Abstract
Next generation sequencing (NGS) is superseding Sanger technology for analysing intra-host viral populations, in terms of genome length and resolution. We introduce two new empirical validation data sets and test the available viral population assembly software. Two intra-host viral population ‘quasispecies’ samples (type-1 human immunodeficiency and hepatitis C virus) were Sanger-sequenced, and plasmid clone mixtures at controlled proportions were shotgun-sequenced using Roche's 454 sequencing platform. The performance of different assemblers was compared in terms of phylogenetic clustering and recombination with the Sanger clones. Phylogenetic clustering showed that all assemblers captured a proportion of the most divergent lineages, but none were able to provide a high precision/recall tradeoff. Estimated variant frequencies mildly correlated with the original. Given the limitations of currently available algorithms identified by our empirical validation, the development and exploitation of additional data sets is needed, in order to establish an efficient framework for viral population reconstruction using NGS.
Next generation sequencing (NGS) is becoming an established experimental framework for genomics and clinical diagnostics1. NGS use covers a broad spectrum of applications, including de novo sequencing, re-sequencing, and metagenomics2. NGS has been employed successfully to characterize highly variable pathogens in different contexts3,4,5,6,7,8,9,10. The combination of huge coverage, deep resolution, primer- or shotgun-based sequencing, along with the possibility to analyze multiple samples in a single run, at a reasonable cost, makes NGS a valuable resource for diagnostic purposes11,12.
Several challenges have arisen concerning the raw NGS data processing, namely mapping to an appropriate reference sequence, error detection/correction13,14, single nucleotide variants (SNV) identification, and genome assembly methods15. In addition to sample processing and library design, problems for computational assembly and SNV detection of long haploid/diploid genomes (several thousands of kilo-bases and above) include genome coverage, repeated regions, and genome rearrangements. Distinct complexities arise with viral populations or so-called ‘quasispecies’16,17. Quasispecies are composed of different proportions of distinct variants, some at very low frequency, that are often subject to recombination. One supposes here that the experimental procedures, including polymerase chain reaction amplification, minimize the chance to generate in vitro recombinants. When processing a quasispecies with NGS, it is vital to correctly recognize minority SNV which can be close to the sequencing error rate. Another challenge is to correctly assemble whole-gene or whole-genome variants, along with their proportions, without generating spurious in silico recombinants.
The current approach for analyzing a whole-gene or whole-genome quasispecies relies on clonal Sanger sequencing. While clonal sequencing has the advantage of an almost perfect variant reconstruction, given an appropriate primer design, the approach is not readily automatable, and cumbersome in terms of bench-work. In principle, NGS will overcome Sanger's limitations. A key step will be when available SNV detection and assembly algorithms are sufficiently reliable. Approaches for SNV analysis that can identify minority variants have been devised8,18,19,20. Several methodologies for quasispecies assembly, all based on reference mapping and overlap-graph paradigm, have been also introduced21,22,23,24,25,26,27,28,29,30, along with software implementations, tested both on simulated and quasi-empirical data, such as pooled inter-host samples18 or artificially created mixtures19. Performance of quasispecies reconstruction algorithms depend on many biological factors: the efficacy of error correction methods, overlap lengths, assembly heuristics, including the quasispecies diversity and of the extent of heterogeneity. It is of vital importance to assess the efficacy of quasispecies assembly methods with real data sets in order to test their use for diagnostics in a clinical context.
This work introduces two new empirical validation data sets and tests quasispecies assembly algorithms. From the plasma samples of two patients infected with type-1 human immunodeficiency or hepatitis C viruses (HIV-1, HCV), using Sanger sequencing, plasmids isolated from transformed bacterial cultures were mixed at controlled proportions, and then the mixtures were shotgun-sequenced with the Roche's 454 sequencing platform. Performance of four different assembly algorithms, including QuRe, PredictHaplo, Geneious™ de novo, and ShoRAH (see Beerenwinkel et al.26 for a comprehensive review) were evaluated in terms of phylogenetic clustering with the original Sanger clones, precision/recall indices, diversity and recombination analysis.
Results
The HCV data set comprised 16 sequences encompassing 669 bases of envelope regions E1–E2. The average diversity was 6.47%. Variant prevalence was approximately [41%, 28%, 13%, 5%, 3%, 3%, 2%, 2%, 1%, 1%, 0.2%, 0.2%, 0.1%, 0.1%, 0.1%, 0.1%]. The optimal nucleotide substitution model, according to the Akaike information criterion (AIC), was the Tamura 3-parameter (1992), with a γ-distributed rate of heterogeneity parameterized on α = 0.28. The HIV-1 data set comprised of 20 env sequences (1,409 bases of the gp120 regions V1–V5), with an average diversity of 4.7%. Variant prevalence was approximately [18%, 13%, 13%, 10%, 10%, 8%, 8%, 5%, 5%, 3%, 3%, 1%, 1%, 0.6%, 0.6%, 0.3%, 0.3%, 0.1%, 0.1%, 0.1%]. The optimal nucleotide substitution model according to AIC was the Tamura-Nei (1993), with a γ-distributed rate of heterogeneity parameterized on α = 0.32.
The Roche's 454 platform (GS FLX Titanium XLR70) yielded 156,655 reads for the HCV and 153,670 for the HIV-1 plasmid mixture, sequenced on two separate lanes. The average (st.dev.) read length was 363 (109) for HCV and 346 (98) for HIV. Using QuRe, 27,912 (17.82%) and 53,930 (35.09%) reads were successfully mapped (and trimmed) to the HCV and HIV-1 reference sequences, respectively. The length ratios between the sequenced regions and the plasmid were ~ 16% for HCV and ~ 39% for HIV-1. The average (st.dev.) coverage of each mapped base was 9,553 (1,881) for HCV and 9,565 (4,216) for HIV. All the reads were also mapped to the plasmid reference sequence and a random subset of 70,000 reads was retained from both experiments to assess sequencing error rates.
Error rate assessment and correction
Out of the two 70,000 random read sets selected from both experiments, 49,866 (71.24%) of the HCV and 62,303 (89.00%) of the HIV-1 mixture were retained when mapped (and trimmed) to the plasmid reference. Note, the observed differences in plasmid mapping and sequence recovery are because two different insertion points were used, whilst the same linear plasmid reference sequence was used for both mappings.
The overall mean error prevalence (standard error, SE) using reads from the HCV mixture was 1.26% (0.010). Insertions, deletions and mismatches accounted for 0.40% (0.005), 0.39% (0.005), and 0.47% (0.006) of the total, respectively. As expected errors were more frequent in homopolymeric regions: 1.33% (0.017) and 1.22% (0.012), respectively. When mapping reads from the HIV-1 mixture, overall error prevalence (SE) was 1.30% (0.006). Base insertions, deletions and mismatches accounted for 0.36% (0.003), 0.47% (0.004), and 0.47% (0.004) of the total, respectively. Error prevalence (SE) in homopolymeric regions was 2.14% (0.016) and 1.05% (0.006) in non-homopolymeric regions.
Using the error correction method embedded in QuRe, the overall error was decreased by 85.29% (6.80 fold) in the HCV sample and by 69.77% (3.54 fold) in the HIV-1 sample. With ShoRAH, the decrease was 85.96% (7.12 fold) for HCV and 83.95% (6.66 fold) for HIV-1. Table 1 reports error rates for the two data sets and correction methods, overall and stratified by error type (insertions, deletions, mismatches and homopolymeric regions).
Table 1. Raw and corrected (QuRE, ShoRAH) estimated error rates for the Roche's GS FLX Titanium XLR70, as estimated by shotgun-sequencing the plasmid TOPO2.1 with HCV/HIV-1 inserts, and mapping the reads against the TOPO2.1 reference using the QuRe read aligner/filter module.
| error prevalence (standard error) by type | ||||||||
|---|---|---|---|---|---|---|---|---|
| data set (n = 70,000) | mapped (n;%) | error correction method | overall error prevalence (standard error) | insertions | deletions | mismatches | homopolymeric | non-homopolymeric |
| HCV | 49,866; 71.24% | non-corrected | 1.26% (0.0096) | 0.40% (0.0054) | 0.39% (0.0054) | 0.47% (0.0059) | 1.33% (0.0168) | 1.22% (0.0117) |
| QuRe | 0.19% (0.0037) | 0.02% (0.0012) | 0.14% (0.0032) | 0.02% (0.0014) | 0.27% (0.0076) | 0.14% (0.0040) | ||
| ShoRAH | 0.18% (0.0037) | 0.00% (0.0000) | 0.12% (0.0031) | 0.06% (0.0021) | 0.24% (0.0073) | 0.14% (0.0041) | ||
| HIV-1 | 62,303; 89.00% | non-corrected | 1.30% (0.0060) | 0.36% (0.0032) | 0.47% (0.0037) | 0.47% (0.0036) | 2.14% (0.0162) | 1.05% (0.0062) |
| QuRe | 0.39% (0.0033) | 0.06% (0.0013) | 0.25% (0.0027) | 0.08% (0.0015) | 0.95% (0.0108) | 0.23% (0.0029) | ||
| ShoRAH | 0.21% (0.0025) | 0.00% (0.0001) | 0.13% (0.0020) | 0.07% (0.0015) | 0.47% (0.0079) | 0.13% (0.0022) | ||
Quasispecies assembly and evaluation of reconstruction performance
QuRe, PredictHaplo, ShoRAH and Geneious™ de novo were applied to both NGS data sets, using the corresponding HIV-1/HCV reference sequences. QuRe used the homopolymeric and non-homopolymeric error rates as estimated from mapping the reads to the plasmid reference (default parameters for the rest). The other programs were run with default parameters.
QuRe, PredictHaplo, ShoRAH and Geneious™ de novo yielded 9, 4, 200, and 81 distinct variants respectively for the HCV data set, and 9, 4, 1,247, and 159 variants for HIV-1 data set. Tables 2 and 3 report precision/recall performance of each reconstruction algorithm, population characteristics and recombination tests. QuRe and PredictHaplo exhibited the highest precision (between 0.55 and 1.00), whilst ShoRAH had the highest recall (0.3 to 0.5). Reconstructed variants were classified as ‘correct’ when clustering with original Sanger clones in a phylogenetic tree at ≥ 75% bootstrap support (see Methods). Of note, when inferring a phylogeny from the sole Sanger data set, the proportion of highly-supported nodes (>75% bootstrap) was 43% and 50%, for HCV and HIV-1, respectively (Figure 1, upper panels). Therefore placement for some variants in the tree was ambiguous (i.e. some variants were probably too similar to distinguish).
Table 2. Performance of quasispecies assembly algorithms.
| data set | Assembly method | # variants output | # correct variants | Precision | Recall |
|---|---|---|---|---|---|
| HCV (n = 16) | ShoRAH | 200 | 8 | 0.04 | 0.50 |
| QuRE | 9 | 8 | 0.89 | 0.50 | |
| PredictHaplo | 4 | 4 | 1.00 | 0.25 | |
| Geneious™ de novo | 81 (18)* | 5 | 0.06 (0.28)* | 0.31 | |
| HIV-1 (n = 20) | ShoRAH | 1,247 | 6 | 0.005 | 0.3 |
| QuRE | 9 | 5 | 0.55 | 0.25 | |
| PredictHaplo | 4 | 3 | 0.75 | 0.15 | |
| Geneious™ de novo | 159 (10)* | 4 | 0.02 (0.40)* | 0.20 |
*considering only contigs > 500 (for HCV) and > 900 (for HIV-1) base pair long.
Table 3. Population characteristics of the original Sanger and of the assembled quasispecies (post-alignment figures after gap-stripping).
| Data set | method | # variants output | PHI-test for recombination (P-value) | Average diversity (p-distance) | Minimal diversity (p-distance) | γ-distributed rate of heterogeneity (α) | effective population size | Segregating sites | Parsimony info |
|---|---|---|---|---|---|---|---|---|---|
| HCV | original Sanger clones | 16 | P = 0.043 | 0.0647 | 0.001 | 0.47 | 0.83 | 19% | 12% |
| ShoRAH | 200 | P = 0.0002 | 0.0531 | 0.000 | 0.22 | 0.16 | 28% | 26% | |
| QuRE | 9 | P = 0.023 | 0.0689 | 0.007 | 0.19 | 0.21 | 17% | 11% | |
| PredictHaplo | 4 | P = 0.03 | 0.0748 | 0.068 | 0.05 | 0.56 | 13% | 3% | |
| Geneious™ de novo | 81 (18)* | P < 0.0001 | 0.078 | 0.000 | 0.52 | 0.31 | 36% | 12% | |
| HIV-1 | original Sanger clones | 20 | P < 0.0001 | 0.047 | 0.002 | 0.09 | 13.26 | 14% | 10% |
| ShoRAH | 1,247 | P < 0.0001 | 0.0109 | 0.000 | 0.17** | 116.77 | 36% | 29% | |
| QuRE | 9 | P < 0.0001 | 0.0425 | 0.000 | 0.10 | 44.72 | 10% | 6% | |
| PredictHaplo | 4 | P < 0.0001 | 0.0685 | 0.016 | 0.05 | 20.09 | 12% | 4% | |
| Geneious™ de novo | 159 (10)* | P < 0.0001 | 0.0524 | 0.000 | 0.57 | 41.64 | 19% | 8% |
*considering only contigs > 500 (for HCV) and > 900 (for HIV-1) base pair long.
**estimated on a random subset of 200 sequences.
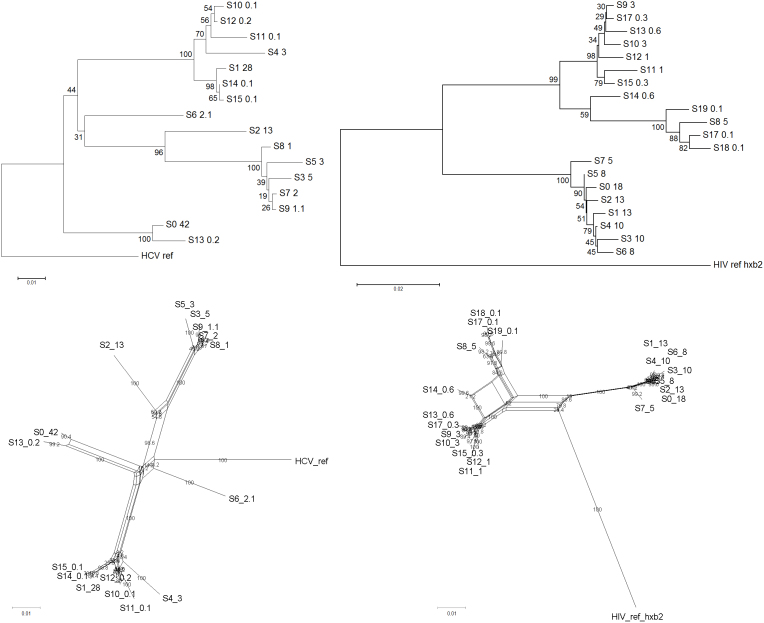
Figure 1. Phylogenetic trees (upper panels) and networks (lower panels) of the original Sanger sequences for the HCV (left panels) and HIV-1 (right panels) data sets.
Neighbor-joining and neighbor-net algorithms were run on optimized models of evolution, over 500 bootstrap runs. Node labels show bootstrap percentages. Numbers after the labels represent variant prevalence (%).
Detection of recombination, using the pairwise homoplasy index test (PHI-test), was marginally significant in the HCV Sanger data set (P = 0.043), and highly significant in the HIV-1 data set (P < 0.0001). Recombination networks are shown in Figure 1 (lower panels). QuRe and PredictHaplo yielded the lowest number of in silico recombinants, showing a moderate decrease of the PHI-test P-value. In terms of variant frequency estimation, all methods correlated significantly with the original distributions, with Pearson's linear correlation ranging from 0.3 to 0.6 (P < 0.0001).
Figure 2 depicts phylogenetic trees for the HCV experiment that compare variants output by each assembler together with the original Sanger sequences. Trees for the HIV-1 experiment are shown in Figure 3. Figure 4 shows trees for the two data sets (left and right panels for HCV and HIV-1, respectively) inferred by combining all variants from all algorithms and original Sanger sequences. Although reconstruction performance was not optimal in most cases, the same –overall- population structure can be found across all trees.
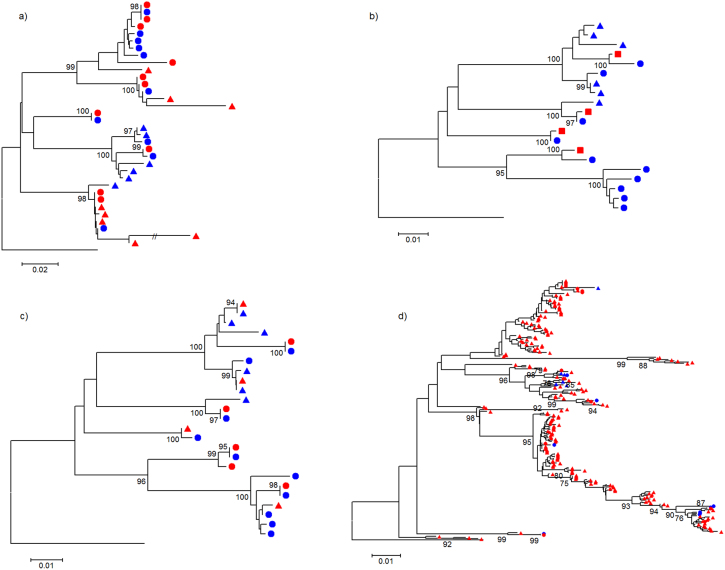
Figure 2. Evolutionary history inferred by neighbor-joining, using an optimized nucleotide substitution model, that compares HCV variants reconstructed by each quasispecies assembler with the original Sanger clones; trees are rooted using the mapping reference sequence.
Panels (a), (b), (c), and (d) show Geneious™ de novo, PredictHaplo, QuRe and ShoRAH, respectively. Node numbers represent% bootstrap replicates (of 500) ≥ 75%. Bullets represent variants at a frequency ≥ 5%, and triangles those < 5% (not available for PredictHaplo, shown by squares). Blue color indicates Sanger isolates, and red reconstructed variants.
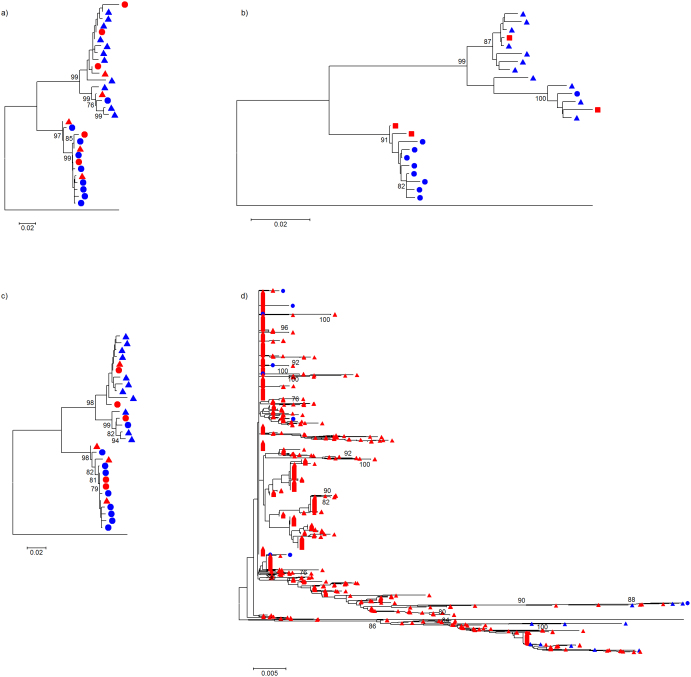
Figure 3. Evolutionary history inferred by neighbor-joining, using an optimized nucleotide substitution model, that compares HIV-1 variants reconstructed by each quasispecies assembler with the original Sanger clones; trees are rooted using the mapping reference sequence.
Panels (a), (b), (c), and (d) show Geneious™ de novo, PredictHaplo, QuRe and ShoRAH, respectively. Node numbers represent% bootstrap replicates (of 500) ≥ 75%. Bullets represent variants at a frequency ≥ 5%, and triangles those < 5% (not available for PredictHaplo, shown by squares). Blue color indicates Sanger isolates, and red reconstructed variants.
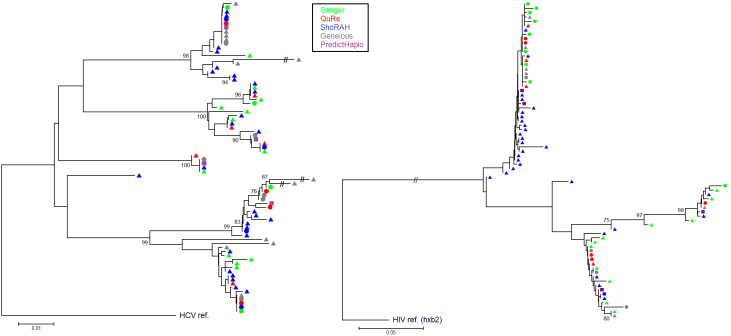
Figure 4. Phylogenetic trees comparing together all reconstructed variants from different assemblers with the original Sanger sequences for the HCV and HIV-1 experiments (left and right panels, respectively).
Trees have been inferred using neighbor-joining on an optimized model of evolution, rooted on the mapping reference sequence, performing 500 bootstraps (nodes with ≥ 75% bootstrap support are shown). For ease of read, only the 30 highest-frequency variants from ShoRAH have been included.
Discussion
This work evaluated the performance of viral quasispecies assembly algorithms in an empirical setting. Comparisons were performed using both an HCV and HIV-1 data sets, sequencing plasmid clones with Sanger technology and mixing them at known proportions. Plasmid mixtures were shotgun-sequenced using Roche's 454 platform.
The estimated error rates were similar in the HIV-1 and HCV samples. The overall error prevalence was consistent with previous estimates10,13,14, as well as the higher prevalence of errors in homopolymeric regions, and the higher prevalence of insertions and deletions as compared to mismatches (although the ratio varied across previous studies). ShoRAH provided a better error reduction as compared to QuRe. Note that the error correction was evaluated only in terms of removing mismatches or indels from the plasmid reference sequence, and not on the ability to distinguish true signal from false signal, especially in presence of minority changes. As a consequence, error correction should be evaluated also on data sets where true mutations exist and assess precision/recall rates at different frequency thresholds. Additionally, the read filtering, mapping, and error counting was carried out with QuRe, which potentially gives QuRe an advantage in the error correction phase. This is because QuRe performed error correction after mapping, while ShoRAH corrected reads before the mapping. Then ShoRAH reads were re-mapped with QuRe (not enabling its error correction module) in order to obtain comparable output.
All of the quasispecies assembly methods published to-date have performed robustly in simulations and empirical experiments18,23,31,32. This study is the first to consider actual intra-patient quasispecies, sequenced with Sanger, mixed at controlled proportions and then processed using NGS. Previously, validation on real data had been performed using inter-patient samples18,23. A recent work also used a controlled proportion approach to assess performance of error correction algorithms, but an artificial quasispecies was generated19. Here, both for HCV and HIV-1 data sets, all assemblers were able to capture a proportion of the most divergent lineages as indicated by the phylogenetic analysis. PredictHaplo was the most conservative algorithm, with the least number of variants reported, while ShoRAH yielded the highest number. QuRe and PredictHaplo exhibited the highest precision, whilst ShoRAH had the highest recall. QuRe showed the best precision/recall tradeoff. Estimated variant frequencies mildly correlated with the original values. In silico recombination was more pronounced in the output by ShoRAH and Geneious™ de novo. Geneious™ de novo was the least effective assembler in terms of precision and recall, but when considering only long contiguous sequences its performance increased substantially. Note that originally Geneious™ de novo has not been designed to assemble a quasispecies. In regards to the parameter set-up of assemblers, PredictHaplo did not require complex tuning and results were similar across different runs. ShoRAH may have benefited from a parameter optimisation (especially the sliding window sizes, but also number of iterations and the hyper-parameter α), but the search space would have required a considerable computational burden. QuRe required only a few parameters (error rates, estimated from the plasmid data, and number of iterations) and the rest was auto-optimised during the process. For all variants reconstructed by the algorithms, the cutoff for clustering with a Sanger clone was set to 75%, without a fixed threshold on nucleotide differences. Results may change when the cutoff or distance criteria are altered.
To determine the performance of reconstruction algorithms, one has to evaluate their output in different contexts. Performance of programs can be also subject to error patterns proper of NGS machinery. Besides the experimental yield, i.e. read length and coverage, factors to consider include: the number of variants in the population, the variant prevalence, the average diversity, the rate of heterogeneity, and the recombination signal. We look forward to testing methodological approaches for quasispecies assembly that specifically incorporate recombination, such as the recent QuasiRecomb, which currently performs only local reconstruction33.
Evaluation of quasispecies assemblers in different scenarios, using both simulated and empirical data, will be an important step in characterizing the clinical relevance of viral populations in infected individuals.
Methods
Sample preparation and processing
Plasmids containing HIV-1 env gp120 V1–V5, or genotype 1 HCV envelope E1–E2 sequences, derived from virus in the plasma of infected patients, previously published by Gray et al.34 and Ho et al.35, isolated from transformed bacterial cultures, were mixed at controlled decreasing proportions (from 42% to 0.1%, see the Results section). Sample preparation and processing procedures were based on the previous work of Zagordi et al.18. The cloning kit used was Invitrogen TOPO® TA Cloning® Kit (with pCR®2.1-TOPO® Vector) with One Shot® Mach1™ T1 Phage-Resistant Chemically Competent E. coli (http://products.invitrogen.com/ivgn/product/K451020).
Study design and sample usage for viral genotyping was approved by University of Florida Institutional Review Board. NGS was performed at University of Florida using the Roche's GS FLX Titanium XLR70 platform. Raw NGS data are available for free upon formal request to the authors.
Mapping to a reference and quasispecies assembly
The HCV reference used for read mapping was the H77 (Genbank NC_004102.1), encompassing 669 bases of the two envelope regions E1–E2 (positions 1,315- 1,984 relative to the full-genome numbering). The HIV-1 reference sequence was the HXB2 (Genbank K03455) env gp160 (positions 6,225–8,792 relative to the full-genome numbering).
Reads were mapped to each reference using QuRe32, a multi-threaded, platform-independent software designed for read mapping, error correction and quasispecies assembly. QuRe uses the JAligner (http://jaligner.sourceforge.net/) implementation of the Smith-Waterman-Gotoh local alignment algorithm36 for mapping reads and discards those that do not exhibit an alignment score sufficiently high as compared to a quasi-random score distribution, according to a z-test as discussed by Bacro and Comet37. For this study, gap open and extension penalties of 23 and 0.3 were chosen, respectively. Reads that spanned both a part of the viral insert and the plasmid were trimmed by the local alignment and retained if the statistical test was below the chosen threshold (P < 0.01 by default, corrected with the Benjamini-Hochberg procedure). Therefore, a read could be mapped to both the plasmid reference and the viral reference.
Error correction was performed using QuRe32 and ShoRAH30. The former implemented the method by Wang et al.8, which assumed a Poisson distribution of errors parameterized differently in homopolymeric and non-homopolymeric regions. A position in the reference sequence was defined as homopolymeric if the substring generated by elongating three bases to the left and three bases to the right of that position contained at least three consecutive bases of the same type. ShoRAH implemented a Bayesian probabilistic clustering over sliding windows across the reference sequence, correcting reads by assigning each one to the closest cluster25. Errors were calculated overall and stratified by type (insertion, deletions, mismatches, and homopolymeric vs. non-homopolymeric region).
Quasispecies assembly was executed employing QuRe v0.9994 (http://sourceforge.net/projects/qure/), ShoRAH v0.5.1 (http://www.bsse.ethz.ch/cbg/software/shorah), PredictHaplo v0.4 (http://bmda.cs.unibas.ch/HivHaploTyper/), and the Geneious™ de novo assembler (http://www.geneious.com/). The quasispecies reconstruction method of QuRe employs a method based on an overlap graph constructed over sliding windows, selecting candidate variants using an algorithm based on overlap consistency and similarity of frequency distributions of variants in each window23,32. ShoRAH perform a parsimony-based reconstruction after the sliding window Bayesian clustering30, whilst PredictHaplo extends the sliding window Bayesian clustering approach to a global quasispecies inference based on a hidden Markov model.
Phylogenetic, recombination analysis and performance assessment
Reconstructed variants were aligned with the original Sanger sequences and a phylogenetic/recombination analysis was carried out using the MEGA software v5.0538 and SplitsTree v439, as follows. In MEGA, a multiple alignment was obtained using MUSCLE40, then an optimal nucleotide substitution model was selected by minimizing the AIC, and finally bootstrapped (n = 500) neighbor-joining trees were inferred. The multiple alignment was used also as input for the recombination analysis in SplitsTree, using the PHI-test by Bruen et al.41, and generating phylogenetic networks with the bootstrapped (n = 500) neighbor-net algorithm.
By looking at the inferred phylogenetic trees, an assembled variant that was clustering with an original Sanger clone with ≥ 75% of bootstrap support was considered as a correct reconstruction. If more than one reconstructed variant was clustering with one or more original Sanger clones, this was counted as a unique correct reconstruction. A minimal nucleotide difference threshold was not fixed. This criterion was chosen to account for the fact that assemblers may output variants of different lengths and a subsequent distances obtained from the multiple alignments may be biased towards longer/shorter sequences. Precision (# of correctly reconstructed variants/# total number of reconstructed variants) and recall (# of correctly reconstructed variants/# number of original variants) were used as performance indicators, as well as the P-value of recombination as obtained by the PHI-test.
Author Contributions
M.C.F.P. conceived the study, executed computational analyses and wrote the manuscript; L.Y. designed the laboratory experiments; D.J.N. and A.D.L. performed experiments; M.M.G. supervised the experiments and reviewed the manuscript; M.S. executed phylogenetic analysis and reviewed the manuscript.
Acknowledgments
MCFP was supported by the University of Manchester's Health Research Center (HeRC) funded by the Medical Research Council (MRC) grant MR/K006665/1. MCFP, DJN and MS were supported by the University of Florida award UL1 RR02989, the 2012 EPIG grant, and by the NIH/NINDS R01 grant NS063897-01A2. MMG and LY were supported from the Stephany W. Holloway University Chair for AIDS Research, and the Center for Research in Human Immune Deficiency and Inflammation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Prof. David Robertson (University of Manchester, UK) for his valuable help in revising the manuscript and improving the English language.
References
- Metzker M. L. Sequencing technologies - the next generation. Nat Rev Genet 11, 31–46 (2010). [DOI] [PubMed] [Google Scholar]
- Pareek C. S., Smoczynski R. & Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet 52, 413–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer J. et al. The evolutionary analysis of emerging low frequency HIV-1 CXCR4 using variants through time--an ultra-deep approach. PLoS Comput Biol 6, e1001022 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M. et al. Characterization of quasispecies of pandemic 2009 influenza A virus (A/H1N1/2009) by de novo sequencing using a next-generation DNA sequencer. PLoS One 5, e10256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon A. F. et al. Phylogenetic analysis of population-based and deep sequencing data to identify coevolving sites in the nef gene of HIV-1. Mol Biol Evol 27, 819–832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozera G. et al. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte sub-populations. Retrovirology 6, 15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer J. et al. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS 23, 1209–1218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Mitsuya Y., Gharizadeh B., Ronaghi M. & Shafer R. W. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome research 17, 1195–1201 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn M. R. et al. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog 8, e1002529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L. et al. High-resolution deep sequencing reveals biodiversity, population structure, and persistence of HIV-1 quasispecies within host ecosystems. Retrovirology 9, 108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch J. R. & Grody W. W. Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. J Mol Diagn 10, 484–492 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore S. F. & Saunders C. J. Deep sequencing of patient genomes for disease diagnosis: when will it become routine? Sci Transl Med 3, 87ps23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles A. et al. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics 12, 245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer J. et al. Analysis of high-depth sequence data for studying viral diversity: a comparison of next generation sequencing platforms using Segminator II. BMC Bioinformatics 13, 47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S. et al. Evaluation of next-generation sequencing software in mapping and assembly. J Hum Genet 56, 406–414 (2011). [DOI] [PubMed] [Google Scholar]
- Holmes E. C. The RNA virus quasispecies: fact or fiction? J Mol Biol 400, 271–273 (2010). [DOI] [PubMed] [Google Scholar]
- Mas A., Lopez-Galindez C., Cacho I., Gomez J. & Martinez M. A. Unfinished stories on viral quasispecies and Darwinian views of evolution. J Mol Biol 397, 865–877 (2010). [DOI] [PubMed] [Google Scholar]
- Zagordi O., Klein R., Daumer M. & Beerenwinkel N. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res 38, 7400–7409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skums P. et al. Efficient error correction for next-generation sequencing of viral amplicons. BMC Bioinformatics 13 Suppl 10, S6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalalad A. R. et al. Highly sensitive and specific detection of rare variants in mixed viral populations from massively parallel sequence data. PLoS Comput Biol 8, e1002417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jojic V., Hertz T. & Jojic N. Population sequencing using short reads: HIV as a case study. Pac Symp Biocomput 114–125 (2008). [PubMed] [Google Scholar]
- Eriksson N. et al. Viral population estimation using pyrosequencing. PLoS Comput Biol 4, e1000074 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosperi M. C. et al. Combinatorial analysis and algorithms for quasispecies reconstruction using next-generation sequencing. BMC Bioinformatics 12, 5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerenwinkel N. & Zagordi O. Ultra-deep sequencing for the analysis of viral populations. Curr Opin Virol 1, 413–418 (2011). [DOI] [PubMed] [Google Scholar]
- Zagordi O., Geyrhofer L., Roth V. & Beerenwinkel N. Deep sequencing of a genetically heterogeneous sample: local haplotype reconstruction and read error correction. J Comput Biol 17, 417–428 (2010). [DOI] [PubMed] [Google Scholar]
- Beerenwinkel N., Gunthard H. F., Roth V. & Metzner K. J. Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front Microbiol 3, 329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso N., Tork B., Skums P., Mandoiu I. & Zelikovsky A. Viral quasispecies reconstruction from amplicon 454 pyrosequencing reads. in Bioinformatics and Biomedicine Workshops (BIBMW), 2011 IEEE International Conference on 94–101 (2011). [Google Scholar]
- Huang A., Kantor R., Delong A., Schreier L. & Istrail S. QColors: An algorithm for conservative viral quasispecies reconstruction from short and non-contiguous next generation sequencing reads. In Silico Biol 11, 193–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrooks K. et al. HCV quasispecies assembly using network flows. Lect N Bioinformat 4983, 159–170 (2008). [Google Scholar]
- Zagordi O., Bhattacharya A., Eriksson N. & Beerenwinkel N. ShoRAH: estimating the genetic diversity of a mixed sample from next-generation sequencing data. BMC Bioinformatics 12, 119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrovskaya I. et al. Inferring viral quasispecies spectra from 454 pyrosequencing reads. BMC Bioinformatics 12 Suppl 6, S1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosperi M. C. & Salemi M. QuRe: software for viral quasispecies reconstruction from next-generation sequencing data. Bioinformatics 28, 132–133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagordi O. et al. Probabilistic Inference of Viral Quasispecies Subject to Recombination. in Research in Computational Molecular Biology Vol. 7262, (ed. Chor, B.) 342–354 (Springer Berlin Heidelberg, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R. R. et al. Unexpected maintenance of hepatitis C viral diversity following liver transplantation. J Virol 86, 8432–8439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. K. et al. Genetic determinants in HIV-1 Gag and Env V3 are related to viral response to combination antiretroviral therapy with a protease inhibitor. Aids 23, 1631–1640 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. An improved algorithm for matching biological sequences. J Mol Biol 162, 705–708 (1982). [DOI] [PubMed] [Google Scholar]
- Bacro J. N. & Comet J. P. Sequence alignment: an approximation law for the Z-value with applications to databank scanning. Comput Chem 25, 401–410 (2001). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H. & Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen T. C., Philippe H. & Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]