Abstract
Leucine-rich repeat kinase 2 (LRRK2) is a gene that, upon mutation, causes autosomal-dominant familial Parkinson's disease (PD). Yeast two-hybrid screening revealed that Snapin, a SNAP-25 (synaptosomal-associated protein-25) interacting protein, interacts with LRRK2. An in vitro kinase assay exhibited that Snapin is phosphorylated by LRRK2. A glutathione-S-transferase (GST) pull-down assay showed that LRRK2 may interact with Snapin via its Ras-of-complex (ROC) and N-terminal domains, with no significant difference on interaction of Snapin with LRRK2 wild type (WT) or its pathogenic mutants. Further analysis by mutation study revealed that Threonine 117 of Snapin is one of the sites phosphorylated by LRRK2. Furthermore, a Snapin T117D phosphomimetic mutant decreased its interaction with SNAP-25 in the GST pull-down assay. SNAP-25 is a component of the SNARE (Soluble NSF Attachment protein REceptor) complex and is critical for the exocytosis of synaptic vesicles. Incubation of rat brain lysate with recombinant Snapin T117D, but not WT, protein caused decreased interaction of synaptotagmin with the SNARE complex based on a co-immunoprecipitation assay. We further found that LRRK2-dependent phosphorylation of Snapin in the hippocampal neurons resulted in a decrease in the number of readily releasable vesicles and the extent of exocytotic release. Combined, these data suggest that LRRK2 may regulate neurotransmitter release via control of Snapin function by inhibitory phosphorylation.
Keywords: kinase, LRRK2, Snapin, SNAP-25, SNARE vesicle, synaptotagmin
Introduction
Leucine-rich repeat kinase 2 (LRRK2) has been identified as a gene corresponding to PARK8, a locus causing familial Parkinson's disease (PD) in an autosomal-dominant manner.1, 2 PD is the second most common neurodegenerative disease and affects more than 1% of people older than 65 years. LRRK2 is a large protein consisting of 2527 amino acids and contains active GTPase and functional kinase domains, with protein interaction domains including leucine-rich repeat and WD40.1, 2, 3, 4 Each of these functional GTPase and kinase domains may be critical for LRRK2 functions and PD pathogenesis, given that the two most prevalent PD-specific mutations among LRRK2 mutations, G2019S and R1441C/G, were mapped in the kinase and GTPase domains, respectively.1, 2, 5 Overexpression of wild-type (WT) or PD-specific mutants of LRKK2 exhibited increased cellular protein aggregations,6, 7, 8 shortening of neurite length, and decrease of neurite branch number during neurite outgrowth,9, 10 as well as enhanced, oxidative stress-induced neurotoxicity.11, 12, 13 In most studies, these phenotypes were intensified in cells expressing mutants, especially G2019S, which showed increased kinase activity, compared to cells expressing WT.10, 14, 15, 16 Therefore, that part of LRRK2 research conducted thus far has focused on identifying its kinase substrates with the goals of elucidating the PD-pathogenic mechanism and developing PD therapeutics.
Several biochemical and genetic methods such as yeast two-hybrid screening and co-immunoprecipitation have been carried out to identify LRRK2-interacting proteins.4, 6, 15, 17, 18, 19, 20, 21, 22, 23 From these studies, moesin, mitogen-activated protein kinase kinase, 4E-BP, eukaryotic initiation factor 4E-binding protein, Drosophila microtubule-binding protein Futsch and ARHGEF7 were identified as substrates for LRRK2 kinase,15, 24, 25, 26, 27 although it remains unclear how their phosphorylation affects PD pathogenesis.
We previously identified Rab5b, an early endosome marker, as an LRRK2-interacting protein by yeast two-hybrid screening and showed that their interaction slows endocytosis of synaptic vesicles.18 From the same screening, we also isolated Snapin as an LRRK2-interacting protein. Snapin was originally reported as a SNAP-25-binding protein that is associated with soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex proteins.28 In Snapin knockout mice, association of SNAP-25 with synaptotagmin-1 was impaired and calcium-dependent exocytosis was significantly reduced.29 Recent studies reported that in Snapin knockout mice, late endocytic proteins were increased30 and that synaptic vesicle fusion was desynchronized.31 In addition, Snapin was also reported to be an important link between the aquaporin water channel and the target SNARE complex,32 and as a regulator of late endosomal transport, which is critical for autophagy–lysosomal function in neurons.33 It was also reported that protein kinase A (PKA) phosphorylated Snapin at its Ser50 residue, and increased both its binding of SNAP-25 and interaction of synaptotagmin-1 with the SNARE complex.34 The results from these studies implied that Snapin is a critical protein to regulate synaptic vesicle trafficking and the fusion of synaptic vesicles to the plasma membrane. A recent study showed that Snapin also functions in fine-tuning of neurite outgrowth via interaction with AC6 (type VI adenylyl cyclase),35 suggesting that Snapin negatively regulates neurite outgrowth.
Materials and methods
Materials
Human Snapin gene (KU35533) was purchased from KUGI (Korean UniGene Information) and cloned into modified pcDNA3.1MycHis vector (Invitrogen, Calsbad, CA, USA), which contained a Flag tag instead of a Myc tag, and into the bacterial expression vector pET29b (Novagen, Darmstadt, Germany) using EcoRI and XhoI restriction enzymes. Vectors containing Snapin T20A, T117A or T117D mutation were synthesized by in vitro site-directed mutagenesis using a mutagenesis kit (Stratagene, La Jolla, CA, USA) with proper primer pairs (Table 1), and the desired mutations were confirmed by sequencing the full length of the cloned open reading frame. GST-Snapin was constructed by cloning the Snapin gene into pGEX4T-1 (GE Healthcare, Seoul, Korea). Construction of LRRK2 and its mutant plasmids was previously described.13, 18 The ΔApaI and ΔXcmI deletion of LRRK2 was carried out by ApaI and XcmI digestion, respectively, and re-ligation of the longer DNA fragment, resulting in deletion of a specific region of the LRRK2 gene generated by the enzyme treatment. Recombinant GST-LRRK2 WT, G2019S or D1994A proteins with truncated N-terminal 970 amino acids were purchased from Invitrogen.
Table 1. Primer sequence.
| Oligomer name | Sequence (5′ to 3′) |
|---|---|
| Snapin T20A-F | GTGGCGGGGCCCGCAGGCCGCGACC |
| Snapin T20A-R | GGTCGCGGCCTGCGGGCCCCGCCAC |
| Snapin T117A-F | GTTGCCAAGGAAGCAGCCCGCAGGAG |
| Snapin T117A-R | CTCCTGCGGGCTGCTTCCTTGGCAAC |
| Snapin T117D-F | GTTGCCAAGGAAGACGCCCGCAGGAG |
| Snapin T117D-R | CTCCTGCGGGCGTCT TCCTTGGCAAC |
GST-SNAP-25 was obtained from Dr Sheng, Z.H. at NIH (Bethesda, MD, USA). Antibodies against Myc and flag tags were purchased from Sigma (St Louis, MO, USA) and Snapin antibodies from SySy (#148002, Goettingen, Germany) or Abnova (H00023557-A01, Taipei City, Taiwan). The HEK293T cell stably expressing flag-tagged LRRK2 WT, G2019S or an empty vector36 was purchased from University of Dundee, UK. The vGlut1-pHluorin was kindly provided by Prof. Rubenstein, John L.R. at UCSF (San Francisco, CA, USA).
Cell culture and transfection
Human embryonic kidney 293T cells were maintained with DMEM containing 10% fetal bovine serum at 37 °C with 5% CO2. To overexpress LRRK2 domains or Snapin, the indicated plasmids were transiently transfected by the standard CaCl2 precipitation method or using Lipofectamine 2000 (Invitrogen). To induce LRRK2 proteins, HEK293T cells stably expressing flag-tagged LRRK2 WT, G2019S or an empty vector were treated with doxycycline (2 μℳ) for 2 days,36 and expression of LRRK2 proteins was confirmed by western blot analysis with flag antibody.
Human SH-SY5Y cells were cultured in DMEM containing 10% fetal bovine serum at 37 °C with 5% CO2 and differentiated in the medium containing all-trans retinoic acid (10 μℳ) for 5 days to obtain neuron-like properties.37
In vitro kinase assay
To measure the kinase activity of LRRK2, two kinds of LRRK2 proteins were used as enzyme sources. One was Myc-LRRK2 WT, G2019S or D1994A proteins overexpressed in HEK293T cells and immunoprecipitated by anti-myc antibody and the other is commercial GST-LRRK2 WT, G2019S or D1994A proteins whose N-terminal 969 amino acids were deleted and fused to GST protein (Invitrogen). For the former, proteins expressed in one well of a 6-well plate were used for one kinase reaction, and for the latter, 30 ng of proteins was used for each kinase reaction. The indicated proteins were incubated in 40 μl of kinase buffer (10 μCi of γ32P-ATP(SBP-501, IZOTOP Budapest, Hungary), 50 μℳ ATP, 25 mℳ Tris–HCl (pH 7.5), 5 mℳ β-glycerol phosphate, 2 mℳ DTT, 0.1 mℳ NA3VO4, 10 mℳ MgCl2) containing approximately 1.5 μg of Snapin WT or mutant proteins purified from Escherichia coli BL21 strain, at 37 °C for 20 min. The samples were analyzed by autoradiography or a PhosphorImager analyzer (Typhoon 9200 imager, GE Healthcare).
Yeast two-hybrid screening, immunoprecipitation, and GST pull-down assay
The yeast two-hybrid screening method has been described in detail.18, 38 For the GST pull-down assay, GST, GST-Snapin or GST-SNAP-25 fusion proteins were expressed in E. coli BL21 strain and isolated using glutathione beads (GE Healthcare), as previously described.39 Lysates of HEK293T cells over-expressing Myc-tagged LRRK2 WT or mutant proteins were incubated with purified GST or GST-Snapin proteins and bound proteins were eluted, subjected to SDS–PAGE (polyacrylamide gel electrophoresis), and detected by anti-myc antibody. To test whether phosphorylation of Snapin by LRRK2 affected Snapin-SNAP-25 interaction, Myc-tagged LRRK2 G2019S protein was overexpressed in 293T cells, immunoprecipitated by anti-Myc antibody and then incubated with Snapin recombinant proteins for phosphorylation. Supernatants containing Snapin phosphorylated by LRRK2 were incubated with GST-SNAP-25 recombinant proteins. Snapin bound to GST-SNAP-25 was detected -by a GST pull-down assay and western blot analysis using anti-Snapin antibody (SySy).
To test the effect of Snapin phosphorylation mediated by LRRK2 on synaptotagmin, we followed the previously described protocol.40 Rat brain lysate (3 mg) was mixed with recombinant Snapin WT or T117D proteins (5 μg) and incubated with anti-syntaxin (Abcam, #ab3265, Cambridge, UK) or mouse control immunoglobulin G (Santa Cruz, Santa Cruz, CA, USA) in a binding buffer (phosphate-buffered saline with 0.1% Triton X-100, 0.1% BSA, 200 μℳ CaCl2, 1 × protease inhibitor cocktail (Roche)) overnight at 4 °C. The mixture was then incubated with protein-G agarose (Peptron, Daejeon, Korea) for 2 h at 4 °C. The purified complex was washed with a washing buffer (phosphate-buffered saline with 0.1% Triton X-100) three times and subjected to SDS–PAGE and western blot analyses using the indicated antibodies (synaptotagmin-1: Santa Cruz, #sc-12466; SNAP-25: BD, #610366; VAMP2: Synaptic Systems, SySy #69.1).
For immunoprecipitation of mouse brain lysate, G2019S transgenic (TG, #009609, purchased from Jackson Laboratory, Bar Harbor, ME, USA) and normal control mice were killed by cervical dislocation. Whole mouse brains were isolated with a scalpel blade. Co-immunoprecipitation analysis was carried out as previously described.41 Briefly, whole brains were disrupted in 1 ml of 50 mℳ Tris, pH 7.5, 50 mℳ NaCl, 0.5% Triton X-100 and 1% protease inhibitor cocktail using a Dounce Homogenizer (10 stroke) and lysed by passing the extracts through a 22-gauge needle five times. Extracts were centrifuged at 12 000 g for 30 min at 4 °C to remove insoluble material, and supernatants were mixed with equivalent volume of binding buffer (50 mℳ Tris, pH 7.5, 50 mℳ NaCl, 0.1% bovine serum albumin (BSA), 0.1% Triton X-100 and 1% protease inhibitor cocktail (PIC)) and then centrifuged at 20 000 g for 30 min at 4 °C. The supernatants were finally immunoprecipitated with anti-SNAP25 or anti-LRRK2 (MJFF2, Epitomics, Burlingame, CA, USA) at 4 °C for 2 h and further incubated with protein-G agarose (Pierce, Rockford, IL, USA) for 2 h. The antibody–protein complexes were washed three times with binding buffer minus BSA and protease inhibitor cocktail, and subjected to western blot analysis using the indicated antibodies (Snapin:Abnova, H00023557, LRRK2:1E11 or UDD 3 30 (Epitomics)).
The flag-tagged LRRK2 protein was induced from HEK293T stable cells36 by treatment of doxycycline (2 μℳ for 2 days). The cell lysates centrifuged at 14 000 g for 20 min at 4 °C and were incubated with the agarose immobilized flag epitope tag antibody (Novus, Littleton, CO, USA, #NB600-351). Then, immunoprecipitation and western blot analysis were performed as above.
Neuron culture and transfection
The hippocampal neurons derived from E-18 pregnant Sprague-Dawley female rats were prepared as described.42 Briefly, the embryonic hippocampi were dissected, dissociated with papain and triturated with a polished, half-bore Pasteur pipette. Dissociated neurons (2.5 × 105) in minimum Eagle's medium supplemented with 0.6% glucose, 1 mℳ pyruvate, 2 mℳ ℒ-glutamine, 10% fetal bovine serum and antibiotics were plated on poly-𝒟-lysine-coated glass coverslips in a 60 mm Petri dish. Four hours after plating, the medium was replaced with Neurobasal media (Invitrogen) supplemented with 2% B-27, 0.5 mℳℒ-glutamine, to which 4 μℳ of 1-β-𝒟-cytosine-arabinofuranoside (Ara-C, Sigma) was added as needed. Neurons were transfected using a modified calcium–phosphate method.18 Briefly, 6 μg of cDNA and 9.3 μl of 2 ℳ CaCl2 were mixed in distilled water to a total volume of 75 μl, and the same volume of 2X BBS was added. Neurobasal medium was completely replaced by transfection medium (minimum Eagle's medium, 1 mℳ pyruvate, 0.6% glucose, 10 mℳ glutamine and 10 mℳ N-2-hydroxyl piperazine-N′-2-ethane sulfonic acid, pH 7.65), and the cDNA mixture was added to the neurons, and incubated in a 5% CO2 incubator for 90 min. Neurons were washed twice with washing medium (minimum Eagle's medium, 1 mℳ pyruvate, 0.6% glucose, 10 mℳ glutamine and 10 mℳ N-2-hydroxyl piperazine-N′-2-ethane sulfonic acid (N-2-hydroxyl piperazine-N′-2-ethane sulfonic acid), pH 7.35) and then returned to the original culture medium. The vGlut-pHluorin (vGpH) and pU6mRFP constructs were co-transfected in a ratio of 5:1. The construct vGpH harbors vesicular glutamate transporter 1 fused with pHluorin, a modified GFP with high pH sensitivity.43 When targeted to the synaptic vesicle lumen, vGpH is quenched and upon exocytosis, the relatively basic pH of the extracellular space allows it to fluoresce. Its fluorescence is again quenched after endocytosis and re-acidification of the synaptic vesicles. Thus, this process accurately reflects the kinetics of exo–endocytic trafficking of synaptic vesicles.43, 44
Synaptic vesicle pool size measurement and image analysis
Coverslips were mounted in a perfusion/stimulation chamber equipped with platinum–iridium field stimulus electrodes (EC-S-10, LCI, Seoul, Korea) on the stage of an Olympus IX-71 inverted microscope with × 40, 1.0 NA oil lens (Olympus, Tokyo, Japan). Neurons were continuously perfused at room temperature with Tyrode solution (136 mℳ NaCl, 2.5 mℳ KCl, 2 mℳ CaCl2, 1.3 mℳ MgCl2, 10 mℳ N-2-hydroxyl piperazine-N′-2-ethane sulfonic acid, 10 mℳ glucose, pH 7.3). To reduce spontaneous activity and prevent recurrent excitation during stimulation, 10 μℳ 6-cyano-7-nitroquinoxaline-2, 3-dione and 50 μℳ of DL-2-amino-5-phosphonovaleric acid were added to the Tyrode solution. Time-lapse images were acquired every 5 s for 4 min using a back-illuminated Andor iXon 897 EMCCD camera (Andor Technologies, Belfast, Northern Ireland) driven by MetaMorph Imaging software (Molecular Devices, Sunnyvale, CA, USA). From the fourth frame, neurons were stimulated (1 ms, 20–50 V, bipolar) using an A310 Accupulser current stimulator (World Precision Instruments, Sarasota, FL, USA) to mimic action potential generation. Quantitative measurement of fluorescence intensity at individual boutons was obtained by averaging a selected area of pixel intensities using MetaMorph software. Individual regions were selected by hand. Then, rectangular regions of interest were drawn around the synaptic boutons, and average intensities were calculated. Large puncta, typically representative of smaller synapse clusters, were rejected during the selection procedure. The center of intensity of each synapse was calculated to correct for any image shift over the course of the experiment. Fluorescence was expressed in intensity units that correspond to fluorescence values averaged over all pixels within the region of interest. Changes in net fluorescence were obtained by subtracting the average intensity of the first four frames (F0) from the intensity of each frame (Ft) for individual boutons. They were then normalized to the maximum fluorescence intensity (Fmax−F0) and averaged. For the exocytosis assays, neurons were pre-incubated with bafilomycin A1 (Calbiochem, San Diego, CA, USA) for 60 s to block re-acidification and stimulated for 30 s at 20 Hz/600 action potentials (APs), a combination, which is known to totally deplete the recycling pool of vesicles.44 Bafilomycin A1 was dissolved in Me2SO at 0.2 mℳ and diluted to a final concentration of 0.5 μℳ prior to the experiments. Bafilomycin A1 was applied throughout the fluorescence measurements.
To estimate the size of each fraction of the synaptic vesicle pool, vGpH-transfected neurons at DIV 18 were stimulated with 40 APs at 20 Hz, which is known to release the store of readily releasable pool of synaptic vesicles in the hippocampal synapses,44 followed by 1800 APs at 20 Hz stimulation in the presence of 1 μℳ bafilomycin A1 to release the entire recycling pool of synaptic vesicles.39, 44 The resting pool that cannot be mobilized by such neuronal activity can be uncovered by applying NH4Cl solution to unquench all acidic vesicles not yet released. Data are presented as means±s.e.m. Statistical analysis was carried out with PASW Statistics 19 (IBM, Armonk, NY, USA). For multiple conditions, we compared means by analysis of variance followed by Tukey's honestly significant difference post hoc test or Fisher's LSD test (depending on the number of groups).
Results
LRRK2 phosphorylates Snapin at Thr-117 residue
To investigate the mechanism for LRRK2 to generate PD, we attempted to elucidate the normal physiological functions of LRRK2. We employed yeast two-hybrid screening to isolate proteins interacting with LRRK2, and tried to deduce LRRK2's function by identifying the functions of the LRRK2-interacting proteins. Snapin (SNAP-25-interacting protein) was originally isolated as an LRRK2-interacting protein by yeast two-hybrid screening using regions from ROC to the kinase (GK) domain or containing the WD40 domain as baits. Both screenings yielded multiple identical Snapin clones whose open reading frame started at the 33rd amino acid. The Snapin clone also interacted with the bait containing the full length LRRK2 gene (data not shown).
As both Snapin and LRRK2 were reported to have roles in vesicle trafficking,18, 29, 31, 45 we first tested the possibility that Snapin was a LRRK2 kinase substrate. An in vitro kinase assay was performed with bacterially expressed, recombinant Snapin and immunoprecipitated, Myc-tagged LRRK2 WT, G2019S or kinase-dead D1994A proteins overexpressed in HEK293T cells. The results showed that LRRK2 phosphorylated Snapin in the order of G2019S>WT, and this order was also observed in LRRK2 autophosphorylation, as previously reported (Figure 1a).3, 15, 24 As expected, the kinase dead mutant D1994A11 showed weaker phosphorylation of Snapin and no autophosphorylation, a pattern similar to the reaction with the vector control (Figure 1a). The weak phosphorylation of Snapin observed in the vector and D1994A lanes might be derived from other kinases non-specifically immunoprecipitated by the myc antibody (Figures 1ai and iii, lanes 1, 4). A similar pattern of Snapin phosphorylation was also observed using commercially available GST-LRRK2 proteins (Invitrogen, Figure 1b). As a negative control, we tested ARF1 proteins that were expressed in the same pET 29 vector as Snapin, as substrates for an in vitro kinase assay. GST-LRRK2 G2019S could not phosphorylate the ARF1 proteins (Figure 1bii; strongly suggesting that phosphorylation of Snapin by LRRK2 is specific.
Figure 1.
Snapin phosphorylation by leucine-rich repeat kinase 2 (LRRK2). (a) Immunoprecipitated LRRK2 phosphorylates Snapin. Recombinant Snapin protein was expressed in E. coli as His-tagged fusion protein and isolated by affinity chromatography with nickel resin. Indicated LRRK2 wild-type (WT) or mutant proteins were expressed in HEK293T cells as Myc-tagged forms and immunoprecipitated by anti-myc antibody. The immunoprecipitates (IP) were incubated with Snapin for an in vitro kinase assay. Autoradiograms of phosphorylated LRRK2 (i) and phosphorylated Snapin (iii) are shown. In addition, the amount of immunoprecipitated LRRK2 (ii) and recombinant Snapin (iv) proteins were detected by anti-myc antibody and Coomassie blue staining, respectively. The empty vector was also transfected to HEK293T cells and the cell lysates were used as a control (vector). (b) Recombinant GST-LRRK2 (Invitrogen) proteins also phosphorylate Snapin, but not ARF1. The purchased LRRK2 WT (WT), G2019S (G) or R1441C (R) proteins were used for an in vitro kinase assay. Autoradiograms showed that LRRK2 and Snapin (i), but not ARF1, proteins (ii) were phosphorylated by LRRK2 (ii). (iii) Positions of both Arf1 and Snapin proteins are indicated by arrows. (c) The amino-acid sequence of human Snapin. The conserved phosphorylation candidate sites at the 20th and 117th threonine residues are indicated as bold letters. (d) LRRK2 phosphorylates Snapin at Thr-117. GST-LRRK2 WT or G2019S (Invitrogen) were subjected to an in vitro kinase assay with recombinant Snapin WT, T20A or T117A protein. Autoradiograms of autophosphorylated GST-LRRK2 (i) and phosphorylated Snapin (ii) are shown. For each kinase assay, use of equal amounts of substrates was confirmed by SDS–PAGE and Coomassie blue staining (aiv, biii, diii).
To identify the phosphorylation site of Snapin by LRRK2, we searched an F/Y-X-T-X-R/K motif (the underlined T is the phosphorylation site), a putative phosphorylation site by LRRK2 that was previously reported.46 Out of four Thr residues in Snapin, two at the 20th (PVAGPTGRD) and 117th (SVAKETARR) amino acid contained a conserved arginine (indicated by an underlined letter) at the +2 position (Figure 1c). We mutated each threonine to alanine (Snapin T20A and T117A), and purified them as bacterially expressed recombinant proteins. Using the purified Snapin proteins, we performed an in vitro kinase assay with the GST-LRRK2 WT or G2019S proteins. The results showed that phosphorylation of T117A by the recombinant GST-LRRK2 G2019S proteins was almost, but not completely, eradicated, whereas that of T20A remained at a level similar to that of WT. This suggests that Thr-117 is at least one of the phosphorylation sites of Snapin by LRRK2 (Figure 1dii).
LRRK2 interacts with Snapin via its ROC domain
To confirm interaction between Snapin and LRRK2, we conducted a GST pull-down assay. Figure 2a shows that LRRK2 overexpressed in 293T cells specifically interacted with bacterially expressed GST-Snapin. As Snapin was reported to interact with many different proteins of various functions by yeast two-hybrid screening,41, 47, 48, 49, 50, 51 and it has even been suggested that its interaction with SNAP-25 is non-specific,52 we used murine-constitutive androstane receptor (CAR, NR1I3), a nuclear receptor, as a nonspecific negative control in the GST pull-down assay. As expected, CAR did not interact with GST-Snapin whereas LRRK2 specifically interacted with GST-Snapin (Figure 2a). Three PD-pathogenic mutants (G2019S, I2020T and R1441C), interacted with GST-Snapin with little differences in their affinities to Snapin in comparison with WT (Figure 2b). To further test the interaction of Snapin with LRRK2, we expressed Myc-tagged Snapin in HEK293T cells stably expressing flag-tagged LRRK2, and performed co-immunoprecipitation of LRRK2 by flag antibody. The result showed that LRRK2 co-immunoprecipitated Snapin (Figure 2c). We also tested the interaction of LRRK2 with Snapin in the differentiated SH-SY5Y cells without any overexpression. The endogenous LRRK2 weakly, but specifically interacted with endogenous Snapin (Figure 2d).
Figure 2.
Leucine-rich repeat kinase 2 (LRRK2) interacts with Snapin. (a) Lysates of 293T cells transfected with myc-LRRK2 (LRRK2 WT) or HA-murine CAR (CAR) were co-incubated with purified GST-Snapin (S) or GST proteins and subjected to a GST pull-down assay and western blot analysis using anti-myc or anti-HA antibody. (b) LRRK2 WT, G2019S, R1441C or I2020T was expressed in HEK293T cells and the cell lysates were used for a GST pull-down assay as (a) Input (I) of each sample is 2.5% of the total proteins. The amount of GST-Snapin or GST proteins used was detected by Coomassie staining, and it was confirmed that similar amounts of GST proteins were used (a, b). (c) Co-immunoprecipitation of flag-LRRK2 with myc-Snapin. The HEK293T cells were induced by doxycycline to stably express flag-LRRK2 and transfected with plasmids expressing myc-Snapin. Cell lysates were immunoprecipitated with flag antibody-agarose. The immunoprecipitate (IP) was washed and prepared for western analysis with anti-flag or anti-myc. 10% of cell lysates were shown as input (Input). * indicates a non-specific band. (d) SH-SY5Y cells differentiated by retinoic acid (10 μℳ for 5 days) were immunoprecipitated by LRRK2 (MJFF2, Epitomics) antibody and the immunoprecipitates were analyzed by antibody against LRRK2 (1E11) or Snapin (SYSY). An input of 2% was not enough to detect endogenous LRRK2, but sufficient for Snapin detection. WB, western blot.
To identify the LRRK2 domain interacting with Snapin, we constructed several LRRK2 deletion mutants (Figure 3a), overexpressed them in HEK293T cells and then repeated the GST pull-down assay using the HEK293T cell lysates and GST-Snapin. The results suggested both ROC and N-terminal (1–937 amino acids) domains as the interaction domains (Figure 3b). As we isolated the Snapin clone from the yeast two-hybrid screening using the GK and WD40 domains as baits, the GST pull-down assay results partially supported the yeast two-hybrid results. However, we were unable to test the interaction of Snapin with the WD40 domain, because transient expression of the WD40 domain in HEK293T cells was too weak to perform a GST pull-down assay.
Figure 3.
Mapping of leucine-rich repeat kinase 2 (LRRK2) domain interacting with Snapin. (a) A scheme of several LRRK2 deletion mutant constructs. LRR: Leucine-rich repeats; Roc, Ras of complex proteins; CoR, C-terminal of Roc domain. (b) GST pull-down assay of LRRK2. The indicated LRRK2-mutant proteins were expressed in 293T cells as a Myc-tagged form by transient transfection and total cell lysates were subjected to a GST pull-down assay using GST (-) or GST-Snapin (S). Input (I) of each sample was 2.5% of the total proteins. The LRRK2 proteins were detected with anti-myc antibody, except for the kinase domain, which was detected by a previously reported LRRK2 antibody, 1E11.67 Because expressions of the ΔApaI, Roc and kinase domains were weaker those of the other domains, exposure times of these domains were extended. * indicates nonspecific protein band.
Snapin phosphorylation by LRRK2 decreases interaction of Snapin with SNAP-25
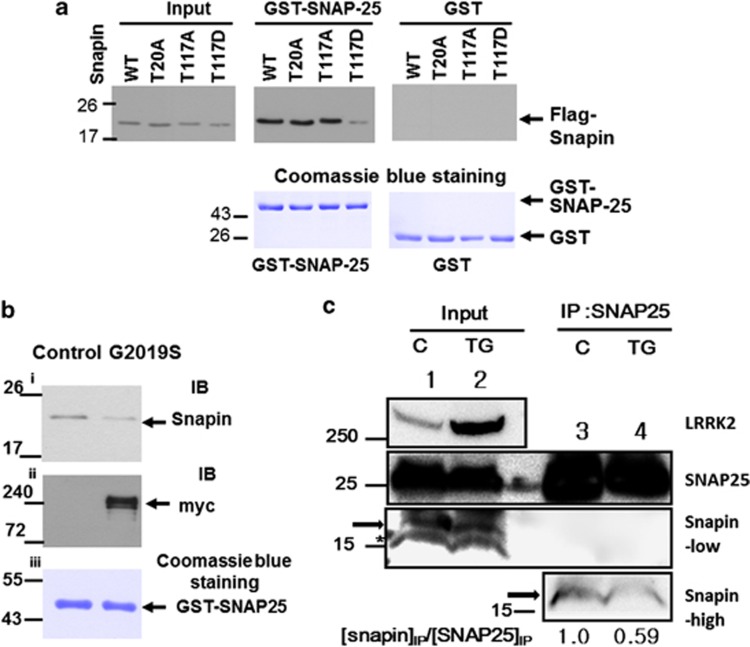
To investigate the physiological function of Snapin phosphorylation by LRRK2, we constructed Snapin T117D, a phosphorylation mimetic mutant, and both Snapin WT and T117D recombinant proteins were expressed in E. coli, purified and tested for specific interaction with GST-SNAP-25. Figure 4a exhibits that Snapin T117D very weakly interacted with GST-SNAP-25 in comparison with interaction of Snapin WT and T117A. This suggested that the phosphorylation of Snapin T117 inhibited the interaction of SNAP-25 with Snapin. To determine whether Snapin phosphorylation by LRRK2 exhibits similar results, Myc-tagged, G2019S proteins expressed in HEK293T cells were immunoprecipitated by anti-myc antibody and the precipitate was incubated with recombinant Snapin WT protein so that Snapin WT could be phosphorylated. The resultant Snapin proteins were tested for interaction with GST-SNAP-25 by a GST pull-down assay. The results showed that Snapin, which might be phosphorylated by LRRK2, considerably decreased the interaction of SNAP-25 with Snapin (Figure 4b), similar to the Snapin T117D mutants. We could not, however, directly confirm the phosphorylation status of Snapin, because no phospho-Snapin-specific antibody is currently available.
Figure 4.
Phosphorylation of Snapin by leucine-rich repeat kinase 2 (LRRK2) causes a decrease of interaction with SNAP-25. (a) Snapin T117D-mutant mimicking phosphorylation by LRRK2 interacts with SNAP-25 more weakly than the wild type (WT). Recombinant Flag-tagged Snapin WT and indicated mutant proteins were subjected to a GST pull-down assay using GST-SNAP-25 and detected by western blot analysis using anti-Flag antibody. Coomassie blue staining of GST and GST-SNAP-25 showed that similar amounts of each protein were used. (b) Snapin WT protein phosphorylated by LRRK2 exhibited weaker interaction with SNAP-25 than the untreated and presumably unphosphorylated Snapin (control). The myc-tagged LRRK2 G2019S was transiently expressed in HEK293T cells and immunoprecipitated by anti-myc antibody. The immunoprecipitated beads were co-incubated with recombinant flag tagged Snapin WT protein with ATP under the in vitro kinase assay conditions. The supernatant was co-incubated with purified GST-SNAP-25 and subjected to a GST pull-down assay and SDS–PAGE. The bound Snapin was detected by anti-flag antibody (Sigma) and the amount was compared to the control transfected with an empty vector (i). The presence of LRRK2 in immunoprecipitates was confirmed by anti-myc antibody (ii). Coomassie blue staining (iii) confirmed that equal amounts of GST-SANP25 protein were used for both incubations (control and G2019S). (c) Interaction of SNAP25 with Snapin was weaker in G2019S transgenic (TG) than in a control (C) mouse brain lysate. Whole brain lysates were prepared from both G2019S TG and normal control mice and immunoprecipitated with SNAP25 antibody. The amount of Snapin in each brain lysates (20% input) was shown with Snapin antibody (Abnova, 1:500 dilution, Snapin-low). The immunoprecipitates, which showed no signal under the diluted condition above, were subjected to western blot analysis with more concentrated Snapin antibody (Abnova, 1:50 dilution, Snapin-high). The amount of specific immunoprecipitated Snapin was calculated as the amount of immunoprecipitated Snapin divided by the amount of immunoprecipitated SNAP25. An arrow or * indicated Snapin or nonspecific protein band, respectively.
To confirm that Snapin phosphorylation by LRRK2 causes its interaction with SNAP25 to be weaker than with non-phosphorylated Snapin, we carried out co-immunoprecipitation of SNAP25 from brain lysates of G2019S TG and control normal mice, and the co-immunoprecipitated complex was subjected to the western blot analysis using Snapin antibody. The result showed that the amount of co-immunoprecipitated Snapin in the TG brain lysates, which contained more LRRK2 kinase activity, was reduced by almost half compared with that of the normal brain lysates (Figure 4c, lanes 3 and 4) although the amount of input Snapin was slightly higher in the control lysates (Figure 4c lanes 1 and 2).
LRRK2-mediated phosphorylation of Snapin decreased interaction of synaptotagmin-1 with SNAP25-syntaxin-VAMP2 complex
To identify the physiological functions of LRRK2-mediated Snapin phosphorylation, we tested Snapin-mediated interaction of synaptotagmin with SNAP-25. It was shown that PKA phosphorylated Snapin at serine 50 and the phosphomimetic Snapin S50D protein, enhanced synaptotagmin association with the core synaptic fusion complex containing SNAP-25, VAMP2 (synaptobrevin-2) and syntaxin.40 We assumed that inhibitory phosphorylation of Snapin by LRRK2 might decrease association of synaptotagmin with the SNARE complex, in contrast to PKA-mediated activatory phosphorylation of Snapin.
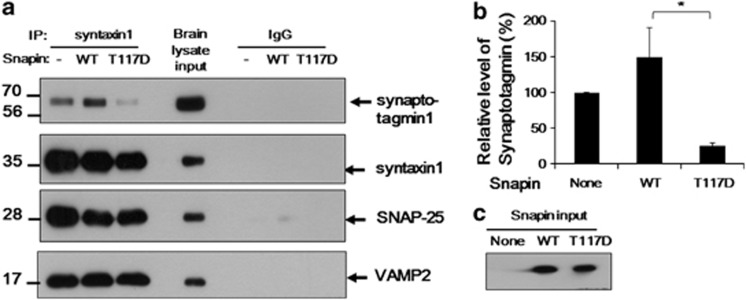
To test our hypothesis, rat brain lysate was incubated with Snapin WT or T117D protein and the complex was co-immunoprecipitated by syntaxin antibody. The immunoprecipitates were analyzed using western blot analysis with synaptotagmin-1 antibody. Figure 5 shows that Snapin WT increased the association of synaptotagmin-1 with the core SNARE complex. In contrast, T117D-mutant protein decreased the association. The amount of each component of the SNARE complex was similar except for SNAP-25, whose interaction with syntaxin might be decreased by the addition of Snapin.
Figure 5.
T117D Snapin, a phosphomimetic mutant, decreased interaction of synaptotagmin with VAMP2-syntaxin-SNAP 25 complex. Rat brain lysate was incubated with none (−), recombinant Snapin wild type (WT) or T117D overnight and immunoprecipitated with syntaxin1 antibody or control rabbit immunoglobulin G antibody. The immunoprecipitate was subjected to SDS–PAGE for western blot analysis. Co-precipitated synaptotagmin-1 was detected by synaptotagmin-1 antibody and each component of the VAMP2-syntaxin-SNAP-25 SNARE complex was also detected by specific antibodies. The experiment was repeated three times and then band densities were analyzed. Data are presented as means±s.e.m. *P⩽0.01 (analysis of variance and Turkey's honestly significant difference post hoc test). A representative image (a) and the resulting graph are shown (b). Five percent of the total brain lysate (a) is shown as input. Use of similar amounts of the recombinant Snapin proteins was confirmed (c). Snapin was detected by the His tag, which was used for purification of the recombinant protein. *P⩽0.05 (analysis of variance and Turkey's honestly significant difference post hoc test).
LRRK2-dependent phosphorylation of Snapin decreased the fraction of readily releasable pool and the extent of exocytotic release.
As we found that LRRK2-induced phosphorylation on Snapin T117 decreased the interaction of synaptotagmin-1 with the SNAP25-syntaxin-VAMP2 complex, we next tested the effect of LRRK2-induced phosphorylation of Snapin on synaptic transmission of the cultured hippocampal neurons. We co-transfected WT Snapin alone, T117D alone, G2019S and WT Snapin or D1994A and WT Snapin with vGpH in cultured rat hippocampal neurons.43 The vesicles of a synapse are organized into three distinct pools, two of which recycle (readily releasable pool (RRP) and a reserve pool), and one which normally does not (resting pool). The vesicles of the RRP are known to be docked at the plasma membrane, ready for immediate release upon stimulation.44 After the RRP has been depleted, continued release occurs from the reserve pool. The amplitude of the response to a train of 40 APs at 20 Hz corresponds to the size of the RRP of the synaptic vesicles. Those recycling vesicles released by a stimulus of 1800 APs at 20 Hz after the addition of bafilomycin A1, a V-type ATPase inhibitor that blocks acidification of endocytosed synaptic vesicles, represent the remainder of the total recycling pool.44, 53 The resting pool of vesicles, which are refractory to stimulation and normally do not participate in recycling, can be uncovered by adding NH4Cl to trap all the vesicles in an alkaline state and hence unquench all acidic vesicles that have not been released (Figure 6a44, 53).
Figure 6.
Leucine-rich repeat kinase 2 (LRRK2)-dependent phosphorylation of Snapin decreased the size of RRP and the extent of exocytotic release. (a) Rat hippocampal neurons were co-transfected with wild-type Snapin alone, T117D alone, G2019S and wild-type Snapin, or D1994A and wild-type Snapin with vGpH. To measure RRP size, neurons were stimulated with 40 APs at 20 Hz. They were then stimulated with 1800 APs at 20 Hz in the presence of bafilomycin (baf). Subtraction of RRP fluorescence from the fluorescence plateau reflected the reserve pool (RP). All remaining acidic vesicles were alkalized by NH4Cl treatment, revealing the size of the resting pool. Fluorescence intensity was normalized to the maximum fluorescence change upon NH4Cl treatment. (b) (left) Average fraction of RRP in wild-type Snapin alone, T117D alone, G2019S and wild-type Snapin, or D1994A and wild-type Snapin (20.7%±2.8% for wild-type Snapin; 16.6%±2.2% for T117D; 12.4%±1.3% for G2019S and wild-type Snapin; 29.2%±3.6% for D1994A and wild-type Snapin). (middle) Average fraction of reserve pool (29.6%±3.0% for wild-type Snapin; 32.8%±3.1% for T117D; 41.6%±2.9% for G2019S and wild-type Snapin; 37.4%±6.4% for D1994A and wild-type Snapin). (right) Average fraction of resting pool (49.7%±3.0% for wild-type Snapin; 50.5%±1.4% for T117D; 46.0%±2.8% for G2019S and wild-type Snapin; 33.8%±6.6% for D1994A and wild-type Snapin). (c) The extent of exocytotic release. Rat hippocampal neurons were co-transfected with wild-type Snapin alone, T117D alone, G2019S and wild-type Snapin, or D1994A and wild-type Snapin with vGpH. The neurons were stimulated with 100 APs at 10 Hz, and then treated with NH4Cl to obtain the total synaptic vesicle pool size. Fluorescence intensity was normalized to the maximum fluorescence change upon NH4Cl treatment. Data are presented as means±s.e.m. *P⩽0.01 (analysis of variance and Turkey's honestly significant difference post hoc test).
We found that LRRK2 G2019S with WT Snapin resulted in a decrease in the RRP fraction at the expense of the reserved fraction (Figure 6b, RRP fraction:reserved fraction=20.7±2.8%:29.6±3.0% for control; 12.4±1.3%:41.6±2.9% for G2019S). Subsequently, the extent of exocytotic release during 100 APs stimulation was also significantly decreased by G2019S (Figure 6c). LRRK2 D1994A with WT Snapin expression showed somewhat opposite effects: the RRP and reserve pool fractions were increased at the expense of the resting pool fraction (Figure 6). The extent of exocytotic release was also significantly increased by D1994A.
Although Snapin T117D itself did not show any effect on the synaptic vesicle pool size and the extent of exocytotic release (Figure 6), we assume that this might be attributed to the compensatory effect of endogenous Snapin or the presence of LRRK2-induced phosphorylation sites other than T117 in Snapin.
Discussion
We reported here that LRRK2 interacts with and phosphorylates Snapin, possibly at threonine 117. Snapin has been reported to interact with numerous proteins including SNAP-25,28 EBAG9,47 cypin,54 casein kinase 1-delta,50 a subunit of Exo70,49 TRPC6,51 EHD1,55 disbindin-1,56 type VI adenylyl cyclase,48 ryanodine receptor57 and UT-A1 urea transporter.41 Most of these Snapin-interacting proteins function in vesicle trafficking. However, the various functions of interaction partners also suggest a possibility that Snapin might non-specifically interact with these proteins.52 However, a Snapin knockout animal model exhibited impaired calcium-dependent exocytosis of large dense-core vesicles in chromaffin cells, confirming specific physiological roles of Snapin in synaptic vesicle trafficking.29
Until now, PKA and casein kinase 1δ were known to phosphorylate Snapin.40, 50 We report here LRRK2 as another Snapin kinase. One of the phosphorylation sites of Snapin by LRRK2 was identified as threonine 117. Although we identified this site based on the conserved sequences for LRRK2's phosphorylation site,46 a recent study on LRRK2's phosphorylation site suggested that LRRK2 phosphorylates amino acids whose surrounding regions have high pI values, rather than the conserved amino-acid sequences.58 The pI of the peptide containing the 113rd amino acid to 121st amino acids (VAKETARRR) of Snapin is 11.7 (calculated by a program from http://web.expasy.org/protparam/), confirming that threonine 117 is acceptable as a phosphorylation site of LRRK2 regardless of the rule that is applied.46, 58 However, LRRK2's phosphorylated motif sequence is not well conserved. Several studies have reported that LRRK2 could phosphorylate threonine or even serine at sites unrelated to the reported F/Y-X-T-X-R/K threonine sites.59, 60 Therefore, it is possible that serines or threonines in Snapin are phosphorylated by LRRK2 in addition to T117, because T117A mutant proteins were still weakly phosphorylated by LRRK2 in the in vitro kinase assay (Figure 1d). We found that Snapin T117D did not significantly affect synaptic vesicle composition and extent of exocytosis, and this might be due to the presence of additional phosphorylation sites in Snapin by LRRK2 (Figure 6b).
We did not observe any significant differences in the interaction of Snapin with LRRK2 WT and its pathogenic mutants (Figure 2). This suggests that interaction of LRRK2 with Snapin is not directly related to PD pathogenicity. However, it is still possible that G2019S phosphorylates Snapin more than WT given G2019S's stronger kinase activity. This may result in stronger inhibitory phosphorylation of Snapin by the G2019S mutant (Figures 1a and b), which may be related to PD pathogenesis.
Previously, we showed that LRRK2 was enriched in the soluble synaptosome fraction, suggesting the possibility that LRRK2 interacts with synaptic proteins.18 A recent report suggested that LRRK2 is part of a presynaptic protein network and controls synaptic vesicle trafficking.61 Silencing of the expression of LRRK2 caused redistribution of vesicles within the bouton and altered recycling dynamics.61 Neurons from Snapin-deficient mice also showed desynchronized fusion of synaptic vesicles and impairment of synaptic efficacy and precision.31
Snapin was reported to promote SNAP-25 binding to synaptotagmin-1 and to stabilize binding of synaptotagmin-1 to the SNARE complex.31 This resulted in an increase in the number of synaptic vesicles in the readily releasable state.29, 31 Thus far, both positive and negative regulators for the interaction between Snapin and SNAP-25 have been reported. PKA phosphorylated Snapin at serine 50, increased the Snapin interaction with SNAP-25, and caused an increase of synaptic vesicle release.34, 40 Two negative regulators, EBAG9 and EHD1, inhibited interaction of Snapin with SNAP-2547, 55 and interaction of SNAP25 with synaptotagmin-1. Consistent with these previous findings, we also found that LRRK2-mediated phosphorylation of Snapin decreased interaction of synaptotagmin with the core SNARE complex (Figure 5). This subsequently resulted in a reduced RRP fraction and extent of exocytosis, suggesting that LRRK2 is another negative regulator of the Snapin and SNAP-25 interaction during synaptic transmission.
Snapin has been reported to directly interact with synaptotagmin31 in addition to indirect interaction via SNAP25. Whether Snapin S50D protein enhances its direct interaction with synaptotagmin is yet unknown. It would be interesting to investigate the effect of LRRK2-mediated phosphorylation of Snapin, on its direct interaction with synaptotagmin.
One of the well-known LRRK2 kinase functions is negative regulation of neurite length. In contrast, SNAP-25 is known as a positive regulator of neurite length.62 Recently, type VI adenylyl cyclase (AC6) was reported as a Snapin-interacting protein and a suppressor of neurite extension.63 Downregulation of Snapin or overexpression of SNAP-25 reversed neurite shortening by AC6.63 Investigation of the effect of LRRK2-mediated Snapin phosphorylation on neurite length could be of further interest.
In addition, it would be interesting to investigate whether the pattern of interaction of Snapin phosphorylated at Thr-117 with various other previously reported Snapin partner proteins is changed.28, 41, 47, 49, 50, 51, 54, 55, 56
Both Snapin and LRRK2 have been reported to regulate trafficking of proteins from late endosomes to lysosomes positively and negatively, respectively.33, 64 In addition, several papers have reported that overexpression of α-synuclein or LRRK2 causes defects in synaptic vesicle trafficking.45, 65, 66 Our data may provide a clue to explain this phenotype in another direction, possibly by inhibitory phosphorylation of Snapin by LRRK2, although this requires further experiments.
Acknowledgments
We thank Drs ZH Sheng (NIH, USA) and CJ Gloeckner (Munich, Germany) for GST-SNAP-25 and LRRK2 antibody (1E11), respectively. This research was supported by the Global Partnership Program (2009-00505 to WS) and Basic Science Research Program (2012R1A1A3008447 to WS) and SRC (MSIP No. 2011-0030775, 2009-0070560, 2009-0070562 to SH) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology, and by a grant from InAm Neuroscience Research Center (Sanbon Hospital, Wonkwang University, Korea). The research herein was also supported by a grant from the Korea Health Technology R&D Project (A092058 to SC) funded by the Ministry of Health & Welfare, and by a grant from the Biomembrane Plasticity Research Center (No. 20100029395 to SC) funded by the NRF of Korea.
The authors declare no conflict of interest.
References
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, et al. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci USA. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol W. Biochemical and molecular features of LRRK2 and its pathophysiological roles in Parkinson's disease. BMB Rep. 2010;43:233–244. doi: 10.5483/bmbrep.2010.43.4.233. [DOI] [PubMed] [Google Scholar]
- Mata IF, Kachergus JM, Taylor JP, Lincoln S, Aasly J, Lynch T, et al. Lrrk2 pathogenic substitutions in Parkinson's disease. Neurogenetics. 2005;6:171–177. doi: 10.1007/s10048-005-0005-1. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, et al. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci USA. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, et al. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- Liou AK, Leak RK, Li L, Zigmond MJ. Wild-type LRRK2 but not its mutant attenuates stress-induced cell death via ERK pathway. Neurobiol Dis. 2008;32:116–124. doi: 10.1016/j.nbd.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo HY, Park JM, Kim CH, Han BS, Kim KS, Seol W. LRRK2 enhances oxidative stress-induced neurotoxicity via its kinase activity. Exp Cell Res. 2010;316:649–656. doi: 10.1016/j.yexcr.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, et al. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzon-Toro B, Rubio de la Torre E, Delgado A, Perez-Tur J, Hilfiker S. Mechanistic insight into the dominant mode of the Parkinson's disease-associated G2019S LRRK2 mutation. Hum Mol Genet. 2007;16:2031–2039. doi: 10.1093/hmg/ddm151. [DOI] [PubMed] [Google Scholar]
- Dachsel JC, Taylor JP, Mok SS, Ross OA, Hinkle KM, Bailey RM, et al. Identification of potential protein interactors of Lrrk2. Parkinsonism Relat Disord. 2007;13:382–385. doi: 10.1016/j.parkreldis.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N, Jeong H, Kwon J, Heo HY, Kwon JJ, Yun HJ, et al. LRRK2 regulates synaptic vesicle endocytosis. Exp Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L, et al. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J Neurosci. 2008;28:3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillardon F. Interaction of elongation factor 1-alpha with leucine-rich repeat kinase 2 impairs kinase activity and microtubule bundling in vitro. Neuroscience. 2009;163:533–539. doi: 10.1016/j.neuroscience.2009.06.051. [DOI] [PubMed] [Google Scholar]
- Gillardon F. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability—a point of convergence in Parkinsonian neurodegeneration. J Neurochem. 2009;110:1514–1522. doi: 10.1111/j.1471-4159.2009.06235.x. [DOI] [PubMed] [Google Scholar]
- Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho RM, Law BM, Harvey K. Mutations in the LRRK2 Roc-COR tandem domain link Parkinson's disease to Wnt signalling pathways. Hum Mol Genet. 2009;18:3955–3968. doi: 10.1093/hmg/ddp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem. 2009;109:959–968. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, et al. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. Embo J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu HP, Lin WY, Guo H, Lu B. LRRK2 kinase regulates synaptic morphology through distinct substrates at the presynaptic and postsynaptic compartments of the Drosophila neuromuscular junction. J Neurosci. 2010;30:16959–16969. doi: 10.1523/JNEUROSCI.1807-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haebig K, Gloeckner CJ, Miralles MG, Gillardon F, Schulte C, Riess O, et al. ARHGEF7 (Beta-PIX) acts as guanine nucleotide exchange factor for leucine-rich repeat kinase 2. PLoS One. 2010;5:e13762. doi: 10.1371/journal.pone.0013762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilardi JM, Mochida S, Sheng ZH. Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat Neurosci. 1999;2:119–124. doi: 10.1038/5673. [DOI] [PubMed] [Google Scholar]
- Tian JH, Wu ZX, Unzicker M, Lu L, Cai Q, Li C, et al. The role of Snapin in neurosecretion: snapin knock-out mice exhibit impaired calcium-dependent exocytosis of large dense-core vesicles in chromaffin cells. J Neurosci. 2005;25:10546–10555. doi: 10.1523/JNEUROSCI.3275-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Cai Q, Tian JH, Sheng ZH. Snapin associates with late endocytic compartments and interacts with late endosomal SNAREs. Biosci Rep. 2009;29:261–269. doi: 10.1042/BSR20090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan PY, Tian JH, Sheng ZH. Snapin facilitates the synchronization of synaptic vesicle fusion. Neuron. 2009;61:412–424. doi: 10.1016/j.neuron.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry AC, Mallick R, Klein JD, Weimbs T, Sands JM, Frohlich O. Syntaxin specificity of aquaporins in the inner medullary collecting duct. Am J Physiol Renal Physiol. 2009;297:F292–F300. doi: 10.1152/ajprenal.00196.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Lu L, Tian JH, Zhu YB, Qiao H, Sheng ZH. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 2010;68:73–86. doi: 10.1016/j.neuron.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur P, Stevens DR, Sheng ZH, Rettig J. Effects of PKA-mediated phosphorylation of Snapin on synaptic transmission in cultured hippocampal neurons. J Neurosci. 2004;24:6476–6481. doi: 10.1523/JNEUROSCI.0590-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Lin JT, Chien CL, Chang WC, Lai HL, Chang CP, et al. Type VI adenylyl cyclase (AC6) regulates neurite extension by binding to Snapin and Snap25. Mol Cell Biol. 2011;31:4874–4886. doi: 10.1128/MCB.05593-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols RJ, Dzamko N, Hutti JE, Cantley LC, Deak M, Moran J, et al. Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson's disease. Biochem J. 2009;424:47–60. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HR, Hu LS, Li GY. SH-SY5Y human neuroblastoma cell line: in vitro cell model of dopaminergic neurons in Parkinson's disease. Chin Med J (Engl) 2010;123:1086–1092. [PubMed] [Google Scholar]
- Lee SJ, Lee JK, Maeng YS, Kim YM, Kwon YG. Langerhans cell protein 1 (LCP1) binds to PNUTS in the nucleus: implications for this complex in transcriptional regulation. Exp Mol Med. 2009;41:189–200. doi: 10.3858/emm.2009.41.3.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Choi JH, Suh PG, Chang JS. Pleckstrin homology domain of phospholipase C-gamma1 directly binds to 68-kDa neurofilament light chain. Exp Mol Med. 2006;38:265–272. doi: 10.1038/emm.2006.32. [DOI] [PubMed] [Google Scholar]
- Chheda MG, Ashery U, Thakur P, Rettig J, Sheng ZH. Phosphorylation of Snapin by PKA modulates its interaction with the SNARE complex. Nat Cell Biol. 2001;3:331–338. doi: 10.1038/35070000. [DOI] [PubMed] [Google Scholar]
- Mistry AC, Mallick R, Frohlich O, Klein JD, Rehm A, Chen G, et al. The UT-A1 urea transporter interacts with snapin, a SNARE-associated protein. J Biol Chem. 2007;282:30097–30106. doi: 10.1074/jbc.M705866200. [DOI] [PubMed] [Google Scholar]
- Chang S, De Camilli P. Glutamate regulates actin-based motility in axonal filopodia. Nat Neurosci. 2001;4:787–793. doi: 10.1038/90489. [DOI] [PubMed] [Google Scholar]
- Voglmaier SM, Kam K, Yang H, Fortin DL, Hua Z, Nicoll RA, et al. Distinct endocytic pathways control the rate and extent of synaptic vesicle protein recycling. Neuron. 2006;51:71–84. doi: 10.1016/j.neuron.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Burrone J, Li Z, Murthy VN. Studying vesicle cycling in presynaptic terminals using the genetically encoded probe synaptopHluorin. Nat Protoc. 2006;1:2970–2978. doi: 10.1038/nprot.2006.449. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, et al. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genet. 2010;6:e1000902. doi: 10.1371/journal.pgen.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pungaliya PP, Bai Y, Lipinski K, Anand VS, Sen S, Brown EL, et al. Identification and characterization of a leucine-rich repeat kinase 2 (LRRK2) consensus phosphorylation motif. PLoS One. 2010;5:e13672. doi: 10.1371/journal.pone.0013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruder C, Reimer T, Delgado-Martinez I, Hermosilla R, Engelsberg A, Nehring R, et al. EBAG9 adds a new layer of control on large dense-core vesicle exocytosis via interaction with Snapin. Mol Biol Cell. 2005;16:1245–1257. doi: 10.1091/mbc.E04-09-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JL, Huang CL, Lai HL, Hung AC, Chien CL, Kao YY, et al. Regulation of type VI adenylyl cyclase by Snapin, a SNAP25-binding protein. J Biol Chem. 2004;279:46271–46279. doi: 10.1074/jbc.M407206200. [DOI] [PubMed] [Google Scholar]
- Bao Y, Lopez JA, James DE, Hunziker W. Snapin interacts with the Exo70 subunit of the exocyst and modulates GLUT4 trafficking. J Biol Chem. 2008;283:324–331. doi: 10.1074/jbc.M706873200. [DOI] [PubMed] [Google Scholar]
- Wolff S, Stoter M, Giamas G, Piesche M, Henne-Bruns D, Banting G, et al. Casein kinase 1 delta (CK1delta) interacts with the SNARE associated protein snapin. FEBS Lett. 2006;580:6477–6484. doi: 10.1016/j.febslet.2006.10.068. [DOI] [PubMed] [Google Scholar]
- Suzuki F, Morishima S, Tanaka T, Muramatsu I. Snapin, a new regulator of receptor signaling, augments alpha1A-adrenoceptor-operated calcium influx through TRPC6. J Biol Chem. 2007;282:29563–29573. doi: 10.1074/jbc.M702063200. [DOI] [PubMed] [Google Scholar]
- Vites O, Rhee JS, Schwarz M, Rosenmund C, Jahn R. Reinvestigation of the role of snapin in neurotransmitter release. J Biol Chem. 2004;279:26251–26256. doi: 10.1074/jbc.M404079200. [DOI] [PubMed] [Google Scholar]
- Kim SH, Ryan TA. CDK5 serves as a major control point in neurotransmitter release. Neuron. 2010;67:797–809. doi: 10.1016/j.neuron.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lucas KG, Akum BF, Balasingam G, Stawicki TM, Provost JM, et al. A novel role for snapin in dendrite patterning: interaction with cypin. Mol Biol Cell. 2005;16:5103–5114. doi: 10.1091/mbc.E05-02-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Xu Y, Shi H, Wong SH, Han W, Talbot K, et al. EHD1 is a synaptic protein that modulates exocytosis through binding to snapin. Mol Cell Neurosci. 2011;45:418–429. doi: 10.1016/j.mcn.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Talbot K, Cho DS, Ong WY, Benson MA, Han LY, Kazi HA, et al. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum Mol Genet. 2006;15:3041–3054. doi: 10.1093/hmg/ddl246. [DOI] [PubMed] [Google Scholar]
- Zissimopoulos S, West DJ, Williams AJ, Lai FA. Ryanodine receptor interaction with the SNARE-associated protein snapin. J Cell Sci. 2006;119:2386–2397. doi: 10.1242/jcs.02936. [DOI] [PubMed] [Google Scholar]
- Webber PJ, Smith AD, Sen S, Renfrow MB, Mobley JA, West AB. Autophosphorylation in the leucine-rich repeat kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities. J Mol Biol. 2011;412:94–110. doi: 10.1016/j.jmb.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Yuan C, Chen R, Dawson TM, Dawson VL. ArfGAP1 is a GTPase activating protein for LRRK2: reciprocal regulation of ArfGAP1 by LRRK2. J Neurosci. 2012;32:3877–3886. doi: 10.1523/JNEUROSCI.4566-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta E, Kawakami F, Kubo M, Obata F. LRRK2 directly phosphorylates Akt1 as a possible physiological substrate: impairment of the kinase activity by Parkinson's disease-associated mutations. FEBS Lett. 2011;585:2165–2170. doi: 10.1016/j.febslet.2011.05.044. [DOI] [PubMed] [Google Scholar]
- Piccoli G, Condliffe SB, Bauer M, Giesert F, Boldt K, De Astis S, et al. LRRK2 controls synaptic vesicle storage and mobilization within the recycling pool. J Neurosci. 2011;31:2225–2237. doi: 10.1523/JNEUROSCI.3730-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osen-Sand A, Catsicas M, Staple JK, Jones KA, Ayala G, Knowles J, et al. Inhibition of axonal growth by SNAP-25 antisense oligonucleotides in vitro and in vivo. Nature. 1993;364:445–448. doi: 10.1038/364445a0. [DOI] [PubMed] [Google Scholar]
- Wu CS, Lin JT, Chien CL, Chang WC, Lai HL, Chang CP, et al. Type VI adenylyl cyclase regulates neurite extension by binding to Snapin and Snap25. Mol Cell Biol. 2011;31:4874–4886. doi: 10.1128/MCB.05593-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson MW, Zhang T, Jiang C, Chen S, Guo M. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum Mol Genet. 2012;21:1350–1363. doi: 10.1093/hmg/ddr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemani VM, Lu W, Berge V, Nakamura K, Onoa B, Lee MK, et al. Increased expression of alpha-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65:66–79. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, et al. LRRK2 controls an endoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- Bauer M, Kinkl N, Meixner A, Kremmer E, Riemenschneider M, Forstl H, et al. Prevention of interferon-stimulated gene expression using microRNA-designed hairpins. Gene Ther. 2009;16:142–147. doi: 10.1038/gt.2008.123. [DOI] [PubMed] [Google Scholar]