Abstract
Simultaneous monitoring the expression, distribution and dynamics of biological molecules in living cells is one of the most challenging tasks in the analytical sciences. The key to effective and successful intracellular imaging is the development of delivery platforms with high efficiency and ultrasensitive molecular probes for specific targets of interest. To achieve these goals, many nanomaterials are widely used as carriers to introduce nucleic acid probes into living cells for real-time imaging of biomolecules. However, limitations on their use include issues of cytotoxicity and delivery efficiency. Herein, we propose a switchable aptamer micelle flare (SAMF), formed by self-assembly of an aptamer switch probe-diacyllipid chimera, for monitoring of ATP molecules inside living cells. Similarity of hydrophobic composition between diacyllipids in the micelle flares and phospholipid bilayers in the dynamic membranes of living cells allows SAMFs to be uptaken by living cells more efficiently than aptamer switch probes without external auxiliary. Switchable aptamers were found to bind target ATP molecules with high selectivity and specificity, resulting in restoration of the fluorescence signal from “OFF” to “ON” state, thus indicating the presence of the analyte. These switchable aptamer micelle flares, which exhibit cell permeability and nanoscale controllability, show exceptional promise for molecular imaging in bioanalysis, disease diagnosis and drug delivery.
Keywords: switchable aptamer, micelle flare nanostructures, ATP imaging, living cells
Investigation of the distribution and dynamics of biological molecules inside cells is critical for understanding physiological processes, diagnosing disease stages and identifying therapeutic targets.1 To fulfill these purposes, many nucleic acid molecular probes for intracellular imaging use have been designed for targets ranging from proteins to RNAs and even small molecules.2-14 Tyagi et al. have reported the use of fluorescent-labeled hybridization probes, e.g., molecular beacons (MBs), to monitor real-time expression levels and distributions of mRNA in vivo.15 To overcome the instability of hybridization probes inside cells, Tan et al. designed and applied a series of modified nucleic acid probes (e.g., locked nucleic acids (LNA),16 hybrid molecular probes17 and 2′-O-methylribonucleotides18) for long-term, real-time and multiple gene imaging. However, the use of molecular beacon-based nucleic acid probes is still challenged by inefficient probe introduction and uneven distribution of probes inside cells.19
Recently, nanomaterials have gained increasing attention for the delivery of nucleic probes into cells by their highly efficient internalization, excellent biostability, and resistance to degradation by nucleases in living cells.20-24 For example, Mirkin et al. developed nucleic acidconjugated gold nanoparticles called “nanoflares” to quantify molecular analytes and regulate mRNA expression in living cells.21, 25-28 Tang et al. reported intracellular simultaneous and multicolor imaging of tumor-related mRNA based on gold nanoparticles modified with molecular beacons.29-31 In addition, Lin et al. employed complexes of graphene oxide nanosheets and fluorescent-labeled aptamers13, 32, 33 for cellular delivery and molecular imaging in living cells.34 However, the cytotoxicity of nanomaterials still cannot be neglected, especially at high concentrations for intracellular use. Even gold nanoparticles, which are considered the safest nanomaterial, are known to cause nephrotoxicity and eryptosis for sizes between 8 and 37 nm.35, 36
By their controllability, excellent biocompatibility and efficient cellular internalization, DNA nanoassemblies have been widely employed to deliver such cargos as antisense DNA and hydrophobic drugs for cancer therapy.37-40 Previously, our group designed and constructed nucleic acid micelles self-assembled through DNA-diacyllipid conjugates, consisting of hydrophobic lipid tails and single-stranded hydrophilic DNA heads (Supporting Information, Figure S1).41-43 Building on this foundation, we present a molecularly engineered switchable aptamer micelle flare nanostructure for intracellular imaging of biological molecules. Each hydrophobic diacyllipid tail is incorporated into a hydrophilic switchable aptamer head through solid phase phosphoramidite chemistry on an automatic DNA synthesizer, followed by self-assembly to form a uniform spherical nanostructure, which we term switchable aptamer micelle flare (SAMF). Because of the similarity between the intermolecular force of the diacyllipid in DNA micelle flares and the dynamic phospholipid bilayers in cell membranes, SAMFs can easily interact with cell membranes and enter cells.42 SAMFs have many advantages over previous approaches, including facile probe modification, capable self-delivery, high signal-to-background ratio, excellent target selectivity, and superior biocompatibility.43 These properties of SAMFs provide an efficient delivery and imaging platform for living cells without using nanomaterials that may cause potential toxic effects.
Results and Discussion
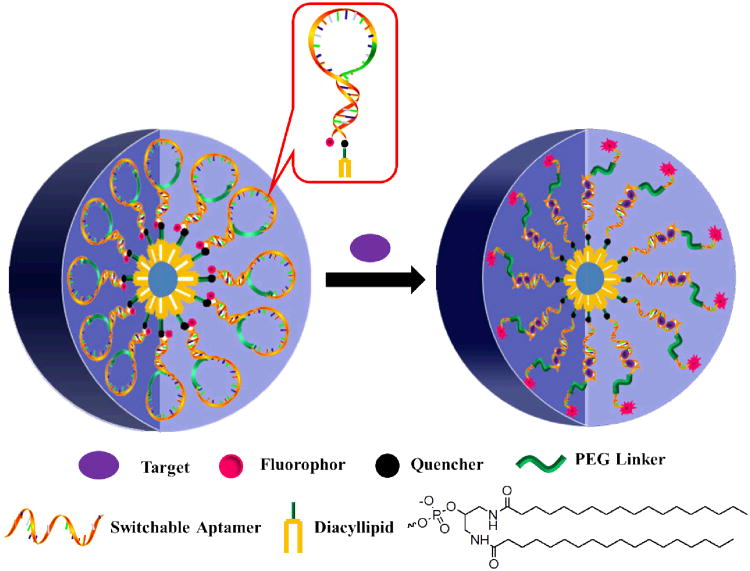
Switchable aptamer micelle flares are self-assembled from conjugates containing an aptamer switch probe,44 a PEG linker to a short DNA sequence complementary to part of the aptamer switch probe, and a diacyllipid tai. In addition, a fluorophore, TAMRA (tetramethylrhodamine), is covalently attached to the 3′-end of the probe, and a quencher, DABYCL (4-(4-dimethylaminophenylazo) benzoic acid), is included between the 5′-end and the diacyllipid tail (Supporting Information, Table S2). In the proof-of-concept experiment, adenosine triphosphate (ATP) was chosen as a target molecule inside cells because of its significant role in cell signaling and cellular reactions. As shown in Figure 1, in the absence of ATP molecules, the aptamer switch probe maintains a loop-stem structure, and fluorescence is quenched as a result of the close proximity between fluorophore and quencher (OFF state). However, the conformation of the probe is altered when the diacyllipid tail interacts with the cell membrane, causing the fluorophore to move far enough away from the quencher to result in restoration of fluorescence signal (ON state).
Figure 1.
Working principle of switchable aptamer micelle flares. Aptamer switch probe and diacyllipid conjugates are self-assembled to form the nanostructure of micelle flares. In the absence of target, the aptamer switch probe maintains a loop-stem structure, and fluorescence is quenched because of the close proximity between fluorophore and quencher. Upon target binding, the conformation of switchable aptamer is altered, resulting in the restoration of fluorescence signal.
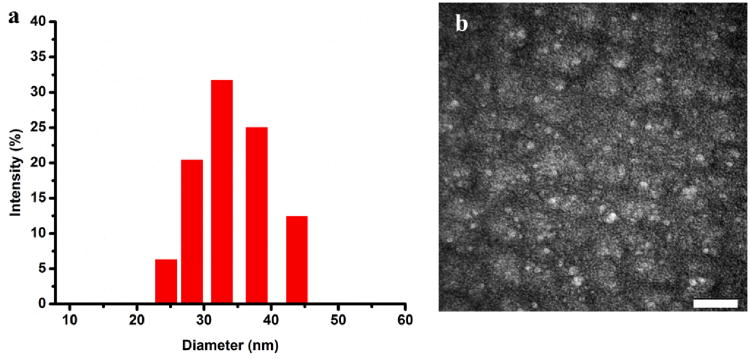
To characterize this design, we first examined the structural formation of SMAFs by agarose gel electrophoresis. As shown in Figure S3 (Supporting Information), in TBE buffer, aptamer switch probes without lipid conjugation move faster than the assembled micelle flares that have larger structures and higher molecular weights, indicating that the micelle flare aggregates were stably formed. Additionally, to provide direct evidence of amphiphilic switchable aptamer-diacyllipid conjugate self-assembly, we employed dynamic light scattering (DLS) to measure the physical size for water-soluble SAMFs (Figure 2a). The hydrodynamic diameter of SAMFs in phosphate buffered saline (PBS) solution was measured as 34.67 nm, which was also demonstrated in transmission electron microscope (TEM) image (27.57 nm, Figure 2b). This highly uniform size originates from the strong hydrophobic interaction between the diacyllipid tails and the precisely designed secondary molecular structure of switchable aptamers.
Figure 2.
Characterization of switchable aptamer micelle flares. (a) Dynamic light scattering (DLS) of SAMFs. (b) TEM image of SAMFs after negative staining by 2% aqueous Uranyl Acetate. Scale bar: 200 nm.
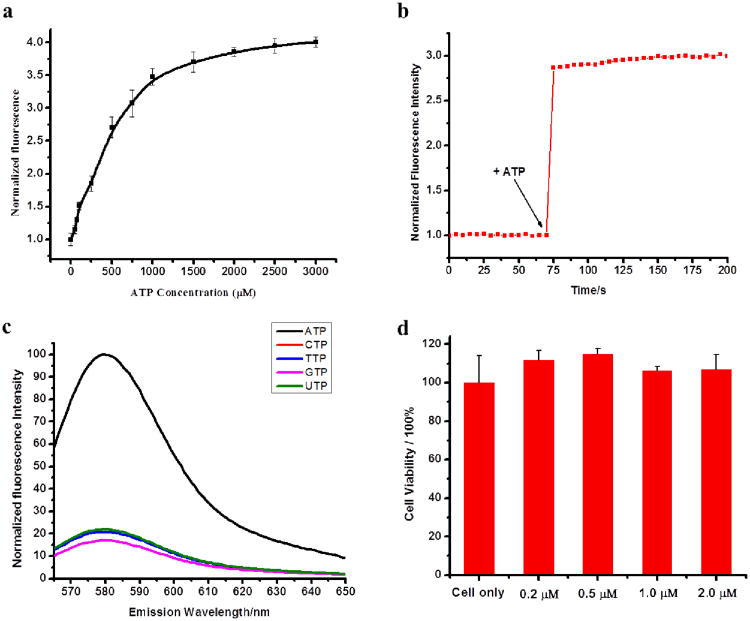
To investigate the response to target molecules, SAMFs (1 μM) in PBS were titrated with a series of ATP concentrations. As shown in Figure 3a, SAMFs showed a significant fluorescence increase upon addition of ATP molecules, with a maximal of 3.9-fold enhancement after adding 2 mM ATP. The cellular ATP concentration is typically in the μM-mM range; thus, our SAMFs were able to sensitively respond to intracellular ATP expression. Furthermore, to test whether the binding affinity of the switchable aptamers was affected after self-assembly, a kinetics study showed a rapid restoration of fluorescence signal after addition of excess amounts of ATP (Figure 3b). This rapid response makes SAMFs feasible for intracellular imaging, essentially because the short detection time avoids unnecessary interferences from cells.
Figure 3.
Switchable aptamer micelle flares respond to target ATP molecules in vitro. (a) Fluorescence titration of SAMF (1 μM) against increased concentrations of ATP (0.1-3.0 mM). (b) Time course of fluorescence associated with the binding of target molecules. The concentration of ATP was 1 mM. (c) Switchable aptamer micelle flares distinguish ATP from its analogs: TTP, GTP, CTP and UTP. The concentration of ATP and its analogs were 2 mM. (d) Cytotoxicity assay of HeLa cells treated with different concentrations of SAMFs (0, 0.2, 0.5, 1, 2 μM).
We next compared the specificity of SAMF to a control switchable DNA micelle flare (CSMF) with two mismatches in the binding domain of the aptamer sequence. SAMF showed more than 4-fold fluorescence increase upon addition of ATP, while the control showed little enhancement, even in a high concentration (2 mM) of ATP, because of the low binding affinity of CSMF to ATP molecules (Supporting Information, Figure S4). Additionally, the selectivity of SAMF was investigated with analogs of ATP, including cytosine triphosphate (CTP), thymidine triphosphate (TTP), guanidine triphosphate (GTP) and uridine triphosphate (UTP). As shown in Figure 3c, these ATP analogs did not induce a significant fluorescence enhancement relative to target ATP molecules, strongly indicating that SAMFs can discriminate between ATP and its analogs in living cells. More significantly, as shown in Figure 3d, switchable aptamer micelle flares showed excellent biocompatibility with living cells based on the cell proliferation assay (MTS). This is most probably attributed to the structure similarity between diacyllipid and phospholipid–the basic component of cell membrane. This result indicates that SAMFs provide a new safe and noncytotoxic nanomaterial for intracellular or in vivo imaging.
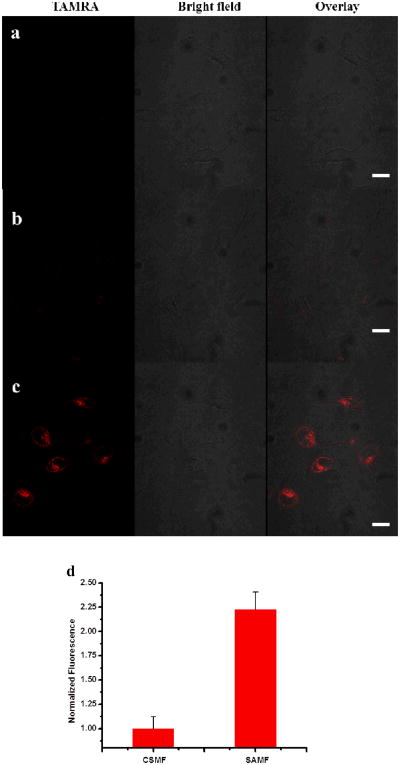
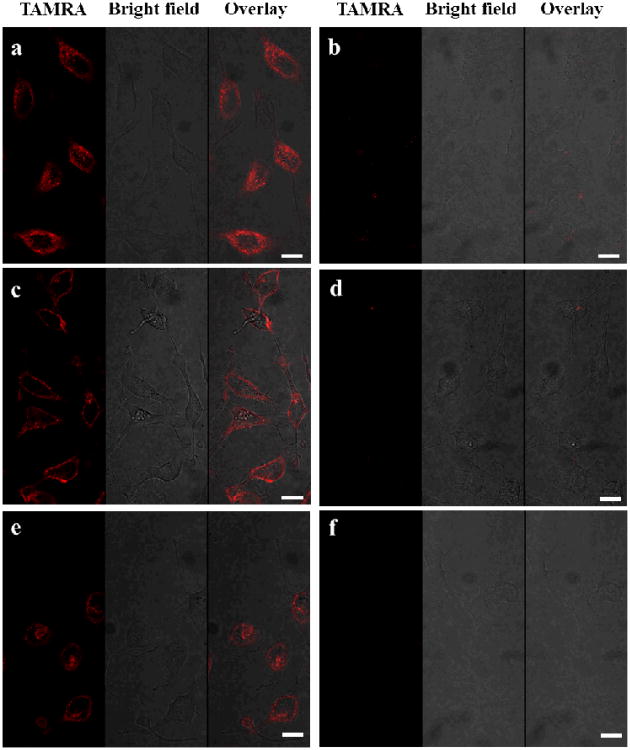
Having demonstrated the specificity and selectivity of SAMFs in vitro, we next verified their cell permeability for imaging of intracellular ATP expression and distribution in HeLa cells (human cervical cancer cwlls). Confocal laser scanning microscopy was used to image the fluorescence signals from HeLa cells incubated with the same amount of SAMF, CSMF and aptamer switch probe without diacyllipid. As shown is Figure 4a, aptamer switch probe without conjugation to the diacyllipid did not result in internalization (no fluorescence signal). However, HeLa cells treated with SAMFs (Figure 4c) showed an intense fluorescence signal compared to cells cultured with CSMFs (Figure 4b), which showed only slight fluorescence intensity, probably resulting from degradation by nucleases or nonspecific opening of the hairpins by protein binding. It is noteworthy that the fluorescence signals were mostly localized in the cytoplasm rather than nucleus. Thus, micelle flares are also able to overcome problems associated with microinjection, a method which, because of nuclear sequestration, delivers most nucleic acid probes into the nucleus. To further quantify the intracellular signal of SAMFs in a high-throughput manner, HeLa cells cultured with equal concentrations of SAMFs or CSMFs were treated with trypsin, followed by characterization by analytical flow cytometry. Samples of 20,000 cells were counted and compared based on cell-associated fluorescence intensity. As shown in Figure 4d, SAMFs showed a 2.3-fold fluorescence enhancement relative to cells treated with CSMFs. The flow cytometry data are consistent with confocal imaging results, and they also demonstrate the cellular internalization and signaling capabilities of SAMF.
Figure 4.
Switchable aptamer micelle flares respond to target molecules in HeLa cells. Confocal microscopy fluorescence imaging of intracellular ATP molecules with 1 μM (a) aptamer switch probe without diacyllipid conjugation, (b) CSMFs and (c) SAMFs. Left panels are TAMRA fluorescence pseudo-colored red, middle panels are the bright-field image and right panels are the overlay of TAMRA fluorescence and the bright-field image. Scale bar: 50 μm. (d) Cell-associated fluorescence (TAMRA) of cell populations treated with 1 μM CSMFs and SAMFs, as determined by flow cytometry.
To reveal the mechanism of SAMFs internalized into HeLa cells, time-dependent fluorescence measurements at 0.5, 1, 2 and 4 hour are carried out. SAMFs were initially found to be distributed on the cell membrane with a weak fluorescence signal at 0.5 h (Supporting Information, Figure S5a). Afterwards, the fluorescence signal became accumulated in cytoplasm from 1 h to 2 h (Supporting Information, Figure S5c, 5e). Finally, SAMFs were mostly distributed in the cytoplasm other than the nucleus at 4 h (Supporting Information, Figure S5g), meanwhile, CSMFs showed a weak increase of fluorescence signal from 0.5 h to 4 h probably resulting from nonspecific opening of the hairpins by protein binding (Supporting Information, Figure S5b, d, f, h). Next, to further investigate the cellular distribution of internalized SAMF, co-localization of SAMFs and lysosomes was examined by co-staining with LysoSensor (lysosome marker). As shown in Figure S6 (Supporting Information), a fraction of SMAFs co-localized with lysosomes, suggesting the minor accumulation of SAMFs in lysosomes by an endocytosis pathway. However, most amphiphilic SAMFs were distributed in the cellular plasma. In light of time-dependent fluorescence measurement and co-localization assay, here we propose the following possible mechanism for internalization of SAMFs into living cells. SAMFs are initially in close proximity to the cell membrane, and then they disintegrate and fuse with the cell membrane. Partial disintegrated SAMFs will permeate the cell through the process of endocytosis, while, most fused SAMF might flip and diffuse from cell membrane to cytoplasm during a process of membrane recycling because of thermodynamically unfavorable structure with hydrophobic tail pointing into aqueous solution.42 The detailed mechanisms will be further investigated with more experiments in the future.
Next, to study the ability of SAMF to respond to changes of ATP expression in living cells, we used two small molecules, etoposide and oligomycin,45, 46 to stimulate or suppress the expression level of intracellular ATP in living HeLa cells, respectively. HeLa cells were first treated with either 100 μM etoposide or 3 μg/ml oligomycin, respectively, followed by culturing with 1 μM SAMF. The confocal results indicate that cells treated with epotoside exhibited increased fluorescence signal compared to untreated HeLa cells, while cells treated with oligomycin showed decreased cell-associated fluorescence (Figure 5a, c, e). Meanwhile, CSMFs showed low fluorescence signals for these drug stimulated HeLa cells, suggesting the change of fluorescence signals induced by variation of ATP expression (Figure 5b, d, f). The imaging data were collected and analyzed by ImageJ software for the measurement of fluorescence signal intensity. The ATP expression in the untreated HeLa cells was normalized as 100 (Supporting Information, Figure S7). The ATP level in HeLa cells treated with 100 μM etoposide showed an increase from 100 to 135, while the expression of intracellular ATP decreased from 100 to 84 in the cells treated with 3 μg/ml oligomycin, in good agreement with previous measurements.27, 47 Furthermore, the average cellular ATP concentration was determined for the bulk HeLa cell samples by using a commercial luciferase ATP assay, which showed the ATP concentrations of 2.5 mM, 3.9 mM and 2.0 mM for untreated, etoposide and oligomycin treated HeLa cells, respectively (Supporting Information, Figure S8). If normalized with untreated HeLa cells, the relatively amount of etoposide treated HeLa cells was 156, and that of oligomycin treated HeLa cells was 80. Thus, the relative fluorescence intensities measured by SAMF were consistent with the cellular average ATP concentrations after treatment with drugs. Since some biological processes are dependent to cell-to-cell variation, the results demonstrate that SAMFs are able to detect the variation of ATP expression inside living cells. In the end, we further estimated cytoplasmic ATP concentration with SAMFs and obtained a value of 2.8 mM following lysis of live HeLa cells (Supporting Information, Figure S9). This value was consistent with the determination of average cellular ATP expression with standard luciferase assay and previous measurement with other aptamer based intracellular approaches, such as aptamers embedded in nanoparticles and nano-flares,23, 27 thus indicating the good sensitivity of SAMF for intracellular quantification of significant biological molecules.
Figure 5.
Confocal microscopy fluorescence imaging of HeLa cell populations treated (a) with 100 μM etoposide, (b) with 3 μg/ml oligomycin and (c) without etoposide or oligomycin, followed by incubation with 1 μM SAMF. Scale bar: 50 μm.
Conclusions
In summary, we assembled switchable aptamers-diacyllipids conjugates at the molecular level to form micelle flares for the measurement of ATP in vitro and further applied the self-assembled nanostructure for molecular imaging inside living cells. The switchable aptamer micelle flares maintain binding affinity to target molecules. They also show potential for the delivery of cargos into living cells without using toxic nanomaterials and for intracellular imaging the expression and distribution of biomolecules. More significantly, switchable aptamer micelle flares can measure the changes of ATP expression in real time when cells are stimulated to increase or decrease ATP levels. Future work will include application of various types of nucleic acid probe micelle flares for ratiometric quantitation of biomolecules in living cells.
Materials and Methods
DNA synthesis
All oligonucleotides were synthesized based on solid phase phosphoramidite chemistry at a 1 μmol scale using an ABI 3400 synthesizer. Lipid phosphoramidite (Figure S2) dissolved in methylene chloride was directly coupled onto the sequence by the DNA synthesizer with an extended coupling time (900s). After synthesis, the DNA sequences were cleaved and deprotected in a solution of methanol: tert-butylamine: water (1:1:2) at 65 °C for 4 h for TAMRA labeling. Deprotected sequences were purified by reverse-phase HPLC using a C4 column (BioBasic-4, 200 mm × 4.6 mm, Thermo Scientific) with 100 mM triethylamine-acetic acid buffer (TEAA, pH 7.5) and acetonitrile (0-30 min, 10-100%) as the eluent. The collected sequences were vacuum dried and stored at -20°C for future use.
Characterization of micelle flares
Each DNA sample was analyzed by electrophoresis for about 90 min at constant 75 V through a 4% agarose gel in 1× TBE (tris(hydroxymethyl)aminomethane (Tris, 89 mM), (EDTA, 2 mM), and boric acid (89 mM), pH 8.0) buffer. The DNA bands were visualized by UV illumination (312 nm) and photographed by a digital camera. The particle sizes (diameters) and their distribution were measured by a ZetaPALS DLS detector (Brookhaven Instruments, Holtsville, NY, USA) at 25°C. The scattering angle was fixed at 90°.
Cell culture
HeLa cervical cancer cells (American Type Culture Collection, Manassas, VA) were maintained in Dulbecco's modification of Eagle's medium (Fisher Scientific) with 10% fetal bovine serum (Invitrogen, Carlsbad, CA) and 0.5 mg/mL penicillin-streptomycin (Sigma, St. Louis, MO) at 37 °C in 5% CO2/air. Cells were plated in 35 mm glass bottomed culture dishes (MatTek Corp., Ashland, MA) and grown to 80% confluency for 48 h before incubation with probes. To stimulate expression of ATP in vivo, the cell medium was immediately mixed with 100 μM etoposide or 3 μg/ml oligomycin (Sigma) before imaging.
Cytotoxicity assay
The cytotoxicity of SAMFs was determined using a CellTiter 96 cell proliferation assay (Promega, Madison, WI, USA). After seeding in 96-well plates and culturing overnight, the cells were incubated with SAMFs (with a final concentration ranging from 0.2 to 2 μM, diluted by culture media) for 2 h, washed with PBS, and then cultured with fresh medium for future cell growth (48 h). After removing the cell medium, CellTiter reagent (20 μL) diluted in fresh medium (100 μL) was added to each well and incubated for 1-2 h. The absorbance (490 nm) was recorded by using a plate reader (Tecan Safire Microplate reader, AG, Switzerland). The cell viability was determined as described by the manufacturer.
Confocal fluorescence microscopy imaging
All cellular fluorescent images were collected with the confocal microscope setup using laser excitation. Switchable and control aptamer micelle flares with TAMRA (1 μM concentration in phosphate buffered saline (PBS) and 5 mM MgCl2) were excited at 543 nm, and the fluorescence was collected at 570 nm. HeLa cells were incubated with SAMF or CSMF for 2 hours, followed by washing twice with PBS to remove unbound probes. To avoid unnecessary dye photobleaching and any damage to the cells, the microscope shutter was opened only long enough to allow the laser to illuminate the bound cells while a fluorescence image was collected. Cells were treated with 3 μg/mL oligomycin or 100 μM etoposide for 0.5 h, followed by incubation with 1 μM switchable aptmaer micelle flares as described above for 2 h.
Flow cytometric assay
HeLa cells incubated with 1 μM SAMF or CSMF for 2 h, respectively, were then washed to remove unbound probes. After treatment with trypsin, cells (200 μL, 1× 106 cells per mL) were suspended in 1× PBS buffer and analyzed by a FACScan cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA).
Cell lysate preparation
HeLa cells were plated in a 35 mm cell culture dish (Corning Incorporated, Corning, NY, USA) and grown around 80% confluency before the experiments. Cells were washed twice with 1 mL PBS and then 0.5 mL of Tris-HCl buffer without proteinase inhibitor was added to the cell culture dish. Finally, cells were scraped off the cell culture dish and ruptured by mechanical shearing using a douncer homogenizer.
Supplementary Material
Acknowledgments
We are grateful to the Interdisciplinary Center for Biotechnology Research (ICBR) at the University of Florida for technical support associated with confocal fluorescence microscopy. We also acknowledge Dr. K. R. Williams for manuscript review. D. Han acknowledges the ACS Division of Analytical Chemistry Fellowship sponsored by the Society for Analytical Chemists of Pittsburgh. This work is supported by grants awarded by the National Institutes of Health (GM066137, GM079359 and CA133086) and by NSF.
Footnotes
Conflict of Interest: The authors declare no competing financial interest
Supporting Information Available: Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org
References and notes
- 1.Spiller DG, Wood CD, Rand DA, White MRH. Measurement of Single-Cell Dynamics. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 2.Tyagi S, Bratu DP, Kramer FR. Multicolor Molecular Beacons for Allele Discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 3.Fang X, Li JJ, Tan W. Using Molecular Beacons To Probe Molecular Interactions between Lactate Dehydrogenase and Single-Stranded DNA. Anal Chem. 2000;72:3280–3285. doi: 10.1021/ac991434j. [DOI] [PubMed] [Google Scholar]
- 4.Li JJ, Fang X, Schuster SM, Tan W. Molecular Beacons: A Novel Approach to Detect Protein – DNA Interactions. Angew Chem Int Ed. 2000;39:1049–1052. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Tan W, Fang X, Li J, Liu X. Molecular Beacons: A Novel DNA Probe for Nucleic Acid and Protein Studies. Chem Eur J. 2000;6:1107–1111. doi: 10.1002/(sici)1521-3765(20000403)6:7<1107::aid-chem1107>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Perlette J, Tan W. Real-Time Monitoring of Intracellular mRNA Hybridization Inside Single Living Cells. Anal Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 7.Li JJ, Fang X, Tan W. Molecular Aptamer Beacons for Real-Time Protein Recognition. Biochem Biophys Res Commun. 2002;292:31–40. doi: 10.1006/bbrc.2002.6581. [DOI] [PubMed] [Google Scholar]
- 8.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the Distribution and Transport of mRNAs in Living Cells. Proc Natl Acad Sci U S A. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake TJ, Medley CD, Sen A, Rogers RJ, Tan W. Stochasticity of Manganese Superoxide Dismutase mRNA Expression in Breast Carcinoma Cells by Molecular Beacon Imaging. ChemBioChem. 2005;6:2041–2047. doi: 10.1002/cbic.200500046. [DOI] [PubMed] [Google Scholar]
- 10.Medley CD, Drake TJ, Tomasini JM, Rogers RJ, Tan W. Simultaneous Monitoring of the Expression of Multiple Genes Inside of Single Breast Carcinoma Cells. Anal Chem. 2005;77:4713–4718. doi: 10.1021/ac050881y. [DOI] [PubMed] [Google Scholar]
- 11.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-Linked Molecular Beacons for Imaging mRNAs in the Cytoplasm of Living Cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen AK, Behlke MA, Tsourkas A. Efficient Cytosolic Delivery of Molecular Beacon Conjugates and Flow Cytometric Analysis of Target RNA. Nucleic Acids Res. 2008;36:e69. doi: 10.1093/nar/gkn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Cao Z, Lu Y. Functional Nucleic Acid Sensors. Chem Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Y, Medley CD, Cao Z, Li J, et al. Molecular Engineering of DNA: Molecular Beacons. Angew Chem Int Ed. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyagi S, Kramer FR. Molecular Beacons: Probes that Fluoresce upon Hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Yang CJ, Medley CD, Benner SA, Tan W. Locked Nucleic Acid Molecular Beacons. J Am Chem Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 17.Yang CJ, Martinez K, Lin H, Tan W. Hybrid Molecular Probe for Nucleic Acid Analysis in Biological Samples. J Am Chem Soc. 2006;128:9986–9987. doi: 10.1021/ja0618346. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Yang CJ, Moroz LL, Tan W. Nucleic Acid Beacons for Long-Term Real-Time Intracellular Monitoring. Anal Chem. 2008;80:3025–3028. doi: 10.1021/ac702637w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyagi S. Imaging Intracellular RNA Distribution and Dynamics in Living Cells. Nat Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 20.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 21.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-Flares: Probes for Transfection and mRNA Detection in Living Cells. J Am Chem Soc. 2007;129:15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Gold Nanoparticles for Biology and Medicine. Angew Chem Int Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen LJ, Olsen LF, Ozalp VC. Aptamers Embedded in Polyacrylamide Nanoparticles: A Tool for in vivo Metabolite Sensing. ACS Nano. 2010;4:4361–4370. doi: 10.1021/nn100635j. [DOI] [PubMed] [Google Scholar]
- 24.Saha K, Agasti SS, Kim C, Li X, Rotello VM. Gold Nanoparticles in Chemical and Biological Sensing. Chem Rev. 2012;112:2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene Regulation with Polyvalent siRNA–Nanoparticle Conjugates. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. Nano-flares for mRNA Regulation and Detection. ACS Nano. 2009;3:2147–2152. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Aptamer Nano-flares for Molecular Detection in Living Cells. Nano Lett. 2009;9:3258–3261. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prigodich AE, Randeria PS, Briley WE, Kim NJ, Daniel WL, Giljohann DA, Mirkin CA. Multiplexed Nanoflares: mRNA Detection in Live Cells. Anal Chem. 2012;84:2062–2066. doi: 10.1021/ac202648w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao G, Gao Y, Li N, Yu Z, Zhuo L, Tang B. Simultaneous Detection of Intracellular Tumor mRNA with Bi-Color Imaging Based on a Gold Nanoparticle/Molecular Beacon. Chem Eur J. 2011;17:11210–11215. doi: 10.1002/chem.201100658. [DOI] [PubMed] [Google Scholar]
- 30.Qiao G, Zhuo L, Gao Y, Yu L, Li N, Tang B. A Tumor mRNA-Dependent Gold Nanoparticle-Molecular Beacon Carrier for Controlled Drug Release and Intracellular Imaging. Chem Commun. 2011;47:7458–7460. doi: 10.1039/c1cc11490e. [DOI] [PubMed] [Google Scholar]
- 31.Li N, Chang C, Pan W, Tang B. A Multicolor Nanoprobe for Detection and Imaging of Tumor-Related mRNAs in Living Cells. Angew Chem Int Ed. 2012;51:7426–7430. doi: 10.1002/anie.201203767. [DOI] [PubMed] [Google Scholar]
- 32.Ellington AD, Szostak JW. In vitro Selection of RNA Molecules that Bind Specific Ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 33.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Li Z, Hu D, Lin CT, Li J, Lin Y. Aptamer/Graphene Oxide Nanocomplex for in situ Molecular Probing in Living Cells. J Am Chem Soc. 2010;132:9274–9276. doi: 10.1021/ja103169v. [DOI] [PubMed] [Google Scholar]
- 35.Chen YS, Hung YC, Liau I, Huang G. Assessment of the in vivo Toxicity of Gold Nanoparticles. Nanoscale Res Lett. 2009;4:858–864. doi: 10.1007/s11671-009-9334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharifi S, Behzadi S, Laurent S, Laird Forrest M, Stroeve P, Mahmoudi M. Toxicity of Nanomaterials. Chem Soc Rev. 2012;41:2323–2343. doi: 10.1039/c1cs15188f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alemdaroglu FE, Ding K, Berger R, Herrmann A. DNA-Templated Synthesis in Three Dimensions: Introducing a Micellar Scaffold for Organic Reactions. Angew Chem Int Ed. 2006;45:4206–4210. doi: 10.1002/anie.200600524. [DOI] [PubMed] [Google Scholar]
- 38.Alemdaroglu FE, Alemdaroglu NC, Langguth P, Herrmann A. DNA Block Copolymer Micelles – A Combinatorial Tool for Cancer Nanotechnology. Adv Mater. 2008;20:899–902. [Google Scholar]
- 39.Kwak M, Herrmann A. Nucleic Acid Amphiphiles: Synthesis and Self-Assembled Nanostructures. Chem Soc Rev. 2011;40:5745–5755. doi: 10.1039/c1cs15138j. [DOI] [PubMed] [Google Scholar]
- 40.Schnitzler T, Herrmann A. DNA Block Copolymers: Functional Materials for Nanoscience and Biomedicine. Acc Chem Res. 2012;45:1419–1430. doi: 10.1021/ar200211a. [DOI] [PubMed] [Google Scholar]
- 41.Liu H, Zhu Z, Kang H, Wu Y, Sefan K, Tan W. DNA-Based Micelles: Synthesis, Micellar Properties and Size-Dependent Cell Permeability. Chem Eur J. 2010;16:3791–3797. doi: 10.1002/chem.200901546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA Aptamer–Micelle as An Efficient Detection/Delivery Vehicle toward Cancer Cells. Proc Natl Acad Sci U S A. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen T, Wu CS, Jimenez E, Zhu Z, Dajac JG, You M, Han D, Zhang X, Tan W. DNA Micelle Flares for Intracellular mRNA Imaging and Gene Therapy. Angew Chem Int Ed. 2013;52:2012–2016. doi: 10.1002/anie.201209440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Z, Mallikaratchy P, Yang R, Kim Y, Zhu Z, Wang H, Tan W. Aptamer Switch Probe Based on Intramolecular Displacement. J Am Chem Soc. 2008;130:11268–11269. doi: 10.1021/ja804119s. [DOI] [PubMed] [Google Scholar]
- 45.Izyumov DS, Avetisyan AV, Pletjushkina OY, Sakharov DV, Wirtz KW, Chernyak BV, Skulachev VP. “Wages of Fear”: Transient Threefold Decrease in Intracellular ATP Level Imposes Apoptosis. BBA - Bioenergetics. 2004;1658:141–147. doi: 10.1016/j.bbabio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Zamaraeva MV, Sabirov RZ, Maeno RZ, Ando-Akatsuka Y, Bessonova SV, Okada Y. Cells Die with Increased Cytosolic ATP During Apotosis: a Bioluminescence Study with Intracellular Luciferase. Cell Death Differ. 2005;12:1390–1397. doi: 10.1038/sj.cdd.4401661. [DOI] [PubMed] [Google Scholar]
- 47.Tan X, Chen T, Xiong X, Mao Y, Zhu G, Yasun E, Li C, Zhu Z, Tan W. Semiquantification of ATP in Live Cells Using Nonspecific Desorption of DNA from Graphene Oxide as the Internal Reference. Anal Chem. 2012;84:8622–8627. doi: 10.1021/ac301657f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.