Abstract
Objective
Oxidative stress is proposed as an important factor in osteoarthritis (OA). We therefore investigated the expression of the three superoxide dismutase (SOD) antioxidant enzymes in OA.
Methods
SOD expression was determined by real-time polymerase chain reaction and immunohistochemistry using human femoral head cartilage. SOD2 expression in Dunkin Hartley guinea pig knee articular cartilage was determined by immunohistochemistry. The DNA methylation status of the SOD2 promoter was determined using bisulfite sequencing. RNA interference was used to determine the consequence of SOD2 depletion on the levels of reactive oxygen species (ROS) using MitoSOX™ and collagenases, matrix metalloproteinase 1 (MMP-1) and MMP-13, gene expression.
Results
All three SOD were abundantly expressed in human cartilage but were markedly down-regulated in end-stage OA cartilage, especially SOD2. In the Dunkin Hartley guinea pig spontaneous OA model SOD2 expression was decreased in the medial tibial chondyle cartilage prior to, and following, the development of OA-like lesions. The SOD2 promoter had significant DNA methylation alterations in OA cartilage. Depletion of SOD2 in chondrocytes gave an increase in ROS but a decrease in collagenase expression.
Conclusion
This is the first comprehensive expression profile of all SOD genes in cartilage and importantly, using an animal model, we show that a reduction in SOD2 is associated with the earliest stages of OA. We found that a decrease in SOD2 associates with an increase in ROS and but a reduction of collagenase gene expression, demonstrating the complexities of ROS function.
Keywords: Osteoarthritis, chondrocytes, superoxide dismutase, gene expression
INTRODUCTION
Osteoarthritis (OA) is a common degenerative joint disease characterised by focal degradation of articular cartilage, centred on load-bearing areas. The primary risk factor for OA development is age, but the mechanisms by which ageing contributes to OA susceptibility and progression remain poorly understood. Articular cartilage extracellular matrix (ECM), which is maintained by chondrocytes, undergoes significant changes in structure and composition with age, closely linked to a decline in chondrocyte function.[1, 2] Many studies have identified molecular characteristics of ageing in OA cartilage or chondrocytes which may contribute to the onset of OA including telomere genomic instability, formation of advanced glycation end-products, increased apoptosis and senescence.[3] Many such changes could be associated with the elevated levels of oxidative stress that occur in OA and aged cartilage [4], leading to an altered chondrocyte phenotype that renders cells unable to respond effectively to normal loading regimes [5] and potentially contributes to disease onset.
Although generally considered a hypoxic environment, oxygen diffuses through articular cartilage and is utilised by chondrocytes to derive approximately 25% of their ATP. Oxidative phosphorylation is a major source of reactive oxygen species (ROS) however, chondrocytes also express NADPH oxidase (NOX) and nitric oxide synthase (NOS) family members along with various oxygenases which principally generate the ROS nitric oxide (NO) and the superoxide anion (O2 ).[4] These ROS generate derivatives including hydrogen peroxide (H2O2), peroxynitrite (ONOO−) and hydroxyl radicals (OH˙). ROS are highly reactive and therefore transient so their production in cartilage has been determined indirectly; an accumulation of lipid peroxidation products [6] and nitrotyrosine residues [7] have been observed in aged and OA cartilage. ROS are able to cause cartilage degradation directly by cleaving collagen, aggrecan and activating matrix metalloproteinases (MMP) [8-10], a family of enzymes that play a key role in cartilage destruction in OA.[11] ROS also act indirectly by modulating redox-sensitive signalling pathways that control MMP expression and in cultured chondrocytes may lead to decreased collagen and aggrecan synthesis.[12, 13]
).[4] These ROS generate derivatives including hydrogen peroxide (H2O2), peroxynitrite (ONOO−) and hydroxyl radicals (OH˙). ROS are highly reactive and therefore transient so their production in cartilage has been determined indirectly; an accumulation of lipid peroxidation products [6] and nitrotyrosine residues [7] have been observed in aged and OA cartilage. ROS are able to cause cartilage degradation directly by cleaving collagen, aggrecan and activating matrix metalloproteinases (MMP) [8-10], a family of enzymes that play a key role in cartilage destruction in OA.[11] ROS also act indirectly by modulating redox-sensitive signalling pathways that control MMP expression and in cultured chondrocytes may lead to decreased collagen and aggrecan synthesis.[12, 13]
To prevent an accumulation of ROS-mediated damage chondrocytes produce a number of antioxidant enzymes including the superoxide dismutases (SOD), catalase and glutathione peroxidase.[4] There are three SOD family members; SOD1 (Cu/Zn-SOD) found principally in the cytosol, SOD2 (Mn-SOD) found in the mitochondrial matrix and SOD3 (EC-SOD) found in the ECM. The SOD family catalyse the dismutation of O2 to O2 and H2O2, thereby limiting the formation of highly aggressive compounds such as ONOO− and OH˙.
to O2 and H2O2, thereby limiting the formation of highly aggressive compounds such as ONOO− and OH˙.
SOD2 has been shown to be down regulated in OA cartilage.[14, 15] SOD3 is also decreased in human OA cartilage and in a mouse model of OA.[16] SOD3 deficient mice show an increased severity of collagen-induced arthritis (CIA).[17] Together, these and other data have led to the proposal that supplementary antioxidants may represent a potential therapy for OA.[18]
We considered that the SOD family may play a key role in controlling ROS levels in cartilage and that any deficiency may account for the elevated oxidative stress observed. Here we report a significant decrease in the expression of all SOD family members in OA cartilage compared to macroscopically normal cartilage (from neck of femur (NOF) fracture patients) at both the RNA and protein levels. We also show a decline in SOD2 expression correlates with the onset of, and importantly precedes, OA-like lesions in an animal model. We also demonstrate that the SOD2 promoter is significantly differentially methylated in OA compared to NOF cartilage, representing a potential mechanism for the reduction in expression observed in OA. Functionally, SOD2 depletion in chondrocytes resulted in a significant increase in ROS but a decrease in the interleukin (IL)-1-induced levels of MMP-1 and MMP-13. Together our data suggest a decrease in SOD2 expression may be presymptomatic of disease and potentially an attempt to reduce the expression of collagenases, enzymes which contribute to cartilage degradation, but concomitantly also contributes to increased oxidative stress.
MATERIALS AND METHODS
Materials
Antibodies against SOD1 and SOD2 were from Stressgen Bioscience (Cambridge, UK) and SOD3 from Abcam (Cambridge, UK). Rabbit IgG isotype control antibody, oligonucleotides (Table 1), PCR Mastermix and 5-aza-2′-deoxycytidine (AZA) were from Sigma-Aldrich (Poole, UK). Anti-rabbit Vectastain ABC Elite kit and Vectashield were from Vector Laboratories Ltd. (Peterborough, UK). Recombinant human IL-1α was a gift from GlaxoSmithKline (Stevenage, UK). Taqman Low Density Arrays (TLDA) were from Applied Biosystems (ABI, Foster City, CA, USA). Sybr-Green Mastermix was from Invitrogen (Paisley, UK). SmartPool™ small interfering RNA (siRNA) against SOD2 (Table 1) and siControl2 were from Dharmacon (Cramlington, UK).
Table 1. Oligonucleotides: real-time RT-PCR, bisulfite PCR primers and SOD2 siGENOME SMART pool siRNA sequences.
| Gene | Method | Primer | Sequence |
|---|---|---|---|
| SOD2 | Sybr Green RT-PCR | Forward Reverse |
5′-CTGGACAAACCTCAGCCCTA 5′-TGATGGCTTCCAGCAACTC |
| SOD2 | DNA Methylation Analysis | Forward Reverse |
5′-GTAATTAAAATTTAGGGGTAGG 5′-AAAAAAAACTACAAACTAACCTC |
| SMARTpool Duplex |
Target Strand | Sequence | |
| 1 | Sense Antisense |
5′-GGACAAACCUCAGCCCUAAUU 5′-PUUAGGGCUGAGGUUUGUCCUU |
|
| 2 | Sense Antisense |
5′-GGAGCACGCUUACUACCUUUU 5′-PAAGGUAGUAAGCGUGCUCCUU |
|
| 3 | Sense Antisense |
5′-AAAGAUACAUGGCUUGCAAUU 5′-PUUGCAAGCCAUGUAUCUUUUU |
|
| 4 | Sense Antisense |
5′-GUAAUCAACUGGGAGAAUGUU 5′-PCAUUCUCCCAGUUGAUUACUU |
RNA Extraction from Cartilage Samples, Chondrocyte Isolation and Cell Culture
Articular cartilage was obtained from patients undergoing joint replacement surgery. Hip cartilage samples from OA patients were compared to lesion-free cartilage from fracture to the neck of femur (NOF) patients with no known history of joint disease. Femoral heads were processed and RNA extracted using established methodology.[19] Human articular chondrocytes (HAC) were isolated from OA cartilage and cultured as previously described.[20] This study was performed with Ethical Committee approval from Norfolk and Norwich University Hospital Trust, Oxford Radcliffe Hospitals NHS Trust and Newcastle and North Tyneside Health Authority and all patients provided informed consent.
Real-Time RT-PCR and siRNA-Mediated Gene Silencing
RNA interference (RNAi) in HAC was as previously described [19] with the siRNA used at 100 nM. Briefly, 2.5 × 104/cm2 (HAC) cells were seeded overnight and the following day transfected with Dharmafect 1 (Dharmacon) and siRNA (SOD2 (Table 1) or non-targeting control, siControl2). Cells were washed 24 hours post-transfection with phosphate buffered saline (PBS) and cultured in serum-free medium for a further 24 hours prior to 24 hour stimulation ± 0.05ng/mL IL-1, or as described. Total RNA was isolated using the Cells-tocDNA II Kit (ABI) and MMP-1 and MMP-13 expression determined by real-time RT-PCR as previously described.[19] Gene depletion was confirmed by real-time RT-PCR and immunoblotting of total cell lysates prepared as described.[19] SOD family gene expression in cartilage cDNA was assessed using a TLDA according to manufacturer’s (ABI) instructions and as previously described.[19] Throughout, mRNA levels were normalised to a housekeeping gene (ABI) using the calculation 2−ΔCt.
Immunohistochemistry
Human Patient Samples
Serial sections (12μm) were prepared and processed from full-depth cartilage taken from intact cartilage of OA and NOF femoral heads as previously described.[21] Serial sections were incubated for 60 minutes at room temperature (RT) with either a polyclonal primary (rabbit) antibody to a SOD or a normal rabbit IgG isotype-matched control. All antibodies were used at a concentration of 1 μg/mL except for SOD3 which was used at a 1:2000 dilution. All antibodies were prepared in PBS containing 1.5% (v/v) normal goat serum.
Dunkin Hartley Guinea Pig Knee Samples
Male Dunkin Hartley guinea pigs (Harlan Olac, UK) aged from 1 to 8 months (five per group) were used for histology. Animals were humanely sacrificed according to UK Home Office regulations. Tibial heads were fixed in 4% (v/v) formaldehyde for 2 days and decalcified in 0.1 M EDTA (pH 7.0) in PBS for 3 weeks. Blocks that included articular cartilage and bone (lateral and medial tibial plateaus) were dehydrated and embedded in paraffin. Five μm-serial, coronal sections were deparaffinised with xylene and rehydrated through a graded ethanol series and finally incubated in 10 mM sodium citrate buffer, pH 6.0, for 2 hours at RT, followed by 3% (v/v) H2O2 in tris-buffered saline (TBS) for 15 minutes. Serial sections were blocked with 1.5% (v/v) normal goat serum in TBS for 30 minutes and then incubated for 90 minutes at RT with an antibody to SOD2 or normal rabbit IgG (1 μg/mL in TBS/1.5% (v/v) normal goat serum). The remainder of the staining procedure and mounting was performed as previously described.[21] Representative sections from each animal were stained with safranin ‘O’/fast green (0.02% (w/v) fast green for 3 minutes, 1% (v/v) acetic acid for 30 seconds, 0.1% (w/v) safranin ‘O’ for 5 minutes).
MitoSOX™ Red staining
HAC were transfected with siRNA against SOD2 or a control (see above) and 24 hr later serum starved overnight. Cells were then stained with 2 μM MitoSOX™ Red (Invitrogen) in serum-free media for 15 min at 37°C, washed twice in PBS for 5 min and fixed in 4% (w/v) cold paraformaldehyde in PBS for 10 min at RT or for SOD2 staining, permeabilised and blocked with 0.5% (v/v) Saponin/PBG (0.5% (w/v) BSA solution/0.2% (v/v) fish skin gelatin) in PBS for 10 mins at RT, incubated with 1μg/ml rabbit anti-human SOD2 antibody in 0.5% Saponin/PBG for 45 mins at RT, washed twice with 0.5% Saponin/PBG and then incubated with secondary antibody (goat anti-rabbit Alexa 488) (1:750 in 0.5% Saponin/PBG) for 45 mins at RT, before fixing as above. All slides were washed twice with PBS and mounted in Vectashield and visualised using confocal microscopy.
Image analysis
Fluorescent and immunohistochemical images were analysed using ImageJ analysis software (Wayne Rasband, NIH, USA). For fluorescent images, individual cells from random fields were selected and the average red (MitoSOX™ Red) and/or green (SOD2) pixels present in the cell (per area) calculated after average background subtraction. Immunological staining from cartilage sections was quantified per section area for SOD3 or on a per cell basis for SOD1 and SOD2.
DNA Methylation Analysis
HAC were cultured ± 10 μM AZA for 35 days prior to RNA isolation with fresh AZA added daily. SOD2 expression was determined by real-time RT-PCR. Genomic DNA (gDNA) was purified from cartilage samples during RNA extraction [22] by ethanol precipitation of the first wash of Qiagen RNeasy columns (Qiagen, Crawley, UK). 200-300ng gDNA was bisulfitetreated using the Epitect Bisulfite kit (Qiagen). PCR was performed to amplify a SOD2 promoter region from 10-15 ng bisulfite-treated gDNA using Titanium Taq (TaKaRa Biomedicals, Wokingham, UK). PCR conditions were: 95°C 1 min, (95°C 15 sec, 64.2°C 30 sec, 68°C 1 min) × 40 cycles, 68°C for 7 min. PCR products were cloned and DNA sequenced until ten unique clones from each patient sample were obtained. DNA methylation analysis was performed using the BiQ analyzer software.[23]
Statistical Analysis
Differences between NOF and OA groups and cultured cell treatments were defined using a two-tailed Mann-Whitney U test. Fisher’s exact test was used for statistical analysis of DNA methylation. Student’s t-tests were performed when quantifying the level of immunological or MitoSOX™ staining.
RESULTS
The SOD gene family are highly but differentially expressed between NOF and OA human cartilage
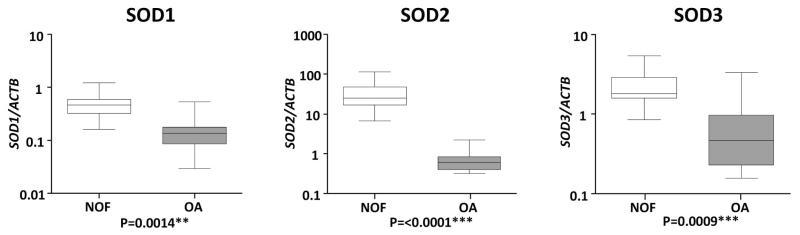
A screen for SOD expression found all three genes to have significantly reduced expression in OA compared to NOF cartilage. The median expression levels of SOD1, SOD2 and SOD3 were reduced 3.4 (P=0.0014), 41.5 (P≤0.0001) and 3.9 (P=0.0009) fold respectively in OA cartilage. This reduction in SOD2 and SOD3 expression compares with previous reports.[14, 16] The quantitative nature of this screen revealed the very high abundance of all the SOD family, particularly SOD2, with the relative abundance of each in NOF samples being: SOD2>>SOD3>ACTB>SOD1 (Fig. 1).
Figure 1.
Comparison of superoxide dismutase (SOD) gene expression between cartilage from neck of femur (NOF) and osteoarthritis (OA) patients. SOD family gene expression was determined by real-time reverse transcriptase polymerase chain reaction (RT-PCR) from cartilage RNA isolated from 12 NOF (open) and 12 OA patients (shaded). Gene expression data are presented as a ratio of SOD gene levels to that of the housekeeping gene Actin B (ACTB) using the calculation 2−ΔCT. Lines within the boxes represent the median, the boxes represent the 25th and 75th percentiles, and the lines outside the boxes correspond to the minimum and maximum values. Presented P values were calculated using the Mann-Whitney U-test, where * represents P<0.05, ** P<0.01 and *** P<0.001.
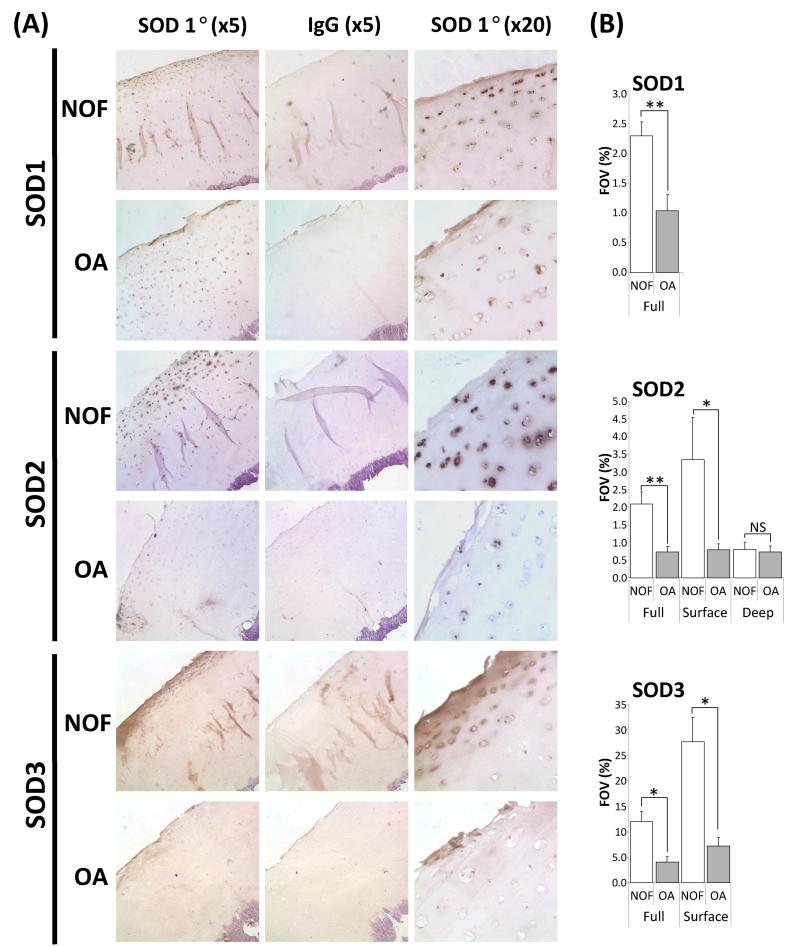
To validate our RNA expression data we performed immunohistochemistry using OA and NOF cartilage (Fig. 2A). As expected SOD1 expression was cellular with staining obvious throughout the sections (i.e. from articular surface to subchondral bone), though staining was more intense at the articular surface. Staining within the full-depth cartilage was significantly reduced in OA sections (Fig. 2B). SOD2 staining was cellular and strongest near the articular surface. There was a clear, significant, decrease in the expression of SOD2 in the surface zones of the diseased tissue though the protein was still detected. A SOD3 antibody gave the expected diffuse staining pattern representative of ECM staining. This was visible throughout the tissue though staining was more intense at the articular surface and again significantly reduced in OA compared to NOF cartilage.
Figure 2.
Immunohistochemical analysis of SOD expression in representative specimens of human articular cartilage from NOF and OA patients. A, Sections were subjected to immunohistochemistry using the labelled anti-SOD or normal rabbit IgG (negative control) and counterstained with haematoxylin. Full-depth sections are shown at × 5 magnification and for clarity the articular surface is shown at × 20. Images are representative of sections from 5 NOF and 5 OA patients. B, immunohistochemical images, of fixed exposure, from 5 NOF and 5 OA patients were quantified using the ImageJ software. For each image the percentage staining in each field of view (FOV) was calculated and the mean with standard error of the mean (SEM) NOF and OA FOV presented. Percentage FOV was calculated for full depth sections or dividing the section into two quadrants ‘deep’ and ‘surface’. P values were calculated using Student’s t-test, where * represents P<0.05 and ** P<0.01.
SOD2 expression declines prior to the appearance of OA erosions
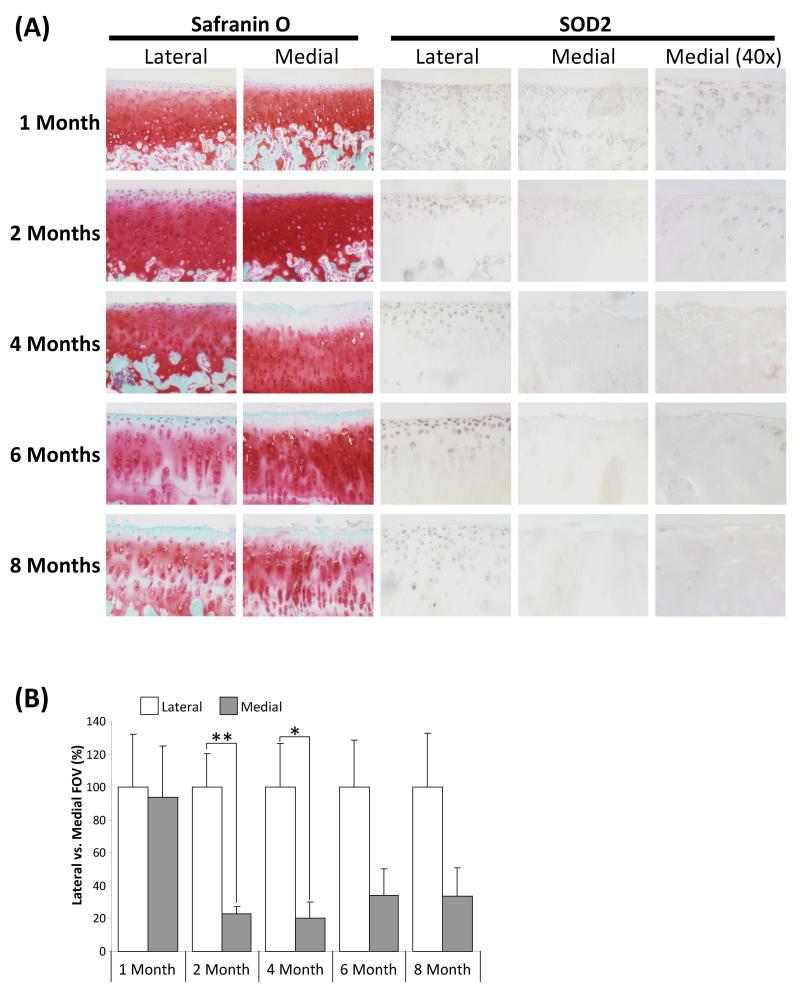
Since the changes in SOD2 expression were most significant we next ascertained its expression during the onset of OA using joint sections from an age-range of Dunkin Hartley guinea pigs; an established model of spontaneous OA [24]. In this model the medial tibial plateau develops OA-like lesions as the animal ages, whilst the lateral tibial plateau develops significantly fewer lesions as highlighted by cellular and surface changes together with a decrease in safranin ‘O’ staining, representative of proteoglycan loss (Fig. 3). In young animals (1 month) SOD2 staining was strongest near the articular surface of both medial and lateral plateaus (Fig. 3A), as observed in human NOF cartilage (Fig. 2). At 4 months of age OA-like lesions could be observed on the medial but not lateral plateau. Concomitantly SOD2 expression was significantly reduced in the diseased medial, but not the normal lateral plateau (Fig. 3A & B). Interestingly, SOD2 expression within the medial plateau significantly declined prior to observation of lesions (i.e. in 2 month old animals) suggesting that this change in expression may be symptomatic of early changes in OA (Fig. 3A & B).
Figure 3.
Immunohistochemical analysis of SOD2 expression in representative specimens of Dunkin-Hartley guinea pig articular cartilage. A, Cartilage sections are representative of 5 animals at each time point (1 to 12 months of age). Sections were either stained with safranin ‘O’ as a marker of proteoglycan or subjected to immunohistochemistry using the labelled anti-SOD2 or normal rabbit IgG (negative control – data not shown). Lateral and medial tibial plateaus are as indicated. Images shown are at × 10 magnification, unless indicated otherwise. B, Immunohistochemical images of lateral and medial tibial plateaus, of fixed exposure, from 5 animals were quantified using the ImageJ software. For each image the percentage staining in each field of view (FOV) was calculated. For each age group the staining in the lateral plateau is arbitrarily 100% + SEM. P values were calculated using Student’s t-test, where * represents P<0.05 and ** P<0.01.
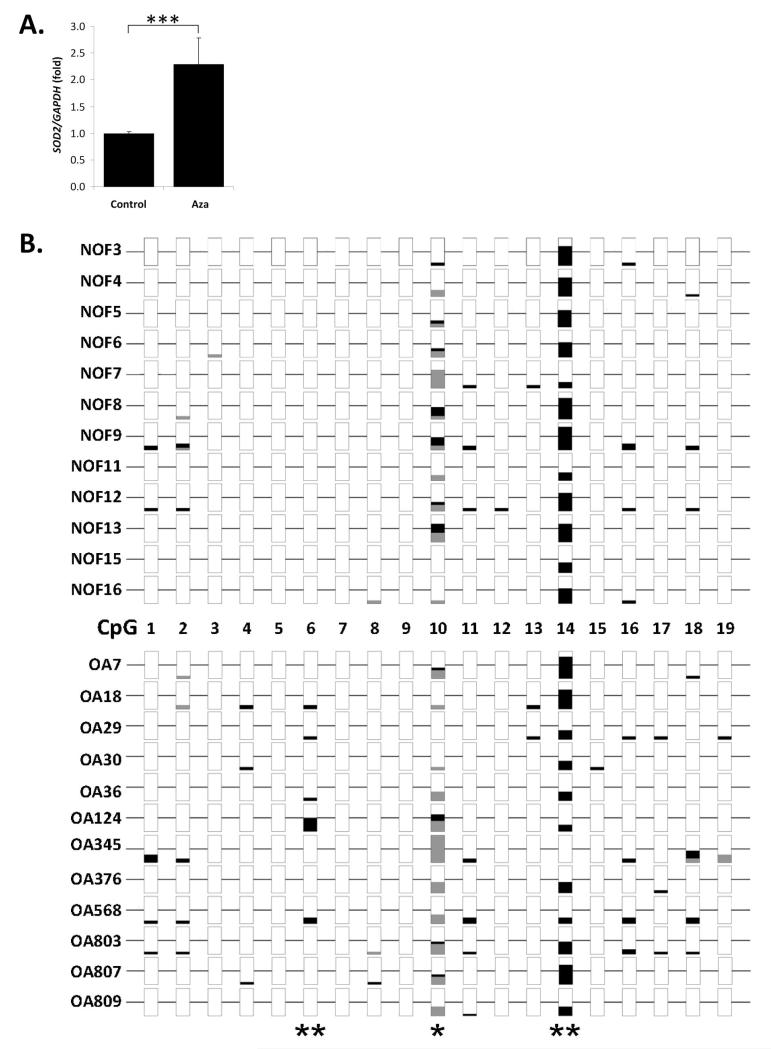
The SOD2 promoter is differentially methylated between NOF and OA cartilage
As SOD2 promoter hypermethylation correlates with decreased gene expression (e.g. [25]), we investigated the role that DNA promoter methylation may play in the SOD2 down-regulation observed in OA chondrocytes. Firstly we found that SOD2 expression increased 2-fold (P≤0.001) in OA HAC treated with the DNA demethylating agent AZA (Fig. 4A), indicating a role for DNA methylation in SOD2 regulation. Next, we analysed a region of the SOD2 promoter upstream of the transcription start site at −297 to −87 (Supp. Fig. 1) known to be subject to differential methylation.[25] Bisulfite sequencing of on average 9.6 clones from each of 12 OA and NOF patient cartilage samples identified three CpG sites, CpG6, 10 and 14, corresponding to nucleotides −222, −183 and −154 respectively, with significant differences in methylation status between NOF and OA cartilage (Fig. 4B). CpG14 and CpG10 showed significantly (P≤0.001 and P≤0.05 respectively) lower methylation in OA cartilage whereas CpG6 was more methylated (P≤0.001). CpG14 is encompassed within a putative AP-2 binding site while CpG10 is located at a single nucleotide polymorphism (SNP) (NCBI reference rs2758343). Overall OA cartilage had, as hypothesised, a significant increase (P≤0.05) in methylated CpG.
Figure 4.
Analysis of the regulation of SOD2 in human articular chondrocytes (HAC) by DNA methylation. A, Primary HAC were treated with the DNA demethylating agent 5-aza-2′deoxycytidine (AZA) for 35 days (2 passages) as described and SOD2 expression measured by real-time RT-PCR and normalised to GAPDH expression levels. Data is presented fold over control and error bars represent SEM. Data is combined from four HAC populations treated in duplicate. Statistical significance was calculated using the Mann-Whitney U-test, where *** represents P<0.001. B, DNA methylation analysis of the SOD2 promoter in cartilage genomic DNA from neck of femur (NOF) and osteoarthritis (OA) patients. Genomic DNA from cartilage of 12 NOF and 12 OA patients was analysed to determine the methylation status of the SOD2 promoter region spanning 19 potential CpG dinucleotides. An average of 9.6 non-clonal DNA sequences per patient sample were analysed and the methylation percentage for each is plotted. Methylation CpG sequences are represented by black bars, open are non-methylated and the bar is grey when the CpG sequence was not detected, generally due to a SNP at that locus (e.g. CpG10). Fisher’s exact test was used for statistical analysis, where * represents P<0.05 and ** P<0.01.
SOD2 depletion modulates MMP-1 and MMP-13 expression in HAC
A reduction in SOD levels are likely to lead to an increase in ROS. Since SOD2 is localised to the mitochondria, the major source of ROS, we used the mitochondria-targeting MitoSOX™ Red, which is oxidized by O2 , but not other ROS or reactive nitrogen species, to confirm that SOD2 depletion increased O2
, but not other ROS or reactive nitrogen species, to confirm that SOD2 depletion increased O2 levels in chondrocytes (Fig 5A & B). A further consequence of a reduction in SOD2 is a decrease in H2O2 which acts as an intracellular signalling moiety.[26] Hence, we tested whether a reduction in SOD2 in chondrocytes would effect basal and pro-inflammatory cytokine (IL-1) induced collagenase (MMP-1 and MMP-13) expression. RNAi depletion of SOD2 in chondrocytes (Fig. 5C) resulted in a significant reduction of basal MMP-1 but not MMP-13 expression (Fig. 5D). OA has an inflammatory component [27] and IL-1, a potent inducer of SOD2 and the collagenases, can be detected within the OA joint. In line with this, IL-1 stimulation led to a robust increase in SOD2 and collagenase expression. However, depletion of SOD2 significantly decreased the IL-1-induced expression of both collagenases (Fig. 5E).
levels in chondrocytes (Fig 5A & B). A further consequence of a reduction in SOD2 is a decrease in H2O2 which acts as an intracellular signalling moiety.[26] Hence, we tested whether a reduction in SOD2 in chondrocytes would effect basal and pro-inflammatory cytokine (IL-1) induced collagenase (MMP-1 and MMP-13) expression. RNAi depletion of SOD2 in chondrocytes (Fig. 5C) resulted in a significant reduction of basal MMP-1 but not MMP-13 expression (Fig. 5D). OA has an inflammatory component [27] and IL-1, a potent inducer of SOD2 and the collagenases, can be detected within the OA joint. In line with this, IL-1 stimulation led to a robust increase in SOD2 and collagenase expression. However, depletion of SOD2 significantly decreased the IL-1-induced expression of both collagenases (Fig. 5E).
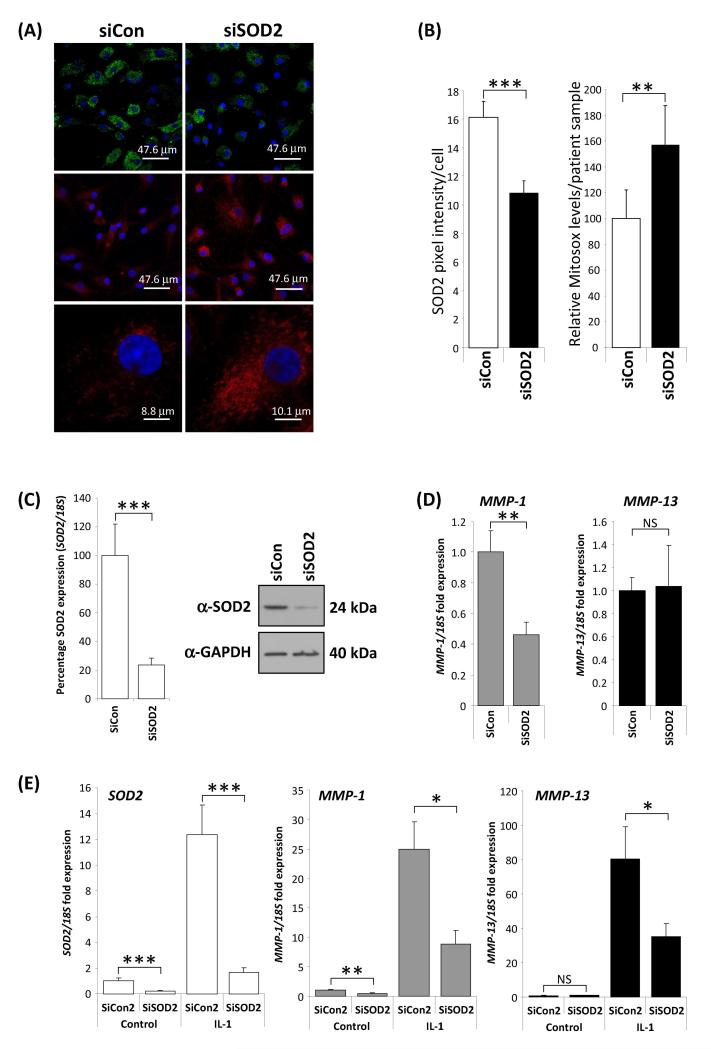
Figure 5.
Functional consequences of SOD2 depletion. A and B, SOD2 depletion (green) by RNAi led to a significant increase in MitoSOX™ Red mitochondrial staining as determined by confocal microscropy. Data are taken from four patient chondrocyte preparations. Images were quantified using ImageJ software on a per cell basis from random field images. P values were calculated using a Student’s t-test, where ** represents P<0.01 and *** P<0.001 C-E, Depletion of SOD2 leads to reduction in collagenase gene expression. C, transfection of primary human articular chondrocytes (HAC) from OA patients with siRNA to SOD2 (siSOD2) for 48 hours under basal conditions (24 hour serum free) compared to transfection with an equivalent amount (100nM) of a non-targeting control siRNA (siCon) resulted in a highly significant approximately 80% reduction in SOD2 RNA level as determined by realtime RT-PCR. Concomitantly a substantial reduction in SOD2 protein expression was measured by immunoblotting (an anti-GAPDH antibody was used as a protein loading control). D, MMP-1 and MMP-13 basal expression levels were measured as described above. SOD2 RNAi resulted in a significant reduction in MMP-1 but no change in the levels of MMP-13. E, interleukin-1 (IL-1, 0.05 ng/ml) stimulation for 24 hours induced the expression of SOD2, MMP-1 and MMP-13, however the induced-expression of all three genes was significantly reduced when SOD2 was depleted by RNAi (siSOD2). Throughout RNA expression levels were normalised to the 18S gene and data plotted as fold over control levels. Data presented are from a single experiment (n=8) using cells from one donor. The experiment was performed three times using different patient cell populations. P values were calculated using the Mann-Whitney U-test, where * represents P<0.05, ** P<0.01 and *** P<0.001. Error bars shown are SEM.
DISCUSSION
Here we found that all of the SOD family of antioxidant enzymes are highly expressed in NOF human cartilage in relation to an established housekeeping gene, which suggests that cartilage must generate significant levels of ROS and that ROS scavenging is necessary for tissue maintenance and homeostasis.[28] Our findings show that the expression of all SOD enzymes is significantly reduced in diseased cartilage, confirming and expanding on previous data for SOD2 and SOD3.[14-16] Regan et al., found that SOD3 was reduced in OA compared to normal (again, hip fracture) human tissue and in the cartilage of the STR/ort mouse spontaneous OA model, interestingly also before histological evidence of disease.[16] SOD3 levels in synovial fluid are also reduced in OA patients compared to patients with injured/painful joints but no cartilage damage.[29] We have now shown not only that SOD2 expression is significantly down regulated in OA chondrocytes in vivo, but that this precedes the development of OA lesions in the Hartley guinea pig, raising the possibility that alterations in SOD2 expression are associated with the earliest stages of OA pathogenesis.
SOD2 expression is subject to epigenetic control via promoter DNA methylation in a number of carcinoma cell-lines.[25] Furthermore, aberrant DNA methylation occurs with disease and ageing.[30] In chondrocytes, SOD2 expression increased following inhibition of DNA methylation (Fig 4A) and analysis of the SOD2 promoter identified a significant overall increase in the number of CpG sites partially methylated in OA compared to NOF cartilage (Fig 4B). Promoter DNA methylation is generally repressive to transcription, thus even the modest increase in CpG methylation observed here may significantly impact on the SOD2 expression level. Re-instigation of SOD2 expression using DNA methyltransferase inhibitors is one way in which they may benefit OA patients, as proposed.[31]
SOD are required for the conversion of reactive O2 to O2 and H2O2 thus a down-regulation of SOD would increase oxidative stress, which is implicated in a number of pathologies including arthritis.[32] As such, research has investigated the consequences of modulating SOD levels in both cells and animal models. SOD1 and SOD3 knockout mice display relatively mild phenotypes [33] but exhibit an increase in oxidative stress markers.[34] SOD2 deletion in mice results in lethality by neonatal day 10.[35] SOD2 is localised to the mitochondria and dismutates O2
to O2 and H2O2 thus a down-regulation of SOD would increase oxidative stress, which is implicated in a number of pathologies including arthritis.[32] As such, research has investigated the consequences of modulating SOD levels in both cells and animal models. SOD1 and SOD3 knockout mice display relatively mild phenotypes [33] but exhibit an increase in oxidative stress markers.[34] SOD2 deletion in mice results in lethality by neonatal day 10.[35] SOD2 is localised to the mitochondria and dismutates O2 produced as a by-product of oxidative phosphorylation. The depletion of SOD2 is proposed to lead to increased mitochondrial DNA mutation and subsequent dysfunction. Interestingly, mitochondrial DNA mutations increase in OA [36], and mitochondrial DNA haplogroups themselves contribute to the pathogenesis of OA.[37]
produced as a by-product of oxidative phosphorylation. The depletion of SOD2 is proposed to lead to increased mitochondrial DNA mutation and subsequent dysfunction. Interestingly, mitochondrial DNA mutations increase in OA [36], and mitochondrial DNA haplogroups themselves contribute to the pathogenesis of OA.[37]
The combined effect of a decrease in all three SOD enzymes could significantly contribute to the increased oxidative stress observed in OA cartilage.[38] Using RNAi we confirmed that depletion of SOD2 from chondrocytes leads to an increase in O2 , which can result in telomere instability and a down regulation of chondrocyte function [38] as well as damage to cartilage macromolecules such as hyaluronan and collagen [9, 39]. A ROS increase could also potentially contribute to the onset of senescence, which may play a role in OA.[1] Together these and other data would suggest that SOD have a protective role in the joint and cartilage. However, the results here demonstrate that down-regulation of SOD2 is in part chondroprotective, reducing the activation of redox-sensitive signalling pathways and thus collagenase expression. Similarly in fibroblasts, SOD2 over-expression increases MMP-1 expression via increased H2O2-dependent activation of ERK1/2 [40] and AP-1 [41], while SOD2-deficient fibroblasts cannot induce MMP-1 and MMP-13 following cytokine stimulation.[40] It is therefore perhaps unsurprising that the depletion of SOD2 in HAC resulted in the decreased basal MMP-1, an observation seen in end-stage OA [22], as well as decreased production of both collagenases following IL-1 stimulation (Fig. 5), presumably by reduced H2O2 levels. H2O2 levels are also controlled by catalase whose expression we found to be significantly increased in OA cartilage (not shown); indeed, increased catalase expression can reverse the H2O2-mediated induction of MMP-1 in fibroblasts.[40] Therefore one potential benefit of a down-regulation in SOD2 would be to reduce the expression of matrix-degrading enzymes by modulation of redox-controlled signalling pathways. However, the role of ROS is complex and increased levels can actually activate latent MMP [8] while more aggressive ROS such as ONOO−, generated from O2
, which can result in telomere instability and a down regulation of chondrocyte function [38] as well as damage to cartilage macromolecules such as hyaluronan and collagen [9, 39]. A ROS increase could also potentially contribute to the onset of senescence, which may play a role in OA.[1] Together these and other data would suggest that SOD have a protective role in the joint and cartilage. However, the results here demonstrate that down-regulation of SOD2 is in part chondroprotective, reducing the activation of redox-sensitive signalling pathways and thus collagenase expression. Similarly in fibroblasts, SOD2 over-expression increases MMP-1 expression via increased H2O2-dependent activation of ERK1/2 [40] and AP-1 [41], while SOD2-deficient fibroblasts cannot induce MMP-1 and MMP-13 following cytokine stimulation.[40] It is therefore perhaps unsurprising that the depletion of SOD2 in HAC resulted in the decreased basal MMP-1, an observation seen in end-stage OA [22], as well as decreased production of both collagenases following IL-1 stimulation (Fig. 5), presumably by reduced H2O2 levels. H2O2 levels are also controlled by catalase whose expression we found to be significantly increased in OA cartilage (not shown); indeed, increased catalase expression can reverse the H2O2-mediated induction of MMP-1 in fibroblasts.[40] Therefore one potential benefit of a down-regulation in SOD2 would be to reduce the expression of matrix-degrading enzymes by modulation of redox-controlled signalling pathways. However, the role of ROS is complex and increased levels can actually activate latent MMP [8] while more aggressive ROS such as ONOO−, generated from O2 and NO, can also modulate redox-sensitive signalling pathways.[42] As a result, the elucidation of the overall consequences of SOD2 down regulation requires more detailed analysis of both the levels of O2
and NO, can also modulate redox-sensitive signalling pathways.[42] As a result, the elucidation of the overall consequences of SOD2 down regulation requires more detailed analysis of both the levels of O2 present in NOF and OA chondrocytes as well as activity of the associated redox-sensitive signalling pathways.
present in NOF and OA chondrocytes as well as activity of the associated redox-sensitive signalling pathways.
Several studies have used antioxidants as treatments for arthritis (reviewed in [18]). Ex vivo SOD3 gene transfer or SOD mimetics (e.g. M40403) can reduce the severity of CIA in models [43, 44] and recombinant SOD1 can inhibit cartilage damage in hens. Intra-articular injections of bovine SOD1 (Orgotein) have also been trialled in OA patients with some success, however the drug was withdrawn due to adverse side effects (reviewed in [18]). In terms of dietary supplements, vitamins have been shown to decrease OA development and increase the expression of antioxidant enzymes in an OA model.[18] However, epidemiological studies examining the benefits of antioxidants in human OA, especially vitamin E (α-tocopherol), are contradictory (reviewed in [4]).
In conclusion, this is the first detailed study to examine the expression of all three SOD antioxidant enzymes in cartilage and OA. Our data clearly shows a decrease in the expression of all three enzymes in OA cartilage, with SOD2 and SOD3 expression decreasing before the appearance of histologically detectable lesions. ROS are normal and useful metabolites, serving important roles in a number of processes, including as a signalling molecule. Depletion of SOD2 reduced the IL-1-induced expression of MMP-1 and MMP-13, indicating that the decrease of SOD2 expression in OA cartilage may represent a chondroprotective mechanism. However, SOD2 depletion also lead to an increase in ROS and such an overproduction, as occurs in injury, disease, or ageing, can lead to cell dysfunction and death.[32] Clearly the balance of ROS levels and the consequences of altered levels on cartilage are complex. Further work is required to determine whether the decrease in antioxidant enzymes reported here are an attempt by the joint to reduce cartilage destruction mediated by modulating the levels of the collagenases but which ultimately leads to an increase in ROS and loss of cartilage homeostasis. Such information is pertinent if antioxidant treatments hold the key to potential OA therapies.
Supplementary Material
The SOD2 promoter. Bioinformatic analysis of the SOD2 promoter was performed using genomatix software. The DNA sequence is numbered with respect to the published transcription start site (+1) of RNA transcript NM_000636 (Genbank ID). Exonic sequence is shown in bold typeface and intronic in lower case. Intron 1 splice site acceptor and donor sequences are underlined. CpG sequences analysed by bisulfite sequencing are shown in bold typeface and numbered consecutively. Dotted arrowed lines indicate the locations of bisulfite primers (Table 1) used to PCR amplify the bisulfite converted SOD2 promoter. Putative binding sites for the factors are shown above (+ orientation) or below (− orientation) the appropriate DNA sequences. SNP (rs2758343) at CpG10 is depicted. This is predicted to cause the loss of a putative HIF-1 binding site to become a FTF site (dashed line). HIF-1, hypoxia inducible factor-1; FTF, alpha fetoprotein transcription factor.
DNA methylation analysis of the SOD2 promoter of neck of femur (NOF) and osteoarthritis (OA) patient cartilage genomic DNA. Figure is essentially as described for Figure 4 but data from all clones sequenced for each patient is presented in the form of a lollipop plot [23]. Open circles show non-methylated CpG dinucleotides, filled represented a non-converted, therefore methylated CpG. A vertical line indicated the lack of a CpG sequence at that locus for the individual clone. A, NOF patient samples. B, OA patient samples.
ACKNOWLEDGMENTS AND AFFILIATIONS
Supported by the Nuffield Foundation (Oliver Bird), Arthritis Research Campaign, the JGW Patterson Foundation the UK NIHR Biomedical Research Centre for Ageing and Age Related Disease Award to the Newcastle upon Tyne Foundation Hospitals NHS Trust. We would like to thank Mark Birch for help with Image quantification and Lorraine Southam and John Loughlin for donating OA genomic DNA samples.
“The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in our licence (http://ard.bmj.com/ifora/licence.pdf).”
REFERENCES
- 1.Martin JA, Buckwalter JA. Aging, articular cartilage chondrocyte senescence and osteoarthritis. Biogerontology. 2002;3(5):257–64. doi: 10.1023/a:1020185404126. [DOI] [PubMed] [Google Scholar]
- 2.Dudhia J. Aggrecan, aging and assembly in articular cartilage. Cell Mol Life Sci. 2005 Oct;62(19-20):2241–56. doi: 10.1007/s00018-005-5217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington JL. Aging bone and cartilage: cross-cutting issues. Biochem Biophys Res Commun. 2005 Mar 18;328(3):700–8. doi: 10.1016/j.bbrc.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 4.Henrotin Y, Kurz B, Aigner T. Oxygen and reactive oxygen species in cartilage degradation: friends or foes? Osteoarthritis Cartilage. 2005 Aug;13(8):643–54. doi: 10.1016/j.joca.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Plumb MS, Aspden RM. The response of elderly human articular cartilage to mechanical stimuli in vitro. Osteoarthritis Cartilage. 2005 Dec;13(12):1084–91. doi: 10.1016/j.joca.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Tiku ML, Shah R, Allison GT. Evidence linking chondrocyte lipid peroxidation to cartilage matrix protein degradation. Possible role in cartilage aging and the pathogenesis of osteoarthritis. J Biol Chem. 2000 Jun 30;275(26):20069–76. doi: 10.1074/jbc.M907604199. [DOI] [PubMed] [Google Scholar]
- 7.Loeser RF, Carlson CS, Del Carlo M, et al. Detection of nitrotyrosine in aging and osteoarthritic cartilage: Correlation of oxidative damage with the presence of interleukin-1beta and with chondrocyte resistance to insulin-like growth factor 1. Arthritis Rheum. 2002 Sep;46(9):2349–57. doi: 10.1002/art.10496. [DOI] [PubMed] [Google Scholar]
- 8.Rajagopalan S, Meng XP, Ramasamy S, et al. Reactive oxygen species produced by macrophage-derived foam cells regulate the activity of vascular matrix metalloproteinases in vitro. Implications for atherosclerotic plaque stability. J Clin Invest. 1996 Dec 1;98(11):2572–9. doi: 10.1172/JCI119076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen SV, Oury TD, Ostergaard L, et al. Extracellular superoxide dismutase (EC-SOD) binds to type i collagen and protects against oxidative fragmentation. J Biol Chem. 2004 Apr 2;279(14):13705–10. doi: 10.1074/jbc.M310217200. [DOI] [PubMed] [Google Scholar]
- 10.Klamfeldt A, Marklund S. Enhanced breakdown in vitro of bovine articular cartilage proteoglycans by conditional synovial medium. The effect of superoxide dismutase and catalase. Scand J Rheumatol. 1987;16(1):41–5. [PubMed] [Google Scholar]
- 11.Rowan AD, Young DA. Collagenase gene regulation by pro-inflammatory cytokines in cartilage. Front Biosci. 2007;12:536–50. doi: 10.2741/2080. [DOI] [PubMed] [Google Scholar]
- 12.Johnson K, Jung A, Murphy A, et al. Mitochondrial oxidative phosphorylation is a downstream regulator of nitric oxide effects on chondrocyte matrix synthesis and mineralization. Arthritis Rheum. 2000 Jul;43(7):1560–70. doi: 10.1002/1529-0131(200007)43:7<1560::AID-ANR21>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 13.Nelson KK, Melendez JA. Mitochondrial redox control of matrix metalloproteinases. Free Radic Biol Med. 2004 Sep 15;37(6):768–84. doi: 10.1016/j.freeradbiomed.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Aigner T, Fundel K, Saas J, et al. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006 Nov;54(11):3533–44. doi: 10.1002/art.22174. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz-Romero C, Calamia V, Mateos J, et al. Mitochondrial dysregulation of osteoarthritic human articular chondrocytes analyzed by proteomics: a decrease in mitochondrial superoxide dismutase points to a redox imbalance. Mol Cell Proteomics. 2009 Jan;8(1):172–89. doi: 10.1074/mcp.M800292-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regan E, Flannelly J, Bowler R, et al. Extracellular superoxide dismutase and oxidant damage in osteoarthritis. Arthritis Rheum. 2005 Nov;52(11):3479–91. doi: 10.1002/art.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross AD, Banda NK, Muggli M, et al. Enhancement of collagen-induced arthritis in mice genetically deficient in extracellular superoxide dismutase. Arthritis Rheum. 2004 Nov;50(11):3702–11. doi: 10.1002/art.20593. [DOI] [PubMed] [Google Scholar]
- 18.Afonso V, Champy R, Mitrovic D, et al. Reactive oxygen species and superoxide dismutases: role in joint diseases. Joint Bone Spine. 2007 Jul;74(4):324–9. doi: 10.1016/j.jbspin.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Hui W, Litherland GJ, et al. Differential Toll-like receptor-dependent collagenase expression in chondrocytes. Ann Rheum Dis. 2008 Feb;7 doi: 10.1136/ard.2007.079574. [DOI] [PubMed] [Google Scholar]
- 20.Shingleton WD, Ellis AJ, Rowan AD, et al. Retinoic acid combines with interleukin-1 to promote the degradation of collagen from bovine nasal cartilage: matrix metalloproteinases-1 and -13 are involved in cartilage collagen breakdown. J Cell Biochem. 2000 Sep 14;79(4):519–31. [PubMed] [Google Scholar]
- 21.Milner JM, Kevorkian L, Young DA, et al. Fibroblast activation protein alpha is expressed by chondrocytes following a pro-inflammatory stimulus and is elevated in osteoarthritis. Arthritis Res Ther. 2006;8(1):R23. doi: 10.1186/ar1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kevorkian L, Young DA, Darrah C, et al. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004 Jan;50(1):131–41. doi: 10.1002/art.11433. [DOI] [PubMed] [Google Scholar]
- 23.Bock C, Reither S, Mikeska T, et al. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005 Nov 1;21(21):4067–8. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 24.Jimenez PA, Glasson SS, Trubetskoy OV, et al. Spontaneous osteoarthritis in Dunkin Hartley guinea pigs: histologic, radiologic, and biochemical changes. Lab Anim Sci. 1997 Dec;47(6):598–601. [PubMed] [Google Scholar]
- 25.Hurt EM, Thomas SB, Peng B, et al. Molecular consequences of SOD2 expression in epigenetically silenced pancreatic carcinoma cell lines. Br J Cancer. 2007 Oct 22;97(8):1116–23. doi: 10.1038/sj.bjc.6604000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock JT. Superoxide, hydrogen peroxide and nitric oxide as signalling molecules: their production and role in disease. Br J Biomed Sci. 1997 Mar;54(1):38–46. [PubMed] [Google Scholar]
- 27.Goldring SR, Goldring MB. The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin Orthop Relat Res. 2004 Oct;(427 Suppl):S27–36. doi: 10.1097/01.blo.0000144854.66565.8f. [DOI] [PubMed] [Google Scholar]
- 28.Henrotin YE, Bruckner P, Pujol JP. The role of reactive oxygen species in homeostasis and degradation of cartilage. Osteoarthritis Cartilage. 2003 Oct;11(10):747–55. doi: 10.1016/s1063-4584(03)00150-x. [DOI] [PubMed] [Google Scholar]
- 29.Regan EA, Bowler RP, Crapo JD. Joint fluid antioxidants are decreased in osteoarthritic joints compared to joints with macroscopically intact cartilage and subacute injury. Osteoarthritis Cartilage. 2008 Apr;16(4):515–21. doi: 10.1016/j.joca.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007 Aug;23(8):413–8. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Roach HI, Aigner T. DNA methylation in osteoarthritic chondrocytes: a new molecular target. Osteoarthritis Cartilage. 2007 Feb;15(2):128–37. doi: 10.1016/j.joca.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 32.McCord JM, Edeas MA. SOD, oxidative stress and human pathologies: a brief history and a future vision. Biomed Pharmacother. 2005 May;59(4):139–42. doi: 10.1016/j.biopha.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Sentman ML, Granstrom M, Jakobson H, et al. Phenotypes of mice lacking extracellular superoxide dismutase and copper- and zinc-containing superoxide dismutase. J Biol Chem. 2006 Mar 17;281(11):6904–9. doi: 10.1074/jbc.M510764200. [DOI] [PubMed] [Google Scholar]
- 34.Reaume AG, Elliott JL, Hoffman EK, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996 May;13(1):43–7. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Huang TT, Carlson EJ, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995 Dec;11(4):376–81. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 36.Grishko VI, Ho R, Wilson GL, et al. Diminished mitochondrial DNA integrity and repair capacity in OA chondrocytes. Osteoarthritis Cartilage. 2008 Jun 17; doi: 10.1016/j.joca.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rego-Perez I, Fernandez-Moreno M, Fernandez-Lopez C, et al. Mitochondrial DNA haplogroups: role in the prevalence and severity of knee osteoarthritis. Arthritis Rheum. 2008 Aug;58(8):2387–96. doi: 10.1002/art.23659. [DOI] [PubMed] [Google Scholar]
- 38.Yudoh K, Nguyen T, Nakamura H, et al. Potential involvement of oxidative stress in cartilage senescence and development of osteoarthritis: oxidative stress induces chondrocyte telomere instability and downregulation of chondrocyte function. Arthritis Res Ther. 2005;7(2):R380–91. doi: 10.1186/ar1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao F, Koenitzer JR, Tobolewski JM, et al. Extracellular superoxide dismutase inhibits inflammation by preventing oxidative fragmentation of hyaluronan. J Biol Chem. 2008 Mar 7;283(10):6058–66. doi: 10.1074/jbc.M709273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranganathan AC, Nelson KK, Rodriguez AM, et al. Manganese superoxide dismutase signals matrix metalloproteinase expression via H2O2-dependent ERK1/2 activation. J Biol Chem. 2001 Apr 27;276(17):14264–70. doi: 10.1074/jbc.M100199200. [DOI] [PubMed] [Google Scholar]
- 41.Wenk J, Brenneisen P, Wlaschek M, et al. Stable overexpression of manganese superoxide dismutase in mitochondria identifies hydrogen peroxide as a major oxidant in the AP-1-mediated induction of matrix-degrading metalloprotease-1. J Biol Chem. 1999 Sep 3;274(36):25869–76. doi: 10.1074/jbc.274.36.25869. [DOI] [PubMed] [Google Scholar]
- 42.Go YM, Patel RP, Maland MC, et al. Evidence for peroxynitrite as a signaling molecule in flow-dependent activation of c-Jun NH(2)-terminal kinase. Am J Physiol. 1999 Oct;277(4 Pt 2):H1647–53. doi: 10.1152/ajpheart.1999.277.4.H1647. [DOI] [PubMed] [Google Scholar]
- 43.Cuzzocrea S, Mazzon E, di Paola R, et al. Synergistic interaction between methotrexate and a superoxide dismutase mimetic: pharmacologic and potential clinical significance. Arthritis Rheum. 2005 Dec;52(12):3755–60. doi: 10.1002/art.21480. [DOI] [PubMed] [Google Scholar]
- 44.Cuzzocrea S, Mazzon E, Paola RD, et al. Effects of combination M40403 and dexamethasone therapy on joint disease in a rat model of collagen-induced arthritis. Arthritis Rheum. 2005 Jun;52(6):1929–40. doi: 10.1002/art.21044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The SOD2 promoter. Bioinformatic analysis of the SOD2 promoter was performed using genomatix software. The DNA sequence is numbered with respect to the published transcription start site (+1) of RNA transcript NM_000636 (Genbank ID). Exonic sequence is shown in bold typeface and intronic in lower case. Intron 1 splice site acceptor and donor sequences are underlined. CpG sequences analysed by bisulfite sequencing are shown in bold typeface and numbered consecutively. Dotted arrowed lines indicate the locations of bisulfite primers (Table 1) used to PCR amplify the bisulfite converted SOD2 promoter. Putative binding sites for the factors are shown above (+ orientation) or below (− orientation) the appropriate DNA sequences. SNP (rs2758343) at CpG10 is depicted. This is predicted to cause the loss of a putative HIF-1 binding site to become a FTF site (dashed line). HIF-1, hypoxia inducible factor-1; FTF, alpha fetoprotein transcription factor.
DNA methylation analysis of the SOD2 promoter of neck of femur (NOF) and osteoarthritis (OA) patient cartilage genomic DNA. Figure is essentially as described for Figure 4 but data from all clones sequenced for each patient is presented in the form of a lollipop plot [23]. Open circles show non-methylated CpG dinucleotides, filled represented a non-converted, therefore methylated CpG. A vertical line indicated the lack of a CpG sequence at that locus for the individual clone. A, NOF patient samples. B, OA patient samples.