Abstract
Uveal melanoma is the most common primary cancer of the eye and often results in fatal metastasis. Here, we describe mutations occurring exclusively at arginine-625 in splicing factor 3B subunit 1 (SF3B1) in low-grade uveal melanomas with good prognosis. Thus, uveal melanoma is among a small group of cancers associated with SF3B1 mutation, and these mutations denote a distinct molecular subset of uveal melanomas.
Uveal melanomas can be divided into prognostically significant subgroups based on their transcriptomic signature. Class 1 tumors rarely metastasize, and they tend be less invasive, more differentiated and occur in younger individuals than class 2 tumors1. In contrast, class 2 tumors frequently metastasize, and they tend to comprise undifferentiated “epithelioid” tumor cells lacking a copy of chromosome 31.
We recently described loss of function mutations in BAP1 (BRCA1-associated protein 1), located at chromosome 3p21.1, in ∼40% of uveal melanomas, virtually all of which were the aggressive class 2 tumors2. In the present study, we searched for additional mutations in uveal melanoma by exome sequencing of 18 primary tumors, including seven class 1 and eleven class 2 tumors. Exome data were filtered down to somatic alterations that were predicted to be deleterious (Supplementary Methods). Only two genes were found to harbor deleterious somatic variants in at least three tumor samples: guanine nucleotide-binding protein G(q) subunit alpha (GNAQ), which is already known to undergo mutation in uveal melanoma3,4, and splicing factor 3B subunit 1 (SF3B1). In the case of SF3B1, the mutation in all three tumors led to a p.R625C alteration (hg19 chr2:198267484G>A) which was confirmed by Sanger sequencing.
We manually examined the entire SF3B1 coding sequence from the other 15 exome samples, but we did not find mutations at any sites other than R625. SF3B1 mutations were recently described in myelodysplastic syndrome (MDS) and chronic lymphocytic leukemia (CLL), clustering within exons 12 to 155,6. Thus, we re-sequenced these exons in a total of 102 primary uveal melanomas and matching blood DNA samples and identified SF3B1 mutations in 19 (18.6%) tumors (Supplementary Table 1). This frequency is similar to that in MDS and CLL5,6, and much higher than that recently reported in breast cancer7. Strikingly, all of the mutations in uveal melanoma occurred atarginine-625, including twelve R625H, five R625C, one R625G and one R625L substitutions (Supplementary Fig. 1). Interestingly, R625 is one of many sites in SF3B1 that are mutated in MDS, but it is the only site that was identified as deleterious by the SIFT algorithm5, which predicts the effect an amino acid substitution has on protein function. SF3B1 mutations were not present in matching blood DNA samples, indicating that they were somatic in origin. In each of the tumors with SF3B1 mutations, wildtype and mutant alleles were present in roughly equal proportions (Supplementary Fig. 1). An evaluation of DNA copy number in 30 of the uveal melanomas, including seven with SF3B1 mutations, revealed no loss of chromosome 2q33.1 where SF3B1 resides (Supplementary Fig. 2),which would have been consistent with a classical tumor suppressor. Rather, these findings are more consistent with SF3B1 mutations functioning as dominant-negative, gain-of-function or haploinsufficient alteration.
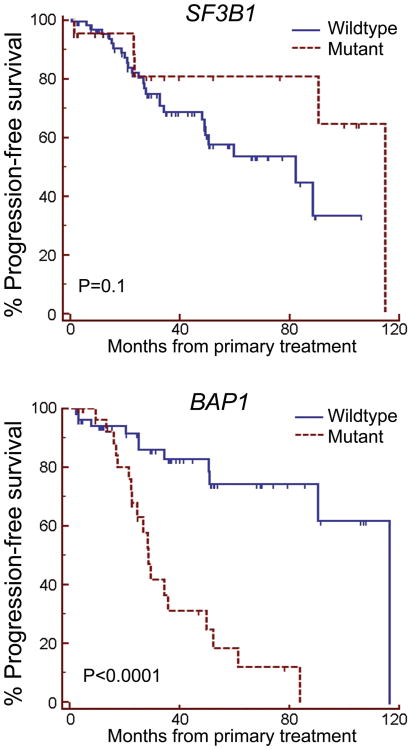
SF3B1 mutations were associated with favorable prognostic features such as younger patient age (P=0.03) and fewer undifferentiated epithelioid cells (P=0.003), and they were inversely associated with poor prognostic features such as the class 2 transcriptomic signature (P = 0.02), loss of chromosome 3 (P = 0.001) and mutation of BAP1 (P=0.002) (Table 1). Patients with SF3B1-mutant tumors trended toward a lower metastatic rate than those with SF3B1-wildtype tumors (P=0.1), which was in striking contrast to the high metastatic rate in patients with BAP1 mutations (P<0.0001)(Fig. 1). Five uveal melanoma samples from distant metastases were available for testing, and none harbored SF3B1 mutations, further supporting the notion that these mutations might be associated with less aggressive tumors. Taken together, these findings indicate that SF3B1 mutations are associated with better prognosis in uveal melanoma, which is similar to findings in MDS5.
Table 1. Associations between SF3B1 mutation and clinical, histopathologic and genetic features.
| Variable | SF3B1 wildtype | SF3B1 mutant | P-value |
|---|---|---|---|
|
| |||
| Patient age | |||
| Mean | 63.0 | 55.3 | 0.03 |
| Median | 65.0 | 60.0 | |
| Minimum, Maximum | 24-87 | 16-76 | |
|
| |||
| Patient sex | |||
| Female | 34 (41%) | 12 (63%) | 0.1 |
| Male | 49 (59%) | 7 (37%) | |
|
| |||
| Tumor diameter (mm) | |||
| Mean | 16.0 | 17.1 | 0.3 |
| Median | 16.0 | 17.2 | |
| Minimum, Maximum | 3-24 | 9-24 | |
|
| |||
| Tumor thickness (mm) | |||
| Mean | 8.8 | 8.6 | 0.8 |
| Median | 9.0 | 8.1 | |
| Minimum, Maximum | 1-16 | 2-15 | |
|
| |||
| Ciliary body involvement | |||
| Yes | 44 (60%) | 9 (47%) | 0.4 |
| No | 29 (40%) | 10 (53%) | |
| Not available | 10 | 0 | |
|
| |||
| Epithelioid cell type | |||
| Yes | 33 (40%) | 1 (5%) | 0.003 |
| No | 49 (60%) | 18 (95%) | |
| Not available | 1 | 0 | |
|
| |||
| Extraocular tumor invasion | |||
| Yes | 17 (21%) | 5 (26%) | 0.8 |
| No | 64 (79%) | 14 (74%) | |
| Not available | 2 | 0 | |
|
| |||
| BAP1 status | |||
| Wildtype | 37 (54%) | 16 (94%) | 0.002 |
| Mutant | 31 (46%) | 1 (6%) | |
| Not available | 15 | 2 | |
|
| |||
| GNAQ status | |||
| Wildtype | 40 (60%) | 10 (53%) | 0.6 |
| Mutant | 27 (40%) | 9 (47%) | |
| Not available | 16 | 0 | |
|
| |||
| GNA11 status | |||
| Wildtype | 28 (43%) | 12 (67%) | 0.1 |
| Mutant | 37 (57%) | 6 (33%) | |
| Not available | 18 | 1 | |
|
| |||
| Gene expression class | |||
| Class 1 | 37 (49%) | 14 (82%) | 0.02 |
| Class 2 | 38 (51%) | 3 (18%) | |
| Not available | 8 | 2 | |
|
| |||
| Chromosome 3 status | |||
| Retention of heterozygosity | 34 (49%) | 14 (93%) | 0.001 |
| Loss of heterozygosity | 36 (51%) | 1 (7%) | |
| Not available | 13 | 4 | |
Figure 1.
SF3B1 mutations in uveal melanoma. (a) Sanger sequence traces of representative tumor samples harboring wild type or mutant SF3B1 alleles at codon 625. (b) Kaplan-Meier survival plots of 102 uveal melanoma patients stratified by SF3B1 mutation status (top) and BAP1 mutation status (bottom).
SF3B1 is one of very few genes that are commonly mutated in uveal melanoma, allowing for a more precise molecular taxonomy of this cancer. Activating oncogenic mutations in GNAQ or GNA11 occur in about 85% of primary uveal melanomas and are thought to represent early events because they are found in uveal melanomas of all stages, including pre-malignant nevi, and they are not associated with prognosis3,8. Consistent with this idea, GNAQ/GNA11 mutations were present in most of our SF3B1-mutant and BAP1-mutant tumors, suggesting that they arise earlier than SF3B1 and BAP1 mutations. In contrast, SF3B1 and BAP1 mutations were almost mutually exclusive, suggesting that they may represent alternative pathways in tumor progression.
SF3B1 encodes subunit 1 of the splicing factor 3b protein complex, which is a component of the U2 small nuclear ribonucleoprotein complex (snRNP) that participates in the splicing of pre-mRNAs9. Splicing factor 3b is also a component of the minor U12-type spliceosome9. To explore the effects of mutant SF3B1 on global RNA expression, we analyzed five SF3B1-mutant and six SF3B1-wildtype class 1 tumors for differentially expressed transcripts using the Illumina Bead Array platform. This analysis was limited to class 1 tumors because most SF3B1 mutations occurred in this subset. Surprisingly, there were only 10 differentially expressed genes, and they provided no insights into the functional significance of the SF3B1 mutations (Supplementary Table 2 and Supplementary Fig. 3). Moreover, none of these genes were the same as those that were differentially expressed in MDS5,10. We therefore investigated if the main consequence of SF3B1 mutations was intron retention rather than differential expression. Three SF3B1-mutant and five SF3B1-wildtype class 1 tumors were analyzed for alterations in splice donor and splice acceptor retention using RNA-Seq (Supplementary Methods). However, no differences in global splice donor or acceptor retention were found between SF3B1-mutant and –wildtype tumors (data not shown). Further, we manually analyzed a set of neural crest regulatory transcripts that are aberrantly spliced in sf3b1-mutant zebrafish (SNAI1, SOX9, TFAP2A, SOX10, ID2, MITF and SF3B1)11, but no splicing abnormalities were found. Despite the known role of SF3B1 in RNA splicing, there have not been consistent results linking SF3B1 mutations to specific splicing errors in MDS or CLL, and the functional consequences of SF3B1 mutations remain elusive despite intensive investigation12. Recent links between SF3B1 and chromatin remodelling complexes13 raise the question of whether the primary effect of SF3B1 mutations on tumor progression involve RNA processing at all. Further investigations are under way to elucidate the effects of SF3B1 mutations on uveal melanoma progression in order to therapeutically target these effects.
Data access
Gene expression micro array and array CGH data have been deposited at the Gene Expression Omnibus (GEO) (accession numbers GSE39717 and GSE42740, respectively). Exome sequences and RNA-seq data are available at the NCBI Sequence Read Archive (accession numbers SRA062369 and SRA062359, respectively).
Supplementary Material
Acknowledgments
This work was supported by NIH R01 CA16187001 (A.M.B. and J.W.H.) and CA12597007 (J.W.H.), Melanoma Research Alliance (J.W.H.), Melanoma Research Foundation (J.W.H.), Tumori Foundation (J.W.H.), Research to Prevent Blindness, Inc. (J.W.H).and by awards to the Department of Ophthalmology and Visual Sciences at Washington University from a Research to Prevent Blindness, Inc. Unrestricted grant, and the NIH Vision Core Grant P30 EY02687c. E.D.O.R. was supported under NIH training grant 5 T32 AR007279-32. We thank Li Cao for technical assistance and Catherine Jordan for exome capture. We thank the patients who volunteered to participate in the study in order to make this work possible.
Footnotes
Author contributions: J.W.H. participated in the conception and design of the study, provided the samples used in the study, performed biostatistical analysis and drafted the manuscript. E.D.O.R. performed the analysis of next generation sequencing data and bioinformatics analysis. H.A. performed Sanger sequencing. M.D.O. performed bioinformatic analysis. L.A.W. managed the tissue bank and clinical database, and prepared DNA and RNA samples. A.M.B. participated in the conception and design of the study and analyzed the data. All authors contributed to the final draft of the manuscript.
Supplementary Information: Supplementary File
• Supplementary Methods, Supplementary Tables 1-2, Supplementary Figures 1-3
Competing financial interests: J.W.H., A.M.B. and Washington University may receive income based on the licensing of related technology by the University to Castle Biosciences, Inc. This work was not supported by Castle Biosciences, Inc.
References
- 1.Harbour JW. Pigment Cell Melanoma Res. 2012;25:171–81. doi: 10.1111/j.1755-148X.2012.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harbour JW, et al. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onken MD, et al. Invest Ophthalmol Vis Sci. 2008;49:5230–4. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Raamsdonk CD, et al. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papaemmanuil E, et al. N Engl J Med. 2011;365:1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, et al. N Engl J Med. 2011;365:2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis MJ, et al. Nature. 2012;486:353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Raamsdonk CD, et al. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Science. 2003;300:980–4. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 10.Visconte V, et al. Blood. 2012;120:3173–86. doi: 10.1182/blood-2012-05-430876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An M, Henion PD. Int J Dev Biol. 2012;56:223–37. doi: 10.1387/ijdb.113383ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visconte V, Makishima H, Maciejewski JP, Tiu RV. Leukemia. 2012 doi: 10.1038/leu.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isono K, Mizutani-Koseki Y, Komori T, Schmidt-Zachmann MS, Koseki H. Genes Dev. 2005;19:536–41. doi: 10.1101/gad.1284605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.