Abstract
C3 glomerulopathy describes glomerular pathology associated with predominant deposition of complement C3 including dense deposit disease and C3 glomerulonephritis. Familial C3 glomerulonephritis has been associated with rearrangements affecting the complement factor H–related (CFHR) genes. These include a hybrid CFHR3-1 gene and an internal duplication within the CFHR5 gene. CFHR5 nephropathy, to date, occurred exclusively in patients with Cypriot ancestry, and is associated with a heterozygous internal duplication of the CFHR5 gene resulting in duplication of the exons encoding the first two domains of the CFHR5 protein. Affected individuals possess both the wild-type nine-domain CFHR5 protein (CFHR512-9) and an abnormally large mutant CFHR5 protein in which the initial two protein domains are duplicated (CFHR51212-9). We found CFHR51212-9 in association with familial C3 glomerulonephritis in a family without Cypriot ancestry. The genomic rearrangement was distinct from that seen in Cypriot CFHR5 nephropathy. Our findings strengthen the association between CFHR51212-9 and familial C3 glomerulonephritis and recommend screening for CFHR51212-9 in patients with C3 glomerulopathy irrespective of ethnicity. Since CFHR51212-9 can result from at least two genomic rearrangements, screening is most readily achieved through analysis of CFHR5 protein.
Aberrant regulation of the complement alternative pathway is associated with C3 glomerulopathy (C3G), a group of glomerular diseases characterized by predominant complement C3 deposition. C3G incorporates entities with distinct EM features: dense deposit disease and C3 glomerulonephritis.1 Complement factor H (CFH) is an important physiological regulator of the complement alternative pathway, and animal and human models have demonstrated a strong link between CFH dysfunction and C3G.2 CFH is part of a family that includes the five complement factor H–related proteins (CFHR1-5). Their contribution to C3G pathogenesis was first revealed by the demonstration that a heterozygous CFHR5 mutation was associated with familial C3G in patients of Cypriot descent (Cypriot CFHR5 nephropathy).3 The mutation resulted in duplication of the first two protein subunits (termed short consensus repeat (SCR) domains resulting in a CFHR5 protein with 11 (CFHR51212-9) rather than the normal nine SCR domains (CFHR512-9). This type of C3G was termed CFHR5 nephropathy. In addition, a novel CFHR3-1 hybrid gene was associated with familial C3G in a family with Irish ancestry.4 Here we report the first case of familial C3G associated with CFHR51212-9 in a family of non-Cypriot descent. In this family, CFHR51212-9 arose as a consequence of a genomic rearrangement distinct from that characterized in Cypriot CFHR5 nephropathy.
RESULTS
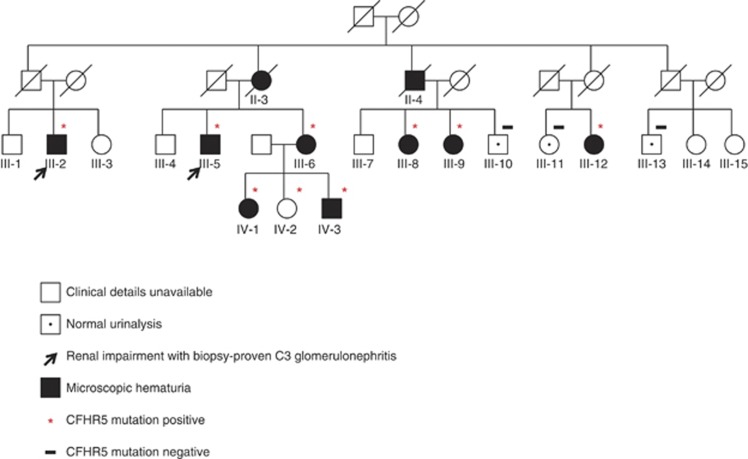
A 46-year-old man (index case, III-5, Figure 1) presented with a several-year history of recurrent intermittent macroscopic hematuria, often associated with symptoms of self-limiting upper respiratory tract infection. There was a family history of renal impairment (Figure 1); his mother (II-3) was dialysis dependent following a history of microscopic hematuria and type 2 diabetes; his maternal uncle (II-4) had had renal failure attributed to diabetes; and one maternal cousin (III-2) had been investigated for microscopic hematuria when 25 years old, was referred for nephrology review at the age of 45 with reduced estimated glomerular filtration rate of 51 ml/min, reached end-stage renal failure in 2005 when 52, and received a living-related renal transplant in 2006. The index case's mother and uncle had not had a renal biopsy. His cousin (III-2) had undergone native renal biopsy in 1998; light microscopy showed mesangial changes, but no glomeruli were present in the sections used for immunofluorescence. In conjunction with the clinical findings at the time, the cousin was diagnosed with probable IgA nephropathy. The cousin (III-2) had received a kidney transplant from his sister (III-3) in 2006, which was functioning; a transplant biopsy at 14 months showed isometric vacuolation only, but there was recent evidence of deteriorating graft functioning with dipstick hematuria. The sister (III-6) of the index case had hematuria with hydronephrosis, and the sister's daughter (IV-1) had episodic macroscopic hematuria. The family originates from a single county in the western part of the United Kingdom and have no known Cypriot ancestry. No other family members suffer from diabetes mellitus. No evidence of immune deficiency has been identified in any family members.
Figure 1.
Pedigree with familial C3 glomerulopathy demonstrating segregation of renal disease with mutant CFHR5 protein. CFHR, complement factor H related.
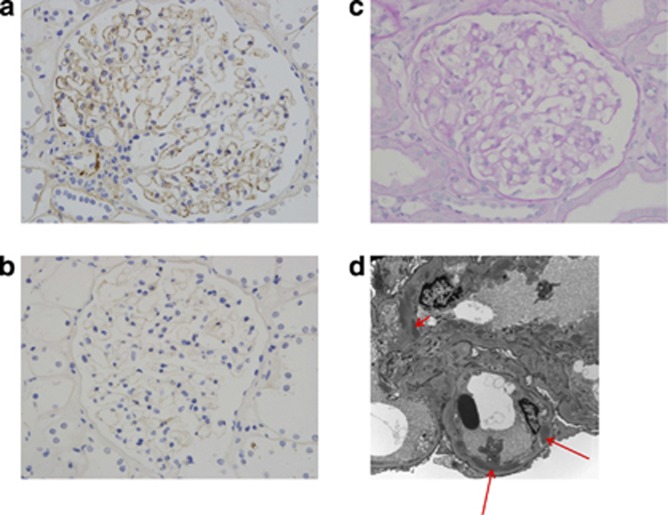
The index case had a protein:creatinine ratio of 15 mg/mmol (normal range <45 mg/mmol) and estimated glomerular filtration rate of 66 ml/min per 1.73 m2. Serum complement C3 and C4 levels were normal on three occasions (C3 1.45, 1.42, 1.26 (normal range 0.65–1.65 g/l); C4 0.25, 0.25, 0.24 (normal range 0.16–0.6 g/l)). Complement hemolytic (CH50) and alternative pathway activity (AP50) assays were in the normal range on two occasions. CFH and complement factor I levels were normal. C3 nephritic factor, measured using a commercial assay (The Binding Site, Birmingham, UK), and anti-CFH antibodies were negative. Renal biopsy contained 10 glomeruli, one of which showed tuft collapse and thickening of Bowman's capsule, whereas the others were normal by light microscopy (Figure 2c). Immunohistochemistry showed granular C3 on glomerular capillary walls (Figure 2a). There was no staining for immunoglobulin A (IgA), IgG, or IgM. Electron microscopy showed segmental sub-endothelial and intra-membranous electron-dense deposits (data not shown). This suggested immunoglobulin-independent complement activation secondary to a complement alternative pathway abnormality. We retrospectively reviewed the native and transplant biopsies of the patient's cousin (III-2). The native kidney showed electron-dense deposits on electron microscopy, similar in morphology to those in cases of CFHR5 nephropathy in Cypriot patients (Figure 2d).3 Although immunostaining was not done on the cousin's native kidney biopsy, transplant renal biopsy 14 months after transplantation showed both C3 staining on immunofluorescence and segmental sub-endothelial and intra-membranous electron-dense deposits on EM. There were no extremely osmiophilic deposits typical of dense deposit disease. The biopsy findings were therefore consistent with C3 glomerulonephritis in the native kidney with recurrence in the transplant, and were similar to those of the index case and the reported Cypriot CFHR5 nephropathy biopsies.
Figure 2.
Renal biopsy images. (a) Complement C3 staining in renal biopsy from index case (III-5). Granular C3 reactivity along the glomerular capillary walls is present. No staining for immunoglobulin A (IgA), IgG, or IgM was seen (data not shown). (b) Glomeruli with negative immunoperoxidase staining for negative control. (c) Light microscopy showing a periodic acid–Schiff–stained glomerulus from the index case (III-5). The glomerulus has a normal appearance by light microscopy. (d) Representative electron microscopic appearances of native renal biopsy of III-2 demonstrating elongated sub-endothelial (long arrows) and mesangial (short arrow) electron-dense deposits.
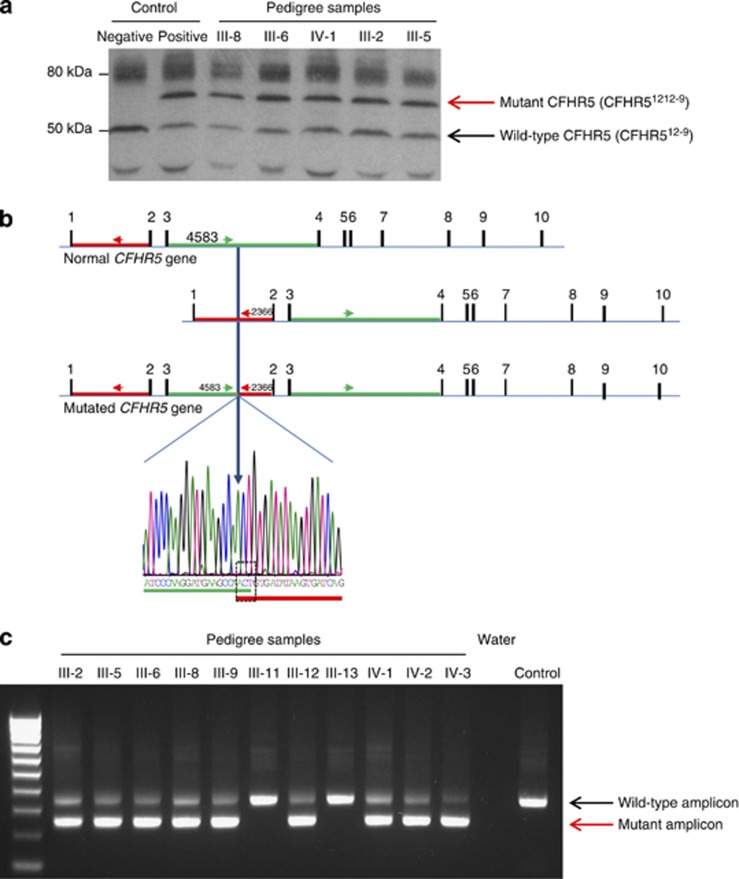
In view of the clinical similarities to CFHR5 nephropathy, we analyzed CFHR5 in the index case. Serum western blotting for CFHR5 revealed an abnormally large CFHR5 protein in addition to the normal CFHR5 protein (Figure 3a). The aberrant band was identical in size to that seen in a Cypriot patient with CFHR5 nephropathy (Figure 3a, lane 2 positive control). No coding or splice site mutations were detected on sequencing the CFHR5 gene. Copy number variation across the CFHR5 gene (ChromGRCh37:1:196946067–196979404:1) demonstrated three copies of CFHR5 exons 2 and 3, a finding identical to that seen in CFHR5 nephropathy previously (Supplementary Figure 1 online).3 Long-range genomic polymerase chain reaction (PCR) and sequencing within this area demonstrated a genomic breakpoint within CFHR5 intron 3 at coordinate 196957850 (nucleotide=A, CFHR5 intron 3), which linked to CFHR5 intron 1 at coordinate 196949652 (nucleotide=G, CFHR5 intron 1) with an intervening ACT triplet (Figure 3b). This breakpoint is close to but distinct from that seen in Cypriot CFHR5 nephropathy (Supplementary Figure 2 online). We next designed a genomic PCR that incorporated primers, which enabled amplification of both the wild-type and mutated locus. Using this PCR and serum CFHR5 western blotting (where serum was available), we screened available family members (Figure 3c). This showed that the two members with biopsy-proven renal disease (Figures 1 and 3; III-2 and III-5) harbored the CFHR51212-9 mutation in heterozygosity. To determine whether CFHR51212-9 segregated with renal disease in the pedigree, we performed dipstick urinalysis from all available consenting family members. We defined microscopic hematuria, with or without macroscopic hematuria, proteinuria, or impaired estimated glomerular filtration rate, as clinical evidence of renal involvement. CFHR51212-9 was present in heterozygosity in all eight members with abnormal urinalysis (Figures 1 and 3c; III-2, III-5, III-6, III-8, III-9, III-12, and IV-1 and IV-3) and one member with no urinalysis available (IV-2), but was absent in the three members with normal urinalysis (III-10, III-11, and III-13).
Figure 3.
Characterization of the abnormal CFHR5 protein in the pedigree. (a) Western blot of serum with a polyclonal anti-human CFHR5 antibody. Both the normal CFHR5 protein and an abnormal higher-molecular-weight protein were detected in the index case and the serum of affected family members. (b) Schematic representation and chromatogram showing genomic breakpoint. The ACT sequence is common to both introns 1 and 3. The abnormal genomic amplicon was generated using the forward primer in intron 3 (green arrow, 5′-TATTGGCTGTGGGTTTGTCA-3′) and the reverse primer in intron 1 (red arrow, 5′-TGACTGATCACTTATATCACAGTTGG-3′). The breakpoint is 4583 bp into CFHR5 intron 3 (A of ACT=4584), where the sequence switches to CFHR5 intron 1 at 2797 (A of ACT=2798). (c) Screening for the genomic breakpoint by polymerase chain reaction. The 337-bp amplicon is amplified from the wild-type CFHR5 gene. If the intronic breakpoint is present, a 239-bp amplicon is generated. CFHR, complement factor H related.
DISCUSSION
The clinical characteristics of the nephropathy in this pedigree were remarkably similar to Cypriot CFHR5 nephropathy: the typical presentation was with microscopic and intermittent macroscopic hematuria, and renal disease was more severe in affected males.5 We identified an abnormal CFHR51212-9 in this kindred identical to the aberrant CFHR5 protein found in Cypriot CFHR5 nephropathy: an internal duplication of SCRs 1 and 2 is caused by duplication of exons 2 and 3 on the CFHR5 gene (CFHR5 exon 1 encodes a leader peptide resulting in exon 2 coding SCR1, and exon 3 encoding SCR2).6 The CFHR51212-9 segregated with renal involvement. The demonstration of familial C3 glomerulonephritis associated with CFHR51212-9 in a demographically unrelated family with a mutation distinct from that described in Cypriot CFHR5 nephropathy strengthens the association of the abnormal CFHR51212-9 and C3G. Although this does not prove causation, it supports the need for a greater understanding of the pathogenic role of CFHR51212-9.
CFHR5 is a member of the CFH family of proteins encoded in the regulators of complement activation gene on chromosome 1.7 CFHR1, CFHR2, and CFHR5 have recently been shown to form homodimers and heterodimers in a head-to-tail configuration via common dimerization domains within SCR1 and SCR2.8 The dimerization of CFHR1, CFHR2, and CFHR5 may enhance the avidity of these proteins for ligand in vivo, enabling them to competitively antagonize CFH and function as complement deregulators.8 The duplication of SCR1 and SCR2, as seen in CFHR51212-9, would theoretically result in the formation of trimers or higher-order complexes. The larger complexes could have enhanced ligand avidity and ability to antagonize CFH. Consistent with this, purified CFHRs from a patient with CFHR51212-9 are more potent in complement deregulation in vitro than healthy controls.8 This phenomenon could apply in vivo particularly where spatial density of C3 is low or when blood flow rate across the site of complement activation is high, such as within the kidney.8 We speculate that CFHR5 nephropathy is a result of a failure of CFH to regulate C3 activation along the glomerular basement membrane owing to enhanced CFH deregulation by CFHR51212-9; however, this requires demonstration in vivo. The deregulation hypothesis could also explain the protective effect of a CFHR3-1 deletion in IgA nephropathy.9 Fewer CFHRs in serum could lead to less CFH deregulation enabling tighter control of complement activation and inflammation. The reason male patients with mutant CFHR51212-9 seem to have more aggressive clinical renal disease remains unclear and warrants further investigation.
Our data demonstrate that an abnormal CFHR5 protein should be looked for in patients with C3G irrespective of Cypriot ancestry and particularly if there is a family history. One approach would be to screen for abnormal CFHR5 protein by serum analysis and then proceed with genetic interrogation of the CFHR locus. This is because we now know that at least two distinct intronic breakpoints within the CFHR5 gene can cause the same mutant CFHR5 protein and C3G. Conversely, protein analysis relies on the fact that the mutant protein is distinguishable from the wild-type protein by mass so would not detect an abnormal CFHR5 protein containing the same number of domains as wild-type CFHR5. Although never reported, if this scenario were suspected, a combination of protein analysis and detailed genetic interrogation would be needed to identify the mutation. In summary, the increasing association between abnormal CFHR proteins and C3G4, 5, 10 indicates that variation across this locus should be looked for in familial C3G.
MATERIALS AND METHODS
The study was conducted according to the Declaration of Helsinki. Informed consent was obtained from all participants.
Genetic analysis of CFHR5
Exon sequencing of CFHR5 was performed in the index case (III-5). Amplicons that targeted the exons and at least 50 bp of contiguous intronic sequence were amplified by PCR using primers (available on request) designed using the Primer3 software (primer3.sourceforge.net). Sequencing reactions were performed using BigDye Terminator 3.1 Cycle Sequencing Kit (Life Technologies, Paisley, UK). Both PCR and sequencing reaction products were purified using Edge Biosystems Gel Filtration plates (Life Technologies). The sequence was run on a 3730 DNA Analyzer (Life Technologies) and analyzed using the Sequencer DNA sequencing analysis software. Genomic DNA was analyzed using the Multiplex Ligation-Dependent Probe Amplification P236-A1 kit according to the manufacturer's instructions (MRC Holland, Amsterdam, The Netherlands, www.mlpa.com). Assay readings were validated using samples from individuals with known copy-number variation across the CFH-CFHR locus. TaqMan probe HS03382618_cn at position chr1:196952738 (Applied Biosystems, Paisley, United Kingdom www.appliedbiosystems.com) was additionally used to validate copy-number variation across the CFHR5 gene. To detect the intronic breakpoint by PCR using genomic DNA, we used the following primers: 5′-TATTGGCTGTGGGTTTGTCA-3′, 5′-GAATTTTAGACCCATATCCCTGAT-3′, and 5′-TGACTGATCACTTATATCACAGTTGG-3′. The 5′-TATTGGCTGTGGGTTTGTCA-3′ and 5′-GAATTTTAGACCCATATCCCTGAT-3′ pair amplify the 337-bp wild-type amplicon, whereas 5′-TATTGGCTGTGGGTTTGTCA-3′ and 5′-TGACTGATCACTTATATCACAGTTGG-3′ pair amplify a 239-bp amplicon if the intronic breakpoint is present. PCR was performed using Qiagen HotStar Taq DNA polymerase (Qiagen, Manchester, UK) with 10 ng of genomic DNA according to the manufacturer's protocol. Both reactions were multiplexed in the same reaction.
Serum analysis of CFHR5
Western blot for CFHR5 was performed using sera from healthy control and affected individuals. The samples were run on standard 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis under nonreducing conditions and probed with a rabbit anti-CFHR5 polyclonal antibody (Abnova, Taipei City, Taiwan, cat. no. H30081494-DO1P).
Acknowledgments
We thank the patients, their families, and their general practitioners for their assistance in providing clinical information and samples for this study. MCP is a Wellcome Trust Senior Fellow in Clinical Science (WT082291MA).
All the authors declared no competing interests.
Footnotes
SUPPLEMENTARY MATERIAL
Figure S1. Copy number variation assay using genomic DNA from the index case and wild type control using multiplex ligation-dependent probe assay (MLPA).
Figure S2. Comparison of the intronic breakpoint reported in this pedigree with that reported in CFHR5 nephropathy in individuals with Cypriot ancestry.
Supplementary material is linked to the online version of the paper at http://www.nature.com/ki
Supplementary Material
References
- Pickering MC, Cook HT. Translational mini-review series on complement factor H: renal diseases associated with complement factor H: novel insights from humans and animals. Clin Exp Immunol. 2008;151:210–230. doi: 10.1111/j.1365-2249.2007.03574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour TD, Pickering MC, Cook HT. Recent insights into C3 glomerulopathy. Nephrol Dialysis Transplantation. 2013;28:1685–1693. doi: 10.1093/ndt/gfs430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale DP, de Jorge EG, Cook HT, et al. Identification of a mutation in complement factor H-related protein 5 in patients of Cypriot origin with glomerulonephritis. Lancet. 2010;376:794–801. doi: 10.1016/S0140-6736(10)60670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik TH, Lavin PJ, Goicoechea de Jorge E, et al. A hybrid CFHR3-1 gene causes familial C3 glomerulopathy. J Am Soc Nephrol. 2012;23:1155–1160. doi: 10.1681/ASN.2012020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou Y, Voskarides K, Gale DP, et al. Familial C3 glomerulopathy associated with CFHR5 mutations: clinical characteristics of 91 patients in 16 pedigrees. Clin J e Am Soc Nephrol. 2011;6:1436–1446. doi: 10.2215/CJN.09541010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JL, Murphy BE, Eyre HJ, et al. Location and structure of the human FHR-5 gene. Genetica. 2002;114:157–161. doi: 10.1023/a:1015114512924. [DOI] [PubMed] [Google Scholar]
- Zipfel PF. Factor H family proteins: on complement, microbes and human diseases. Biochem Soc Trans. 2002;30:971–978. doi: 10.1042/bst0300971. [DOI] [PubMed] [Google Scholar]
- Goicoechea de Jorge E, Caesar JJE, Malik TH, et al. Dimerization of complement factor H-related proteins modulates complement activation in vivo. Proc Natl Acad Sci USA. 2013;110:4685–4690. doi: 10.1073/pnas.1219260110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharavi AG, Kiryluk K, Choi M, et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43:321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortajada A, Yebenes H, Abarrategui-Garrido C, et al. C3 glomerulopathy-associated CFHR1 mutation alters FHR oligomerization and complement regulation. J Clin Invest. 2013;123:2434–2446. doi: 10.1172/JCI68280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.