Summary
We report a novel method for obtaining simultaneous images from multiple vantage points of a microscopic specimen using size-matched microscopic mirrors created from anisotropically etched silicon. The resulting pyramidal wells enable bright-field and fluorescent side-view images, and when combined with z-sectioning, provide additional information for 3D reconstructions of the specimen. We have demonstrated the 3D localization and tracking over time of the centrosome of a live Dictyostelium discoideum. The simultaneous acquisition of images from multiple perspectives also provides a five-fold increase in the theoretical collection efficiency of emitted photons, a property which may be useful for low-light imaging modalities such as bioluminescence, or low abundance surface-marker labelling.
Keywords: Confocal microscopy, fluorescence microscopy, PSF, tilted views, 3D microscopy, 3D reconstruction
Introduction
Microscopy is normally conducted within a single focal plane (XY) at a specific axial depth (Z) providing a single 2D image or stack of images. Because biological systems are highly dynamic and often undergo coordinated inter- and intracellular motion in all three spatial dimensions, it becomes very difficult to capture these movements with standard wide-field or confocal microscopy. We have explored the use of cell-sized mirrors formed by the angled sidewalls of anisotropically etched pyramidal wells in silicon for conducting microscopy from multiple vantage points (Fig. 1A). The mirrored pyramidal wells (MPWs) provide multiple, nearly orthogonal views of the specimen, increase the collection efficiency of the imaging system, and are compatible with biomicroelectromechanical systems devices. Trupke et al. (2006) describe MPWs for particle tracking but not imaging extended objects within the MPW. Although many excellent studies on side-view (Boocock et al., 1985) and mirrored microscopy (Bell & Jeon, 1963; Gustaffson & Wolpert, 1967; Harris, 1969; Ingram, 1969; Hlinka & Sanders, 1972; Sanders & Prasad, 1979; Boocock et al., 1985; Roth et al., 1988; Cao et al., 1997; Dong et al., 1999; Leyton-Mange et al., 2006) can be found in the literature, we feel that a strong advantage offered by MPW microscopy over other methods is the ability to mass fabricate mirrored wells of various controlled sizes. Standard photolithography methods enable feature sizes ranging from 2 μm to as large as several hundred micrometres, allowing MPWs to be custom sized for a particular specimen. MPWs enable interrogation with short working-distance objectives that are physically obstructed by larger mirror systems (Stelzer & Lindek, 1994; Sheppard, 1995).
Fig. 1.
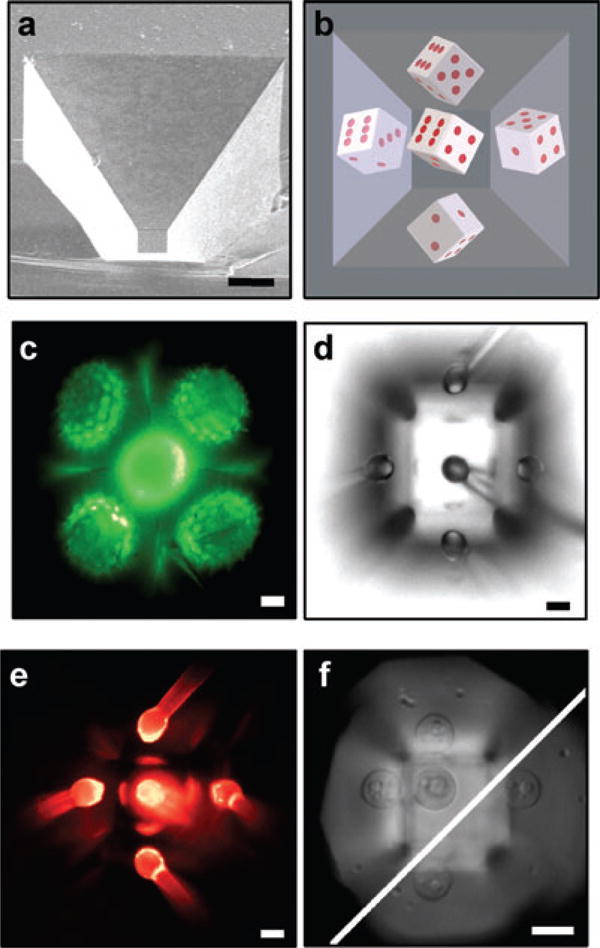
MPW and simultaneous, multiple vantage point and side-view images. (A) SEM image of MPW, (B) 3D rendering of 45° MPW with pipped die, (C) Autofluorescent Helianthus annuus pollen grain in a 54.7° MPW, (D) Pulled glass pipette with 25-μm bulbous tip in optical centre of MPW demonstrating five simultaneous views from multiple vantage points, (E) Fluorescent image of pipette tip of panel D with rhodamine coating and (F) 100× composite image of Saccharomyces cerevisiae cell. Upper left of diagonal are cell and two reflections at the first focal plane. Lower left of diagonal are two reflections at a deeper focal plane. Scale bar: 10 μm.
Methods
The MPWs are fabricated using techniques developed for semiconductor and microelectromechanical systems (MEMS) fabrication more than 30 years ago (Burden & Walley, 1969; Lee, 1969; Williams & Muller, 1996; Kovacs et al., 1998; Madou, 2002; Trupke et al., 2006), and now described in widely available texts (Madou, 2002). Briefly, silicon wafers (P-Type <100>) were wet oxidized at 1100°C to produce an oxide mask layer approximately 600 nm thick. Photoresist was then patterned on the surface to make openings of the top dimension of the completed wells. The wafers were then dipped in a 6:1 buffered oxide etch solution (J.T. Baker, Inc., Phillipsburg, NJ, USA) for ∼ 30 s to selectively etch the oxide and expose the underlying silicon. They were promptly removed and rinsed in deionized water, and acetone was used to remove the photoresist. The wafers were placed in 50% KOH at 80°C for a time appropriate to produce the desired etch depth (etch rate was ∼ 1.4 μm min−1). The wafers were then rinsed in deionized water and returned to the buffered oxide etchant to remove the SiO2 mask layer. Finally, they were coated with a reflective metal film using electron-beam thermal evaporation. The biocompatibility of platinum has been demonstrated in other studies by our group (Werdich et al., 2004), and we have found aluminium to be highly reflective and suitable for repeated cellular studies as well. It may also be possible to use other materials, such as rhodium. Wafers may be used whole, cut into small pieces that can be put in culture dishes, or attached to microscope slides.1
Results
For our single-step etch protocol, the etch angle is fixed at 54.7° and the dimensions of the top, bottom and depth were sculpted as desired consistent with the angle constraint. The coated wells provided up to five simultaneous views of the specimen from nearly orthogonal vantage points (Fig. 1B–E), a high-magnification side-view image from a single mirror (Fig. 1F), or six views using z-sectioning and imaging the reflection off the bottom of the well (data not shown). When a 25-μm glass pipette is placed at the optical centre of the MPW, five reflected images are formed in a single, reflected focal plane (Fig. 1D and E). The central, reflected image in 1D is less intense due to obstruction of the collection cone by the real object, whereas it is more intense in 1E, possibly due to more efficient fluorescence excitation. The mirrors (Fig. 1A) can faithfully reproduce the fine features of a specimen without aberrations. Figure 1(C) illustrates the fine, spiky projections of a Helianthus annuus pollen grain in profile from four sides (see Supplementary Fig. 1C); however, had the grain been elevated in a slightly larger well, the central image would have been in focus with the other four. At higher numerical apertures consideration must be given to the possibility of mismatch between the angles of the collection cone and the MPW wall. However, high-magnification image formation is possible, as illustrated in a reflected bright-field five-way image of a single Saccharomyces cerevisiae cell using a 100× objective (NA 0.75) in an MPW (Fig. 1F). The five images shown in Fig. 1(E) illustrate a theoretical five-fold increase in the collection efficiency of MPW microscopy over conventional means because a greater portion of the total available solid angle (4π steradians) is redirected into the objective. This equates to an increase in detection sensitivity and a higher signal-to-noise ratio for imaging fluorescent or luminescent samples.
Biology exists and functions in all three spatial dimensions simultaneously, and dynamic 3D in silico reconstructions will be useful to biologists. The 3D information in MPW images is evident by direct observation. However, rapid optical sectioning through reflection-space provides a thoroughly sampled dataset for 3D reconstruction. A sample anywhere in the imaging volume of the well (within working distance limits) can be seen in the reflections of each mirror simply by adjusting the focal depth. Figure 2 illustrates the orientation and location of the multiple single-reflection tilted views of a sample specimen denoted β, γ, δ, ε and φ (secondary and tertiary reflections are not shown). The figure shows that all six faces of the die are visible, and at any die orientation five of the six faces will be simultaneously visible in the reflections. The sixth can be observed directly with no reflection at a different focal depth, consistent with detailed measurements of reflections and mirror angles by Trupke et al. (2006).
Fig. 2.
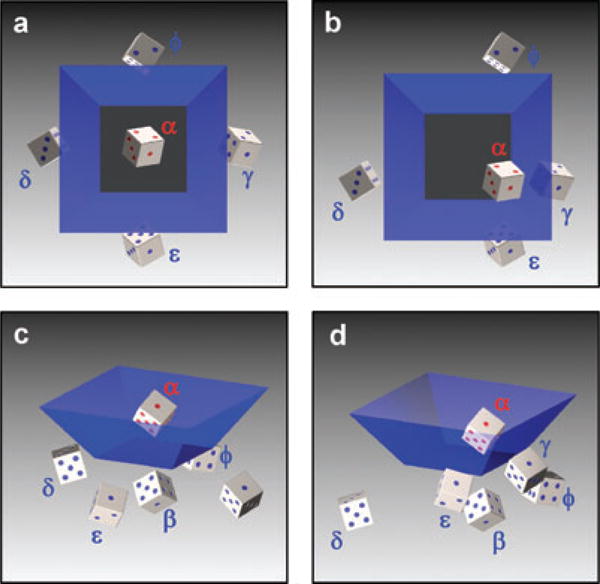
MPW image nomenclature. Red pips and text indicate real object whereas blue pips indicate reflections. Perspective (A, B) and top (C, D) view of MPW reflections for optically centred (A, C) and eccentric (B, D) specimen. α – primary image, β – bottom reflection, γ, δ – first set of opposing mirrors, ε, φ – second set of opposing mirrors (β images are obscured in C and D by α image). Secondary reflections such as a gamma-mirror image of a beta mirror image are denoted by mirror labels (γ-β), or an epsilon mirror image of a gamma (ε-γ), and similarly for tertiary reflections. Scale bar: 10 μm.
Existing deconvolution techniques and 3D reconstruction methods applied to MPW z-sectioned data are likely to provide very accurate 3D models. This is possible because each image acquisition gives multiple tilted views of the sample at the same point in time. A model that incorporates accurate spatial information over time may contain information that is not evident in simple 2D images. Figure 3(A) and (B) show fluorescent images using a 40× oil objective (NA 1.3) at zero and 54 s of a D. discoideum cell stably transfected with green fluorescent protein (GFP)-tubulin (Neujahr et al., 1998). The bright centrosome was visible in the alpha (α) image (Fig. 3Ai) as well as in the primary (γ,ε) and secondary (γ-β, ε-β) (not shown) reflected images. The centrosome migrated from the lower to upper hemisphere of the cell during acquisition (Fig. 3A and B and see Supplementary movie Fig. 3A). Note that the movement of the centrosome in the z-axis was detected without collecting a z-stack, demonstrating the power of this approach. A bright-field series of images at varying focal depths of another D. discoideum cell show what cells look like from varying perspectives (Fig. 3C–E). Given today's sensitive cameras, fast acquisition rates, and rapid microscope stepper motors, rapid z-sectioning should provide simultaneous tilted-view data for 3D reconstructions of live specimens.
Fig. 3.
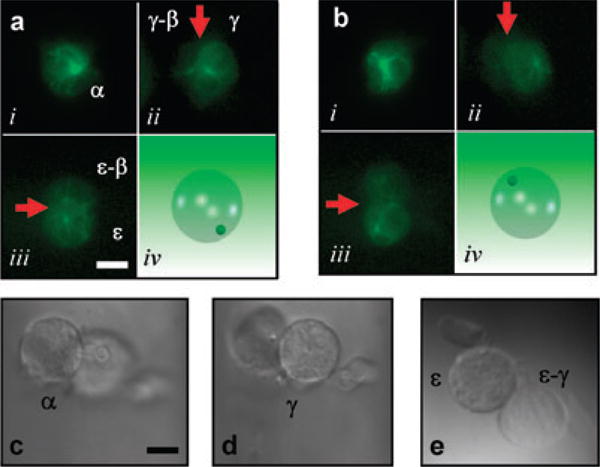
Composite timed-acquisition MPW fluorescent images of GFP-tubulin in a Dictyostelium cell at zero (A) and 54 s (B) as the centrosome migrates from the lower to upper hemisphere of the cell (cartoon insets lower right). Red arrows indicate the mirror plane separating primary from secondary reflections. (C–E) Bright-field α, γ, ε and ε-γ composite images of Dictyostelium cell with pseudopodial extension arising from the dorsal surface of the cell. Scale bar: 10 μm.
Discussion
Three-dimensional microscopy with MPWs offers advantages over other methods because the mirrors reorient the point-spread function (PSF) of the objective. The PSF is the complex 3D intensity pattern in and near the image plane resulting from a point source of light in or near the object plane of a microscope objective. The PSF is also the 3D distribution of points in physical space near the object plane that can contribute to the intensity of a single point in the image plane of a microscope objective. The accuracy of 3D microscopy is ultimately limited by the PSF, which is roughly hourglass shaped, axially elongated and symmetric about the image plane of the microscope (Abbe, 1873; Born & Wolf, 1965). The shape of the PSF and the location of its minima determine the in-plane resolution limit (approximately 200 nm) as well as the axial resolution of 3D imaging by optical sectioning microscopy (Strutt, 1879; Strutt, 1880). Each optical sectioning microscopy ‘slice’ arises from a finite slab of the specimen and contains out of focus light from adjacent slices. The 3D-PSF must be deconvolved from the optical sectioning microscopy image stacks to recover a more accurate 3D structure of the specimen. Iterative, constrained deconvolution is an excellent way to interpolate the missing axial information but requires time for acquisition and implementation (Agard & Sedat, 1983). Tilted view microscopy reorients fixed samples relative to the optical axis, acquiring different cross-sectional images with high in-plane resolution that are recombined into a 3D model (Shaw et al., 1989; Shaw, 1990; Bradl et al., 1994). Tilted view microscopy is limited by precise alignment requirements and the necessity for fixed specimens. Other methods for very high resolution imaging such as theta, 4pi, and I5M all share the common feature of collecting more light from more directions around the sample by multiple objectives or macrosized mirrors (Hell & Stelzer, 1992; Stelzer & Lindek, 1994; Cogswell et al., 1996; Hell et al., 1997; Lindek et al., 1997, 1999; Gustafsson et al., 1999). MPWs offer an exciting and unprecedented opportunity to obtain fast and accurate 3D information of live specimens from several nearly orthogonal directions simultaneously.
We describe a new and simple method for obtaining 3D image data from microscope specimens. The MPWs can be made with any size opening, rectangular or square. The depth of the well and the dimensions of the bottom are then related by the wall angle and etch time. The MPWs should be compatible with many microscopy techniques, including fluorescence lifetime imaging and fluorescence recovery after photobleaching, and will provide precise optical access to any part of the cell. In addition to providing aesthetically pleasing side-view and 3D images, this technique should provide more accurate information for quantitative studies in numerous processes including cell division, immune cell synapse formation, cell motility, and embryo development. Thousands of MPWs can be fabricated in an area of a few square millimetres, allowing MPW technology to be compatible with biomicroelectromechanical systems devices. This technology can ultimately be used for high-throughput screening and drug discovery.
A variety of reconstruction techniques can be used to convert the multiple images acquired with an MPW into either a 3D wire frame or transparent-solid image of the object. We have considered directly populating voxels of a space containing the specimen with combinations (addition and averaging) of the intensities of all constituent reflected images in a transparent-solid. We have developed software for ImageJ and matlab that performs the affine reflection transformation on selected points in simultaneous reflected images, resulting in a swarm of data points in the specimen volume, representing the membrane, for instance. If the sample can be rotated within the MPW, for example by rotation of a micropipette that supports the object or rotation of the MPW around a fixed sample, existing algorithms can be used to create a convex surface representation from the observed silhouettes (Bray et al., 2000; Evertson et al., 2008). Surface features might be used to properly represent concavities (Brodland & Veldhuis, 1998). The key is that multiple images could improve the accuracy of a 3D reconstruction.
The wells can be etched through the full thickness of the silicon wafer to form a mirrored well with four sides and an open bottom. This would provide wells that could be attached to the objective of an upright microscope and lowered down over a small specimen that was attached to a slide. Open-bottom wells could be incorporated into a microfluidic device where the flow is through the open bottom, allowing the possibility of 3D tracking or even dynamic trapping, as cells or particles move through the system. Single atom and molecule imaging methods may also benefit from the increased signal and the precise, 3D spatial localization afforded by the use of the MPWs (Trupke et al., 2006; Betzig et al., 2006).
The MPW in Fig. 1(A) is geometrically concave and introverted, meaning that the optical centre is within the well, due to the etch angle of silicon. Extroverted wells are also geometrically concave but have their optical centre outside of the cavity formed by the well. We have begun exploring the construction of extroverted MPWs with angles other than 54.7° by two methods: a two-step silicon etching process and replica moulding of faceted and polished stainless steel rods. These extroverted MPWs can successfully image specimens that are physically located outside of the well (data not shown). Extroverted MPWs will extend the use of this technology to specimens on planar substrates that cannot be easily positioned within an introverted well, such as highly motile single cells and cell monolayers on a cover slip.
In addition to 3D location and tracking of microscopic particles, the MPWs are useful as simple amplifiers of samples that are scattering or emitting low levels of light. We are using the mirrors for studying bioluminescent specimens and chemical reactions, and for reducing the cytosolic level of standard fluorophores such as calcium-sensitive dyes, with the possibility of reducing dye-associated side effects. We are excited by the possibility of birefringent coatings on each mirror (as a group or individually) for near-sample filtering of excitation and emission wavelengths for multispectral imaging. Because the natural etch angle of silicon is not 45°, true periscope construction as in Boocock et al. (1985) is not straightforward. However, we are exploring aperture functions that take advantage of the steeper angle to illuminate the specimen with one mirror while imaging from another. We are also encouraged by the high density at which the mirrors can be fabricated. We envision miniature MPW devices coupled to microfluidics and charge-coupled device (CCD) image sensors for static dynamic measurements on large numbers of individual cells (Faley et al., 2008); for example, the measurement of a complete blood count with differential white cell populations by labelling of membrane CD markers for HIV prognosis and treatment decisions in resource-poor settings. With the addition of dynamic calcium fluxing measurements in leukocytes, such devices may prove useful for new clinical diagnostics in hospitals and other point of care locations.
Conclusions
We have fabricated MPWs with controllable dimensions over a wide range from macroscopic to sub-cellular. We have studied MPWs microfabricated by silicon etching with an optical centre at which five simultaneous microscope images can be obtained with bright-field and fluorescent microscopy. We demonstrate the use of the MPWs for obtaining 3D geometric localization of sub-cellular features in both bright-field and fluorescent imaging modalities. We have begun extending mirror fabrication beyond silicon etching to construct extroverted mirrors with an optical centre that lies outside of the well and providing up to seven simultaneous images of a specimen. Finally, we are utilizing the wells to increase the collection efficiency and sensitivity of microscopes for bioluminescent and other low-light imaging, and we are beginning to explore the coupling of MPWs to microfluidics and CCD image sensors for imaging device miniaturization.
Supplementary Material
Figure1C_BrightField.jpg A bright field image of the pollen grain from manuscript Figure 1C.
Figure1C_Fluorescent.jpg A larger version of the fluorescent pollen grain image in manuscript Figure 1C.
Figure3A.mov A timelapse movie of the alpha, gamma and epsilon images of a single D. discoideum cell. The bright centrosome migrates from the bottom half of the cell to the top half through the course of the movie.
Acknowledgments
This research has been funded in part by AFOSR grant FA9550-05-1-0349, the Vanderbilt Institute for Integrative Biosystems Research and Education (VIIBRE), and the Systems Biology and Bioengineering Undergraduate Research Experience (SyBBURE). K.T.S. wishes to acknowledge helpful conversations with Mats Gustafsson at the University of California, San Francisco, Sanford Simon at The Rockefeller University, Sandy Rosenthal and Sean Schaffer at Vanderbilt University, and the Vanderbilt Cell Imaging Resource. We thank Allison Price and Don Berry for their editorial assistance.
Footnotes
Prefabricated mirrors can be can be custom fabricated at MEMS exchange (http://www.mems-exchange.org/) or other microfabrication foundries, or obtained from VIIBRE at modest cost. Details for mirror purchase as well as a specific recipe for their production are available at http://www.vanderbilt.edu/viibre/technologies.html.
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Author contributions This work was made possible by the cooperation of all authors, whose contributions are roughly as follows. K.T.S., R.S.R., C.W. and C.J. planned and conducted all imaging experiments. D.A.M. and I.A.G. designed and fabricated the MPW devices. J.P.W. provided significant theoretical and mathematical insight leading to the images of Fig. 1, and all of the work was completed in the laboratories of J.P.W. or C.J. K.T.S. had the primary responsibility for designing and drafting the manuscript, and all other authors contributed to its design and editing.
References
- Abbe E. Beiträge zur theorie des mikroskops und der mikroskopischen wahrnehmung (Contributions to the theory of the microscope and microscopic observations) Arkiv Mikroskop Anat. 1873;9:413–468. [Google Scholar]
- Agard DA, Sedat JW. 3-dimensional architecture of a polytene nucleus. Nature. 1983;302:676–681. doi: 10.1038/302676a0. [DOI] [PubMed] [Google Scholar]
- Bell L, Jeon K. Locomotion of Amoeba proteus. Nature. 1963;198:675–676. [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Boocock CA, Brown AF, Dunn GA. A simple chamber for observing microscopic specimens in both top and side views. J Microsc – Oxf. 1985;137:29–34. doi: 10.1111/j.1365-2818.1985.tb02558.x. [DOI] [PubMed] [Google Scholar]
- Born M, Wolf E. Principles of Optics. Pergamon Press; London: 1965. [Google Scholar]
- Bradl J, Housmann M, Schneider B, Rinke B, Cremer C. A versatile 2-pi-tilting device for fluorescence microscopes. J Microsc – Oxf. 1994;176:211–221. [Google Scholar]
- Bray MA, Lin SF, Wikswo JP. Three-dimensional surface reconstruction and fluorescent visualization of cardiac activation. IEEE Trans Biomed Eng. 2000;47:10, 1382–1391. doi: 10.1109/10.871412. [DOI] [PubMed] [Google Scholar]
- Brodland GW, Veldhuis JH. Three-dimensional reconstruction of live embryos using robotic macroscope images. IEEE Trans Biomed Eng. 1998;45(9):1173–1181. doi: 10.1109/10.709561. [DOI] [PubMed] [Google Scholar]
- Burden MSJ, Walley PA. Evaporation of metals and elemental semiconductors using a work-accelerated electron beam source. Vacuum. 1969;19:397–402. [Google Scholar]
- Cao J, Usami S, Dong C. Development of a side-view chamber for studying cell-surface adhesion under flow conditions. Ann Biomed Eng. 1997;25:573–580. doi: 10.1007/BF02684196. [DOI] [PubMed] [Google Scholar]
- Cogswell CJ, Larkin KG, Klemm HU. Fluorescence microtomography: multiangle image acquisition and 3D digital reconstruction. Proc SPIE. 1996;2655:109–115. [Google Scholar]
- Dong C, Jian C, Struble EJ, Lipowsky HH. Mechanics of leukocyte deformation and adhesion to endothelium in shear flow. Ann Biomed Eng. 1999;27:298–312. doi: 10.1114/1.143. [DOI] [PubMed] [Google Scholar]
- Evertson DW, Holcomb MR, Eames M, et al. High-resolution high-speed panoramic cardiac imaging system. IEEE Trans Biomed Eng. 2008;55:1241–1243. doi: 10.1109/TBME.2007.912417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faley S, Seale K, Hughey J, et al. Microfluidic platform for real-time signaling analysis of multiple single T cells in parallel. Lab on A Chip. 2008 doi: 10.1039/b719799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustaffson T, Wolpert L. Cellular movement and contact in sea urchin morphogenesis. Biolo Rev Camb Philos Soc. 1967;42:85–90. doi: 10.1111/j.1469-185x.1967.tb01482.x. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL, Agard DA, Sedat JW. (IM)-M-5: 3D widefield light microscopy with better than 100 nm axial resolution. J Microsc – Oxf. 1999;195:10–16. doi: 10.1046/j.1365-2818.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- Harris A. Initiation and propagation of ruffle in fibroblast locomotion. J Cell Biol. 1969;43:165a–166a. [Google Scholar]
- Hell S, Stelzer EHK. Properties of a 4pi confocal fluorescence microscope. J Opt Soc Am A – Opt Image Sci Vis. 1992;9:2159–2166. [Google Scholar]
- Hell SW, Schrader M, van der Voort HTM. Far-field fluorescence microscopy with three-dimensional resolution in the 100-nm range. J Microsc –Oxf. 1997;187:1–7. doi: 10.1046/j.1365-2818.1997.2410797.x. [DOI] [PubMed] [Google Scholar]
- Hlinka J, Sanders FK. Real and reflected images of cells in profile .1. method for study of cell movement and adhesion. J Cell Sci. 1972;11:221–231. doi: 10.1242/jcs.11.1.221. [DOI] [PubMed] [Google Scholar]
- Ingram VM. A side view of moving fibroblasts. Nature. 1969;222:641–644. doi: 10.1038/222641a0. [DOI] [PubMed] [Google Scholar]
- Kovacs GTA, Maluf NI, Petersen KE. Bulk micromachining of silicon. Proc IEEE. 1998;86:1536–1551. [Google Scholar]
- Lee DB. Anisotropic etching of silicon. J Appl Phys. 1969;40:4569–4574. [Google Scholar]
- Leyton-Mange J, Yang S, Hoskins MH, Kunz RF, Zahn JD, Dong C. Design of a side-view particle imaging velocimetry flow system for cell-substrate adhesion studies. J Biomech Eng - Trans ASME. 2006;128:271–278. doi: 10.1115/1.2165689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindek S, Stefany T, Stelzer EHK. Single-lens theta microscopy – a new implementation of confocal theta microscopy. J Microsc – Oxf. 1997;188:280–284. [Google Scholar]
- Lindek S, Swoger J, Stelzer EHK. Single-lens theta microscopy: resolution, efficiency and working distance. J Mod Opt. 1999;46:843–858. [Google Scholar]
- Madou MJ. Fundamentals of Microfabrication: The Science of Miniaturization. 2. CRC Press; Boca Raton, FL: 2002. [Google Scholar]
- Neujahr R, Albrecht R, Kohler J, Matzner M, Schwartz JM, Westphal M, Gerisch G. Microtubule-mediated centrosome motility and the positioning of cleavage furrows in multinucleate myosin II null cells. J Cell Sci. 1998;111:1227–1240. doi: 10.1242/jcs.111.9.1227. [DOI] [PubMed] [Google Scholar]
- Roth KE, Rieder CL, Bowser SS. Flexible-substratum technique for viewing cells from the side – some in vivo properties of primary (9 + 0) cilia in cultured kidney epithelia. J Cell Sci. 1988;89:457–466. doi: 10.1242/jcs.89.4.457. [DOI] [PubMed] [Google Scholar]
- Sanders EJ, Prasad S. Observation of cultured embryonic epithelial-cells in side view. J Cell Sci. 1979;38:305–314. doi: 10.1242/jcs.38.1.305. [DOI] [PubMed] [Google Scholar]
- Shaw PJ. 3-dimensional optical microscopy using tilted views. J Microsc – Oxf. 1990;158:165–172. doi: 10.1111/j.1365-2818.1990.tb02989.x. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Agard DA, Hiraoka Y, Sedat JW. Tilted view reconstruction in optical microscopy – 3-dimensional reconstruction of drosophila-melanogaster embryo nuclei. Biophys J. 1989;55:101–110. doi: 10.1016/S0006-3495(89)82783-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard CJR. Fundamental reduction of the observation volume in far-field light-microscopy by detection orthogonal to the illumination axis – confocal theta-microscopy. Opt Commun. 1995;119:693–695. [Google Scholar]
- Stelzer EHK, Lindek S. Fundamental reduction of the observation volume in far-field light-microscopy by detection orthogonal to the illumination axis – confocal theta microscopy. Opt Commun. 1994;111:536–547. [Google Scholar]
- Strutt JW. Investigations in optics, with special reference to the spectroscope. Philos Mag Lett. 1879;8:261–274. 403–411, 477–486. [Google Scholar]
- Strutt JW. Investigations in optics, with special reference to the spectroscope. Philos Mag Lett. 1880;9:40–55. [Google Scholar]
- Trupke M, Ramirez-Martinez F, Curtis EA, et al. Pyramidal micromirrors for microsystems and atom chips. Appl Phys Lett. 2006;88:071116-1–071116-2. [Google Scholar]
- Werdich AA, Lima EA, Ivanov B, Ges I, Wikswo JP, Baudenbacher FJ. A microfluidic device to confine a single cardiac myocyte in a sub-nanoliter volume on planar microelectrodes for extracellular potential recordings. Lab on A Chip. 2004;4:357–362. doi: 10.1039/b315648f. [DOI] [PubMed] [Google Scholar]
- Williams KR, Muller RS. Etch rates for micromachining processing. J Microelectromech Syst. 1996;5(4):256–269. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure1C_BrightField.jpg A bright field image of the pollen grain from manuscript Figure 1C.
Figure1C_Fluorescent.jpg A larger version of the fluorescent pollen grain image in manuscript Figure 1C.
Figure3A.mov A timelapse movie of the alpha, gamma and epsilon images of a single D. discoideum cell. The bright centrosome migrates from the bottom half of the cell to the top half through the course of the movie.


