Abstract
System-wide approaches are now being applied to study vaccine responses, whose mechanisms of action, and failure, are not well understood. These works have repeatedly shown vaccine response to be an orchestrated process involving multiple arms of immunity most noticeable sensing and innate components. Prediction of vaccine responses based on system-wide measures is achievable, but challenges remain for robust population wide predictions based only on pre-vaccination measures, especially in partially efficacious vaccines such as influenza. This is especially true in older adults, who are often less responsive to vaccination and exhibit high level of variation compared to young in many components of immunity. Despite this increase in variation, most of the studies on aging use group averages of immune phenotypes to model immune system behavior. Using systems approaches, it is possible to exploit this variation to form distinguishable clusters of phenotypes within and across individuals to discover underlying immune states.
Introduction
Despite the widespread use of vaccines and success of vaccination in eradicating devastating infectious diseases, how vaccines work and why they fail, remains a mystery. Thus, understanding mechanisms of vaccine response is an important goal for both fundamental immunology and public health. This is particularly true in the context of aging where the immune responses to vaccination are often defective[1-4]. Recently, integrated measurements of an individual's immune profile has become a feasible reality, as technological advances have enabled accurate, rapid and relatively low cost enumeration of multiple components of immune system determinants and in a broad manner, including: 35 parameter cell subset phenotypes and responses by mass cytometry, multiplex (currently 51) serum cytokine abundances, comprehensive affordable sequencing of HLAs, Ig repertoires and antigen specificity, as well as more established technologies such as whole-genome gene expression[5-8]. Each such individual measurement may be considered as a new dimension on which the immune system may be probed. This system-wide approach to immunology has recently been used in vaccination as a means of exploring the immune response in an unbiased manner in humans and it is starting to yield insights involving multiple components of immunity.
Learning the ropes – Application of Systems-wide measurement to highly effective vaccines
Initial work from Gaucher et al.[9] monitored a group of volunteers over a period of a year following their administration of a live attenuated yellow fever vaccine (YF17D). Multiple immune parameters and expression of genes from whole blood were measured at different time points. This unbiased approach identified for the first time, a gene signature induced early after vaccination (days 3 and 7) composed of several transcription factors with functionality spanning multiple arms of immunity. Most prominently these included genes associated with a number of pathways from the innate immune response such as the molecular sensor for single-stranded RNA, Toll-like receptor 7 (TLR7) and its downstream adaptor molecule MyD88, as well as other several molecules with direct antiviral activity such as ISG20 and OAS1, 2, and 3. Genes controlling important innate responses such as type I IFNs, inflammasome, complement system and cytokine signaling, such as IRF7, STAT1, C1QA and C1QB were also significantly induced by the YF17D. A similar time series analysis of YF17D administration, conducted by Querec et al.[10], showed many overlapping genes signatures to those observed by Gaucher et al. and used computational models of an individual's gene expression measurement from early time points (up to day 7), to predict the subsequent magnitude of neutralizing antibody titers and antigen specific CD8+ T cell responses to YF17D. Importantly, as YF17D yields strong immune responses in the wide majority (90%) of individuals[11] and it is logistically easy to recruit study subjects with no prior exposure, its choice as the first vaccine in which to apply an integrative system-wide approach was sensible, and offered good grounds for learning. From the perspective of this review, the strength of response is such that the variation observed between individuals is relatively small compared with other cohorts, for example in aging, where one can expect a much larger variation. Nonetheless, a recent re-analysis by gender of yellow fever vaccination data[9], highlighted the fact that the identified gene signatures induced by YF17D, occurred almost exclusively in females[12], which are known clinically to have a stronger response to vaccination in general[12-15]. This indicates that the factors determining the variation in the immune responses and phenotypes must be taken into account especially in studies conducted in humans where a significant variation is often observed.
Getting complex - Systems-wide vaccination responses in vaccines with variable response and partial efficacy
In more challenging applications, several groups and large consortia [16,17] have been using such integrative system-wide measurement approaches to understanding molecular mechanisms predictive of seasonal influenza vaccine response[18,19,20,21], HIV vaccines[22] and others[20,23-25]. Interestingly, these unbiased profiling analyses are enabling the discovery of new molecules involved in vaccine response, and are generating hypotheses for mechanisms of action that may be tested in animal models. For example, Pulendran and colleagues studied influenza vaccine response[18] in a study design parallel to their initial efforts in yellow fever[10], and showed that the gene expression of the calcium-calmodulin-dependent protein kinase IV (CaMKIV) is predictive at day 3 post-vaccination of end outcome influenza specific antibody titers. CaMKIV was known to have a role in T cell development, inflammatory responses and hematopoietic stem cell maintenance[26-29], but not in the B-cell response. Returning to animal models, they showed that vaccination of CaMKIV-deficient (Camk4-/-) mice with TIV, induced enhanced antigen-specific antibody titers, demonstrating an unappreciated role for CaMKIV in the regulation of antibody responses.
More recently, we have adopted an integrative system-wide approach to find baseline (i.e. prevaccination) correlates of vaccine responsiveness in older adults (>60 years). Results of these studies, which also included young controls, yielded new age-dependent and age-independent predictors of vaccine response encompassing several ‘layers’ of the immune system including the proportion of various cell subsets, levels of serum cytokines, the functional status of immune cells, and expression of genes known to regulate apoptosis. Integration of such different aspects of immunity supported a critical role for apoptosis in the response to the influenza vaccine such that the presence of soluble anti-apoptotic molecules from blood (e.g. sFasL) and sets of genes with pro-apoptotic function such as GSTP1, FCGR2A, ZPB89, and others were positive predictors of the antibody response against influenza vaccination. This new role of the apoptosis machinery in vaccine response was confirmed in apoptosis-deficient animal models. From such studies, we suggested that the observed deficits in apoptosis can cause accumulation of memory cells (also known as memory inflation[30]) that are specific for diverse chronic infectious agents[31]. Due to the limited niche availability, such accumulation may restrict the number of cells able to respond to novel antigenic challenges and vaccination.
Our studies also show that prediction of vaccine response is less accurate and more sensitive in the elderly than in young individuals, indicating that we have not yet exhausted examining the variation across human populations. Moreover, as shown above, the immune predictors of vaccine responsiveness differ depending on the vaccine formulation (live-attenuated versus inactivated vaccines), cohort demographics (age, gender, race, others), exposure history (to the same or other antigens), etc. These findings also mesh with results from Chaussabel and colleagues, who performed a detailed time course investigating immune responses to influenza and pneumococcal vaccines, in a set of adults aged 18-64, and identified both shared and differing gene expression signatures for the different vaccines. Thus, we are experiencing the early days for the application of system-wide approaches for the identification of immunological features involved in the response to vaccination, and more generally, to the understanding of immune variation in humans,
Increased Variation in Older Adults and the Possibility of Immune System States
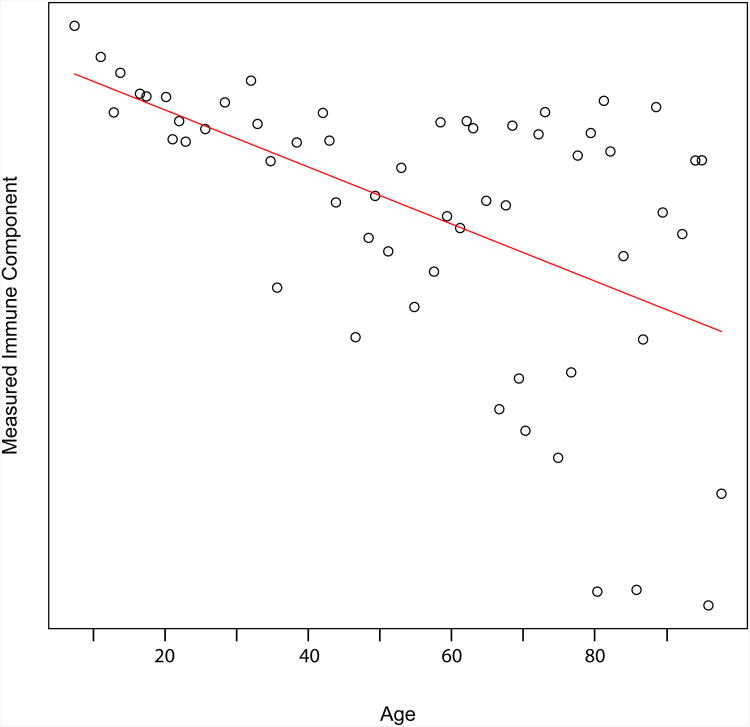
From a predictive point of view, our studies were challenging given the increased variation observed in the elderly as compared with young adults in many of the features (gene, cytokine, cell subset abundance or response) assayed. For example, we observed CD8 Naïve relative cell subset whose mean proportion in blood declines with age, show age dependent increase in coefficient of variation. This is likely the underlying reason why finding predictive models that explain vaccine responsiveness based on immunological features seems to be more difficult in older adults[19]. Indeed, a detailed inspection of the literature reveals that the variation in immune phenotypes in humans increases with aging (measureable as increased CV), such that the elderly exhibit a higher degree of diversity than is seen in young individuals [3,32-38]. Yet, most studies modeling immune behavior and clinical phenotypes with age have reported group mean dysregulations (Fig. 1). This increase in variance observed in older adults, is likely due to an individual's life-history (i.e. footprints of prior exposure and disease), with younger adults starting off from more similar ‘initial conditions’. A notable outlier to test may be the exceptionally old who are a select group of individuals who have buffered the effects of aging and thus may be expected to show lower variance [39]. Importantly though, even in young individuals, the immune system does not start off identical, heritability studies (performed mostly in twins) have pointed towards genetic determinants influencing various leukocyte cell-frequencies[40], abundance of pro-inflammatory cytokines in plasma[41], and gene expression transcripts levels [42] and methylation patterns[43] from blood derived cells.
Figure 1. A characteristic plot of showing the measurement of an immune component as a function of age.
Quiet commonly, especially in humans, the variation in phenotypes increases with age, such that older adults exhibit a higher degree of diversity (increased CV) than is seen in young individuals[3,32-3732-37]. Yet, most studies modeling immune behavior and clinical phenotypes with age have reported group mean dysregulations. Shown in red is a least square regression of the plotted simulated data, the ages on the x-axis vary by immune component assayed.
This raises the question as to whether sets of events (e.g. a low frequency of a certain cell-type in peripheral blood or a high abundance of a certain cytokine in serum) occur independently of one another or co-occur within the same individual. If the latter is the case, it implies the existence of ‘immune states’, likely the result of the ‘network’ nature of the immune system and detectable by analyzing groups of individuals sharing related traits, and/or by following single individuals over time. What would such states look like? They would be a combination of immune factors that co-vary, more so than would be expected in a loosely coupled system where each factor is independently, or close to independently, varying. Implicitly, experienced clinical immunologists have knowledge of such immune system states, stemming from having observed multiple laboratory tests performed on the same patient and for hundreds of patients; they learn to expect to observe abnormalities in a test assaying one factor of the immune system, when abnormal values are observed in another test; assaying a different immune system factor. Such behavior is indicative of a network connection between the two factors. However, the complexity of the immune system is so high that the ability to make such observations quickly decays the more factors are involved, and knowledge is limited primarily to extreme conditions of disease.
We explicitly make a distinction between immune states inferred from individuals with clinically described disease (from hereon: disease immune states) and those derived from those who are considered clinically healthy (or non-diseased) by today's clinical criteria (from hereon: healthy immune states). The distinction between disease and healthy immune states are blurred to some degree by the definition of what exactly is considered a ‘healthy immune system’, a definition likely to shift with time. As a working definition we would propose that healthy immune states are those detected in a healthy population and capturing much of the underlying variation due to differing life history events or genetics, whereas, disease immune states, sit ‘on top’ of healthy immune states, and represent a breakaway from an individual natural immune system alterations. We note, that by this definition the identification and clinical implications of disease immune states are relatively straight forward (for the identification of disease sub-group subsets, progression and treatment, discussion of which we leave for others) whereas the clinical significance of healthy immune states rests in how they affect disease susceptibility and trajectory, and is likely harder to detect. Hence, evidence as to the existence of differing immune states in those we currently consider healthy is scarce, as is the ability to distinguish those that are of good immune health versus those that are at risk within them.
First glimpse of Immune States Inferred from those Considered “Clinically Healthy”
A prime example of what could be considered a healthy immune state with clinical implications is the ‘immune risk profile’ (IRP) established from longitudinal studies of very elderly Swedish cohorts and characterized by an inverted CD4/CD8 ratio (<1), increase in late-differentiated CD8 T cells, as well as low B cells and CMV seropositivity[44-48]. This cluster of measures has been associated with mortality in 85 year-olds, with 2, 4 and 6 year follow-up, and more recent studies suggest that it can be applied to subjects over 65 years of age. We note that the IRP did not investigate any correlations of immune parameters with response to vaccination, and it is important to define whether it can also be applied to other cases and populations. Yet, it is an example that highlights the importance of standardized multi-parameter measurements for the discovery of immune states with clinical implications.
At a more detailed level, system-wide studies of vaccines, such as influenza, where immune response is highly variable and efficacy partial, are identifying segregated populations of responders and non-responders. Beyond their direct usefulness to understanding the conditions in which vaccines fail, these studies are informative for identifying immune states present in what is generally considered a healthy population. For example, in the work of Furman et al., the age-related decay in apoptosis function may well represent a cluster of measurements including genes, serum cytokines, etc. that identify individuals at risk of a weak vaccine response and maybe to other immune responses[19]. Similarly, Pulendran and colleagues were able to identify in young individuals gene signatures early after vaccination that could allow for early identification of subjects at risk of having poor responses to influenza vaccination[18]. Of note, of the 133 genes used to predict the antibody response to the seasonal influenza vaccine, 7 were also predictors of the antibody response to vaccination with the YF-17D vaccine[10,18]. Key genes in the predictive signatures were TNFRSF17, which encodes BCMA, (known to have a key role in B cell differentiation), and CD38, which encodes a surface protein important in lymphocyte development. Interestingly, BCMA belongs to a family of molecules (BAFF, APRIL, BAFF-R and TACI) that regulate plasma cell differentiation and the authors found strong correlations between the expression of genes encoding APRIL, BAFF-R and TACI and the response to both influenza and YF-17D vaccines. These results strongly suggest that the players involved in this network regulating the antibody responses to different vaccines could be also used to identify immune states representing poor versus good immune health.
Systems-wide approaches can identify underlying immune states and their trajectories with age
Immunologists may learn from other scientific fields where complex system theory ideas have taken hold and system level approaches have shown utility in revealing underlying complex system organization. These range from relatively close fields such as cancer genomics, to far flung ones such as gut microbiome interactions and complex ecosystems. A commonality to the approaches taken in probing these systems is the harnessing of the variance exhibited in the system towards an improved understanding. Yet, the view for much of ongoing research in immunology is that high variance is abhorred, considered as anywhere between a necessary nuisance to a reason to stop an experiment short. The identification of immune states necessitates a high level of variation between samples needed to detect coherent co-variation of multiple immune system components. System-wide studies of older adults, where the data generated is both of high dimensionality and highly variant between samples, notwithstanding controlling for confounding factors, may offer an ideal discovery ground for immune states while simultaneously answering clinically relevant questions regarding the variation in immune response.
A dynamic system with many components exerting regulation on one another is expected to gravitate towards a stable state[49,50]. Immune system homeostasis is considered as such a stable condition. Yet both the long term alterations in the immune compartments with aging and the large variation in measures observed between individuals would suggest that more than one stable state exists, both within an individual over time and between individuals. If so, this brings about fascinating and important questions: Within an individual we may ask when did these multiple states appear? Were they present at the onset of an individual's development or appeared as ‘sink holes’ later in life? The low variance observed in the young, compared to the old, would suggest that they are life history dictated, yet minor differences present in ‘initial conditions’ may grow in magnitude to be detectable with age. If the latter is the case, the increase in the variance of immunological parameters could therefore be considered as different immune trajectories of aging. Whichever is the case, sudden events or long term trajectories, it would be important to identify the possible system states, the biological mechanisms which drive an individual's immune system towards a specific state and the clinical implication of different immune system states to disease.
Highlights.
System-wide approaches are now applied to study vaccine response in healthy adults.
Vaccine response involves multiple arms of immunity and can be predicted.
Robust prediction of response from pre-vaccination measures remains a challenge.
Phenotypic variance rises with age; suggest use of individual data over group means.
Variation within and between individuals, may reveal underlying immune states.
Acknowledgments
The authors would like to thank M.M. Davis, A. Butte and B. Kidd for illuminating discussions. This work was supported by US National Institutes of Health (NIH) (U19 AI057229) and (in part) by the Israel Science Foundation (1365/12). SSSO is a Taub Fellow. DF was supported by Stanford Center of Longevity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Oxman MN, Levin MJ, Johnson GR, Schmader KE, Straus SE, Gelb LD, Arbeit RD, Simberkoff MS, Gershon AA, Davis LE, et al. A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N Engl J Med. 2005;352:2271–2284. doi: 10.1056/NEJMoa051016. [DOI] [PubMed] [Google Scholar]
- 2.Schmader KE, Levin MJ, Gnann JW, Jr, McNeil SA, Vesikari T, Betts RF, Keay S, Stek JE, Bundick ND, Su SC, et al. Efficacy, safety, and tolerability of herpes zoster vaccine in persons aged 50-59 years. Clin Infect Dis. 2012;54:922–928. doi: 10.1093/cid/cir970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodwin K, Viboud C, Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24:1159–1169. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 4.Leggat DJ, Thompson RS, Khaskhely NM, Iyer AS, Westerink MA. The elderly immune response to pneumococcal polysaccharides 14 and 23F consists predominantly of switched memory B cells. J Infect Dis. 2013 doi: 10.1093/infdis/jit139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Krishnakumar S, Wilhelmy J, Babrzadeh F, Stepanyan L, Su LF, Levinson D, Fernandez-Vina MA, Davis RW, Davis MM, et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc Natl Acad Sci U S A. 2012;109:8676–8681. doi: 10.1073/pnas.1206614109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Maecker HT, Lindstrom TM, Robinson WH, Utz PJ, Hale M, Boyd SD, Shen-Orr SS, Fathman CG. New tools for classification and monitoring of autoimmune diseases. Nat Rev Rheumatol. 2012;8:317–328. doi: 10.1038/nrrheum.2012.66. A review of novel high-bandwidth technologies aimed at probing immune system components and computational algorithms aimed at extracting the most out of that data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS, Dekker CL, Zheng NY, Huang M, Sullivan M, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med. 2013;5:171ra119. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soon WW, Hariharan M, Snyder MP. High-throughput sequencing for biology and medicine. Mol Syst Biol. 2013;9:640. doi: 10.1038/msb.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, 3rd, Castro E, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monath TP. Yellow fever vaccine. In: Plotkin S, Orenstein W, editors. InVaccines. WB Saunders; Philadelphia: 2004. pp. 1095–1176. [Google Scholar]
- 12*.Klein SL, Jedlicka A, Pekosz A. The Xs and Y of immune responses to viral vaccines. Lancet Infect Dis. 2010;10:338–349. doi: 10.1016/S1473-3099(10)70049-9. This paper presents evidence that vaccine-induced expression of immune genes signatures occurs predominantly in females, which indicates that sources of variation can be detected in simple demographic factors. These must be considered in studies involving human populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennell LM, Galligan CL, Fish EN. Sex affects immunity. J Autoimmun. 2012;38:J282–291. doi: 10.1016/j.jaut.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook IF. Sexual dimorphism of humoral immunity with human vaccines. Vaccine. 2008;26:3551–3555. doi: 10.1016/j.vaccine.2008.04.054. [DOI] [PubMed] [Google Scholar]
- 16.Consortium THIP: Human Immunology Project Consortium. Edited by. 2013 [Google Scholar]
- 17.Inflammation CfHIAa: Center for Human Immunology Autoimmunity and Inflammation. 2013 doi: 10.1111/nyas.12101. [DOI] [PubMed] [Google Scholar]
- 18**.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. Here, the authors present data of vaccine-induced changes in expression of genes that predict the subsequent influenza-specific antibody and T cell responses. An elegant example of how, using systems approaches on blood from clinically healthy individuals, we can identify populations at risk and learn the mechanisms involved in vaccine response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Molecular systems biology. 2013;9:659. doi: 10.1038/msb.2013.15. The paper makes use of aging populations as a source of immune variation and analyses pre-vaccine blood samples in a systems-wide fashion to find metrics of vaccine responsiveness. This paper identifies a common defect in apoptosis by integrating multiple immune system measurements. It also constitutes the first work in predicting vaccine response prior to its administration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Obermoser G, Presnell S, Domico K, Xu H, Wang Y, Anguiano E, Thompson-Snipes L, Ranganathan R, Zeitner B, Bjork A, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. A detailed timecourse primairly analyzing changes in blood gene expression following vaccination and highlighting the dynamics of immune response over time and the differences in response between vaccines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Nino D, Arden N, Quarles JM, Couch RB, Belmont JW. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. The Journal of infectious diseases. 2011;203:921–929. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zak DE, Andersen-Nissen E, Peterson ER, Sato A, Hamilton MK, Borgerding J, Krishnamurty AT, Chang JT, Adams DJ, Hensley TR, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3503–3512. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy RB, Oberg AL, Ovsyannikova IG, Haralambieva IH, Grill D, Poland GA. Transcriptomic profiles of high and low antibody responders to smallpox vaccine. Genes and immunity. 2013 doi: 10.1038/gene.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haralambieva IH, Oberg AL, Ovsyannikova IG, Kennedy RB, Grill DE, Middha S, Bot BM, Wang VW, Smith DI, Jacobson RM, et al. Genome-wide characterization of transcriptional patterns in high and low antibody responders to rubella vaccination. PLoS One. 2013;8:e62149. doi: 10.1371/journal.pone.0062149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haralambieva IH, Oberg AL, Dhiman N, Ovsyannikova IG, Kennedy RB, Grill DE, Jacobson RM, Poland GA. High-dimensional gene expression profiling studies in high and low responders to primary smallpox vaccination. The Journal of infectious diseases. 2012;206:1512–1520. doi: 10.1093/infdis/jis546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krebs J, Wilson A, Kisielow P. Calmodulin-dependent protein kinase IV during T-cell development. Biochem Biophys Res Commun. 1997;241:383–389. doi: 10.1006/bbrc.1997.7823. [DOI] [PubMed] [Google Scholar]
- 27.Anderson KA, Means AR. Defective signaling in a subpopulation of CD4(+) T cells in the absence of Ca(2+)/calmodulin-dependent protein kinase IV. Mol Cell Biol. 2002;22:23–29. doi: 10.1128/MCB.22.1.23-29.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Illario M, Giardino-Torchia ML, Sankar U, Ribar TJ, Galgani M, Vitiello L, Masci AM, Bertani FR, Ciaglia E, Astone D, et al. Calmodulin-dependent kinase IV links Toll-like receptor 4 signaling with survival pathway of activated dendritic cells. Blood. 2008;111:723–731. doi: 10.1182/blood-2007-05-091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitsos CM, Sankar U, Illario M, Colomer-Font JM, Duncan AW, Ribar TJ, Reya T, Means AR. Calmodulin-dependent protein kinase IV regulates hematopoietic stem cell maintenance. J Biol Chem. 2005;280:33101–33108. doi: 10.1074/jbc.M505208200. [DOI] [PubMed] [Google Scholar]
- 30.Karrer U, Sierro S, Wagner M, Oxenius A, Hengel H, Koszinowski UH, Phillips RE, Klenerman P. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J Immunol. 2003;170:2022–2029. doi: 10.4049/jimmunol.170.4.2022. [DOI] [PubMed] [Google Scholar]
- 31.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 32.Stiasny K, Aberle JH, Keller M, Grubeck-Loebenstein B, Heinz FX. Age affects quantity but not quality of antibody responses after vaccination with an inactivated flavivirus vaccine against tick-borne encephalitis. PLoS One. 2012;7:e34145. doi: 10.1371/journal.pone.0034145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, Nayak L, Moss PA. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol. 2002;169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 34.Gibson KL, Wu YC, Barnett Y, Duggan O, Vaughan R, Kondeatis E, Nilsson BO, Wikby A, Kipling D, Dunn-Walters DK. B-cell diversity decreases in old age and is correlated with poor health status. Aging Cell. 2009;8:18–25. doi: 10.1111/j.1474-9726.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Effros RB, Boucher N, Porter V, Zhu X, Spaulding C, Walford RL, Kronenberg M, Cohen D, Schachter F. Decline in CD28+ T cells in centenarians and in long-term T cell cultures: a possible cause for both in vivo and in vitro immunosenescence. Exp Gerontol. 1994;29:601–609. doi: 10.1016/0531-5565(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 36.Comin F, Speziali E, Martins-Filho OA, Caldas IR, Moura V, Gazzinelli A, Correa-Oliveira R, Faria AM. Ageing and Toll-like receptor expression by innate immune cells in chronic human schistosomiasis. Clin Exp Immunol. 2007;149:274–284. doi: 10.1111/j.1365-2249.2007.03403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buffa S, Bulati M, Pellicano M, Dunn-Walters DK, Wu YC, Candore G, Vitello S, Caruso C, Colonna- Romano G. B cell immunosenescence: different features of naive and memory B cells in elderly. Biogerontology. 2011;12:473–483. doi: 10.1007/s10522-011-9353-4. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann J, Spyridopoulos I. Telomere length in cardiovascular disease: new challenges in measuring this marker of cardiovascular aging. Future cardiology. 2011;7:789–803. doi: 10.2217/fca.11.55. [DOI] [PubMed] [Google Scholar]
- 39**.Bergman A, Atzmon G, Ye K, MacCarthy T, Barzilai N. Buffering mechanisms in aging: a systems approach toward uncovering the genetic component of aging. PLoS computational biology. 2007;3:e170. doi: 10.1371/journal.pcbi.0030170. This work raises the conjecture that the population of the very old is non-representative of other older-adults as their favorable genotypes act as mechanisms that buffer the deleterious effect of age-related disease genes, which may result in their the frequency of deleterious phenotype increasing amongst them. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans DM, Frazer IH, Martin NG. Genetic and environmental causes of variation in basal levels of blood cells. Twin research: the official journal of the International Society for Twin Studies. 1999;2:250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- 41.de Maat MP, Bladbjerg EM, Hjelmborg J, Bathum L, Jespersen J, Christensen K. Genetic influence on inflammation variables in the elderly. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:2168–2173. doi: 10.1161/01.ATV.0000143856.01669.e7. [DOI] [PubMed] [Google Scholar]
- 42.Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nature genetics. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nature biotechnology. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wikby A, Maxson P, Olsson J, Johansson B, Ferguson FG. Changes in CD8 and CD4 lymphocyte subsets, T cell proliferation responses and non-survival in the very old: the Swedish longitudinal OCTO-immune study. Mech Ageing Dev. 1998;102:187–198. doi: 10.1016/s0047-6374(97)00151-6. [DOI] [PubMed] [Google Scholar]
- 45**.Strindhall J, Skog M, Ernerudh J, Bengner M, Lofgren S, Matussek A, Nilsson BO, Wikby A. The inverted CD4/CD8 ratio and associated parameters in 66-year-old individuals: the Swedish HEXA immune study. Age (Dordr) 2012 doi: 10.1007/s11357-012-9400-3. An important paper showing 15% of adults aged 66 meet the Immune Risk Profile criteria as first identified in older adults. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsson J, Wikby A, Johansson B, Lofgren S, Nilsson BO, Ferguson FG. Age-related change in peripheral blood T-lymphocyte subpopulations and cytomegalovirus infection in the very old: the Swedish longitudinal OCTO immune study. Mech Ageing Dev. 2000;121:187–201. doi: 10.1016/s0047-6374(00)00210-4. [DOI] [PubMed] [Google Scholar]
- 47.Hadrup SR, Strindhall J, Kollgaard T, Seremet T, Johansson B, Pawelec G, thor Straten P, Wikby A. Longitudinal studies of clonally expanded CD8 T cells reveal a repertoire shrinkage predicting mortality and an increased number of dysfunctional cytomegalovirus-specific T cells in the very elderly. J Immunol. 2006;176:2645–2653. doi: 10.4049/jimmunol.176.4.2645. [DOI] [PubMed] [Google Scholar]
- 48.Ferguson FG, Wikby A, Maxson P, Olsson J, Johansson B. Immune parameters in a longitudinal study of a very old population of Swedish people: a comparison between survivors and nonsurvivors. J Gerontol A Biol Sci Med Sci. 1995;50:B378–382. doi: 10.1093/gerona/50a.6.b378. [DOI] [PubMed] [Google Scholar]
- 49.Bar-Yam Y, Harmon D, de Bivort B. Systems biology. Attractors and democratic dynamics. Science. 2009;323:1016–1017. doi: 10.1126/science.1163225. [DOI] [PubMed] [Google Scholar]
- 50.Waddington CH. Principles of Embryology. London: Allen and Unwin; 1956. [Google Scholar]