Abstract
This study is aimed at gaining insights into the brain site-specific proteomic senescence signature while comparing physiologically aged brains with aging-related dementia brains (for example, Alzheimer's disease (AD)). Our study of proteomic differences within the hippocampus (Hp), parietal cortex (pCx) and cerebellum (Cb) could provide conceptual insights into the molecular mechanisms involved in aging-related neurodegeneration. Using an isobaric tag for relative and absolute quantitation (iTRAQ)-based two-dimensional liquid chromatography coupled with tandem mass spectrometry (2D-LC-MS/MS) brain site-specific proteomic strategy, we identified 950 proteins in the Hp, pCx and Cb of AD brains. Of these proteins, 31 were significantly altered. Most of the differentially regulated proteins are involved in molecular transport, nervous system development, synaptic plasticity and apoptosis. Particularly, proteins such as Gelsolin (GSN), Tenascin-R (TNR) and AHNAK could potentially act as novel biomarkers of aging-related neurodegeneration. Importantly, our Ingenuity Pathway Analysis (IPA)-based network analysis further revealed ubiquitin C (UBC) as a pivotal protein to interact with diverse AD-associated pathophysiological molecular factors and suggests the reduced ubiquitin proteasome degradation system (UPS) as one of the causative factors of AD.
Keywords: aging, Alzheimer's disease, proteasome, proteomics, ubiquitin
Introduction
Understanding the unique alterations in the brain proteomic signature induced by cellular senescence as a result of normal and pathological aging is essential for identification of a specific cure for various neurodegenerative disorders. Alzheimer's disease (AD) is a progressive, aging-related neurodegenerative disorder of the central nervous system and is the most common cause of dementia in the elderly worldwide. It is characterized by impaired memory and the deterioration of higher cognitive functions.1, 2 The main pathological characteristics of AD are accumulations of senile plaques that comprise amyloid-β (Aβ) peptides produced from the β-amyloid precursor protein (APP) following sequential processing by β- and γ-secretases.3, 4, 5, 6 The second major pathological hallmark of AD is neurofibrillary tangles (NFTs) caused by intracellular accumulations of the hyperphosphorylated microtubule-associated protein tau (MAPT).7, 8, 9 Neurodegeneration in AD progresses sequentially, starting first in predisposed induction sites from the entorhinal cortex in the medial temporal lobe and then gradually spreading to the entire hippocampus (Hp) and the limbic system, before advancing in topographically predictable sequences and ultimately expanding to the temporal association cortex. Although the occipital lobe cortex retains nearly normal function, frequently even in terminal-stage patients, the temporal lobe cortex, in contrast, as one of the most fragile parts of the brain, is extremely vulnerable to neuronal death.10, 11, 12, 13 Thus, it might be possible to identify those biomarkers that cause aging-related AD in the temporal lobe. Biomarkers that are capable of preventing neurodegenerative processes within the occipital lobe at early stages of AD may also be identified.14, 15 Because the prevailing assumption is that the cerebellar proteome of the central nervous system area is relatively unaffected by AD, this is a less investigated region and often neglected in AD studies. However, it must be noted that several studies indicated that in AD the cerebellum (Cb) also shows some signs of morphological and metabolic dysfunctions.16 A number of pathological changes, such as widespread deposits of diffuse amyloid, ubiquitin-immunoreactive dystrophic neurites and increased microglia, have been revealed in the AD-affected Cb by immunocytochemical studies. However, tau-immunoreactive NFTs have not been seen. Although the observed changes may be merely epiphenomenal to the pathological processes occurring in the AD neocortex and Hp, the morphological and immunocytochemical differences between cerebral and cerebellar cortices of AD patients may nonetheless give insights into the molecular factors involved in the development of the neuropathological lesions of the AD brain.16 Consequently, more attention has also been given to this brain area in recent years, though with little focus at the molecular level.17, 18, 19, 20, 21, 22, 23, 24
Thus far, proteomic analyses have been considered worldwide with greater emphasis on improving the diagnosis of early stages of AD by identifying novel specific biomarkers from the cerebrospinal fluid or plasma to complement the existing diagnostic methods that are based on autopsy or neuroimaging techniques. Isobaric tags for relative and absolute quantitation (iTRAQ) is a widely accepted approach for quantitative proteomics.25 Accordingly, in this study, we applied the two-dimensional (2D) liquid chromatography coupled with tandem mass spectrometry-based iTRAQ (2D-LC-MS/MS-iTRAQ) technique for quantitative profiling of aging-related brain site-specific proteome changes. We sought to investigate the mechanisms of senescence and pathological aging-related neurodegeneration at the proteomic level in different areas of the brain, including the Hp, the parietal cortex (pCx) and the Cb, by comprehensively identifying the proteins with altered expression levels in response to aging and aging-related dementia (for example, AD). Subsequently, we exploited publically accessible bioinformatic databases to infer the functional role and networks linked to the proteins that we found differentially expressed.
Our data also reveal protein pathway modulations acquired by the Cb to counteract neurodegeneration and thus could eventually become a lucrative target for the identification of plausible therapeutic drugs.
Materials and methods
Reagents
Unless indicated, all reagents used for biochemical methods were purchased from Sigma-Aldrich (St Louis, MO, USA). Materials and reagents for sodium dodecyl sulfate-polyacrylamide gel electrophoresis were from Bio-Rad (Bio-Rad Laboratories, Hercules, CA, USA). The iTRAQ reagent eight-plex kit was bought commercially (Applied Biosystems, Foster City, CA, USA).
Human subjects
Brain tissues were obtained from the brain bank of the Choju Medical Institute of the Fukushimura hospital (Toyohashi, Aichi, Japan), and the protocols utilized were approved by the local ethics committee of the Fukushimura hospital. The scientific use of human material was conducted in accordance with the Declaration of Helsinki, and informed consent was obtained from the guardians of the patients. Brain tissues were weighed at the time of autopsy, snap frozen with liquid nitrogen and stored at –80 °C. Patients with sporadic AD (early stage, low incidence, Table 1) received a pathological diagnosis according to the criteria of the Consortium to Establish a Registry for Alzheimer's Disease (CERAD) and the Braak stage13, 26 as described previously.27, 28 Controls were elderly patients who were age matched but without significant neurological disorders. Patients were also cognitively evaluated by neuropsychological tests using the mini-mental state examination (MMSE) and Hasegawa's dementia scale (HDS, or the HDS revised version (HDS-R)), which is commonly utilized in Japan.14, 15, 27, 28, 29
Table 1. Characteristics of human brain subject tissue samples.
| Subject (Patient) | Pathological diagnosis | Gender | Age (years) | Stage of amyloid deposits (none, A, B, C) | NFT stage (I–VI) | PMI (h) |
|---|---|---|---|---|---|---|
| Control | ||||||
| 1 | Physiological aging | F | 82 | A | I | 34 |
| 2 | Physiological aging | F | 88 | A | I | 36 |
| 3 | Physiological aging | F | 87 | A | II | 7 |
| 4 | Physiological aging | F | 95 | B | II | 24 |
| AD | ||||||
| 1 | SDAT | F | 80 | B | VI | 3.3 |
| 2 | SDAT | F | 88 | C | V | 11 |
| 3 | SDAT | F | 87 | C | VI | 6.5 |
| 4 | SDAT | F | 98 | C | V | 12 |
Abbreviations: AD, Alzheimer's disease; F, female; M, male; NFT, neurofibrillary tangle; PMI (h), post-mortem interval in hours; SDAT, senile dementia/Alzheimer's type. Stages of amyloid deposits: A, rare or a few; B, mild or moderate; C, numerous or marked.
Autopsy, neuropathological diagnostic criteria and immunohistochemistry
Brains were removed at the time of autopsy, weighed, cut mid-sagittally and examined for vascular and other macroscopically detectable lesions. Specimens for diagnostic examination were obtained from the hemisphere showing abnormal findings based on computed tomography scanning or from the left hemisphere when no difference was observed between the left and the right hemispheres. Specimens were fixed in 4% paraformaldehyde (Sigma, Tokyo, Japan) as a hemisphere block. Samples used for diagnostic purposes were taken from the frontal, temporal, parietal and occipital lobes; hippocampal formation; amygdala; basal ganglia; thalamus; and midbrain, including the substantia nigra, pons, medulla and the Cb. The specimens were embedded in paraffin and processed into 5-mm sections for conventional histological and immunohistochemical examination. For standard tissue characterization, specimens were stained using hematoxylin–eosin (to display morphology) and Klüver–Barerra staining methods. Methenamine silver and modified Gallyas–Braak staining were used to detect senile plaques, cerebral amyloid angiopathy and NFTs. Further characterization by immunohistochemistry was performed with specific antibodies against ubiquitin, α-synuclein, Aβ (1–40/42), and MAPT (IBL, Fujioka, Japan). Immunostaining methods were applied using the antibodies in a dilution of 1:1000 and a standard Avidin–Biotin Complex (ABC) method using a VectaStain Elite ABC kit (Vector Laboratories, Burlingame, CA, USA) and the protocols provided by the supplier. After being extensively washed, immunoreactive products were detected with DAKO Envision+ (Dako, Kyoto, Japan) and visualized after the addition of diaminobenzidine as the chromogen.14, 15, 27, 28, 29
Human brain tissue protein extraction from Hp, pCx and Cb
Briefly, brain tissues from Hp, pCx and Cb (Table 1) were excised, snap-frozen in liquid nitrogen and then powdered using a mortar and pestle. Upon addition of lysis buffer (2% sodium dodecyl sulfate, 0.5 ℳ triethyl ammonium bicarbonate buffer (TEAB), 1 Complete protease inhibitor cocktail tablet (Roche, Mannheim, Germany) and 1 PhosSTOP phosphatase inhibitor cocktail tablet (Roche)), the samples were vortexed for 1 min and incubated on ice for an additional 45 min before homogenization (sonication parameters: amplitude, 23% pulse: 5 s/5 s for 5 min) using a Vibra Cell high intensity ultrasonic processor (Jencons Scientific, Leighton Buzzard, Bedfordshire, UK). After centrifugation (20 000 × g/4 °C/30 min), supernatant was collected and stored at −80 °C until further use. The protein was quantified by a ‘2-D Quant' kit (Amersham, Piscataway, NJ, USA) according to the manufacturer's protocol.14, 29
iTRAQ protocol
A detailed description of the 2D-LC-MS/MS-iTRAQ procedures,30, 31 including postproteomic data verification by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blot analyses (Supplementary Figures S1–S3),32, 33, 34 can be found in the Supplementary document.
Experimental design
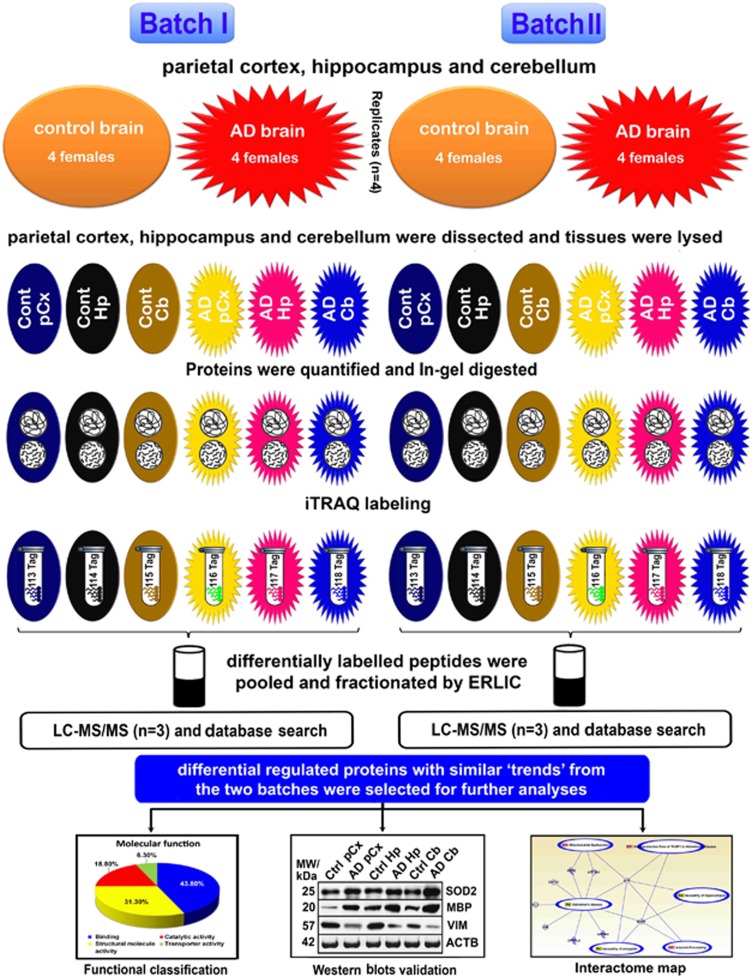
Hp, pCx and Cb tissues were isolated from four AD females and four age-matched non-demented females (controls) using the above-mentioned protocol. Detailed diagnostic features used in this study, such as stages of plaques and NFTs of AD patients and control patients, are described in Table 1. Proteomic analysis was conducted in two batches (B-I and B-II). For each batch, we used four (four biological replicates) different Hp, pCx and Cb tissues pooled from four AD and four control patients, respectively. Each batch was then repeated six times to ensure high confidence in the detection of brain region-specific proteins regulated by aging-related neurodegeneration. The quality of the data set and instrumental reproducibility was achieved by comparing and combining three technical replicates after the samples were labeled with 113–118 isobaric tags and processed in 2D-LC-MS/MS (Figure 1).
Figure 1.
Schematic representation of the experimental design showing biological and technical replicates. Brain tissues (hippocampus (Hp), parietal cortex (pCx) and cerebellum (Cb)) were isolated from four Alzheimer's disease (AD) and four age-matched non-demented control subjects. The quantitative proteomics analyses of AD and control brains were performed by labeling with multi eight-plex isobaric tag for relative and absolute quantitation (iTRAQ) reagents (113–118) (followed by electrostatic repulsion-hydrophilic interaction chromatography (ERLIC)-based fractionation, and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS)-based multidimensional protein identification technology (2D-LC-MS/MS-iTRAQ). The obtained data were analyzed using ProteinPilot software, classified by the Panther database and validated by western blots.
To verify the tissue specificity and that the protein samples were covering the whole human brain tissue proteome, all the identified proteins were uploaded into JVirGel (http://www.jvirgel.de/), a database that creates a virtual 2D-gel picture (Supplementary Figure S1).35
In addition, we used online databases (for example, Panther (Protein Analysis Through Evolutionary Relationship) at www.pantherdb.org, UniProt, NCBI and ‘softberry' http://linux1.softberry.com/berry.phtml) to classify the functions of the iTRAQ-identified proteins modulated in the AD brains. Further biocomputational network analysis of the iTRAQ-identified proteins using the Ingenuity Pathways Analysis (IPA, http://www.ingenuity.com) offered us additional valuable clues about the complex interactions between these proteins in the AD brain.
Following the database search and classification of proteins, western blots were performed on randomly selected proteins to verify the iTRAQ values. The western blot images correlated well and thus validated the obtained iTRAQ values (Supplementary Figures S2 and S3).
Statistical analysis
The data obtained by the western blot analyses are illustrated as the mean±s.d. Student's t-tests were used to determine statistical significance. SPSS 19.0 (Statistical Products and Service Solutions, IBM Corporation, Armonk, NY, USA) for Windows was used to perform an analysis of variance followed by Fisher's protected least significant difference post hoc tests, when warranted. For the iTRAQ analysis, ProteinPilot Software 3.0 (Applied Biosystems) was used. To be considered statistically significant, we required a probability value to be <0.05 (95% confidence limit, *P<0.05).29, 32, 33
Results
Identification of brain region-specific proteome changes in the Hp, pCx and Cb of AD
We identified a total of 950 proteins in the Hp, pCx and Cb through the use of iTRAQ. Of those identified proteins, 825 were quantified with a strict cutoff of an unused ProtScore ⩾2 as the qualification criteria. These results correspond to a peptide confidence level of 99% and an applied false discovery rate of 0.33% (<1.0%). A total of 31 proteins showed an altered expression level in the investigated brain sites (Figure 1 and Tables 2, 3, 4, 5, 6, 7, 8, with the cutoff for up- and down-regulation predefined at 1.2 and 0.83, respectively (P-values are the average P-values of batch-I and batch-II; s.d. values are s.d. of batch-I and batch-II)).
Table 2. List of regulated proteins in the Alzheimer's disease (AD) hippocampus (Hp).
| Protein IDs | Gene symbols | Protein names | Biological process | No. of peptides | AD Hp/Control | P-value | s.d. value | Subcellular location |
|---|---|---|---|---|---|---|---|---|
| Structural constituent of cytoskeleton | ||||||||
| IPI00219301.7 | MARCKS | Myristoylated alanine-rich C-kinase substrate | Calcium-mediated signaling | 30 | 2.05 | 4.43E–05 | 0.7725 | Plasma membrane |
| IPI00001453.2 | INA | α-Internexin | Cellular component morphogenesis | 24 | 1.86 | 0.0349 | 0.1334 | Intermediate filament |
| Nucleic acid binding | ||||||||
| IPI00645192.3 | MECP2 | Isoform B of methyl-CpG-binding protein 2 | Regulation of transcription from RNA polymerase II promoter | 11 | 3.35 | 0.0209 | 0.6794 | Nucleus |
| IPI00217467.3 | HIST1H1E | Histone H1.4 | Nucleotide and nucleic acid metabolic process | 12 | 7.65 | 0.0392 | 1.2124 | Nucleus |
| Transfer | ||||||||
| IPI00745872.2 | ALB | Isoform 1 of serum albumin | Transport | 31 | 1.67 | 0.0155 | 0.3689 | Secreted |
| GTPase activity/ATP binding | ||||||||
| IPI00026268.3 | GNB1 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β-1 | Ras protein signal transduction | 6 | 0.71 | 0.0302 | 0.1165 | Nucleus |
| IPI00640817.1 | AK1 | Adenylate kinase 1 | ATP metabolic process | 7 | 1.67 | 0.0307 | 0.2267 | Cytoplasm |
| Fructose binding/nervous system development | ||||||||
| IPI00796333.1 | ALDOA | Fructose-bisphosphate aldolase A | Glycolysis | 26 | 0.57 | 0.0315 | 0.0111 | Cytoplasm |
| IPI00160552.3 | TNR | Tenascin-R | Signal transduction | 10 | 0.67 | 0.0004 | 0.0238 | Secreted |
| Chaperone-mediated protein folding | ||||||||
| IPI00400826.1 | CLU | Isoform 2 of Clusterin | Apoptosis | 3 | 0.51 | 0.0479 | 0.0168 | Cytoplasm |
Table 3. List of regulated proteins in the Alzheimer's disease (AD) parietal cortex (pCx).
| Protein IDs | Gene symbols | Protein names | Biological process | No. of peptides | AD pCx/Control | P-value | s.d. value | Subcellular location |
|---|---|---|---|---|---|---|---|---|
| Neurological system process/hydrolase activity | ||||||||
| IPI00251507.2 | SYN1 | Isoform IB of Synapsin-1 | Synaptic transmission | 106 | 0.72 | 0.0334 | 0.0406 | Golgi apparatus |
| IPI00440493.2 | ATP5A1 | ATP synthase subunit α | Cation transport | 48 | 1.30 | 0.0424 | 0.0231 | Mitochondrial |
Table 4. List of regulated proteins in the Alzheimer's disease (AD) cerebellum (Cb).
| Protein IDs | Gene symbols | Protein names | Biological process | No. of peptides | AD Cb/Control | P-value | s.d. value | Subcellular location |
|---|---|---|---|---|---|---|---|---|
| Neurological system process | ||||||||
| IPI00219661.2 | PLP1 | Isoform 1 of myelin proteolipid protein | Cellular component morphogenesis | 6 | 5.77 | 0.0039 | 0.7291 | Plasma membrane |
| IPI00015964.4 | GAP43 | Neuromodulin | Activation of protein kinase C activity by G-protein-coupled receptor protein signaling pathway | 21 | 2.75 | 9.32E–05 | 0.1643 | Cell membrane |
| Hydrogen ion transporting ATP synthase activity/oxidoreductase activity | ||||||||
| IPI00257508.4 | DPYSL2 | Dihydropyrimidinase-related protein 2 | Nucleotide and nucleic acid metabolic process | 47 | 1.69 | 0.0328 | 0.0542 | Cytoplasm |
| IPI00014439.4 | QDPR | Dihydropteridine reductase | Cellular amino acid catabolic process | 11 | 1.59 | 0.0386 | 0.6406 | Cytoplasm |
| Nucleotide binding/lyase activity | ||||||||
| IPI00789551.1 | MATR3 | Matrin-3 | Structural molecule activity | 6 | 0.66 | 0.0504 | 0.0304 | Nucleus |
| IPI00465248.5 | ENO1 | α-Enolase | Glycolysis | 42 | 0.75 | 0.0175 | 0.0980 | Cytoplasm |
| Structural constituent of cytoskeleton | ||||||||
| IPI00026314.1 | GSN | Isoform 1 of Gelsolin | Cellular component morphogenesis | 10 | 0.69 | 0.0569 | 0.0540 | Cytoplasm |
Table 5. List of common regulated proteins in the Alzheimer's disease (AD) hippocampus (Hp) and parietal cortex (pCx).
| Protein IDs | Gene symbols | Protein names | Biological process | No. of peptides | AD pCx/Control | P-value | s.d. value | No. of peptides | AD Hp/Control | P-value | s.d. value | Subcellular location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alzheimer disease-amyloid secretase pathway | ||||||||||||
| IPI00219186.1 | APP | APP Isoform APP714 of amyloid β A4 protein (Fragment) | Signal transduction | 13 | 2.82 | 0.0128 | 0.0707 | 10 | 2.39 | 0.0136 | 0.2291 | Membrane |
| Calcium ion binding | ||||||||||||
| IPI00221226.7 | ANXA6 | Annexin A6 | Intracellular protein transport | 17 | 1.83 | 0.0632 | 0.0487 | 18 | 2.64 | 0.0134 | 0.3616 | Cytoplasm |
Table 6. List of common regulated proteins in the Alzheimer's disease (AD) hippocampus (Hp) and cerebellum (Cb).
| Protein IDs | Gene symbols | Protein names | Biological process | No. of peptides | AD Hp/Control | P-value | s.d. value | No. of peptides | AD Cb/Control | P-value | s.d. value | Subcellular location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Structural molecule activity | ||||||||||||
| IPI00220175.5 | MAPT | Isoform Tau-E of Microtubule-associated protein tau | Cellular component morphogenesis | 10 | 2.01 | 0.0013 | 0.2714 | 11 | 1.49 | 0.0371 | 0.0931 | Plasma membrane |
| IPI00020356.4 | MAP1A | MAP1A 331 kDa protein | Microtubule associated complex | 12 | 1.51 | 0.0107 | 0.0183 | 17 | 1.70 | 0.0472 | 0.0222 | Cytoplasm |
| Protein binding | ||||||||||||
| IPI00021812.2 | AHNAK | Neuroblast differentiation-associated protein AHNAK | Nervous system development | 8 | 2.73 | 4.84E–07 | 0.1066 | 6 | 0.55 | 0.0216 | 0.0239 | Nucleus |
| IPI00295601.1 | CEND1 | Cell cycle exit and neuronal differentiation protein 1 | Involved in neuroblastoma cell differentiation | 6 | 5.7 | 0.0398 | 0.2243 | 2 | 0.52 | 0.0428 | 0.0205 | Membrane |
| Oxidoreductase activity | ||||||||||||
| IPI00219018.7 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis | 33 | 0.39 | 0.0027 | 0.0649 | 184 | 0.7955 | 2.12E–05 | 0.0621 | Cytoplasm |
Table 7. List of common regulated proteins in the Alzheimer's disease (AD) parietal cortex (pCx) and cerebellum (Cb).
| Protein IDs | Gene symbols | Protein names | Biological process | No. of peptides | AD pCx/Control | P-value | s.d. value | No. of peptides | AD Cb/Control | P-value | s.d. value | Subcellular location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA binding/hydrolase activity | ||||||||||||
| IPI00217466.3 | HIST1H1D | Histone H1.3 | Nucleotide and nucleic acid metabolic process | 7 | 4.12 | 0.0220 | 0.8771 | 13 | 0.57 | 0.0423 | 0.0852 | Nucleus |
| IPI00289159.3 | GLS | Isoform KGA of Glutaminase kidney isoform | Cellular amino acid metabolic process | 3 | 0.35 | 0.0443 | 0.0229 | 14 | 0.81 | 0.059 | 0.0370 | Cytoplasm |
Table 8. List of common regulated proteins in the Alzheimer's disease (AD) parietal cortex (pCx), hippocampus (Hp) and the cerebellum (Cb).
| Protein IDs | Gene symbols/protein names | No. of peptides | AD pCx/Control | P-value | s.d. value | No. of peptides | AD Hp/Control | P-value | s.d. value | No. of peptides | AD Cb/Control | P-value | s.d. value | Subcellular location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oxidoreductase activity | ||||||||||||||
| IPI00022314.1 | SOD2; superoxide dismutase (Mn) | 4 | 2.19 | 0.0406 | 0.6278 | 4 | 1.64 | 0.0393 | 0.2031 | 4 | 1.53 | 0.0452 | 0.1132 | Mitochondrial |
| Structural constituent of myelin sheath/structural constituent of cytoskeleton | ||||||||||||||
| IPI00021907.2 | MBP; isoform 1 of myelin basic protein | 61 | 2.05 | 0.0250 | 0.6187 | 30 | 1.53 | 9.31E–06 | 0.4068 | 61 | 3.40 | 2.6466E–05 | 0.4032 | Cell membrane |
| IPI00418471.6 | VIM; Vimentin | 24 | 0.49 | 0.0039 | 0.0303 | 42 | 0.74 | 0.0241 | 0.0209 | 24 | 0.59 | 0.0213 | 0.0303 | Cytoplasm |
Specifically, 11 proteins (APP, MARCKS, INA, MECP, HIST1H1E, ALB, GNB1, AK1, ALDOA, TNR and CLU) in the Hp (Table 2), 2 proteins (SYN1 and ATP5A1) in the pCx (Table 3), and 7 proteins (PLP1, GAP43, DPYSL2, QDPR, MATR3, ENO1 and GSN) in the Cb (Table 4) were modulated. In addition, ANXA6 was modulated in both Hp and pCx (Table 5). MAPT, MAP1A, AHNAK, CEND1 and GAPDH were altered in both Hp and Cb (Table 6) and HIST1H1D and GLS were modulated in both pCx and Cb (Table 7). SOD2, MBP and VIM were modulated in all brain regions investigated (Table 8).
As expected, APP was found to be upregulated in the AD Hp, whereas MAPT and MAP1A were upregulated in both AD Hp and Cb. It is noteworthy that Clusterin (CLU) was downregulated in the Hp, AHNAK was upregulated in the Hp and downregulated in the Cb, the extracellular matrix protein Tenascin-R (TNR) was downregulated in the Hp, HIST1H1D/E were upregulated in the pCx and the Hp and downregulated in the Cb and MECP2 was upregulated in the Hp of AD brains. In addition, the antiapoptotic protein SOD2 was upregulated in all three areas (Hp, pCx and Cb) of AD subjects.
Biocomputational classification of the regulated proteins in the Hp, pCx and Cb of AD brains
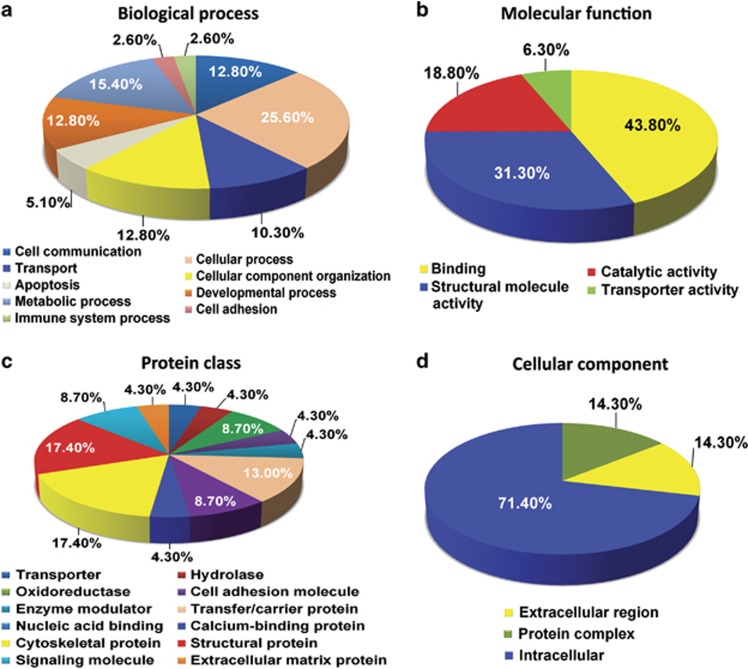
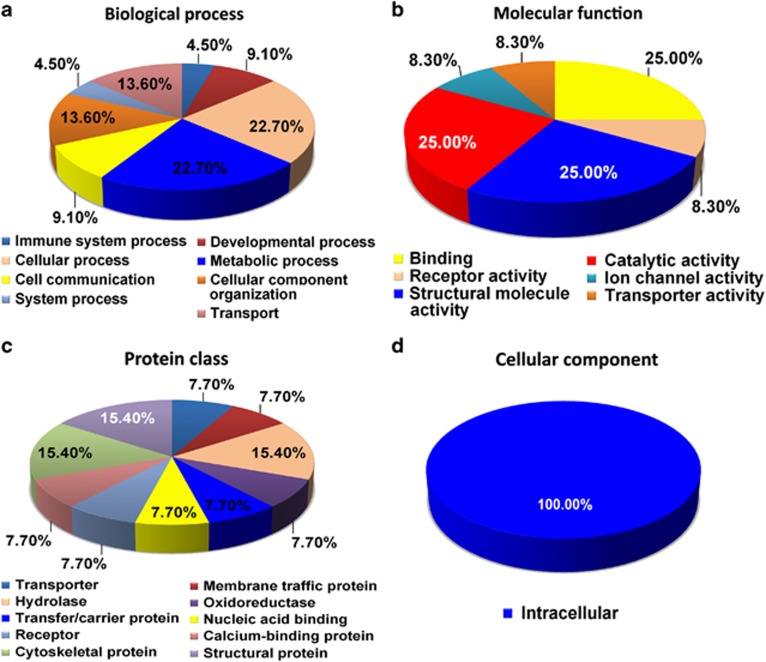
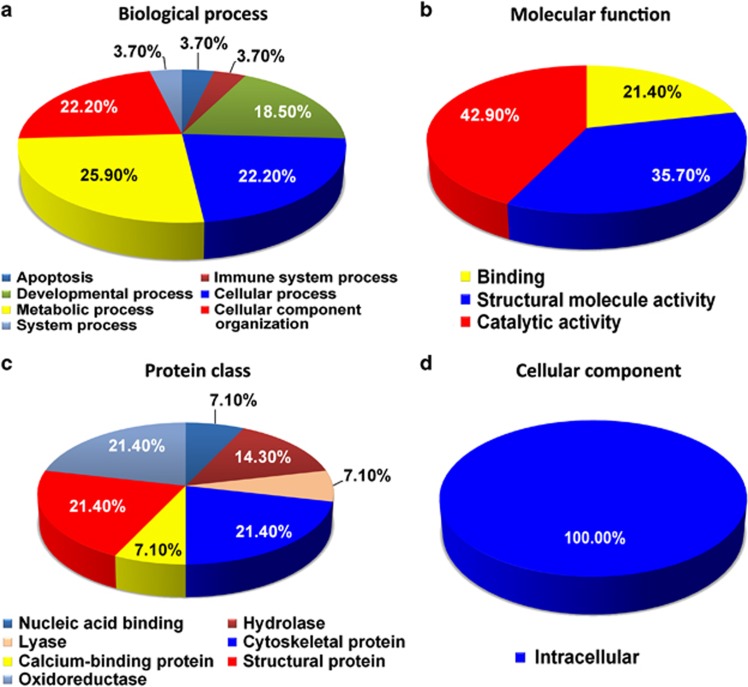
Next, we classified all of the 31 differentially regulated proteins from the Hp, pCx and Cb of AD brains based on their classes as well as cellular and molecular functions using the Panther database (Figures 2, 3, 4 and Tables 2, 3, 4, 5, 6, 7, 8). As predicted, both molecular and cellular functions of these proteins, as well as their associated biological pathways, were significantly altered in the Hp and the pCx compared with the Cb (Figures 2, 3, 4). All investigated regions (Hp, pCx and Cb) revealed severe alterations in metabolic, cellular and developmental processes. The most interesting observation was that the percentage of alterations in developmental processes was highest in the Cb (35.7% it was 29% for Hp and 25% for pCx), whereas the percentage of alterations in cellular processes was equivalent in both the pCx (62.5%) and the Hp (58%) (Figures 2, 3, 4a). As part of these changes, binding proteins (43.8% HIST1H1E and APP) were significantly altered in the Hp as well as the structural proteins (31.3% MAPT, MARCKS, MBP, INA and vimentin) and catalytic regulators (18.8% GAPDH and GNB1). Alterations in binding proteins (25% HIST1H1D and ANNEXIN A6), catalytic proteins (25% GLS, ATP5A1 and SOD2) and structural proteins (25% MBP, synapsin1 and vimentin) were similar in the pCx. Catalytic proteins (42.9% GLS, GAPDH, ENO1 and SOD2) in the Cb were the most affected group of proteins. Structural proteins (35.7% MAPT, GSN, PLP1, MBP and Vimentin) and binding proteins (21.4% HIST1H1D and GSN) were also significantly affected (Figures 2, 3, 4b). It is of interest to note that a substantial proportion of extracellular proteins (for example, TNR) were significantly downregulated exclusively in the Hp, whereas downregulation was only observed among intracellular proteins in the pCx and the Cb (Figures 2, 3, 4).
Figure 2.
Pie chart depicting the functional classification of differentially regulated proteins in the hippocampus (Hp) of Alzheimer's disease (AD) brains. The isobaric tag for relative and absolute quantitation (iTRAQ)-identified hippocampal proteome was characterized within the molecular function Gene Ontology (GO) category. Subcellular and functional categories were based on the annotations of GO using the online tool at www.pantherdb.org in the following categories: (a) biological process, (b) molecular function, (c) protein class and (d) cellular component.
Figure 3.
Pie chart depicting the functional classification of differentially regulated proteins in the parietal cortex (pCx) of Alzheimer's disease (AD) brains. The isobaric tag for relative and absolute quantitation (iTRAQ)-identified parietal cortical proteome was characterized within the molecular function Gene Ontology (GO) category. Subcellular and functional categories were based on the annotations of GO using the online tool at www.pantherdb.org in the following categories: (a) biological process, (b) molecular function, (c) protein class and (d) cellular component.
Figure 4.
Pie chart depicting the functional classification of differentially regulated proteins in the cerebellum (Cb) of Alzheimer's disease (AD) brains. The isobaric tag for relative and absolute quantitation (iTRAQ)-identified cerebellar proteome was characterized within the molecular function Gene Ontology (GO) category. Subcellular and functional categories were based on the annotations of GO using the online tool at www.pantherdb.org in the following categories: (a) biological process, (b) molecular function, (c) protein class and (d) cellular component.
Biocomputational network analysis of the proteins regulated in the Hp, pCx and Cb of AD brains
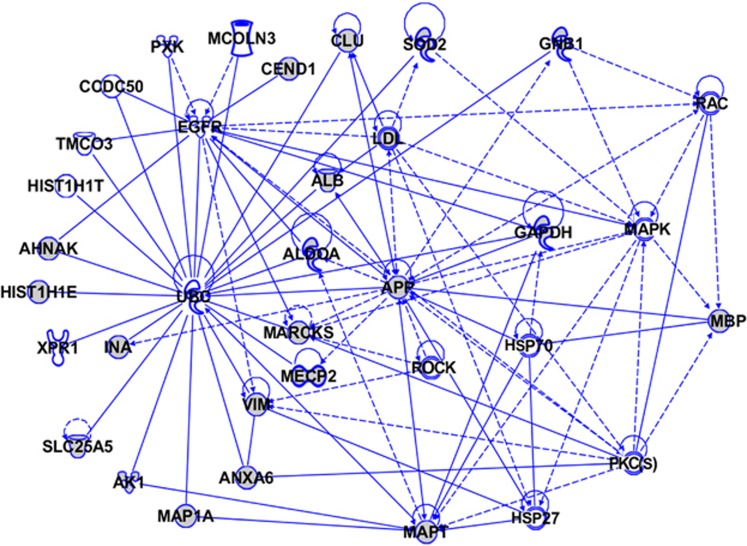
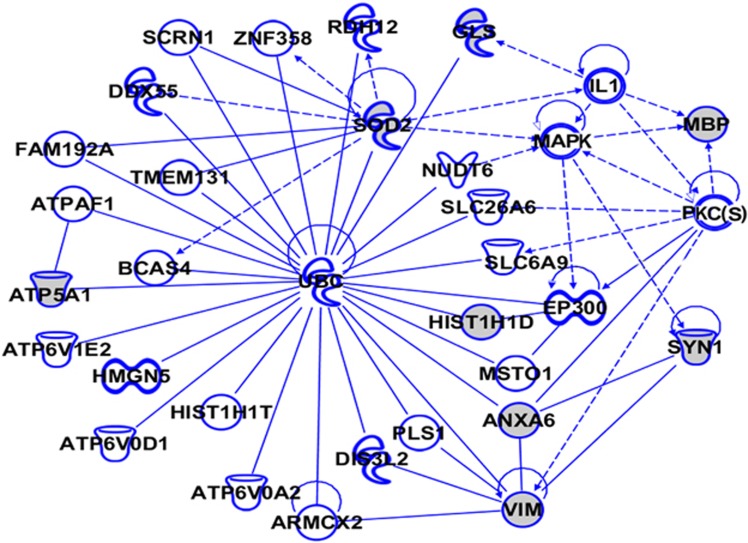
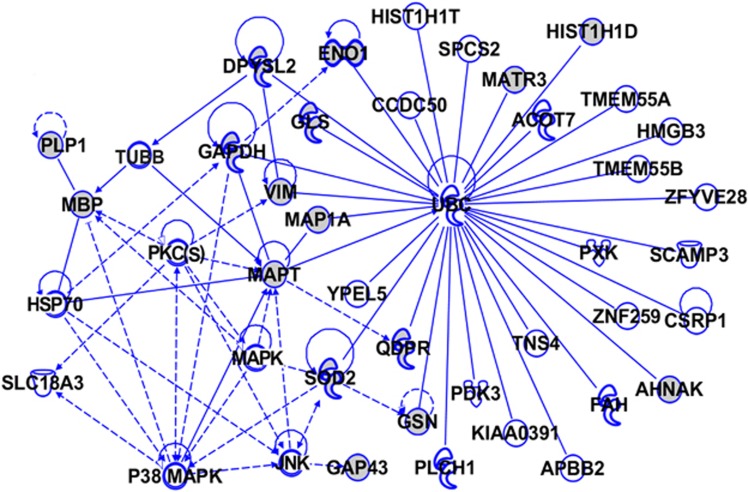
To gain further insight into the potential biological mechanisms involving the iTRAQ-identified differentially regulated proteins in the AD brain, we used IPA (http://www.ingenuity.com). IPA identifies protein networks based on the known interactions (either direct or indirect) between proteins. In addition, IPA defines common functional and canonical pathways, thereby offering additional information about the complex interactive links between these proteins in the brain (Figures 5, 6, 7). The Hp network comprises 35 proteins (AHNAK, AK1, ALB, ALDOA, ANXA6, APP, CCDC50, CEND1, CLU, EGFR, GAPDH, GNB1, HIST1H1E, HIST1H1T, HSP27, HSP70, INA, LDL, MAP1A, MAPK, MAPT, MARCKS, MBP, MCOLN3, MECP2, PKC(s), PXK, RAC, ROCK, SLC25A5, SOD2, TMCO3, UBC, VIM and XPR1), of which 22 proteins were quantified by iTRAQ. The remaining 13 proteins were found by IPA to interact with the proteins quantified by iTRAQ. These proteins mediate biological processes such as glucose metabolism, signal transduction and apoptosis (Figure 5). The pCx network contained 33 proteins (ATP6V0D1, ATPAF1, ATP5A1, ATP6V1E2, ATP6V0A2, ANXA6, ARMCX2, BCAS4, DDX55, DIS3L2, EP300, FAM192A, GLS, HIST1H1D, HIST1H1T, HMGN5, IL1, MAPK, MBP, MSTO1, NUDT6, PKC(s), PLS1, RDH12, SYN1, SCRN1, SLC26A6, SLC6A9, SOD2, TMEM131, UBC, VIM and ZNF358), of which 8 proteins were quantified by iTRAQ. The remaining 25 proteins were found by IPA to interact with the proteins quantified by iTRAQ (Figure 6). The Cb network comprises 43 proteins (ACOT7, AHNAK, APBB1, CCDC50, CSRP1, DPYSL2, ENO1, FAH, GAPDH, GLS, GAP43, GSN, HIST1H1D, HIST1H1T, HMGB3, HSP70, JNK, KIAA0391, MAP1A, MAPK, MAPT, MATR3, MBP, P38MAPK, PDK3, PLCH1, PLP1, PKC(s), PXK, QDPR, SCAMP3, ZNF259, SLC18A3, SOD2, SPCS2, TUBB, TMEM55A, TMEM55B, TNS4, UBC, VIM, YPEL5 and ZFYVE28), of which 16 proteins were quantified by iTRAQ. The remaining 27 proteins were found by IPA to interact with the proteins quantified by iTRAQ. These proteins are mainly involved in developmental processes, and their downregulation is critically associated with various neurological, skeletal and muscular disorders (Figure 7). Interestingly, our IPA networks of differentially regulated proteins from Hp, pCx and Cb revealed a strong interaction between most of the AD-associated proteins (such as AHNAK, CLU, CEND1, SOD2, GNB1, ANXA6, ATP5A1, TNR and HIST1H1 variants) with ubiquitin C (UBC), a type of post-translational modification system. UBC binds to target proteins either as a monomer or as a polymer and forms a polyubiquitin chain to label those proteins for the ubiquitin proteasome degradation system (UPS).
Figure 5.
The Ingenuity Pathway Analysis (IPA)-generated network of isobaric tag for relative and absolute quantitation (iTRAQ)-identified proteins and their interacting partners in Alzheimer's disease (AD) hippocampus (Hp). The IPA construct networks based on the differentially regulated proteins and their potential link with other known proteins in human AD Hp. Solid and dashed connecting lines indicate the presence of direct and indirect interactions, respectively. Modulatory roles of proteins on the expression of other proteins are indicated by arrows.
Figure 6.
The Ingenuity Pathway Analysis (IPA)-generated network of isobaric tag for relative and absolute quantitation (iTRAQ)-identified proteins and their interacting partners in Alzheimer's disease (AD) parietal cortex (pCx). The IPA construct networks based on the differentially regulated proteins and their potential link with other known proteins in human AD pCx. Solid and dashed connecting lines indicate the presence of direct and indirect interactions, respectively. Modulatory roles of proteins on the expression of other proteins are indicated by arrows.
Figure 7.
The Ingenuity Pathway Analysis (IPA)-generated network of isobaric tag for relative and absolute quantitation (iTRAQ)-identified proteins and their interacting partners in Alzheimer's disease (AD) cerebellum (Cb). The IPA construct networks based on the differentially regulated proteins and their potential link with other known proteins in human AD Cb. Solid and dashed connecting lines indicate the presence of direct and indirect interactions, respectively. Modulatory roles of proteins on the expression of other proteins are indicated by arrows.
Discussion
Aging-related neurodegenerative disorders such as AD are multifactor disorders. These factors include a variety of brain changes that begin as many as 20 years before symptoms appear and involve the disruption of the most intricate neuronal networks regulating synaptic plasticity, signal transduction and transport. In this study, we provide for the first time a comprehensive quantitative brain site-specific proteome study of aging and aging-related AD-type dementia brains.
In recent years, several proteomic studies have described the differential protein expression profile in the brains of AD patients, with great emphasis on the cortical and hippocampal proteomes. Surprisingly, none of the reports focused on the cerebellar proteome despite its unique behavior of staying unaffected in AD. However, several reports have confirmed that the Cb also reveals molecular changes in response to AD, but this area of the brain likely counteracts those changes more efficiently.17, 18, 19, 20, 21, 22, 23, 24 Therefore, we aimed to compare the Cb proteome with the Hp and pCx proteomes to provide critical information on brain region-specific mechanisms that are counteracting the development and progression of aging and AD-related neurodegeneration and may illuminate how the Cb sequesters itself from neuronal death in AD. We identified 31 proteins that were altered significantly in the investigated brain areas, including Hp, pCx and Cb. The biocomputational protein classification analysis using the Panther database suggests that these proteins are involved in diverse biological processes but with different configurations in the Hp, pCx and Cb. The IPA indicates that the proteins that were altered in the AD brains had a strong interaction with UBC signaling in all three of the investigated brain regions. UBC mediates the polyubiquitination of proteins and consequently targets them to be transported to the proteasome for degradation back to the basic building blocks of cells so that they may be recycled elsewhere.
In addition, our data are in agreement with the expression levels of several proteins altered in the brains of AD-based neuropathology. For instance, ATP5A1 was suggested to contribute to neurodegeneration as it accumulates in the cytosol at early stages of NFT-based neurodegenerative processes.36 Furthermore, the protein levels of ALDOA were reported to be increased in AD hippocampal tissue.37 Our data are in line with previous studies of ATP5A1, which was downregulated in the Hp whereas ALDOA expression was upregulated.38 Impairment of brain metabolism has been recognized as a hallmark of AD, and the reduction of glucose utilization is paralleled by a decrease in the expression of glycolytic enzymes. We observed a significant downregulation of ALDOA in the Hp, ENO1 in the Cb and GAPDH in both the Hp and the Cb, thus explaining the impaired glucose metabolic system in AD.
Hyperphosphorylation of MAPT (tau) is closely related to various neurodegenerative diseases, including aging-related dementia such as AD.39, 40 For instance, deposition of hyperphosphorylated tau in the Cb of PS1 E280A AD has recently been reported.18 Many mechanisms could be involved in tau hyperphosphorylation, including the upregulation of tau kinases, the downregulation of phosphatases and other covalent modifications of tau.7, 41, 42 Although the MAPT hyperphosphorylation in the Hp and the pCx has been reported in neurodegenerative tauopathies,43 no thorough data have been reported yet regarding its changes in the Cb. Our data reveal high protein expression levels of MAPT in both AD Hp and AD Cb tissues. Recently, it has been hypothesized that a prion-like transmission of misfolded hyperphosphorylated MAPT or Aβ aggregates between neurons is one possible explanation for AD-associated anatomical irregularity and progression that appears in the absence of cortical Aβ pathology and MAPT lesions in the transentorhinal region. Misfolded MAPT in the neuronal cytoplasm may function as a seed that triggers hyperphosphorylation and misfolding of the natively unfolded MAPT protein. Disease progression is therefore associated with the intercellular transfer of pathogenic proteins, such as hyperphosphorylated MAPT aggregates.44, 45, 46, 47 This sheds further light on the current discussion that an impairment of the UPS is affected at early stages of AD,48, 49 and is in agreement with our current and previous data showing that pivotal proteins of the chaperone/proteasomal pathways are changed at early stages of AD.14, 15 It is possible that these abnormally high expressions of MAPT and the disturbed UPS could contribute to the hyperphosphorylation of MAPT in the Hp and eventually in the Cb too. In addition, Sepulveda-Falla et al.18 reported that the deposition of hyperphosphorylated MAPT in the Cb was found in AD and was caused by a presenilin-1 mutation E280A. Thus, combined with our proteomic results, the high expression of (hyperphosphorylated) MAPT in the Cb could also be involved in aging-related neuropathologies such as AD. However, this suggests that the Cb exploits cellular tools to inhibit hyperphosphorylated MAPT-based NFT formation. Furthermore, we identified several other proteins critically regulated in AD brains that could eventually become potential therapeutic targets for various neurodegenerative disorders. In the following sections, we describe some of these proteins with respect to AD.
CLU and AD
CLU is another important protein involved in neurodegenerative diseases.50 Association of CLU gene polymorphisms with late-onset AD has been reported recently,51, 52, 53, 54, 55 and additional data provide a possible link between the CLU and APOE genotypes in the etiology of AD.56, 57 Some in vitro studies have demonstrated that at certain concentrations, purified CLU can interact with Aβ and result in an inhibition of fibril formation and thus functions as an extracellular chaperone that prevents the aggregation of nonnative proteins.58, 59, 60 In contrast, in a mouse model of AD, CLU was found to cause neuritic dystrophy by promoting Aβ plaque formation.61 This effect of CLU varied according to the amount of the prefibrillar substrate.62 Regardless, CLU could play an antiapoptotic function by interfering with the activity of the proapoptotic protein Bax.63 Overexpression of CLU has also been found in several human gland cancers.64 Thus, our data support the hypothesis that the strict downregulation of CLU in the AD Hp could contribute to neurodegenerative processes because of a reduced chaperone function and decreased antiapoptotic activity.
TNR and AD
TNR is an extracellular glycoprotein known to be primarily involved in signal transduction and cell–matrix adhesion. Loss of TNR impairs cognition, synaptic plasticity and motor abilities in mice. Homozygous deletion of TNR is associated with intellectual disability and cognitive deficits.65, 66 Our data are consistent with these reports, indicating that TNR was significantly downregulated. However, this TNR downregulation only occurred in the Hp, and not in the pCx or Cb. Thus, we postulate that the downregulation of TNR expression might be one specific direct cause of impaired cognitive abilities in AD. Interfering with the TNR protein expression level could potentially be useful for novel AD treatment strategies especially through an improvement of Hp function.
AHNAK and AD
Recent findings have disclosed the crucial role of AHNAK in myelination processes during development, neuronal plasticity and neuro-re-/de-generation events;67, 68 because the development of tau lesions in AD is traceable to differences between early- versus late-maturing oligodendrocytes and to the exceptionally protracted myelination of late-developing portions of the human brain,69, 70, 71 AHNAK becomes of pivotal interest for future investigations.
Gelsolin (GSN) and AD
Previous studies have identified the antiamyloidogenic role of gelsolin in AD. Gelsolin can reduce the amyloid burden by acting as an inhibitor of Aβ fibrillization and as an antioxidant and antiapoptotic protein.72, 73 However, the expression level of GSN in the brain of AD patients has been discussed controversially.74, 75 Our data indicate that GSN is significantly downregulated in the AD Cb, which is consistent with a recent report that a reduced plasma GSN level in AD patients was found. This suggests that GSN may function as an additional plasma biomarker candidate that could contribute to the diagnosis of early-stage aging-related AD.75
MECP2 and AD
Environmental factors, including metals and dietary factors, operate by interfering with the interaction of methylated CpG clusters and binding proteins, such as MeCP2 and SP1. The impact of these factors on AD has been discussed previously.76 MECP2 was found to be involved in various neurodevelopmental disorders77 and to promote neuronal death.78 Thus, the extremely high expression of MECP2 in the AD Hp could contribute to neurodegenerative pathways.
Histones and AD
The post-translational modifications of histones, such as acetylation and deacetylation, have been increasingly recognized as critical factors affecting gene activation and silencing in the brain of individuals with neurodegenerative disorders.79, 80, 81, 82, 83 Aside from its post-translational modifications, the expression levels of histones are also important for cell cycle control.84 Here, we show that the expression of HIST1H1E was increased in the AD Hp, whereas the expression of HIST1H1D was increased in the AD pCx but decreased in the AD Cb. In general, increased levels of histone variants were found in affected areas (pCx and Hp) in AD, whereas the Cb reflected reduced levels of another cell cycle protein, CEND1. The different changes in expression levels of histones in different brain regions indicate their brain site-specific roles in the pathology of AD. However, further studies are required to unravel the link between the expression levels and cellular functions of these proteins to accurately evaluate their specific pathological roles in AD and other neurodegenerative disorders.
CEND1 and AD
CEND1 is another neural stem cell-specific protein involved in cell cycle exit and neuronal differentiation.85 The correlation of neurodegeneration and deregulation of the cell cycle in AD have been frequently discussed over the past few years.86, 87, 88, 89, 90, 91, 92 Brain site-specific CEND1 expression changes (increased in the Hp and decreased in the Cb) may cause neurodegeneration by promoting abnormal cell cycle re-entry.93 Thus, increased CEND1 expression in the Hp could be responsible for a reduced regeneration capacity, whereas its downregulation in the Cb might be a survival strategy acquired by this area to induce dynamic regeneration. Thus, our study suggests a potential therapeutic usage of CEND1 in neurodegenerative disorders.
UPS and AD
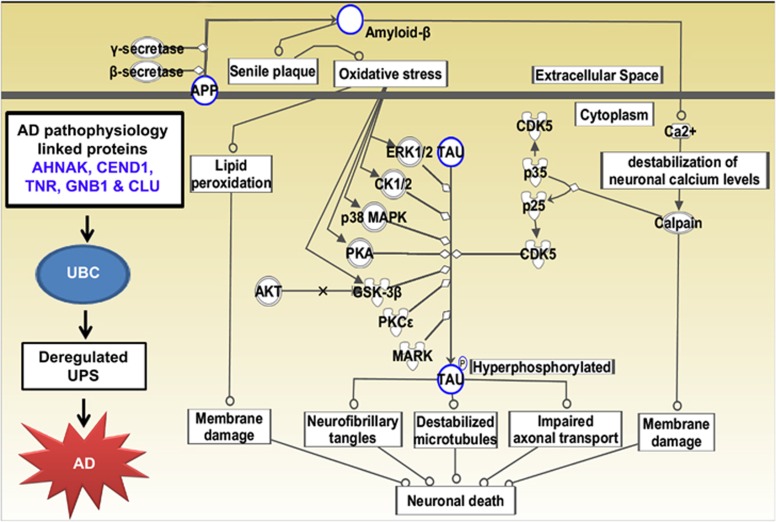
Our iTRAQ data provide information about various proteins with an altered brain site-specific expression pattern, and it is intriguing to notice that many of them functionally interact with the pivotal UPS regardless of their resources. Our data are consistent with previous reports and reinforce that the function of the UPS is impaired in AD,94, 95, 96, 97 and thus emphasize the importance of the unfolded protein response and proteostasis in AD (Figure 8).98
Figure 8.
Ingenuity Pathway Analysis (IPA)-derived neurodegenerative disease-specific network of isobaric tag for relative and absolute quantitation (iTRAQ)-identified differentially regulated proteomes of hippocampus (Hp), parietal cortex (pCx) and cerebellum (Cb) of Alzheimer's disease (AD) brains. IPA analysis identified a group of proteins modulated in aging demented brain and their potential interactive links in the context of various neurodegenerative diseases. The nine different pathways identified by IPA are as follows: (1) amyloid processing (APP, MAPT), (2) Huntington's disease signaling (GNB1, GLS), (3) axonal guidance signaling (DPYSL2, GNB1), (4) mitochondrial dysfunction (SOD2, APP, ATP5A1), (5) glycolysis/gluconeogenesis (ENO1, ALDOA, GAPDH), (6) clathrin-mediated endocytosis signaling (CLU, ALB), (7) protein kinase A signaling (HIST1H1D, HIST1H1E), (8) signaling Rho family GTPases (VIM, GNB1) and (9) phospholipase C signaling (AHNAK, GNB, MARCKS).
The UPS is the major proteolytic pathway that degrades intracellular proteins in a regulated manner, usually essential for protein repair, turnover and degradation. Deregulation of the UPS has been implicated in the pathogenesis of many neurodegenerative disorders such as Parkinson's disease, Huntington disease, Pick's disease and prion diseases. In addition, deregulation of the UPS has also been implicated in several other genetic diseases, including cystic fibrosis, Angelman's syndrome and Liddle syndrome, suggesting that activation of similar mechanisms must occur in neurodegeneration as a basic phenomenon.99, 100, 101, 102, 103
Recently, it has also become clear that the UPS is perturbed in aging-related dementia such as AD.96, 97, 104, 105, 106, 107, 108 Pathological aging, with respect to protein repair and degradation via the UPS, and selective protein oxidation may cause protein damage, or protein mutations may induce a dysfunction of the proteasome and a deficit of the UPS in AD. Such events eventually lead to activation of cell death pathways and to an aberrant aggregation or incorporation of ubiquitinated proteins into hallmark structures of AD such as neuritic amyloid plaques or NFTs. In AD, insoluble Aβ peptide aggregates or inclusion bodies within nerve cells are commonly observed with ubiquitin immunoreactivity.109, 110 The accumulation of ubiquitin–protein conjugates in neuropathological lesions was first detected in NFTs isolated from human brain tissue,111, 112, 113 and it is these NFTs that best correlate with the degree of dementia.13 A more direct relationship between the ubiquitin system and pathogenesis of AD was established with the discovery of a frameshift mutation in the ubiquitin transcript, which results in an ubiquitin protein with 20 extra amino-acid residues at its C-terminus (UBB+1).95, 108, 114 However, it is not fully understood why ubiquitin is accumulated in intra- and extra-cellular deposits or how it is involved in AD pathogenesis. Our iTRAQ analysis provides compelling evidence that many of the altered molecular factors in AD brains interact with UBC. Thus, the interconnection of AD-related proteins with UBC implicates a pivotal role of ubiquitin UPS in aging-related neurodegeneration such as AD.
Although our proteomic data are in agreement with others,115 quantitative proteomics analyzing aging brain tissue has yet to be evaluated and reviewed with critical considerations. There are currently a combination of different methods used, such as (1) 2D difference gel electrophoresis (2D-DIGE) combined with MALDI-TOF-MS/MS (matrix-assisted laser desorption/ionization time-of-flight MS/MS) proteomics17, 37, 38, 116 or (2) 2D-LC-MS/MS-iTRAQ/ICAT (isotope-coded affinity tag); some proteins appear more frequently (for example, 14-3-3 proteins, VDACs, VIM, creatine kinase B, Aldolase, GFAP and HSP70 proteins), whereas others are hardly detected (for example, STAT3, p60TRP, NF-κB, NTRK1 and α-, β- and γ-secretases).117, 118 Despite improved sensitivity of our 2D-LC-MS/MS analyses, limitations of ‘undersampling' in LC-MS/MS still restrict the number of peptides that can be identified in a specific sample. Though this technical limitation can be assessed by the inclusion of technical replicates or identical samples that are processed in parallel,119 further technical improvements of quantitative 2D-DIGE/LC-MS/MS (iTRAQ/ICAT) proteomics are required to make more significant translational research-based contributions that may provide more clinically relevant data.
Further studies using higher numbers of control and AD patients, shorter post-mortem intervals and more individual patient-to-patient comparisons may certainly improve our knowledge regarding the neurodegenerative processes in the aging brain.
In conclusion, our current study demonstrates a brain site-specific aging-related proteome pattern and emphasizes the importance of UPS in the AD brain. We report higher MAPT expression levels in the AD Cb suggesting transmission of neurodegeneration signals to this area. Our study also indicates a crucial role of AHNAK in processes of aging-related dementia, and we hypothesize that the Cb exploits proteins such as CEND1 or GSN to fight against neurodegeneration. An even more comprehensive study of the cerebellar proteome would most likely shed further light on how this area counteracts NFT formation and AD-induced neurodegeneration.
In conclusion, our findings have unraveled the complex proteome changes that occur in aging brains and open a new avenue for drug discovery in aging-related brain disorders such as AD.
Acknowledgments
We thank Mr T Kanesaka, Mr N Ogawa, Mr Y Tani and Ms C Taniguchi (all at the Choju Medical Institute, Fukushimura Hospital) for technical assistance, patient care, sampling and tissue data acquisition. This work was supported by the research fund of Hanyang University (HY-2012-N).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Mattson MP. Pathways towards and away from Alzheimer's disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese K, Akatsu H. Alzheimer's disease--an interactive perspective. Curr Alzheimer Res. 2006;3:109–121. doi: 10.2174/156720506776383022. [DOI] [PubMed] [Google Scholar]
- Holscher C. Development of beta-amyloid-induced neurodegeneration in Alzheimer's disease and novel neuroprotective strategies. Rev Neurosci. 2005;16:181–212. doi: 10.1515/revneuro.2005.16.3.181. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ, Schenk D. Alzheimer's disease: molecular understanding predicts amyloid-based therapeutics. Annu Rev Pharmacol Toxicol. 2003;43:545–584. doi: 10.1146/annurev.pharmtox.43.100901.140248. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Claeysen S, Cochet M, Donneger R, Dumuis A, Bockaert J, Giannoni P. Alzheimer culprits: cellular crossroads and interplay. Cell Signal. 2012;24:1831–1840. doi: 10.1016/j.cellsig.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Goedert M. Tau protein pathology in neurodegenerative diseases. Trends Neurosci. 1998;21:428–433. doi: 10.1016/s0166-2236(98)01337-x. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Grundke-Iqbal I. Metabolic/signal transduction hypothesis of Alzheimer's disease and other tauopathies. Acta Neuropathol. 2005;109:25–31. doi: 10.1007/s00401-004-0951-y. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Braak H, Markesbery WR. Neuropathology and cognitive impairment in Alzheimer disease: a complex but coherent relationship. J Neuropathol Exp Neurol. 2009;68:1–14. doi: 10.1097/NEN.0b013e3181919a48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyckaerts C, Delatour B, Potier MC. Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 2009;118:5–36. doi: 10.1007/s00401-009-0532-1. [DOI] [PubMed] [Google Scholar]
- Lace G, Savva GM, Forster G, de Silva R, Brayne C, Matthews FE, et al. Hippocampal tau pathology is related to neuroanatomical connections: an ageing population-based study. Brain. 2009;132:1324–1334. doi: 10.1093/brain/awp059. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Yokota T, Mishra M, Akatsu H, Tani Y, Miyauchi T, Yamamoto T, et al. Brain site-specific gene expression analysis in Alzheimer's disease patients. Eur J Clin Invest. 2006;36:820–830. doi: 10.1111/j.1365-2362.2006.01722.x. [DOI] [PubMed] [Google Scholar]
- Yokota T, Akatsu H, Miyauchi T, Heese K. Characterization of the novel protein P9TLDR (temporal lobe down-regulated) with a brain-site-specific gene expression modality in Alzheimer's disease brain. FEBS Lett. 2012;586:4357–4361. doi: 10.1016/j.febslet.2012.10.050. [DOI] [PubMed] [Google Scholar]
- Larner AJ. The cerebellum in Alzheimer's disease. Dement Geriatr Cogn Disord. 1997;8:203–209. doi: 10.1159/000106632. [DOI] [PubMed] [Google Scholar]
- Ciavardelli D, Silvestri E, Del Viscovo A, Bomba M, De Gregorio D, Moreno M, et al. Alterations of brain and cerebellar proteomes linked to Abeta and tau pathology in a female triple-transgenic murine model of Alzheimer's disease. Cell Death Dis. 2010;1:e90. doi: 10.1038/cddis.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda-Falla D, Matschke J, Bernreuther C, Hagel C, Puig B, Villegas A, et al. Deposition of hyperphosphorylated tau in cerebellum of PS1 E280A Alzheimer's disease. Brain Pathol. 2011;21:452–463. doi: 10.1111/j.1750-3639.2010.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K, Andersen BB, Pakkenberg B. Stereological quantification of the cerebellum in patients with Alzheimer's disease. Neurobiol Aging. 2012;33:e111–e120. doi: 10.1016/j.neurobiolaging.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Zheng W, Wang T, Yu D, Feng WY, Nie YX, Stoltenberg M, et al. Elevation of zinc transporter ZnT3 protein in the cerebellar cortex of the AbetaPP/PS1 transgenic mouse. J Alzheimers Dis. 2010;20:323–331. doi: 10.3233/JAD-2010-1363. [DOI] [PubMed] [Google Scholar]
- Kusbeci OY, Bas O, Gocmen-Mas N, Karabekir HS, Yucel A, Ertekin T, et al. Evaluation of cerebellar asymmetry in Alzheimer's disease: a stereological study. Dement Geriatr Cogn Disord. 2009;28:1–5. doi: 10.1159/000228544. [DOI] [PubMed] [Google Scholar]
- Du J, Sun B, Chen K, Zhang L, Liu S, Gu Q, et al. Metabolites of cerebellar neurons and hippocampal neurons play opposite roles in pathogenesis of Alzheimer's disease. PLoS One. 2009;4:e5530. doi: 10.1371/journal.pone.0005530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann PA, Schlafer C, Seidl U, Santos VD, Essig M, Schroder J. The cerebellum in mild cognitive impairment and Alzheimer's disease - a structural MRI study. J Psychiatr Res. 2008;42:1198–1202. doi: 10.1016/j.jpsychires.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Lomoio S, Lopez-Gonzalez I, Aso E, Carmona M, Torrejon-Escribano B, Scherini E, et al. Cerebellar amyloid-beta plaques: disturbed cortical circuitry in AbetaPP/PS1 transgenic mice as a model of familial Alzheimer's disease. J Alzheimers Dis. 2012;31:285–300. doi: 10.3233/JAD-2012-112198. [DOI] [PubMed] [Google Scholar]
- Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- Akatsu H, Takahashi M, Matsukawa N, Ishikawa Y, Kondo N, Sato T, et al. Subtype analysis of neuropathologically diagnosed patients in a Japanese geriatric hospital. J Neurol Sci. 2002;196:63–69. doi: 10.1016/s0022-510x(02)00028-x. [DOI] [PubMed] [Google Scholar]
- Akatsu H, Yamagata H, Chen Y, Miki T, Kamino K, Takeda M, et al. TAFI polymorphisms at amino acids 147 and 325 are not risk factors for cerebral infarction. Br J Haematol. 2004;127:440–447. doi: 10.1111/j.1365-2141.2004.05219.x. [DOI] [PubMed] [Google Scholar]
- Mishra M, Akatsu H, Heese K. The novel protein MANI modulates neurogenesis and neurite-cone growth. J Cell Mol Med. 2011;15:1713–1725. doi: 10.1111/j.1582-4934.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Manavalan A, Sze SK, Heese K. Neuronal p60TRP expression modulates cardiac capacity. J Proteomics. 2012;75:1600–1617. doi: 10.1016/j.jprot.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Sundaramurthi H, Manavalan A, Ramachandran U, Hu JM, Sze SK, Heese K. Phenotyping of tianma-stimulated differentiated rat neuronal b104 cells by quantitative proteomics. Neurosignals. 2012;20:48–60. doi: 10.1159/000331492. [DOI] [PubMed] [Google Scholar]
- Shen Y, Inoue N, Heese K. Neurotrophin-4 (ntf4) mediates neurogenesis in mouse embryonic neural stem cells through the inhibition of the signal transducer and activator of transcription-3 (stat3) and the modulation of the activity of protein kinase B. Cell Mol Neurobiol. 2010;30:909–916. doi: 10.1007/s10571-010-9520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam O, Loo TX, Heese K. Brain-derived neurotrophic factor (BDNF) has proliferative effects on neural stem cells through the truncated TRK-B receptor, MAP kinase, AKT, and STAT-3 signaling pathways. Curr Neurovasc Res. 2009;6:42–53. doi: 10.2174/156720209787466028. [DOI] [PubMed] [Google Scholar]
- Manavalan A, Feng L, Sze SK, Hu JM, Heese K. New insights into the brain protein metabolism of Gastrodia elata-treated rats by quantitative proteomics. J Proteomics. 2012;75:2468–2479. doi: 10.1016/j.jprot.2012.02.029. [DOI] [PubMed] [Google Scholar]
- Hiller K, Schobert M, Hundertmark C, Jahn D, Munch R. JVirGel: Calculation of virtual two-dimensional protein gels. Nucleic Acids Res. 2003;31:3862–3865. doi: 10.1093/nar/gkg536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant N, Wattez A, Galvan-valencia M, Ghestem A, David JP, Lemoine J, et al. Association of ATP synthase alpha-chain with neurofibrillary degeneration in Alzheimer's disease. Neuroscience. 2003;117:293–303. doi: 10.1016/s0306-4522(02)00747-9. [DOI] [PubMed] [Google Scholar]
- Sultana R, Boyd-Kimball D, Cai J, Pierce WM, Klein JB, Merchant M, et al. Proteomics analysis of the Alzheimer's disease hippocampal proteome. J Alzheimers Dis. 2007;11:153–164. doi: 10.3233/jad-2007-11203. [DOI] [PubMed] [Google Scholar]
- Muller T, Jung K, Ullrich A, Schrotter A, Meyer HE, Stephan C, et al. Disease state, age, sex, and post-mortem time-dependent expression of proteins in AD vs. control frontal cortex brain samples. Curr Alzheimer Res. 2008;5:562–571. doi: 10.2174/156720508786898488. [DOI] [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci USA. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Klug A, Crowther RA. Tau protein, the paired helical filament and Alzheimer's disease. J Alzheimers Dis. 2006;9:195–207. doi: 10.3233/jad-2006-9s323. [DOI] [PubMed] [Google Scholar]
- Billingsley ML, Kincaid RL. Regulated phosphorylation and dephosphorylation of tau protein: effects on microtubule interaction, intracellular trafficking and neurodegeneration. Biochem J. 1997;323 (Pt 3:577–591. doi: 10.1042/bj3230577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzori C, Ghetti B, Piva R, Srinivasan AN, Zolo P, Delisle MB, et al. Activation of the JNK/p38 pathway occurs in diseases characterized by tau protein pathology and is related to tau phosphorylation but not to apoptosis. J Neuropathol Exp Neurol. 2001;60:1190–1197. doi: 10.1093/jnen/60.12.1190. [DOI] [PubMed] [Google Scholar]
- Himmelstein DS, Ward SM, Lancia JK, Patterson KR, Binder LI. Tau as a therapeutic target in neurodegenerative disease. Pharmacol Ther. 2012;136:8–22. doi: 10.1016/j.pharmthera.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11:301–307. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Del Tredici K. Alzheimer's pathogenesis: is there neuron-to-neuron propagation. Acta Neuropathol. 2011;121:589–595. doi: 10.1007/s00401-011-0825-z. [DOI] [PubMed] [Google Scholar]
- Goedert M, Clavaguera F, Tolnay M. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci. 2010;33:317–325. doi: 10.1016/j.tins.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Lee VM. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J Biol Chem. 2011;286:15317–15331. doi: 10.1074/jbc.M110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Jomary C. Clusterin. Int J Biochem Cell Biol. 2002;34:427–431. doi: 10.1016/s1357-2725(01)00155-8. [DOI] [PubMed] [Google Scholar]
- Ma JF, Liu LH, Zhang Y, Wang Y, Deng YL, Huang Y, et al. Association study of clusterin polymorphism rs11136000 with late onset Alzheimer's disease in Chinese Han population. Am J Alzheimers Dis Other Demen. 2011;26:627–630. doi: 10.1177/1533317511432735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Wei X, Chen S, Kurz A, Muller U, Gasser T, et al. Association of clusterin gene polymorphisms with late-onset Alzheimer's disease. Dement Geriatr Cogn Disord. 2011;32:198–201. doi: 10.1159/000331276. [DOI] [PubMed] [Google Scholar]
- Yu JT, Li L, Zhu QX, Zhang Q, Zhang W, Wu ZC, et al. Implication of CLU gene polymorphisms in Chinese patients with Alzheimer's disease. Clin Chim Acta. 2010;411:1516–1519. doi: 10.1016/j.cca.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Pankratz ND, Choi Y, Rothstein JH, Faber KM, Cheng R, et al. Genome-wide association of familial late-onset Alzheimer's disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet. 2011;7:e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, et al. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol. 2011;68:320–328. doi: 10.1001/archneurol.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golenkina SA, Gol'tsov A, Kuznetsova IL, Grigorenko AP, Andreeva TV, Reshetov DA, et al. [Analysis of clusterin gene (CLU/APOJ) polymorphism in Alzheimer's disease patients and in normal cohorts from Russian populations] Mol Biol (Mosk) 2010;44:620–626. [PubMed] [Google Scholar]
- Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara E, Soto C, Governale S, Frangione B, Ghiso J. Apolipoprotein J and Alzheimer's amyloid beta solubility. Biochem J. 1996;316 (Pt 2:671–679. doi: 10.1042/bj3160671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Wals P, Osterburg HH, Johnson SA, Pasinetti GM, Morgan TE, et al. Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1-42) and forms slowly sedimenting A beta complexes that cause oxidative stress. Exp Neurol. 1995;136:22–31. doi: 10.1006/exnr.1995.1080. [DOI] [PubMed] [Google Scholar]
- Matsubara E. [Apolipoprotein J and Alzheimer's amyloid beta solubility] Nihon Ronen Igakkai Zasshi. 1999;36:110–115. doi: 10.3143/geriatrics.36.110. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, O'Dell MA, Parsadanian M, Taylor JW, Harmony JA, Bales KR, et al. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer's disease. Proc Natl Acad Sci USA. 2002;99:10843–10848. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerbury JJ, Poon S, Meehan S, Thompson B, Kumita JR, Dobson CM, et al. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 2007;21:2312–2322. doi: 10.1096/fj.06-7986com. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol. 2005;7:909–915. doi: 10.1038/ncb1291. [DOI] [PubMed] [Google Scholar]
- Gleave M, Jansen B. Clusterin and IGFBPs as antisense targets in prostate cancer. Ann NY Acad Sci. 2003;1002:95–104. doi: 10.1196/annals.1281.020. [DOI] [PubMed] [Google Scholar]
- Dufresne D, Hamdan FF, Rosenfeld JA, Torchia B, Rosenblatt B, Michaud JL, et al. Homozygous deletion of Tenascin-R in a patient with intellectual disability. J Med Genet. 2012;49:451–454. doi: 10.1136/jmedgenet-2012-100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag-Sallaz M, Montag D. Severe cognitive and motor coordination deficits in tenascin-R-deficient mice. Genes Brain Behav. 2003;2:20–31. doi: 10.1034/j.1601-183x.2003.00003.x. [DOI] [PubMed] [Google Scholar]
- von Boxberg Y, Salim C, Soares S, Baloui H, Alterio J, Ravaille-Veron M, et al. Spinal cord injury-induced up-regulation of AHNAK, expressed in cells delineating cystic cavities, and associated with neoangiogenesis. Eur J Neurosci. 2006;24:1031–1041. doi: 10.1111/j.1460-9568.2006.04994.x. [DOI] [PubMed] [Google Scholar]
- Salim C, Boxberg YV, Alterio J, Fereol S, Nothias F. The giant protein AHNAK involved in morphogenesis and laminin substrate adhesion of myelinating Schwann cells. Glia. 2009;57:535–549. doi: 10.1002/glia.20782. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K. Alzheimer's disease: pathogenesis and prevention. Alzheimers Dement. 2012;8:227–233. doi: 10.1016/j.jalz.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K. Where, when, and in what form does sporadic Alzheimer's disease begin. Curr Opin Neurol. 2012;25:708–714. doi: 10.1097/WCO.0b013e32835a3432. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici-Braak K. Evolutional aspects of Alzheimer's disease pathogenesis. J Alzheimers Dis. 2012;33 (Suppl 1:S155–S161. doi: 10.3233/JAD-2012-129029. [DOI] [PubMed] [Google Scholar]
- Carro E. Gelsolin as therapeutic target in Alzheimer's disease. Expert Opin Ther Targets. 2010;14:585–592. doi: 10.1517/14728222.2010.488222. [DOI] [PubMed] [Google Scholar]
- Chauhan V, Ji L, Chauhan A. Anti-amyloidogenic, anti-oxidant and anti-apoptotic role of gelsolin in Alzheimer's disease. Biogerontology. 2008;9:381–389. doi: 10.1007/s10522-008-9169-z. [DOI] [PubMed] [Google Scholar]
- Ji L, Chauhan A, Wegiel J, Essa MM, Chauhan V. Gelsolin is proteolytically cleaved in the brains of individuals with Alzheimer's disease. J Alzheimers Dis. 2009;18:105–111. doi: 10.3233/JAD-2009-1127. [DOI] [PubMed] [Google Scholar]
- Guntert A, Campbell J, Saleem M, O'Brien DP, Thompson AJ, Byers HL, et al. Plasma gelsolin is decreased and correlates with rate of decline in Alzheimer's disease. J Alzheimers Dis. 2010;21:585–596. doi: 10.3233/JAD-2010-100279. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Basha MR, Ge YW, Zawia NH. How and when environmental agents and dietary factors affect the course of Alzheimer's disease: the “LEARn” model (latent early-life associated regulation) may explain the triggering of AD. Curr Alzheimer Res. 2007;4:219–228. doi: 10.2174/156720507780362164. [DOI] [PubMed] [Google Scholar]
- Gonzales ML, LaSalle JM. The role of MeCP2 in brain development and neurodevelopmental disorders. Curr Psychiatry Rep. 2010;12:127–134. doi: 10.1007/s11920-010-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastidar SG, Bardai FH, Ma C, Price V, Rawat V, Verma P, et al. Isoform-specific toxicity of Mecp2 in postmitotic neurons: suppression of neurotoxicity by FoxG1. J Neurosci. 2012;32:2846–2855. doi: 10.1523/JNEUROSCI.5841-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arosio B, Bulbarelli A, Bastias Candia S, Lonati E, Mastronardi L, Romualdi P, et al. Pin1 contribution to Alzheimer's disease: transcriptional and epigenetic mechanisms in patients with late-onset Alzheimer's disease. Neurodegener Dis. 2012;10:207–211. doi: 10.1159/000333799. [DOI] [PubMed] [Google Scholar]
- Daniilidou M, Koutroumani M, Tsolaki M. Epigenetic mechanisms in Alzheimer's disease. Curr Med Chem. 2011;18:1751–1756. doi: 10.2174/092986711795496872. [DOI] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer's disease. Neurobiol Aging. 2011;32:1161–1180. doi: 10.1016/j.neurobiolaging.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Sancho M, Diani E, Beato M, Jordan A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 2008;4:e1000227. doi: 10.1371/journal.pgen.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsimpardi L, Gaitanou M, Malnou CE, Lledo PM, Charneau P, Matsas R, et al. BM88/Cend1 expression levels are critical for proliferation and differentiation of subventricular zone-derived neural precursor cells. Stem Cells. 2008;26:1796–1807. doi: 10.1634/stemcells.2007-0921. [DOI] [PubMed] [Google Scholar]
- Herrup K. The contributions of unscheduled neuronal cell cycle events to the death of neurons in Alzheimer's disease. Front Biosci (Elite Ed) 2012;4:2101–2109. doi: 10.2741/527. [DOI] [PubMed] [Google Scholar]
- Keeney JT, Swomley AM, Harris JL, Fiorini A, Mitov MI, Perluigi M, et al. Cell cycle proteins in brain in mild cognitive impairment: insights into progression to Alzheimer disease. Neurotox Res. 2011;22:220–230. doi: 10.1007/s12640-011-9287-2. [DOI] [PubMed] [Google Scholar]
- Sultana R, Butterfield DA. Regional expression of key cell cycle proteins in brain from subjects with amnestic mild cognitive impairment. Neurochem Res. 2007;32:655–662. doi: 10.1007/s11064-006-9123-x. [DOI] [PubMed] [Google Scholar]
- Moh C, Kubiak JZ, Bajic VP, Zhu X, Smith MA, Lee HG. Cell cycle deregulation in the neurons of Alzheimer's disease. Results Probl Cell Differ. 2011;53:565–576. doi: 10.1007/978-3-642-19065-0_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonda DJ, Lee HP, Kudo W, Zhu X, Smith MA, Lee HG. Pathological implications of cell cycle re-entry in Alzheimer disease. Expert Rev Mol Med. 2010;12:e19. doi: 10.1017/S146239941000150X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JP, Oliveira CR, Agostinho P. Cell cycle re-entry in Alzheimer's disease: a major neuropathological characteristic. Curr Alzheimer Res. 2009;6:205–212. doi: 10.2174/156720509788486590. [DOI] [PubMed] [Google Scholar]
- Lee HG, Casadesus G, Zhu X, Castellani RJ, McShea A, Perry G, et al. Cell cycle re-entry mediated neurodegeneration and its treatment role in the pathogenesis of Alzheimer's disease. Neurochem Int. 2009;54:84–88. doi: 10.1016/j.neuint.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Esiri MM, Smith AD. The cell division cycle and the pathophysiology of Alzheimer's disease. Neuroscience. 1998;87:731–739. doi: 10.1016/s0306-4522(98)00293-0. [DOI] [PubMed] [Google Scholar]
- Ambegaokar SS, Jackson GR. The downward spiral of tau and autolysosomes: a new hypothesis in neurodegeneration. Autophagy. 2012;8:1144–1145. doi: 10.4161/auto.20515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick L, Gentle L, Strachan J, Layfield R. Review: unchained maladie - a reassessment of the role of Ubb(+1) -capped polyubiquitin chains in Alzheimer's disease. Neuropathol Appl Neurobiol. 2012;38:118–131. doi: 10.1111/j.1365-2990.2011.01236.x. [DOI] [PubMed] [Google Scholar]
- Riederer BM, Leuba G, Vernay A, Riederer IM. The role of the ubiquitin proteasome system in Alzheimer's disease. Exp Biol Med (Maywood) 2011;236:268–276. doi: 10.1258/ebm.2010.010327. [DOI] [PubMed] [Google Scholar]
- Upadhya SC, Hegde AN. Role of the ubiquitin proteasome system in Alzheimer's disease. BMC Biochem. 2007;8 (Suppl 1:S12. doi: 10.1186/1471-2091-8-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper W, Nijholt DA, Hoozemans JJ. The unfolded protein response and proteostasis in Alzheimer disease: preferential activation of autophagy by endoplasmic reticulum stress. Autophagy. 2011;7:910–911. doi: 10.4161/auto.7.8.15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. Bioessays. 2008;30:1172–1184. doi: 10.1002/bies.20852. [DOI] [PubMed] [Google Scholar]
- Huang Q, Figueiredo-Pereira ME. Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications. Apoptosis. 2010;15:1292–1311. doi: 10.1007/s10495-010-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijholt DA, De Kimpe L, Elfrink HL, Hoozemans JJ, Scheper W. Removing protein aggregates: the role of proteolysis in neurodegeneration. Curr Med Chem. 2011;18:2459–2476. doi: 10.2174/092986711795843236. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Tanaka K. Does impairment of the ubiquitin-proteasome system or the autophagy-lysosome pathway predispose individuals to neurodegenerative disorders such as Parkinson's disease. J Alzheimers Dis. 2010;19:1–9. doi: 10.3233/JAD-2010-1231. [DOI] [PubMed] [Google Scholar]
- Lowe J, Blanchard A, Morrell K, Lennox G, Reynolds L, Billett M, et al. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson's disease, Pick's disease, and Alzheimer's disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J Pathol. 1988;155:9–15. doi: 10.1002/path.1711550105. [DOI] [PubMed] [Google Scholar]
- Cecarini V, Bonfili L, Cuccioloni M, Mozzicafreddo M, Rossi G, Buizza L, et al. Crosstalk between the ubiquitin-proteasome system and autophagy in a human cellular model of Alzheimer's disease. Biochim Biophys Acta. 2012;1822:1741–1751. doi: 10.1016/j.bbadis.2012.07.015. [DOI] [PubMed] [Google Scholar]
- de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM. Protein quality control in Alzheimer's disease by the ubiquitin proteasome system. Prog Neurobiol. 2004;74:249–270. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Oddo S. The ubiquitin-proteasome system in Alzheimer's disease. J Cell Mol Med. 2008;12:363–373. doi: 10.1111/j.1582-4934.2008.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Jung YK. Alzheimer's disease meets the ubiquitin-proteasome system. Trends Mol Med. 2004;10:565–570. doi: 10.1016/j.molmed.2004.09.005. [DOI] [PubMed] [Google Scholar]
- van Leeuwen FW, Hol EM, Fischer DF. Frameshift proteins in Alzheimer's disease and in other conformational disorders: time for the ubiquitin-proteasome system. J Alzheimers Dis. 2006;9:319–325. doi: 10.3233/jad-2006-9s336. [DOI] [PubMed] [Google Scholar]
- Layfield R, Lowe J, Bedford L. The ubiquitin-proteasome system and neurodegenerative disorders. Essays Biochem. 2005;41:157–171. doi: 10.1042/EB0410157. [DOI] [PubMed] [Google Scholar]
- Forman MS, Farmer J, Johnson JK, Clark CM, Arnold SE, Coslett HB, et al. Frontotemporal dementia: clinicopathological correlations. Ann Neurol. 2006;59:952–962. doi: 10.1002/ana.20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Kondo J, Ihara Y. Ubiquitin is a component of paired helical filaments in Alzheimer's disease. Science. 1987;235:1641–1644. doi: 10.1126/science.3029875. [DOI] [PubMed] [Google Scholar]
- Perry G, Friedman R, Shaw G, Chau V. Ubiquitin is detected in neurofibrillary tangles and senile plaque neurites of Alzheimer disease brains. Proc Natl Acad Sci USA. 1987;84:3033–3036. doi: 10.1073/pnas.84.9.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G, Mulvihill P, Fried VA, Smith HT, Grundke-Iqbal I, Iqbal K. Immunochemical properties of ubiquitin conjugates in the paired helical filaments of Alzheimer disease. J Neurochem. 1989;52:1523–1528. doi: 10.1111/j.1471-4159.1989.tb09203.x. [DOI] [PubMed] [Google Scholar]
- van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- Martin B, Brenneman R, Becker KG, Gucek M, Cole RN, Maudsley S. iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer's mice: understanding the interface between physiology and disease. PLoS One. 2008;3:e2750. doi: 10.1371/journal.pone.0002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana R, Robinson RA, Di Domenico F, Abdul HM, Clair DK, St, Markesbery WR, et al. Proteomic identification of specifically carbonylated brain proteins in APP(NLh)/APP(NLh) x PS-1(P264L)/PS-1(P264L) human double mutant knock-in mice model of Alzheimer disease as a function of age. J Proteomics. 2011;74:2430–2440. doi: 10.1016/j.jprot.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn BC, Wang J, Markesbery WR, Lovell MA. Quantitative changes in the mitochondrial proteome from subjects with mild cognitive impairment, early stage, and late stage Alzheimer's disease. J Alzheimers Dis. 2010;19:325–339. doi: 10.3233/JAD-2010-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorobogatko YV, Deuso J, Adolf-Bryfogle J, Nowak MG, Gong Y, Lippa CF, et al. Human Alzheimer's disease synaptic O-GlcNAc site mapping and iTRAQ expression proteomics with ion trap mass spectrometry. Amino Acids. 2011;40:765–779. doi: 10.1007/s00726-010-0645-9. [DOI] [PubMed] [Google Scholar]
- Gozal YM, Duong DM, Gearing M, Cheng D, Hanfelt JJ, Funderburk C, et al. Proteomics analysis reveals novel components in the detergent-insoluble subproteome in Alzheimer's disease. J Proteome Res. 2009;8:5069–5079. doi: 10.1021/pr900474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.