the Supplementary Online Materials]
Author Contributions
References and Notes
- 1.Cone RD. Studies on the physiological functions of the melanocortin system. Endocrine reviews. 2006 Dec;27:736. doi: 10.1210/er.2006-0034. [DOI] [PubMed] [Google Scholar]
- 2.Hay DL, Poyner DR, Sexton PM. GPCR modulation by RAMPs. Pharmacology & therapeutics. 2006 Jan;109:173. doi: 10.1016/j.pharmthera.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Hinkle PM, Sebag JA. Structure and function of the melanocortin2 receptor accessory protein (MRAP) Molecular and cellular endocrinology. 2009 Mar 5;300:25. doi: 10.1016/j.mce.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooray SN, Clark AJ. Melanocortin receptors and their accessory proteins. Molecular and cellular endocrinology. 2011 Jan 15;331:215. doi: 10.1016/j.mce.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Metherell LA, et al. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nature genetics. 2005 Feb;37:166. doi: 10.1038/ng1501. [DOI] [PubMed] [Google Scholar]
- 6.Chan LF, Metherell LA, Clark AJ. Effects of melanocortins on adrenal gland physiology. Eur J Pharmacol. 2011 Jun 11;660:171. doi: 10.1016/j.ejphar.2010.11.041. [DOI] [PubMed] [Google Scholar]
- 7.Chan LF, et al. MRAP and MRAP2 are bidirectional regulators of the melanocortin receptor family. Proc Natl Acad Sci U S A. 2009 Apr 14;106:6146. doi: 10.1073/pnas.0809918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebag JA, Hinkle PM. Regulation of G protein-coupled receptor signaling: specific dominant-negative effects of melanocortin 2 receptor accessory protein 2. Science signaling. 2010;3:ra28. doi: 10.1126/scisignal.2000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tschop MH, et al. A guide to analysis of mouse energy metabolism. Nature methods. 2012 Jan;9:57. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balthasar N, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005 Nov 4;123:493. doi: 10.1016/j.cell.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 11.Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocrine reviews. 2006 Dec;27:710. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 12.Johansson LE, et al. Genetic variance in the adiponutrin gene family and childhood obesity. PLoS One. 2009;4:e5327. doi: 10.1371/journal.pone.0005327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altarejos JY, et al. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nature medicine. 2008 Oct;14:1112. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halley JT, Nelson TS, Kirby LK, Johnson ZB. Relationship between dry matter digestion and metabolizable energy. Poultry science. 1985 Oct;64:1934. doi: 10.3382/ps.0641934. [DOI] [PubMed] [Google Scholar]
- 15.Adams DJ, et al. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics. 2005 Dec;86:753. doi: 10.1016/j.ygeno.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome research. 2003 Mar;13:476. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, et al. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci. 2003 Aug 6;23:7143. doi: 10.1523/JNEUROSCI.23-18-07143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.