Abstract
MYC alterations influence the survival of patients with diffuse large B-cell lymphoma. Most studies have focused on MYC translocations but there is little information regarding the impact of numerical alterations and protein expression. We analyzed the genetic alterations and protein expression of MYC, BCL2, BCL6, and MALT1 in 219 cases of diffuse large B-cell lymphoma. MYC rearrangement occurred as the sole abnormality (MYC single-hit) in 3% of cases, MYC and concurrent BCL2 and/or BCL6 rearrangements (MYC double/triple-hit) in 4%, MYC amplifications in 2% and MYC gains in 19%. MYC single-hit, MYC double/triple-hit and MYC amplifications, but not MYC gains or other gene rearrangements, were associated with unfavorable progression-free survival and overall survival. MYC protein expression, evaluated using computerized image analysis, captured the unfavorable prognosis of MYC translocations/amplifications and identified an additional subset of patients without gene alterations but with similar poor prognosis. Patients with tumors expressing both MYC/BCL2 had the worst prognosis, whereas those with double-negative tumors had the best outcome. High MYC expression was associated with shorter overall survival irrespectively of the International Prognostic Index and BCL2 expression. In conclusion, MYC protein expression identifies a subset of diffuse large B-cell lymphoma with very poor prognosis independently of gene alterations and other prognostic parameters.
Introduction
MYC is a pleiotropic transcription factor involved in many different cellular processes. The oncogenic activation of MYC may occur by direct gene alterations such as translocations and amplifications or by dysregulation of upstream signaling pathways. MYC translocation is a recurrent genetic alteration in aggressive B-cell lymphomas such as Burkitt’s lymphoma (BL), diffuse large B-cell lymphoma (DLBCL) and B-cell lymphoma, unclassifiable with features intermediate between DLBCL and BL (BCLU). BCLU is a provisional category recently introduced in the World Health Organization (WHO) classification to identify a group of poorly characterized, very aggressive lymphomas. These tumors have variable morphology, most are CD10-positive and carry secondary MYC translocations frequently associated with additional genetic alterations.1–3 Approximately, 40 to 80% of these cases have additional rearrangements of BCL2 and/or BCL6 genes, and have been referred to as double- or triple-hit lymphomas.4–6 However, the precise borders of BCLU are not well-defined yet.2
DLBCL is a heterogeneous disease with varied clinical, morphological and genetic features. MYC translocations have been observed in 6% to 14% of cases.7–15 The addition of rituximab (R) to the classic CHOP (cyclophosphamide, doxorubicin, vincristine and prednisone) chemotherapy has significantly improved the outcome of these patients. Nevertheless, 30% to 40% of them die of the disease usually within 1–2 years after the diagnosis and it is necessary to identify such poor-risk patients who may benefit from alternative treatment strategies. The International Prognostic Index (IPI) as well as most of the gene-signatures that classify DLBCL into prognostically significant groups have retained their significance after the use of R-CHOP schemes.16,17 However, to date a molecular classification is not feasible in routine clinical practice and translational results of gene expression profiling (GEP) data are necessary.18
GEP studies also recognized a subset of DLBCL with molecular signatures resembling BL and an unfavorable outcome if treated with non-intensive chemotherapy. Most of these cases carried a MYC translocation but corresponded to classical DLBCL by morphology and immunophenotype and were unidentifiable by these methods, increasing the need for methods to recognize such tumors.11,19 Furthermore, studies using gene set enrichment analysis showed that high MYC transcriptional activity confers a poorer survival to patients with DLBCL independently of the presence of MYC translocations.20,21 Other studies have demonstrated higher levels of MYC mRNA in DLBCL with increased gene copy numbers and correlated these results with an unfavorable prognosis.22,23 In brief, all these studies indicate the importance of identifying MYC genetic changes in DLBCL. However, most of these studies evaluated only the impact of isolated changes of the MYC gene, were performed in small series of patients, or considered patients treated with CHOP and R-CHOP chemotherapy.15,22,23 At the protein level, high expression of MYC detected by immunohistochemistry may be useful to identify cases with MYC translocations.20,24,25
In this study we investigated the clinical impact of the spectrum of MYC gene alterations and MYC protein expression in DLBCL in comparison to that of other gene alterations.
Methods
The methods are fully described in the Online Supplementary Appendix. Briefly, data regarding 219 patients (125 males, 94 females; median age, 61 years) consecutively diagnosed with de novo DLBCL between 2002 and 2007 were retrieved from the files of five institutions of the Grup per l’Estudi dels Limfomes de Catalunya i Balears (GELCAB). All tumors were classified as DLBCL according to the current WHO classification and no immunodeficiency-associated lymphomas or transformed low-grade lymphomas were included. Informed consent was obtained from all the patients according to the guidelines of the different Ethic Committees.
The diagnostic samples were reviewed by expert hematopathologists (LC, FC, JLM, SS, IE, EC) from the five hospitals involved in the study. Tissue microarrays were constructed and the immunohistochemical studies included CD10 (clone 56C6), MUM1/IRF4 (clone MUM1p), BCL2 (clone 124), and Ki-67 (clone MIB-1) (all from Dako), BCL6, kindly provided by Dr. Roncador (Centro Nacional de Investigaciones Oncologicas, Madrid, Spain), and MYC (clone Y69, Epitomics, USA). The conditions for all these antibodies and their evaluation were as previously described and followed the guidelines recommended for their interpretation by the Luneburg Lymphoma Biomarker Consortium26,27 The cut-off used to determine BCL2 expression was 50%, similar to that used in other studies.12 MYC was evaluated using computerized image analysis with Aperio ImageScope software, version 9.0.0.1521 (Aperio Technologies, Vista, CA, USA). A mean number of 7000 cells were evaluated per case (range, 850–50000 cells). To select the optimal cut-off of the quantitative MYC assessment for predicting survival, a maximally selected rank statistics test was performed using the Maxstat package (R statistical package, version 2.8.1, Vienna, Austria).28 The best threshold obtained was 10% of positive cells, as shown in Online Supplementary Figure S1.
Fluorescence in situ hybridization (FISH) using split signal probes for BCL2, MALT1, BCL6 and MYC genes was performed as previously described29 and the cut-off values for the interphase FISH analyses were established following the criteria of Ventura.30 Gains were considered when three or four copies of the gene studied were identified, whereas more than four copies were considered as amplifications.31 However, as we did not use a centromer probe in this study, we cannot distinguish between true polysomies and partial chromosomal gains.
Categorical data were compared using Fisher’s exact test and a two-sided P value, whereas non-parametric tests were used for ordinal data. Standard definitions of complete response, progression-free survival and overall survival were used.32 The actuarial survival analysis was carried out according to the method described by Kaplan and Meier and the curves compared by the log-rank test.33 The multivariate analyses for survival were performed using the stepwise proportional hazards model (Cox).34
Results
Clinical features
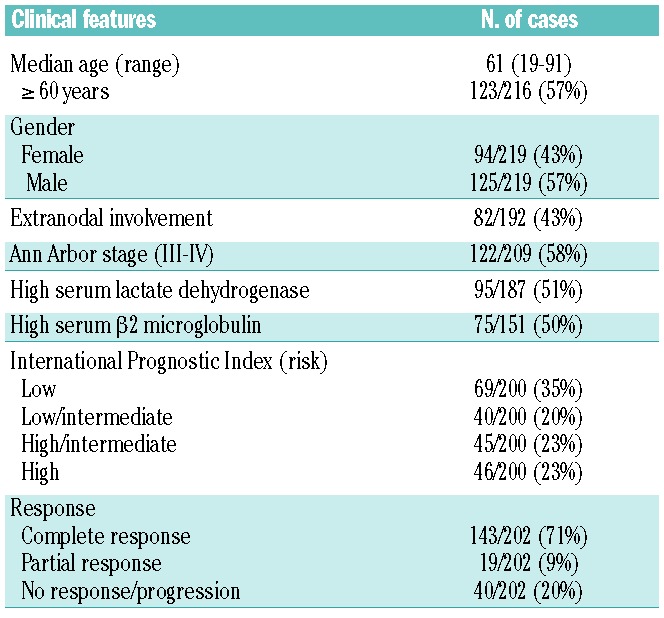
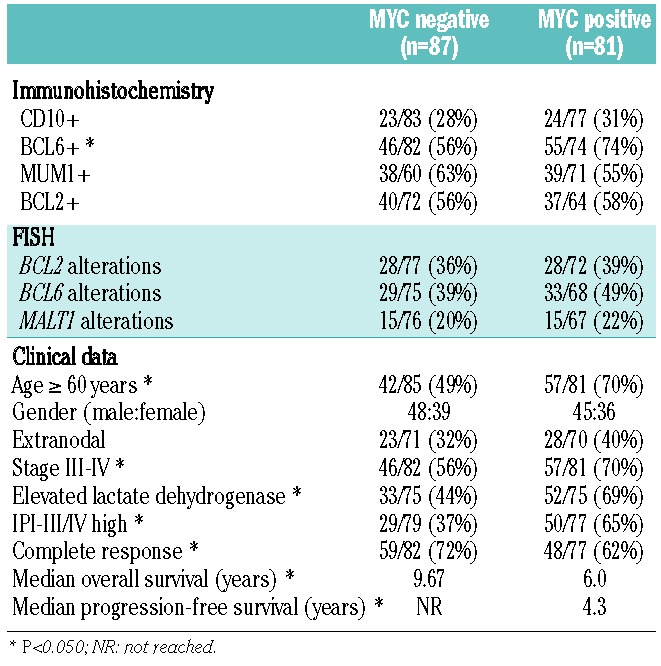
The patients’ main clinical features at diagnosis are presented in Table 1. Eleven patients who received no treatment were excluded from the survival analyses. All the remaining patients received immunochemotherapy, including adriamycin-containing regimens in 196 cases (185 received R-CHOP, and 11 patients received R-high dose-CHOP/R-ESHAP schemes; etoposide, methylprednisolone, high-dose cytarabine and cisplatin). One hundred forty-three of 202 patients (71%) with assessable response reached a complete response. The median progression-free survival was 7.5 years. After a median follow-up of 5.2 years for surviving patients, 87 patients had died, with a 5-year overall survival rate of 60% [95% confidence interval (CI): 53–67.
Table 1.
Clinical features of 219 patients with de novo DLBCL.
Morphology and immunohistochemistry
One hundred fifty-three out of the 219 cases (70%) were classified as centroblastic, 20 (9%) as immunoblastic and 46 (21%) as other variants. The phenotype of these tumors included CD10-positivity in 54/199 (27%), BCL6-positivity in 112/197 (57%) and MUM1/IRF4-positivity in 93/185 (50%). BCL2 was expressed in 95/164 cases (58%).
MYC protein immunostaining was studied by a computerized method in 168 cases and the protein was found to be expressed (cut-off 10%) in 81/168 (48%) cases. In all positive cases, MYC was expressed only in the nucleus. Since the cut-off for MYC immunohistochemistry was 40% in other studies, we also performed the same analyses using this value; with this cut-off, only 21 cases (13%) showed MYC overexpression (Online Supplementary Material).9,10,12 In addition we also evaluated MYC immunostaining in a semi-quantitative manner finding a good correlation with the digital method. All the information on the semi-quantitative approach, the relationship with the digital method and the impact on the outcome of the patients is shown in the Online Supplementary Material (Online Supplementary Table S1, Online Supplementary Figures S2, S3C and S3D, and Online Supplementary Methods).
MYC, BCL2, BCL6 and MALT1 genetic alterations
Table 2 presents a summary of the genetic alterations. The highest incidence of alterations was detected for BCL6, 71/165 (43%) cases, followed by BCL2 65/172 (38%), MYC 49/176 (28%) and MALT1 37/164 (23%). In 117 of 167 (70%) cases with complete information at least one genetic alteration was recognized using these probes.
Table 2.
Genetic alterations detected by FISH.
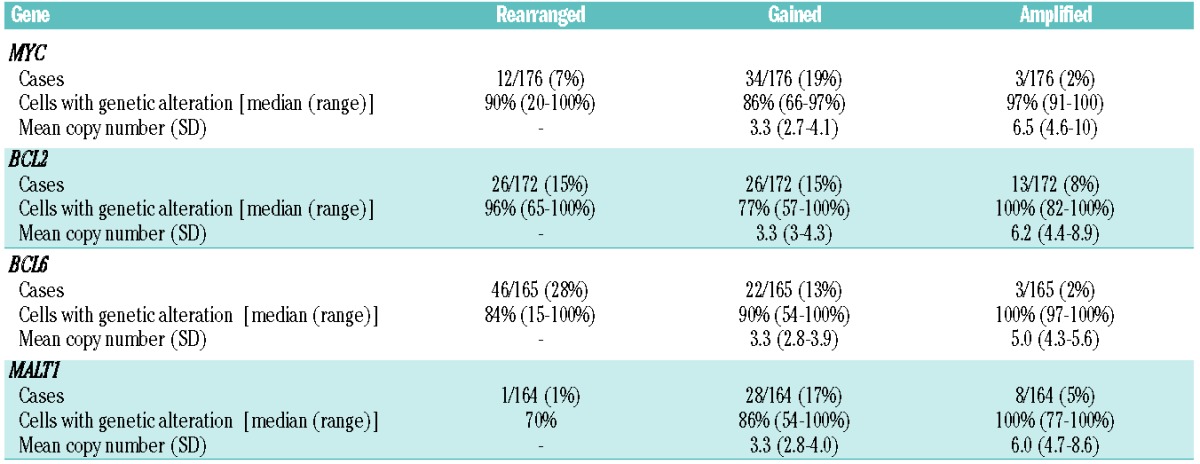
Twelve of 176 (7%) evaluable cases had MYC rearrangements, present in a median of 90% of the neoplastic cells (range, 20–100%). Four cases had MYC rearrangement as the sole abnormality (MYC single-hit). One additional case with MYC rearrangement had only gains of BCL6 and was also included in this group. Simultaneous rearrangements of MYC and BCL2 or BCL6 (MYC double-hit) were identified in five cases: four had MYC and BCL2 rearrangements and one had MYC and BCL6. Two additional cases presented triple-hit MYC-BCL2–BCL6; one case had a non-MYC double-hit, simultaneously involving the BCL2 and BCL6 genes. Among ten evaluable MYC rearranged tumors, eight cases were IGH/MYC and two non-IGH/MYC. Gains and amplifications of MYC were identified in 34/176 (19%) and 3/176 (2%) cases, respectively.
Based on the type of alterations of MYC we classified the tumors into five groups which had particular clinic-pathological and genetic characteristics, summarized in Tables 3 and 4. These groups were MYC negative (MYC without alterations), MYC gained, MYC amplified, MYC single-hit, and MYC double/triple-hit. The correlations between morphological and immunophenotypic features are shown in Table 3. The 12 tumors with MYC breaks were classified as centroblastic in eight cases (5 MYC double/triple-hit and 3 MYC single-hit), immunoblastic in two cases (both MYC single-hit) and not otherwise specified in the other two cases (MYC double/triple-hit). MYC double/triple-hit lymphomas were more frequently CD10 and BCL6-positive (5/7, 71%) and MUM1-negative (5/7, 71%). Of note, all double/triple-hit cases overexpressesd BCL2, usually very strongly, whereas MYC single-hit cases had lower or no expression of BCL2.
Table 3.
Morphological and immunophenotypic features related to MYC alterations.
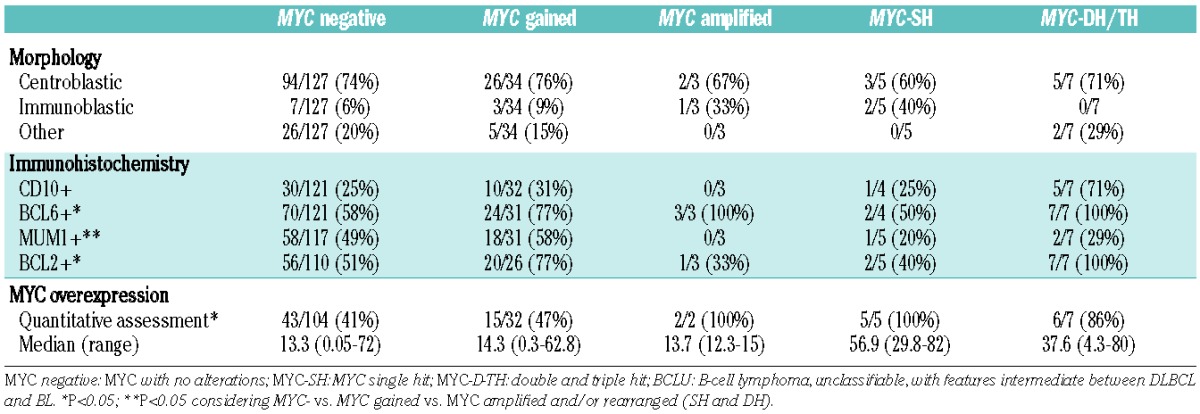
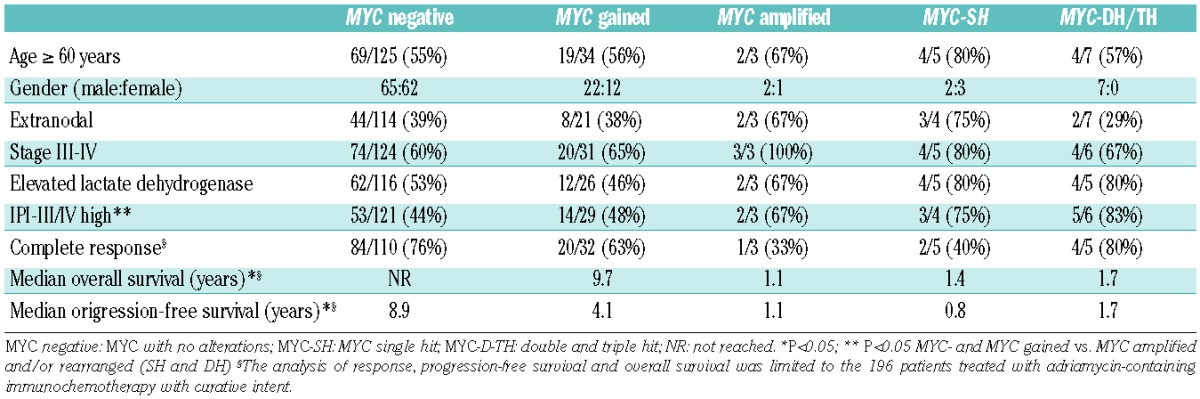
Table 4.
Clinical features and survival of patients according to MYC alterations.
Cases with MYC rearrangements or amplifications had MYC overexpression more frequently than the remaining cases (13/14 versus 58/136; P<0.005; Table 3). Ten of 12 cases with MYC breaks had at least 30% or more positive cells, whereas only two cases expressed MYC protein in 19% and 4% of the neoplastic cells. The mean percentage of cells expressing MYC in the former cases was 53% (range, 30–82%). Among cases with amplifications or gains of MYC gene, MYC protein expression was observed in an average of 14% of cells in each group (range, 12–15% for amplifications and 0–63% for gains). However, there were cases with no MYC gene alterations that had high expression of MYC protein. Thus, 20/104 (19%) MYC-negative cases and 6/32 (19%) MYC-gained cases had more than 30% positive cells expressing MYC protein (Online Supplementary Table S1).
Rearrangements of BCL2 were as common as gains of the gene (15%). A significant correlation was observed between gene alteration and protein expression: 91% cases with rearranged BCL2 and 69% with gains/amplifications had high expression of BCL2 (P<0.005). Among BCL2-translocated cases 19/26 (73%) were CD10-positive (P<0.005). BCL6 was more commonly rearranged than gained or amplified (28%, 13% and 2%, respectively). BCL6 protein was expressed in 91% of translocated cases and 74% of gained and amplified tumors (P<0.005). Gains were the most common gene alterations detected for MALT1. Notably, 28/36 (78%) cases with gains or amplification of BCL2 also presented gains or amplifications of MALT1.
Clinical impact of MYC genetic alterations and MYC expression
The correlations of MYC genetic abnormalities and the main clinical data are detailed in Table 4. Patients with MYC rearrangements or amplifications more frequently had high/intermediate- or high-risk IPI scores than the other patients (77% versus 46%; P=0.03). The initial clinical features of patients with MYC gains were completely similar to those with no MYC alterations. Table 5 shows the clinico-pathological features of the patients according to MYC expression. Patients with MYC overexpression were more frequently older and more frequently had advanced stage disease, high serum lactate dehydrogensase (LDH) concentration and high-risk IPI score. Online Supplementary Tables S2 and S3 and Online Supplementary Figure S3 show the results considering the 40% cut-off for MYC expression.
Table 5.
Main clinico-pathological data according to MYC expression.
Genetic alterations or MYC expression were not taken into consideration to decide the patients’ therapy. Among the 196 patients treated with curative intent, the median progression-free and overall survival were 7.5 and 9.7 years, respectively. All the following results refer to these patients.
Progression-free survival according to MYC genetic alterations is detailed in Table 4 and plotted in Figure 2A. The 5-year progression-free survival rates for patients with no alterations, MYC gains, and MYC rearrangements were 65%, 41%, and 15 %, respectively (P=0.003). In addition, 5-year progression-free survival rates according to MYC protein expression were 65% and 49% for negative versus positive cases, respectively (P=0.003), as shown in Figure 2C. Other variables predicting poor progression-free survival were advanced stage, age >60 years, high serum LDH, and intermediate/high or high IPI score (P<0.05 in all cases). In the multivariate analysis, including MYC gene status, MYC protein expression and IPI, the Cox model with 141 cases showed that IPI (relative risk: 1.5; P<0.001) was the only variable predicting progression-free survival.
Figure 2.
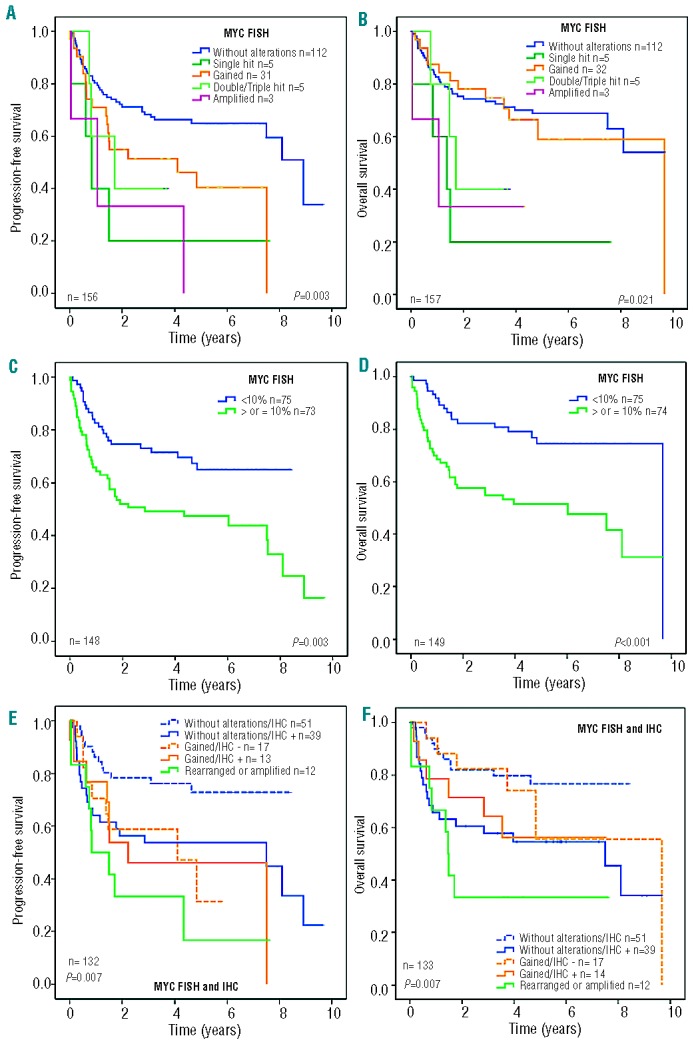
Kaplan-Meier analysis of de novo DLBCL patients treated with immunochemotherapy with curative intent in different settings. (A) Progression-free survival (PFS) and (B) overall survival (OS) according to MYC gene alterations as assessed by FISH. (C) PFS and (D) OS of patients according to MYC expression as assessed by quantitative immunohistochemistry (IHC); a threshold of 10% was obtained by the Maxstat test. (E) PFS and (F) OS of patients according to MYC gene alterations and MYC expression.
Overall survival according to MYC genetic alterations is shown in Table 4 and plotted in Figure 2B. Five-year overall survival rates for patients with no alterations, MYC gains, and MYC rearrangements were 69%, 59%, and 31%, respectively (P=0.021). Of note, no differences were found in overall survival rates between patients who were MYC negative and those with MYC gains. Moreover, 5-year overall survival rates according to MYC protein expression were 75% and 52% for negative versus positive cases, respectively (P<0.001), as shown in Figure 2D. Other variables predicting overall survival were stage, age, serum LDH and IPI (P<0.05 in all cases). In the multivariate analysis, including MYC gene status, MYC expression and IPI, the Cox model with 141 cases showed that IPI (relative risk: 1.5; P=0.001) and MYC protein expression (relative risk: 1.95; P=0.023) were the most important variables for predicting overall survival.
MYC protein expression was analyzed along with the presence of MYC genetic alterations. Figures 2E and 2F show the progression-free and overall survival curves according to the combination of MYC gene alterations and MYC expression. We observed that the presence of MYC protein expression had an unfavorable impact on both progression-free survival and overall survival (P=0.007) (Figure 2E and 2F, respectively).
Clinical impact of BCL2, BCL6 and MALT1 genetic alterations and protein expression
Genetic changes involving BCL2, BCL6 and MALT1, as well as BCL6 protein expression did not influence the outcome of the patients (data not shown). With regards to BCL2, patients with tumors expressing BCL2 had shorter progression-free survival and overall survival than those who were BCL2-negative (5-year progression-free survival 49% versus 69%, respectively; P=0.009 and 5-year overall survival: 57% versus 73%, respectively; P=0.09).
Figure 1.
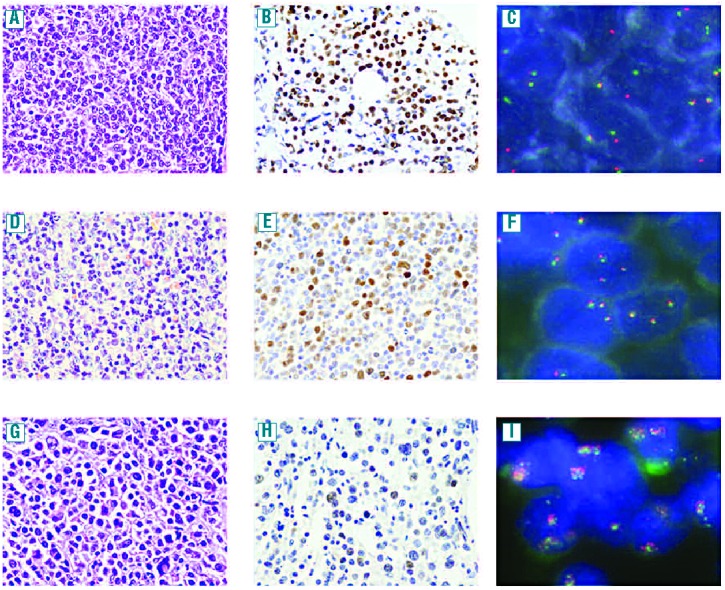
Hematoxylineosin stain (x400), MYC immunohistochemistry (x400) and FISH in DLBCL cases (x1000). DLBCL with MYC protein expression in 82% of tumor cells and MYC rearrangements (1A–1C); MYC expression in 33% of cells in a case with MYC gains (1D–1F); MYC expression in 15% tumor cells and MYC amplifications (1G–1I).
The impact of MYC and BCL2 expression was also evaluated in the present series, since recent studies have shown a remarkable impact of the expression of both proteins on the outcome of patients with DLBCL.10,12,24Figures 3A and 3B illustrate the progression-free and overall survival curves plotted according to the different combinations of MYC and BCL2 protein expression. As can be seen, patients with tumors positive for both MYC and BCL2 had the worst outcome, whereas those with double-negative tumors had the best outcome. Isolated BCL2 or MYC expression conferred an intermediate prognosis. A multivariate analysis was performed including BCL2 and MYC expression as co-variables. Both BCL2 (HR: 2.1; P=0.009) and MYC (HR: 2.1; P=0.009) maintained prognostic importance for progression-free survival in a model with 120 cases, whereas only MYC expression (HR: 3.0; P<0.001) showed a prognostic impact on overall survival. Finally, when BCL2 expression was included in the multivariate analyses for progression-free survival and overall survival along with the main clinico-pathological variables, it did not reach independent prognostic value.
Figure 3.
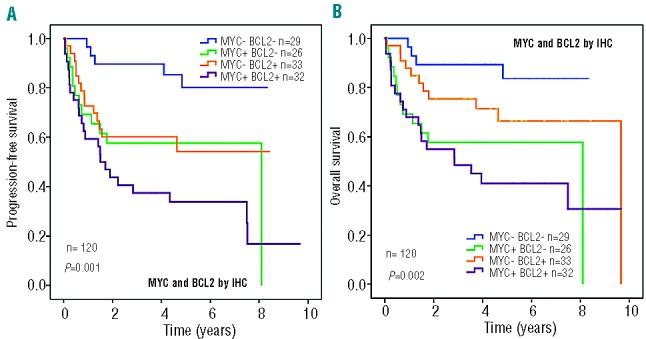
Progression-free survival (A) and overall survival (B) of cases grouped according to MYC and BCL2 expression.
Discussion
In this study we analyzed the clinical impact of MYC genetic alterations and protein expression in a large cohort of patients with DLBCL treated with immunochemotherapy. Since MYC dysregulation can occur by different mechanisms, our study focused on the analysis of both gene alterations and protein expression, and their influence on the clinical behavior of the tumors. Using FISH we observed that changes involving MYC, as well as those of BCL2, BCL6 and MALT1, were events occurring in most tumor cells, independently of the type of alteration. MYC alterations occurred either at gene or protein level in 41% cases (18 and 43%, respectively). The incidence of the genetic changes is similar to that found in other series in which the same methods of detection were used.7,9,10,35,36 The relative higher number of cases overexpressing MYC protein in our study is due to the computerized method used to quantify the expression and the different cut-off obtained using a statistical method.28 With this approach we obtained the most significant cut-off value at 10%.
Recent studies have focused on the significance of MYC rearrangements in DLBCL, but there is very little information regarding the clinical impact of gains and amplifications in series of patients homogeneously treated with regimens including immunotherapy. In our study the incidence of gains and amplifications was 19% and 2%, respectively. Patients with MYC gains had similar clinical features as those with no detectable MYC alterations. Gains were usually associated with additional genetic alterations in the other genes studied and only 13% cases had MYC gains as the unique alteration. Two previous studies have evaluated the impact of gains of MYC gene in DLBCL. Testoni et al. studied 166 patients treated with R-CHOP by array comparative genetic hybridization and found that cases with gains had additional alterations and expressed high levels of MYC mRNA. MYC gains only had an impact on overall survival when they were associated with deletions of 8p.37 Yoon et al. studied 156 patients and observed that cases with increased copy number and translocations had a shorter survival. However, in that study only 23 of 129 patients with available follow-up were treated with immunochemotherapy. Moreover, in the same study cases with gains were analyzed along with the cases with MYC amplification.23 In our study we considered the presence of three or four copies as gains, and more than four gene copies as amplifications: with this approach we identified a small group of patients with MYC amplifications who had a very unfavorable outcome similar to that of patients with double/triple-hit DLBCL. Mossafa et al. also reported that MYC amplification was an unfavorable alteration in a group of patients with high-grade B-lymphomas, including 12 with transformed low-grade B-cell lymphomas and three DLBCL.31
MYC rearrangements have been detected in 6 to 14% of de novo DLBCL. In our study, 12 (7%) cases showed MYC rearrangement and 7/12 (58%) had additional rearrangements of BCL2 and/or BCL6. This is consistent with previous FISH studies showing that 40–80% of DLBCL with MYC rearrangements have concurrent BCL2 and/or BCL6 translocations, and also with conventional genetic studies indicating that MYC genetic alterations are usually associated with additional chromosomal changes in these tumors.5 Recent studies have shown a dismal prognosis for double/triple-hit DLBCL, even in patients treated with immunochemotherapy. However, only a small number of these studies analyzed the impact of concurrent double/triple-hit compared to MYC single-hit cases. Green et al. studied a series of 191 DLBCL by FISH for MYC and BCL2, as well as the protein expression of these markers by immunohistochemistry.9 After excluding concurrent double-hit lymphomas, they did not find that MYC gene breaks had a significant impact on the survival of their patients. Similarly, Johnson et al. observed a very unfavorable impact of MYC rearrangements only when they were associated with either BCL2 breaks or protein overexpression.12 In our series the behavior of single-hit MYC and double/triple-hit lymphomas was similar. These findings suggest that single-hit MYC should not be underestimated, particularly when associated with high protein expression. It seems clear that BCL2 and/or BCL6 breaks confer a very unfavorable behavior to MYC rearrangements. The differences between studies may be due to the very low number of cases showing single-hit MYC. Further studies are needed to clarify the prognostic value of single-hit MYC.
MYC protein expression was detected in 81/168 (48%) of cases. In our series a high average of cells (over 30%) with MYC protein expression was observed in 83% of tumors with MYC rearrangement, 19% of cases with MYC gains and in 19% of cases with no MYC gene alterations. Thus, the presence of MYC rearrangement correlated with MYC protein expression in this series and our results were similar to those of other studies using the same antibody.24,25 The MYC protein expression studies also identified a subset of tumors with levels of expression over 30% without gene alterations (19%) indicating that mechanisms other than gene alterations may cause overexpression of the protein.10,20,24,25
We observed that high MYC protein expression had an unfavorable prognostic impact in patients with DLBCL. Maximally selected log-rank statistics were applied for MYC protein expression to determine the most appropriate cut-off values capable of separating two subgroups with different survival distributions. Establishing the most appropriate cut-off is of critical importance for the translation of new biomarkers into the clinical practice, and this approach has been found to be useful by us and other groups in the identification of the most suitable cut-off points for markers as prognosticators.38–41 The threshold that best captured the unfavorable impact of MYC alterations in our series was 10%. Using a digitally determined cut-off of 40% and an observational cut-off of 25% MYC alterations still had a significant prognostic impact on overall survival but not on progression-free survival. Moreover, high MYC protein expression but not the genetic alteration maintained the prognostic impact on survival in the multivariate analysis, together with the IPI, when the cut-off value was 10%, but not the 40% by digital analysis or the 25% by the manual approach (Online Supplementary Table S2). Three recent studies evaluated the impact of MYC protein expression in patients with DLBCL treated with R-CHOP. Notably, a cut-off of 40% was coincidentally used in all, and only in the study by Horn et al. did MYC protein expression have a significant prognostic impact in the univariate survival analyses.9,10,12 The impact of MYC and BCL2 co-expression was also evaluated in these studies. Green et al. and Horn et al. described scores predicting outcome and observed that patients with MYC+/BCL2+ scores had an unfavorable prognosis.9,10 Johnson et al. found that patients with simultaneous expression of both markers had inferior overall and progression-free survival.12 We performed the same analysis and obtained similar results, emphasizing the impact of MYC and BCL2 proteins in DLBCL. Notably, none of the previous studies used the same cut-off value for BCL2. Differently from these studies, in our study MYC-positive cases with no BCL2 overexpression retained an unfavorable prognosis. This difference may be attributed to the different cut-offs used for BCL2 in the four studies, and the different threshold for MYC in ours, in addition to the different methodological approaches used to evaluate MYC and BCL2. During the process of the review of our paper a new study reinforcing the unfavorable impact of MYC+/BCL2+ cases has been pre-published.42
In summary, in this study we have shown the prognostic impact of MYC gene rearrangements, amplifications and protein overexpression in DLBCL, particularly when these alterations are associated with BCL2/BCL6 rearrangements or BCL2 protein overexpression. The immunohistochemical detection of MYC protein may be a screening method to identify a subgroup of DLBCL patients with poor prognosis. However, further studies with larger cohorts of patients are needed to clarify whether the immunohistochemical detection may substitute the genetic analysis of MYC, BCL2 and BCL6 for the identification of “double-hit” genetic tumors.
Acknowledgments
The authors would like to thank Elena Gonzalvo, Ingrid Victoria, Mònica Marin and Laura Gelabert for their excellent technical assistance.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
This study was supported by “Comisión Interministerial de Ciencia y Tecnología Española” (CICYT) SAF08/3630 SAF12/38432, Red Temática de Investigación Cooperativa del Cáncer (RTICC) (RD06/0020/0039). RD12/0036/0023, and Fondo de Investigación Sanitaria (PI12/01536), Spanish Ministry of Health, and grant “Feno/genotipatge DLBCL”, La Caixa.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117(19):5019–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaffe ES, Pittaluga S. Aggressive B-cell lymphomas: a review of new and old entities in the WHO classification. Hematology Am Soc Hematol Educ Program. 2011; 2011:506–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slack GW, Gascoyne RD. MYC and aggressive B-cell lymphomas. Adv Anat Pathol. 2011;18(3):219–28 [DOI] [PubMed] [Google Scholar]
- 4.Aukema SM, Siebert R, Schuuring E, van Imhoff GW, Kluin-Nelemans HC, Boerma EJ, et al. Double-hit B-cell lymphomas. Blood. 2011;117(8):2319–31 [DOI] [PubMed] [Google Scholar]
- 5.Salaverria I, Siebert R. The gray zone between Burkitt’s lymphoma and diffuse large B-cell lymphoma from a genetics perspective. J Clin Oncol. 2011;29(14):1835–43 [DOI] [PubMed] [Google Scholar]
- 6.Snuderl M, Kolman OK, Chen YB, Hsu JJ, Ackerman AM, Dal CP, et al. B-cell lymphomas with concurrent IGH-BCL2 and MYC rearrangements are aggressive neoplasms with clinical and pathologic features distinct from Burkitt lymphoma and diffuse large B-cell lymphoma. Am J Surg Pathol. 2010;34(3):327–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akyurek N, Uner A, Benekli M, Barista I. Prognostic significance of MYC, BCL2, and BCL6 rearrangements in patients with diffuse large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone plus rituximab. Cancer. 2012;118(17):4173–83 [DOI] [PubMed] [Google Scholar]
- 8.Barrans S, Crouch S, Smith A, Turner K, Owen R, Patmore R, et al. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J Clin Oncol. 2010;28(20):3360–5 [DOI] [PubMed] [Google Scholar]
- 9.Green TM, Young KH, Visco C, Xu-Monette ZY, Orazi A, Go RS, et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30(28):3460–7 [DOI] [PubMed] [Google Scholar]
- 10.Horn H, Ziepert M, Becher C, Barth TF, Bernd HW, Feller AC, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121(12):2253–63 [DOI] [PubMed] [Google Scholar]
- 11.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354(23):2419–30 [DOI] [PubMed] [Google Scholar]
- 12.Johnson NA, Slack GW, Savage KJ, Connors JM, Ben-Neriah S, Rogic S, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30(28):3452–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klapper W, Stoecklein H, Zeynalova S, Ott G, Kosari F, Rosenwald A, et al. Structural aberrations affecting the MYC locus indicate a poor prognosis independent of clinical risk factors in diffuse large B-cell lymphomas treated within randomized trials of the German High-Grade Non-Hodgkin’s Lymphoma Study Group (DSHNHL). Leukemia. 2008;22(12):2226–9 [DOI] [PubMed] [Google Scholar]
- 14.Nitsu N, Okamoto M, Miura I, Hirano M. Clinical significance of 8q24/c-MYC translocation in diffuse large B-cell lymphoma. Cancer Sci. 2009;100(2):233–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savage KJ, Johnson NA, Ben-Neriah S, Connors JM, Sehn LH, Farinha P, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114(17):3533–7 [DOI] [PubMed] [Google Scholar]
- 16.Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziepert M, Hasenclever D, Kuhnt E, Glass B, Schmitz N, Pfreundschuh M, et al. Standard International prognostic index remains a valid predictor of outcome for patients with aggressive CD20+ B-cell lymphoma in the rituximab era. J Clin Oncol. 2010;28(14):2373–80 [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez-Garcia G, Cardesa-Salzmann T, Climent F, Gonzalez-Barca E, Mercadal S, Mate JL, et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood. 2011;117 (18):4836–43 [DOI] [PubMed] [Google Scholar]
- 19.Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354(23):2431–42 [DOI] [PubMed] [Google Scholar]
- 20.Kluk MJ, Chapuy B, Sinha P, Roy A, Dal CP, Neuberg DS, et al. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PLoS One. 2012;7(4): e33813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrader A, Bentink S, Spang R, Lenze D, Hummel M, Kuo M, et al. High Myc activity is an independent negative prognostic factor for diffuse large B cell lymphomas. Int J Cancer. 2012;131(4):E348–61 [DOI] [PubMed] [Google Scholar]
- 22.Stasik CJ, Nitta H, Zhang W, Mosher CH, Cook JR, Tubbs RR, et al. Increased MYC gene copy number correlates with increased mRNA levels in diffuse large B-cell lymphoma. Haematologica. 2010;95(4):597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon SO, Jeon YK, Paik JH, Kim WY, Kim YA, Kim JE, et al. MYC translocation and an increased copy number predict poor prognosis in adult diffuse large B-cell lymphoma (DLBCL), especially in germinal centre-like B cell (GCB) type. Histopathology. 2008;53(2):205–17 [DOI] [PubMed] [Google Scholar]
- 24.Green TM, Nielsen O, de SK, Xu-Monette ZY, Young KH, Moller MB. High levels of nuclear MYC protein predict the presence of MYC rearrangement in diffuse large B-cell lymphoma. Am J Surg Pathol. 2012;36 (4):612–9 [DOI] [PubMed] [Google Scholar]
- 25.Tapia G, Lopez R, Munoz-Marmol AM, Mate JL, Sanz C, Marginet R, et al. Immunohistochemical detection of MYC protein correlates with MYC gene status in aggressive B cell lymphomas. Histopathology. 2011;59(4):672–8 [DOI] [PubMed] [Google Scholar]
- 26.Colomo L, Lopez-Guillermo A, Perales M, Rives S, Martinez A, Bosch F, et al. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood. 2003;101(1):78–84 [DOI] [PubMed] [Google Scholar]
- 27.de JD, Xie W, Rosenwald A, Chhanabhai M, Gaulard P, Klapper W, et al. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications (a study from the Lunenburg Lymphoma Biomarker Consortium). J Clin Pathol. 2009;62(2):128–38 [DOI] [PubMed] [Google Scholar]
- 28.Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Computational Statistics & Data Analysis. 2003;43(2):121–37 [Google Scholar]
- 29.Valera A, Balague O, Colomo L, Martinez A, Delabie J, Taddesse-Heath L, et al. IG/MYC rearrangements are the main cytogenetic alteration in plasmablastic lymphomas. Am J Surg Pathol. 2010;34(11): 1686–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ventura RA, Martin-Subero JI, Jones M, McParland J, Gesk S, Mason DY, et al. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn. 2006;8(2):141–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mossafa H, Damotte D, Jenabian A, Delarue R, Vincenneau A, Amouroux I, et al. Non-Hodgkin’s lymphomas with Burkitt-like cells are associated with c-Myc amplification and poor prognosis. Leuk Lymphoma. 2006;47(9):1885–93 [DOI] [PubMed] [Google Scholar]
- 32.Cheson BD, Pfistner B, Juweid ME, Gascoyne RD, Specht L, Horning SJ, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25(5):579–86 [DOI] [PubMed] [Google Scholar]
- 33.Kaplan EL, Meier P. Non-parametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–81 [Google Scholar]
- 34.Cox D. Regression models and life tables. J R Stat Assoc. 1972;34:187–220 [Google Scholar]
- 35.Copie-Bergman C, Gaulard P, Leroy K, Briere J, Baia M, Jais JP, et al. Immunofluorescence in situ hybridization index predicts survival in patients with diffuse large B-cell lymphoma treated with R-CHOP: a GELA study. J Clin Oncol. 2009;27(33):5573–9 [DOI] [PubMed] [Google Scholar]
- 36.Foot NJ, Dunn RG, Geoghegan H, Wilkins BS, Neat MJ. Fluorescence in situ hybridisation analysis of formalin-fixed paraffinembedded tissue sections in the diagnostic work-up of non-Burkitt high grade B-cell non-Hodgkin’s lymphoma: a single centre’s experience. J Clin Pathol. 2011;64(9):802–8 [DOI] [PubMed] [Google Scholar]
- 37.Testoni M, Kwee I, Greiner TC, Montes-Moreno S, Vose J, Chan WC, et al. Gains of MYC locus and outcome in patients with diffuse large B-cell lymphoma treated with R-CHOP. Br J Haematol. 2011;155(2):274–7 [DOI] [PubMed] [Google Scholar]
- 38.Bomben R, Dal BM, Zucchetto A, Zaina E, Nanni P, Sonego P, et al. Mutational status of IgV(H) genes in B-cell chronic lymphocytic leukemia and prognosis: percent mutations or antigen-driven selection? Leukemia. 2005;19(8):1490–2 [DOI] [PubMed] [Google Scholar]
- 39.Gine E, Martinez A, Villamor N, Lopez-Guillermo A, Camos M, Martinez D, et al. Expanded and highly active proliferation centers identify a histological subtype of chronic lymphocytic leukemia (“accelerated” chronic lymphocytic leukemia) with aggressive clinical behavior. Haematologica. 2010;95(9):1526–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krober A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100(4): 1410–6 [PubMed] [Google Scholar]
- 41.Zucchetto A, Sonego P, Degan M, Bomben R, Dal BM, Bulian P, et al. Surface-antigen expression profiling of B cell chronic lymphocytic leukemia: from the signature of specific disease subsets to the identification of markers with prognostic relevance. J Transl Med. 2006;4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, et al. MYC/BCL2 protein co-expression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2013; 121(20):4021–31; 5 [DOI] [PMC free article] [PubMed] [Google Scholar]


