Abstract
Motivation: 3′ end processing is important for transcription termination, mRNA stability and regulation of gene expression. To identify 3′ ends, most techniques use an oligo-dT primer to construct deep sequencing libraries. However, this approach can lead to identification of artifactual polyadenylation sites due to internal priming in homopolymeric stretches of adenines. Although heuristic filters have been applied in these cases, they typically result in a high proportion of both false-positive and -negative classifications. Therefore, there is a need to develop improved algorithms to better identify mis-priming events in oligo-dT primed sequences.
Results: By analyzing sequence features flanking 3′ ends derived from oligo-dT-based sequencing, we developed a naïve Bayes classifier to classify them as true or false/internally primed. The resulting algorithm is highly accurate, outperforms previous heuristic filters and facilitates identification of novel polyadenylation sites.
Contact: nathan.lawson@umassmed.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
3′ end processing of pre-mRNAs influences transcription termination, mRNA stability and localization and dynamic regulation of translation. In plants, yeast and metazoans, sequence elements in the 3′ untranslated region (3′UTR) direct cleavage and polyadenylation (reviewed in Millevoi and Vagner, 2010; Proudfoot, 2011). Among these elements is the polyadenylation signal (PAS), a defined hexameric sequence located 10–30 nucleotides (nt) upstream of the cleavage and polyadenylation site (pA site), which binds Cleavage and Polyadenylation Specificity Factor complex (Millevoi and Vagner, 2010; Proudfoot, 2011). The PAS predominantly comprises the sequence AAUAAA (Proudfoot and Brownlee, 1976), although single nucleotide variants are also functional (Beaudoing et al., 2000; Sheets et al., 1990). In addition to the PAS, a guanine/uracil- or uracil-rich downstream sequence element can be found 20–40 nt downstream of the pA site that is recognized by Cleavage Stimulatory Factor (Millevoi and Vagner, 2010; Proudfoot, 2011). In some instances, a uracil-rich sequence element is present upstream of the PAS, which may also act to enhance usage of a specific PAS by recruiting Cleavage Factor I (Millevoi and Vagner, 2010; Proudfoot, 2011). In combination, these sequence elements help define the site of cleavage and polyadenylation at the 3′ end of a pre-mRNA.
Most efforts to identify 3′ ends of mRNAs have relied on priming with an oligonucleotide of deoxythymines (oligo-dT). These efforts include early studies relying on expressed sequence tags (ESTs; Beaudoing et al., 2000; Tian et al., 2005; Zhang et al., 2005), as well as more recent work using deep sequencing (reviewed in Mueller et al., 2013). These include Poly(A) Site sequencing (PAS-Seq; Shepard et al., 2011) and PolyA-Seq (Derti et al., 2012), which rely on oligo-dT containing primers for first strand cDNA synthesis. While these approaches are technically straightforward, oligo-dT binding can occur in internal homopolymeric stretches of adenines (Nam et al., 2002) leading to identification of false-positive pA sites. A more selective method is poly(A)-position profiling by sequencing (referred to as 3pseq), where a splint RNA:DNA oligonucleotide with overhanging thymines is hybridized and ligated to the polyadenylated tail of mRNAs to prevent internal priming (Jan et al., 2011). However, 3pseq is technically demanding and most laboratories are more likely to use oligo-dT-primed approaches. In these latter cases, internal priming events are generally filtered from datasets based on the number of adenines in the genomic sequence downstream of the cleavage site (Beaudoing et al., 2000; Brockman et al., 2005; Fu et al., 2011; Haenni et al., 2012; Liu et al., 2007; Shen et al., 2011; Shepard et al., 2011; Smibert et al., 2012; Tian et al., 2005; Wilkening et al., 2013; Wu et al., 2011; Zhang et al., 2005). However, the strict definition of these heuristic filters inevitably misses some internal priming events (false positives) and also excludes true 3′ ends (false negatives; Sherstnev et al., 2012). Thus, additional methods are needed to easily analyze oligo-dT primed deep sequencing data to identify true pA sites.
A naïve Bayes classifier, based on Bayes theorem, is a supervised learning algorithm in which the features used to predict the class are considered conditionally independent (Alpaydın, 2010). Naïve Bayes classifiers are computationally efficient, require relatively small training datasets, handle both continuous and discrete features and ignore non-relevant features (Kotsiantis et al., 2006; Alpaydın, 2010). Here, we demonstrate the effectiveness of a naïve Bayes classifier to identify internal priming events in oligo-dT primed sequencing data. We find that our trained algorithm outperforms heuristic filters and enriches for 3′ ends in oligo-dT sequencing data that bear canonical motifs important for cleavage and polyadenylation. Biological validation shows that our method is highly accurate, facilitating identification of novel 3′UTRs and 3′ ends in multiple animal species.
2 METHODS
2.1 Zebrafish care and staging
Zebrafish were maintained as described in (Westerfield, 1993) and staged as described in (Kimmel et al., 1995). Studies were performed under the approval of the University of Massachusetts Medical School Institutional Animal Care and Usage Committee.
2.2 RNA purification
Total RNA was purified from either 6 or 24 hpf wild-type CF zebrafish and treated with DNase I (Qiagen RNeasy Mini Kit, Qiagen RNase-Free DNase Set). Polyadenylated RNA was selected using magnetic oligo-dT beads (Invitrogen mRNA Direct Kit).
2.3 RNA-seq libraries and data analysis
The 24 hpf zebrafish RNA-seq library was built using an Illumina mRNA-seq protocol (Part # 1004898 Rev. D) and paired-end sequenced on an Illumina Genome Analyzer II (76 nt reads) and an Illumina Hi-Seq (101 nt reads). Sanger 6 hpf RNA-seq data were downloaded from the European Bioinformatics Institute (run ERR022485). RNA-seq reads from both developmental stages starting with at least five thymines (the reverse complement of a polyadenylated mRNA) or ending with at least five adenines were mapped to the zebrafish genome (Zv9) using Bowtie (Langmead et al., 2009; Supplementary Fig. S1A). Those that mapped to the genome were taken as sites for potential internal oligo-dT priming and included in the True Negative training set (Supplementary Fig. S1A). The site of internal priming was assigned to the single nucleotide immediately upstream of the last mapped 3′ adenine in this set (referred to as RNA-seq internally primed sites). Sequence fragments that did not initially map were trimmed of terminal adenines (or thymines) and re-mapped (Supplementary Fig. S1A). Mapped reads (referred to as RNA-seq putative pA sites) were combined with the PAS-Seq data for establishment of the True Positive training sets (Supplementary Fig. S1A; see Training Sets).
2.4 3′ end deep sequencing datasets
We constructed PAS-Seq libraries as described in (Shepard et al., 2011), using barcoded adapters, and paired-end sequenced on an Illumina Hi-Seq (101 nt reads) with a custom sequencing primer described in (Shepard et al., 2011) designed to exclude the remainder of the poly(A) tail from sequencing. Libraries were de-convoluted using Perl scripts and mapped to the zebrafish genome (Zv9) using Tophat (Trapnell et al., 2009). Zebrafish 6 and 24 hpf 3pseq (Ulitsky et al., 2011) and mammalian polyA-seq alignments (Derti et al., 2012) were downloaded from the Gene Expression Omnibus (accession numbers GSE32880, GSE30198). cleanUpdTSeq (see below) was used to classify putative sites from unfiltered polyA-seq as true or false, using a probability assignment cutoff = 0.5. No additional filtering was performed on the 3pseq or the originally filtered polyA-seq datasets.
2.5 RNA-Seq Transcriptome Analysis
To assess the utility of our classifier on an annotated transcriptome, we used it on previously published RNA-seq models for zebrafish embryos described in Pauli et al. (2012). Unique 3′ transcript ends were classified using cleanUpdTSeq as described below.
2.6 pA site builds
A custom Perl script clustered mapped sequencing reads into putative pA sites. Mapped reads were trimmed 3′ terminal nt, which corresponds to the site of cleavage. Reads were clustered first for identically matching sites. An iterative process was used to cluster adjacent sites within ±5 nt, starting with the site with the highest number of reads. Within a cluster, the putative pA site was defined as the location with the most reads and the total reads were combined to give the height. Mann–Whitney test was performed to assess height differences between datasets (Hollander and Wolfe, 1999). Concordance between datasets was defined as being within ±10 nt using a Perl script. The distance from the putative PAS to the pA site was determined as the distance from the 3′ end of the PAS to the pA site.
2.7 Training Sets
RNA-seq putative pA sites were combined with the PAS-Seq putative pA sites and clustered as described above (Supplementary Fig. S1A). Sites concordant between the PAS-Seq and the 3pseq data sets were assigned to the True Positive training set (Supplementary Fig. S1A). 3pseq coordinates were used if there was not an exact match. RNA-seq internally primed sites not concordant with 3pseq were assigned to the True Negative training set (Supplementary Fig. S1A). Only sites that were present in both the 6 and 24 hpf datasets were used for training (Supplementary Fig. S1B). We did not take the number of sequencing reads that composed a putative pA site into account.
2.8 cleanUpdTSeq
The function buildFeatureVector in the cleanUpdTSeq package was used to build feature vectors for training dataset and test dataset. Features include presence/absence of 4096 hexamers in the upstream of the pA sites, downstream mononucleotide count, downstream dinucleotide count and average distance of downstream adenines to the pA site (Supplementary Fig. S2B). The upstream features are modeled as binomial variables and the downstream features are modeled as normal variables. A naïve Bayes classifier was built using the training data and the function buildClassifier, which leverages the R package e1071 with laplace set to 1. To classify the test dataset, the predictClass function was applied. These functions along with sequence fetching utilities and training data are available on our website (lawsonlab.umassmed. edu/cleanupdtseq.html). The package cleanUpdTSeq is available at Bioconductor.org.
2.9 Performance Metrics
Precision, recall, true negative rate (TNR), false discovery rate (FDR), false positive rate (FPR), accuracy, F-score and Matthew’s correlation coefficient (MCC) were calculated using the following equations. TP = true positive, TN = true negative, FP = false positive, FN = false negative.
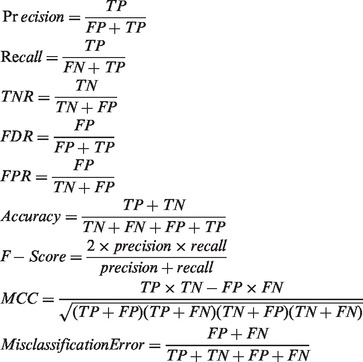 |
Pearson’s correlation coefficient was used to assess nucleotide profile correlation (Pietrokovski, 1996; R Core Team, 2013). To apply heuristic filters (8A and 8A plus top 10 PAS), Perl scripts were used to classify all putative pA sites from the training set (True Positives and True Negatives).
2.10 Model selection, training set size and relative feature importance
For performance evaluation, the training datasets were randomly split (70% used for training and 30% used for cross-validation) in 10 trials, each with a range of probability cutoffs from 0 to 1 at an interval of 0.1 for each combination of upstream (20–50 nt in increments of 10 nt) and downstream (30–50 nt in increments of 10 nt) sequence. We calculated average precision, recall, F-score, accuracy, true negative rate, false discovery rate, false positive rate and MCC from 10 cross-validations of each model, using a probability of true cutoff of 0.5 (Supplementary Table S1). We chose 40 nt of upstream and 30 nt of downstream sequence for subsequent PAS identification.
To evaluate training set size, we trained the classifier with 15 995 (50%), 19 194 (60%), 22 393 (70%), 25 592 (80%) or 28 791 (90%) peaks and used the remainder for cross validation. Average precision, recall, F-score, accuracy, true negative rate, false discovery rate, false positive rate and MCC from 10 cross-validation trials were calculated as above.
To evaluate the relative importance of each feature, P-values using prop.test in R and odds ratio were calculated for the binary features (Mladenic and Grobelnik, 1999). For the continuous features, P-values were calculated using t-test. Features were ordered by P-values, with lowest values indicating greatest importance. Upstream features (binary) and downstream features (continuous) are listed separately in Supplementary Tables S2A and B, respectively. In addition, the top 200 positive upstream features are listed in Supplementary Table S2C, which only include top upstream features with odds ratio <0. The presence of positive features is associated with increased probability of being a true polyadenylation site.
2.11 PAS flanking sequence characterization
Commonly used zebrafish PASs were identified by applying Multiple Em for Motif Elicitation (MEME) (Bailey and Elkan, 1994) to 50 nt upstream of 3′ ends annotated in Ensembl (v61). These were used to build a Perl script to search for a canonical or variant PASs. For mammals, a Perl script was used to search for a canonical or variant PAS in order of decreasing usage in polyA-seq data (Derti et al., 2012). For the True Positive and True Negative training sets, 50 nt upstream for all of the sites was examined using MEME (Bailey and Elkan, 1994) with the following settings: -minw 5 -maxw 10 –oops. 50 nt downstream of all of the sites was examined using the options: -minw 5 -maxw 50 –oops. For the other datasets, 40 nt upstream of the pA site and 30 nt downstream of 10 000 randomly chosen sites within the dataset were used for analysis (upstream: -minw 5 -maxw 10 –oops; downstream: -minw 5 -maxw 30 – oops).
2.12 Poly(A) tail length assays
Total RNA was purified from 24 hpf wild-type CF zebrafish (Qiagen RNeasy Mini Kit). For the G-tail assay, we used the Affymetrix Poly(A) Tail-Length Assay Kit to add guanosines and inosines to the 3′ end of the polyadenylated mRNAs (Martin and Keller, 1998). Subsequently, reverse transcription was performed with a poly-cytosine anchored primer. Alternatively, we used an oligo-dT(10) primer to make cDNA (Murray and Schoenberg, 2008). In both cases cDNAs were used as a template in a 20 cycle primary PCR with Hot Master Taq DNA polymerase (5′) to amplify the 3′ end with poly(A) tail with a forward primer and assay-specific reverse primer (G-Tail: Affymetrix Poly(A) Tail-Length Assay Kit Universal Primer, oligo-dT: GGGGATCCGCGGTTTTTTTTTT; Murray and Schoenberg, 2008). Nested PCR was performed for 20 to 35 cycles using 1 µl of a 1:50 dilution of the primary PCR as template, a nested forward primer and the assay-specific reverse primer. PCR products were run on a 2% agarose gel. Gene-specific oligonucleotides were also used to help estimate the size of the 3′UTR without any poly(A) tail. The lower part of the smear or single band were excised from the gel, column purified (Qiagen MinElute Gel Extraction Kit), shotgun cloned (Promega pGEM-T Easy Vector System I) and sequence verified.
3 RESULTS AND DISCUSSION
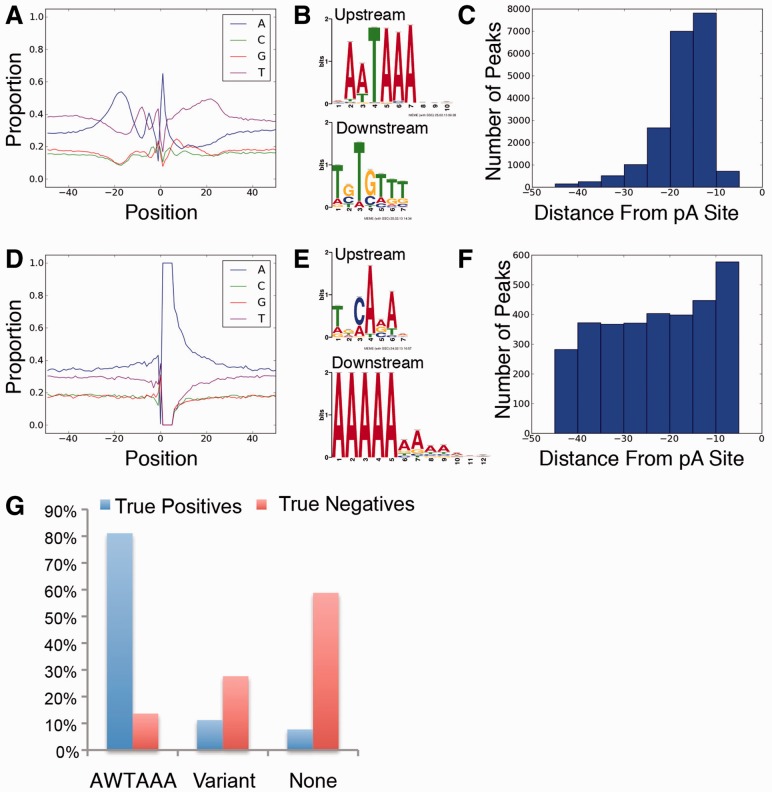
To distinguish between true and false pA sites in oligo-dT primed deep sequencing data, we trained a naïve Bayes classifier using defined True Positive and True Negative sites. Given the demonstrated technical rigor of 3pseq, True Positives were defined as the intersection of 3′ ends identified by both 3pseq and PAS-Seq datasets from the same stage of zebrafish embryos (Supplementary Fig. S1A). True Negatives were derived from oligo-dT primed RNA-Seq reads with at least five genomically templated terminal adenines or proximal thymines and were not present in 3pseq (Supplementary Fig. S1A). The training set consists of 22 770 True Positives and 9 219 True Negatives, representing the majority of zebrafish-coding genes (data not shown). As expected of pA sites (Graber et al., 1999), True Positives exhibited a prevalence of adenines and thymines upstream of the cleavage site (Fig. 1A) and a canonical PAS clustered near the 3′ end (Fig. 1B and C). Downstream of the pA site we noted thymine and a slight enrichment of guanine (Fig. 1A). By contrast, True Negative sites failed to exhibit characteristics associated with pA sites (Fig. 1D and E), and the small fraction of PASs in these sequences did not cluster near the 3′ end (Fig. 1F and G).
Fig. 1.
Training sets display characteristics of true pA sites and internally oligo-dT primed sites. (A) Nucleotide composition of pA site flanking True Positives. (B) Over-represented motifs upstream and downstream of True Positives. (C) Distribution of cleavage distance for consensus PASs of True Positive training set. (D) Nucleotide composition flanking True Negatives. (E) Over-represented motifs upstream and downstream of True Negatives. (F) Distribution of cleavage distance for canonical or variant PASs of True Negative training set. (G). PAS distribution upstream of True Positives and True Negatives. Hexamers included in ‘variant’ are AGTAAA, TATAAA, AATACA, CATAAA, AATGAA, TTTAAA, AACAAA, GATAAA
3.1 Algorithm training and performance
To delineate the True Positives from the True Negatives, we chose algorithm features to represent sequence elements known to direct cleavage and polyadenylation (Supplementary Fig. S2A). In the upstream sequence region, this included all hexamer permutations to allow for self-discovery of potential canonical and variant PASs and uracil-rich elements (Supplementary Fig. S2B). Downstream of a putative cleavage site, guanine/uracil- or uracil-rich elements may signify a true pA site (Proudfoot, 2011), while adenine richness may indicate internal oligo-dT priming. Therefore, we also included mono- and di-nucleotide counts, as well as the average distance of the adenines to the pA site, as features (Supplementary Fig. S2B). The upstream features were modeled as a binomial distribution and the downstream features were modeled as a normal distribution. The relative importance of each feature is listed in Supplementary Table S2A. As expected, canonical PASs, AATAAA and ATTAAA, are among the top four most important binary features (Supplementary Table S2A), while downstream T/GT rich elements, adenine richness and proximity to the pA site are among the most important continuous features (Supplemental Table S2B). Variant PASs also aid in identification of true pA sites (Supplemental Table S2C: top 200 positive hexamers upstream). Variation of up- (20–50 nt) and downstream (30–50 nt) sequence lengths for these features (Supplementary Fig. S2C) demonstrated low variability between the different models (Supplementary Table S1). We chose 40 nt of upstream sequence, as not to miss any possible PASs in the upstream region due to variations in cleavage site usage, and 30 nt downstream for subsequent training (Pauws et al., 2001).
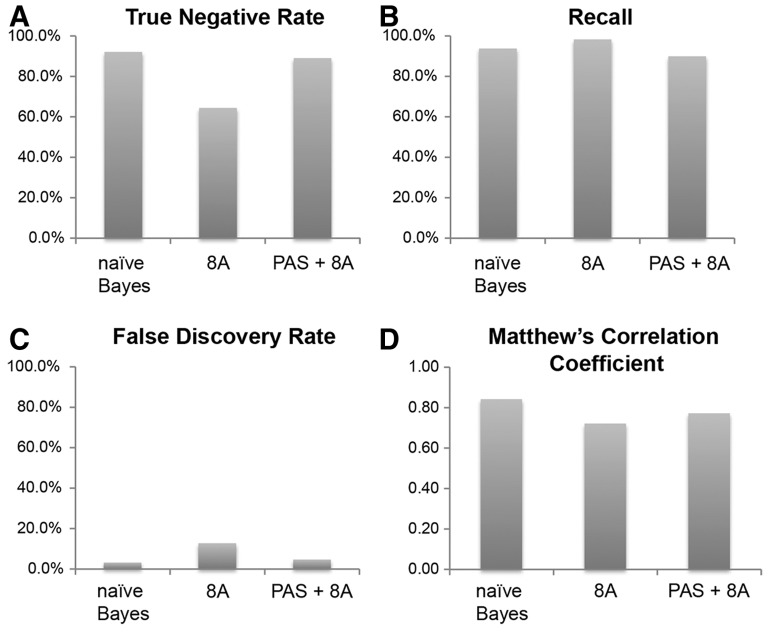
To develop and test the naïve Bayes classifier, we randomly sampled 70% of the training set to build the classifier (training) and the remaining 30% to evaluate performance (cross-validation) and averaged the results of 10 trials. Following training, we found that the naïve Bayes classifier recalled 92.2% of True Negatives (true negative rate, Table 1 and Fig. 2A) and 93.8% of True Positives (recall, Table 1 and Fig. 2B), while it incorrectly categorized only 3.2% of predicted positives (false discovery rate, Table 1 and Fig. 2C). By contrast, a heuristic filter defined as 8 or more adenines in 10 nt downstream of the pA site (referred to hereafter as 8A) generally performed worse at identifying True Negatives and False positives, although recall with this filter was quite good (Fig. 2 and Table 1). While removing all sites without a putative PAS in combination with the 8A filter (PAS + 8A) improved performance (Fig. 2A–C), the naïve Bayes classifier generally outperformed both of these heuristic filters, as determined by MCC, a balanced measure of true positives, false positives, true negatives and false negatives (Matthew’s correlation coefficient, Table 1 and Fig. 2D; Matthews, 1975). While the size of the training set may lead to over-fitting due to biased sequence composition, algorithm performance was similar using 50, 60, 70, 80 or 90% of the True Positives and True Negatives for training (Supplementary Fig. S3), demonstrating that our initial training set was of sufficient size. Taken together, the naive Bayes classifier outperforms the heuristic filters on these initial training and cross-validation sets. Furthermore, the increased specificity appears to come with little cost to sensitivity.
Table 1.
Performance measurement from naïve Bayes classifier and indicated heuristic filters
| naïve Bayes | 8A only | PAS + 8A | |
|---|---|---|---|
| True-negative rate | 0.922 | 0.645 | 0.891 |
| Recall | 0.938 | 0.984 | 0.899 |
| False discovery rate | 0.032 | 0.127 | 0.047 |
| Matthew’s correlation coefficient | 0.843 | 0.722 | 0.773 |
| Precision | 0.968 | 0.873 | 0.953 |
| F-score | 0.953 | 0.925 | 0.926 |
| Accuracy | 0.934 | 0.886 | 0.897 |
| False-positive rate | 0.078 | 0.355 | 0.109 |
Note: naive Bayes classifier outperforms heuristic filters based on the number of adenines downstream of a putative site and polyadenylation consensus signal upstream of a putative pA site.
See methods for a description of filters and equations of the performance metrics.
Fig. 2.
The trained algorithm outperforms heuristic filters. Performance metrics for naïve Bayes classification compared with 8A or PAS + 8A filters (see text for description of filters). (A) True Negative Rate. (B) Recall. (C) False Discovery Rate. (D) Matthew’s Correlation Coefficient
3.2 Application to PAS-Seq data
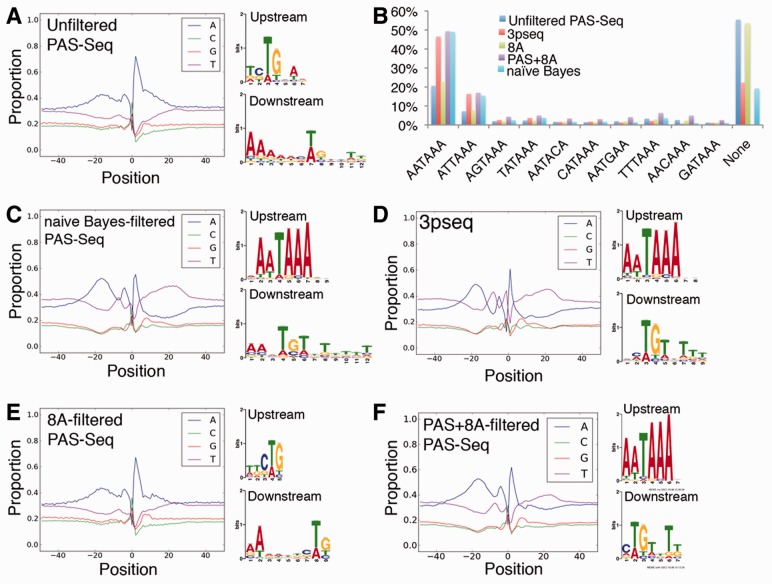
To more generally test the performance of the naïve Bayes classifier, we used all True Positives and True Negatives to build the classifier and used it to categorize unfiltered oligo-dT primed 3′ end deep sequencing (PAS-Seq) data from 24 hpf zebrafish embryos. Genomic sequence flanking 3′ ends from unfiltered PAS-Seq data shows enrichment for adenines upstream and downstream of the pA site (Fig. 3A), similar to our True Negative training set (see Fig. 1D and E; rA = 0.89, rC = 0.78, rG = 0.79, rT = 0.76). Furthermore, we failed to identify a canonical PAS as an over-represented motif upstream of putative pA sites in unfiltered PAS-Seq data (Fig. 3A). Indeed, only 20.6% of the putative pA sites contain an upstream AATAAA and 55.4% have no identifiable PAS (Fig. 3B). Thus, this dataset likely contains a high proportion of sequences derived from internal oligo-dT priming. Filtering these data with the trained naïve Bayes classifier calls 65.4% of putative pA sites from PAS-Seq as false, and the nucleotide profile of pA sites called true resembled that of pA sites identified at the same developmental stage by the more technically rigorous 3pseq approach (compare Fig. 3C and D; rA = 0.83, rC = 0.68, rG = 0.85, rT = 0.86). Similar upstream (canonical PAS) and downstream sequence (guanine/thymine-rich) elements were also easily identified in both 3pseq and filtered PAS-Seq datasets (Fig. 3C and D). In all, 49% of the true pA sites called by the classifier contain AATAAA and only 19.2% have no PAS, in agreement with genome-wide PAS distributions (Li et al., 2012; Ulitsky et al., 2012) and similar to 3pseq data (Fig. 3B). In contrast to the naïve Bayes classifier, categorizing PAS-Seq using the 8A heuristic filter classified only 18.1% of putative pA sites as false, leading to only slightly better correlation of sequence composition of the remaining sites with those from 3pseq data (rA = 0.45, rC = 0.12, rG = 0.65, rT = 0.61) compared with the unfiltered PAS-Seq. However, the adenine richness in the downstream sequence region and lack of identifiable consensus PAS suggest a large number of internally oligo-dT primed sites are called as positives after applying the 8A filter (Fig. 3B and E). The PAS + 8A heuristic filter performs better than the 8A filter, likely by excluding more false positives (rA = 0.76, rC = 0.57, rG = 0.83, rT = 0.82; Fig. 3F). However, based on its strict definition, the PAS + 8A filter eliminates all true pA sites that do not contain a consensus PAS (Fig. 3B). Together, these results demonstrate that our naïve Bayes classifier performs better than heuristic filters, resulting in a set of putative pA sites that closely resembles 3pseq. Importantly, our classifier is also able to identify 3′ ends that do not bear a consensus PAS. Further examination of this subset reveals 53% contain the top 50 positive hexamers upstream, and 74% contain the top 100 positive hexamers upstream (Supplementary Table S2C).
Fig. 3.
Algorithm-filtered PAS-Seq 3′ ends resemble those identified by 3pseq. Nucleotide composition, and over-represented motifs up- and downstream of pA sites in (A) 24 hpf unfiltered PAS-Seq, (C) 24 hpf PAS-Seq filtered by naïve Bayes classifier, (D) 24 hpf 3pseq, (E) 8A filtered 24 hpf PAS-Seq, (F) PAS + 8A filtered 24 hpf PAS-Seq. (B) PAS distribution for unfiltered 24 hpf PAS-Seq, 24 hpf 3pseq, 8A filtered 24 hpf PAS-Seq, PAS + 8A filtered 24 hpf PAS-Seq, naïve Bayes classified 24 hpf PAS-Seq
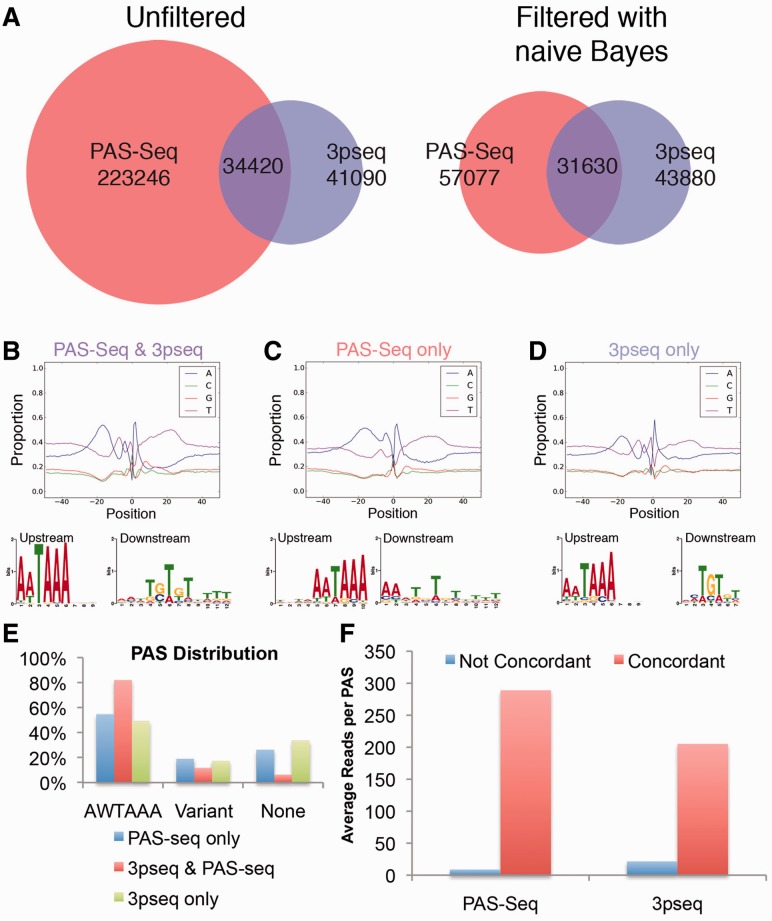
Comparison of the proportion of pA sites common to PAS-Seq and 3pseq data in 24 hpf zebrafish embryos revealed an increase from 13.0 to 35.7% after filtering PAS-Seq data with the naïve Bayes classifier (Fig. 4A). Interestingly, both common and unique pA sites exhibit characteristics typical of true PAS (Fig. 4B–D), although those unique to only 3pseq or PAS-Seq datasets show a higher proportion of variant PAS usage (Fig. 4E). The occurrence of these sites in only 3pseq or PAS-Seq datasets may be due to low levels of expression that are not consistently detected at this sequencing depth. Accordingly, pA sites common to PAS-Seq and 3pseq comprise significantly (P < 2.2e-16) more sequencing reads than those unique to either PAS-Seq (mean of 288.69 versus 8.85) or 3pseq (mean of 204.94 versus 21.4; Fig. 4F). Other technical issues may also contribute to a pA site being uniquely found in either dataset. For example, internal oligo-dT binding may block extension from an oligo-dT bound to the poly(A) tail (Nam et al., 2002), thus inhibiting identification of true 3′ ends in adenine-rich genomic loci in PAS-Seq. Variable PAS usage due to polymorphisms between the zebrafish strains used to generate the PAS-Seq and 3pseq datasets may also be possible (Howe et al., 2013).
Fig. 4.
Comparison of raw and filtered PAS-Seq 3′ ends with those from 3pseq. (A) Overlap of 24 hpf zebrafish putative pA sites from PAS-Seq and 3pseq before and after filtering of PAS-Seq by the naïve Bayes classifier. (B–D) Nucleotide composition graphs, and sequence logos for over-represented motifs 40 nt upstream and 30 nt downstream of pA sites (B) common to PAS-Seq and 3p seq, or uniquely found in (C) PAS-Seq or (D) 3pseq datasets only. (E) PAS distribution. (F) Mean number of sequencing reads contributing to a putative pA site
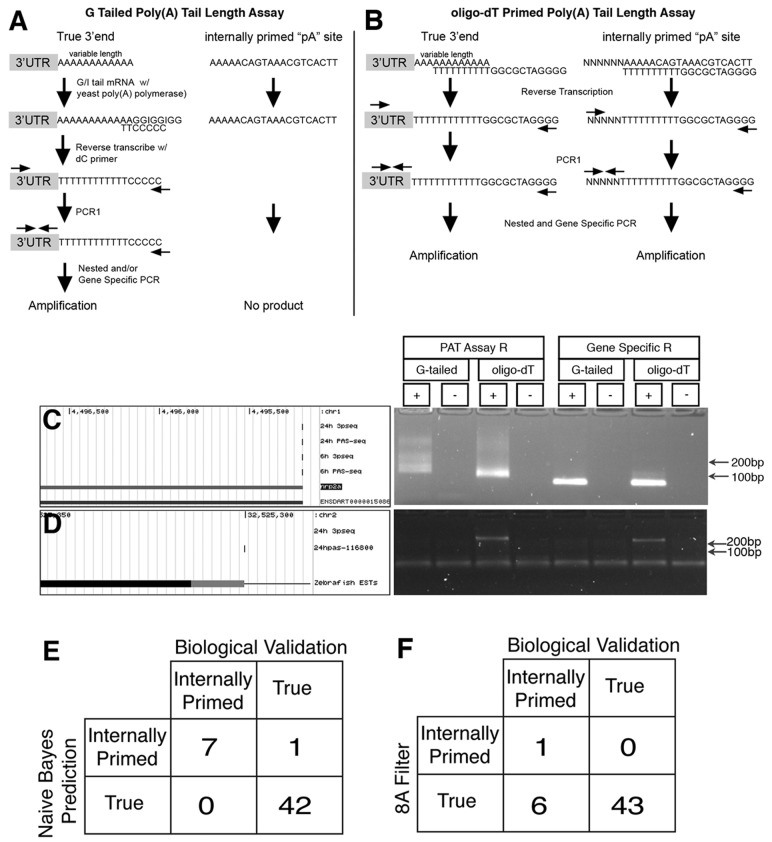
To biologically cross-validate the in silico predictions, we conducted two different poly(A) tail length (PAT) assays to verify a putative pA site. In the G-tailed PAT assay (GPAT), yeast poly(A) polymerase is used to ligate guanosines and inosines to the 3′ end of polyadenylated RNA, followed by reverse transcription with an anchored poly-cytosine primer (Fig. 5A). Alternatively, an oligo-dT containing primer was used for reverse transcription (dtPAT; Fig. 5B) (Murray and Schoenberg, 2008). In both assays, nested PCR was performed using a gene-specific forward primer and an assay-specific reverse primer to amplify the 3′ end of the transcript including the poly(A) tail, as well as gene-specific forward and reverse primers to amplify fragments without the poly(A) tail (Fig. 5A and B). Due to different poly(A) tail lengths or variable oligo-dT binding along the poly(A) tail, validation of a true 3′ end resulted in a smear on an agarose gel in both assays (Fig. 5A–C). Conversely, an internally primed site will result in no product in the GPAT assay and a single product in the dtPAT assay (Fig. 5A, B and D). We applied GPAT and dtPAT assays to 50 putative pA sites in the zebrafish genome defined by our classifier (Supplementary Table S3). Forty-two of these sites were called True by the classifier, of which 22 corresponded to annotated 3′UTRs (Zv9, ENSEMBL v68) and 20 represented novel 3′ ends. All of these True sites were amplified using the GPAT assay, indicating these are true polyadenylated 3′ ends (Fig. 5C and E; Supplementary Table S3), including 11 novel 3′ ends that were identified by 24 hpf PAS-Seq but not 24 hpf 3pseq. Thirteen validated True sites contained PASs other than the canonical AAUAAA within 40 nt upstream of the cleavage site and one lacked any consensus motif, demonstrating that our classifier can identify 3′ ends without a consensus PAS (Supplementary Table S3).
Fig. 5.
Biological validation of filtered 3′ ends. (A and B) Schematics depicting (A) G Tailed poly(A) Tail Length Assay and (B) oligo-dT Primed poly(A) Tail Length Assay. (C) Left, UCSC genome browser screenshot of 3′end of nrp2a annotated by Ensembl (v68) and RefSeq in 6 and 24 hpf PAS-Seq and 3pseq datasets. Right, GPAT and dtPAT assays for 3′ end of nrp2a. (D) Left, UCSC genome browser (reversed to show negative strand in same orientation as C) shows a putative false pA site in an EST expressed in 24 hpf PAS-Seq but not 24 hpf 3pseq. Right, GPAT and dtPAT assay for 3′ indicated at left. (C and D) ‘+’: reaction included reverse transcriptase; ‘−’ : no reverse transcriptase. ‘PAT Assay R’ denotes use of assay-specific reverse primer. ‘Gene Specific R’ denotes use of gene-specific reverse primer. ‘G-tailed’ or ‘oligo-dT’ indicate the method by which the initial cDNA template was made, and which assay-specific reverse primer was used for the lanes labeled ‘PAT Assay R’. Total RNA from 24 hpf whole embryos was used for biological validation. Confusion matrices for biologically validated sites compared with (E) naïve Bayes classifier or (F) 8A filter
Along with the putative True set, we assayed eight sites that were classified as False, only one of which was annotated as a 3′ end in ENSEMBL (Supplementary Table S3). Half of these sites displayed a variant PAS near the putative 3′ end, while the remaining sites contained no PAS 40 nt upstream. Seven out of the eight sites classified as False failed to amplify in the GPAT assay, but were detected by dtPAT suggesting that they arise from internal oligo-dT priming (Supplementary Table S3; Fig. 5D, E). Furthermore, six of these sites contained fewer than eight adenines in the downstream region and were called true by the 8A heuristic filter (Fig. 5F; Supplementary Table S3). One False site, which did not possess a consensus PAS and contained only three downstream adenines, was amplified by the GPAT assay (Fig. 5E). Together, our biological cross-validation of putative pA sites demonstrates the high accuracy of the naïve Bayes classifier. Importantly, our classifier facilitated the identification of novel pA sites from PAS-Seq allowing the discovery of new 3′UTRs in the zebrafish transcriptome.
3.3 Naïve Bayes classifier displays utility in other species
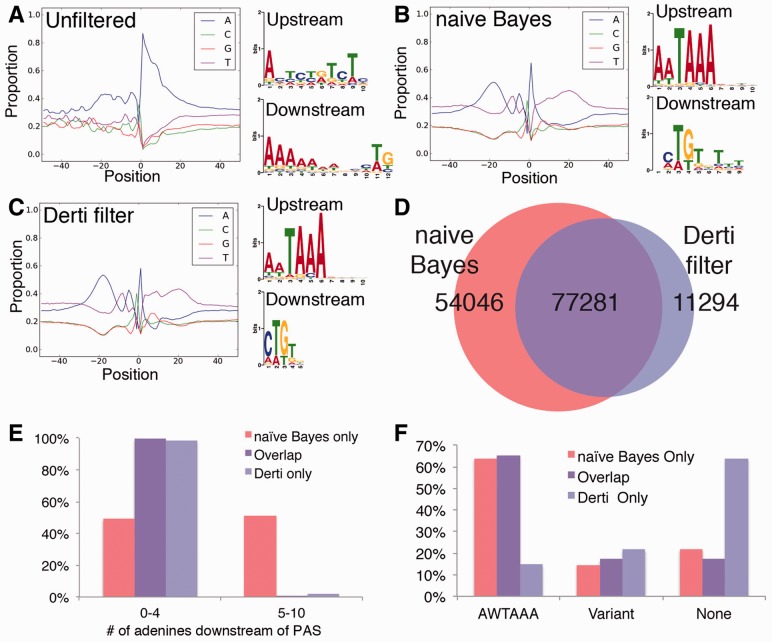
To determine whether our algorithm, which was trained using zebrafish datasets, could be applied to other species, we used it to filter mammalian datasets generated using polyA-seq, an alternative oligo-dT primed 3′ end sequence method (Derti et al., 2012; see following and data not shown). In this previous study, data were filtered using an empirically derived threshold of a log ratio, calculated as a product of mono-nucleotide frequencies in the 10 nt downstream of putative 3′ ends relative to those from internal priming sites (referred to hereafter as the ‘Derti filter’). As a metric to assess the efficacy of our classifier, we compared the results of filtering polyA-seq data with the naïve Bayes classifier to the output of the Derti filter. For example, unfiltered polyA-seq data from human kidney exhibited similarities to our negative training set, consistent with internal oligo-dT priming (Fig. 6A). From approximately half million putative 3′ ends in the unfiltered data, application of the naïve Bayes classifier identified >130 000 pA sites, which exhibited the expected characteristics for true polyadenylated 3′ ends (Fig. 6B). In comparison, the Derti filter (Derti et al., 2012) classified 94 945 as true pA sites, which also exhibited the expected sequence patterns (Fig. 6C). Of these, approximately 77 000 were commonly assigned by the two filtering methods (Fig. 6D). Closer inspection revealed that nearly all pA sites identified uniquely by the Derti filter have fewer than 5 adenines in the 10 nt downstream (Fig. 6E), consistent with the focus of this filter on nucleotide frequencies in the downstream region (Derti et al., 2012). In comparison, our naïve Bayes classifier identifies pA sites with all proportions of adenines in the downstream region, including 54 046 true pA sites called false by the Derti filter (Derti et al., 2012). Importantly, the majority of sites uniquely identified by our classifier possess a canonical PAS, suggesting that they are true 3′ ends (Fig. 6F). By contrast, the majority of true sites uniquely identified by the Derti filter did not display a PAS in the upstream region (Fig. 6F), suggesting that many may be false-positive calls. However, without biological cross-validation, it is difficult to assess the false-positive rate within this group. In any event, these observations suggest that our naïve Bayes classifier, trained on zebrafish 3′ end sequencing data, performs well in the identification of pA sites from mammalian species. Furthermore, our classifier discovered many more likely true positive pA sites from unfiltered data than the Derti filter. This is likely due to the interrogation and analysis of multiple sequence elements during the training of this classifier, while the Derti filter is restricted to consideration of only mononucleotide frequencies immediately downstream of the pA.
Fig. 6.
Naïve Bayes classifier shows utility in filtering human 3′ end sequencing datasets. Nucleotide profiles and sequence logos of over-represented motifs 40 nt upstream and 30 nt downstream of putative pA sites that were (A) unfiltered, (B) assigned as true by naïve Bayes classifier, or (C) called true by Derti et al. positional discriminant function (Derti et al., 2012). (D) Overlap of putative pA sites called true by naïve Bayes classifier and Derti et al. positional discriminant function (Derti et al., 2012). (E) Number of downstream adenines or (F) PAS distribution for putative pA sites called true by naïve Bayes, both naïve Bayes and Derti et al. positional discriminant function, or just Derti et al. positional discriminant function
Based on our work, a trained naïve Bayes classifier is clearly beneficial to identify true pA sites from oligo-dT primed 3′ end sequencing data from both zebrafish and other animal species. Additionally, this approach may also be helpful to assess and improve the quality of 3′ ends of RNA-seq transcript models built from standard transcript annotation software, such as Cufflinks or Scripture (Guttman et al., 2010; Trapnell et al. 2010). Indeed, application of our classifier to previously published RNA-seq transcript models from zebrafish (Pauli et al., 2012) suggests a significant number of transcript ends may be due to internal oligo-dT priming, while filtering enriches for canonical and variant PAS hexamers (Supplementary Fig. S4). The usage of our classifier could also be extended to RefSeq and other sequence databases, as these gene models have been largely built from oligo-dT primed cDNAs and likely contain a significant number of incorrect 3′ end annotations. Indeed, an estimated 12% of ESTs labeled as 3′ ends in dbEST human (release October 04, 2001) are due to internal oligo-dT priming (Nam et al., 2002), and our naive Bayes classifier correctly predicted that the 3′ end of vegfc, as annotated by ENSEMBL in the zebrafish genome (Zv9), is due to mis-priming (Supplementary Table S3). Thus, naïve Bayes filtering of annotated sequences in available databases, in addition to previously published genome-wide oligo-dT primed sequencing data, will likely lead to identification of new pA sites and eliminate false internally primed sites. Further studies are needed to assess the performance of the naïve Bayes classifier, trained on zebrafish data, in yeast and plants. In conclusion, the naïve Bayes classifier developed in these studies will facilitate the identification of novel pA sites in combination with simple oligo-dT primed 3′ end sequencing.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jianhong Ou for his help with the Bioconductor Package. We thank George Kourkoulis and Ira Male for fish care and maintenance. We thank the Sanger center for use of unpublished data. We thank the UMASS Medical School Deep Sequencing Core Facility, the Genomics Tufts University Core Facility and the University of California Riverside HT Sequencing Core Facility. Additionally, we would like to thank Victor Ambros, Jeffrey Bailey, Andrea D’Ambrogio, John Keaney, Charles Sagerström and Marian Walhout for helpful discussion about this project.
Funding: S.S. was supported by a pre-doctoral fellowship from the American Heart Association (10PRE4170016). N.L. and L.J.Z. were supported by R01HL101374, from the National Heart, Lung, and Blood Institute for this work.
Conflict of Interest: none declared.
REFERENCES
- Alpaydın E. Introduction to Machine Learning. Second Edition. 2010 The MIT Press Cambridge, Massachusetts. [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Beaudoing E, et al. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman JM, et al. PACdb: PolyA cleavage site and 3′-UTR database. Bioinformatics. 2005;21:3691–3693. doi: 10.1093/bioinformatics/bti589. [DOI] [PubMed] [Google Scholar]
- Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, et al. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber JH, et al. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni S, et al. Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 3′-end-seq. Nucleic Acids Res. 2012;40:6304–6318. doi: 10.1093/nar/gks282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M, Wolfe DA. Nonparametric Statistical Methods. New York, NY: John Wiley & Sons; 1999. [Google Scholar]
- Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, et al. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, et al. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kotsiantis SB, et al. Machine learning: a review of classification and combining techniques. Artif. Intell. Rev. 2006;26:159–190. [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Dynamic landscape of tandem 3′ UTRs during zebrafish development. Genome Res. 2012;22:1899–1906. doi: 10.1101/gr.128488.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, et al. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W. Tailing and 3′-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA. 1998;4:226–230. [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenic D, Grobelnik M. Proceedings of the 16th International Conference on Machine Learning. Morgan Kaufmann Publishers, San Francisco, CA; 1999. Feature selection for unbalanced class distribution and Naive Bayes; pp. 258–267. [Google Scholar]
- Mueller AA, et al. All's well that ends well: alternative polyadenylation and its implications for stem cell biology. Curr. Opin. Cell Biol. 2013;25:222–232. doi: 10.1016/j.ceb.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EL, Schoenberg DR. Assays for determining poly(A) tail length and the polarity of mRNA decay in mammalian cells. Methods Enzymol. 2008;448:483–504. doi: 10.1016/S0076-6879(08)02624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam DK, et al. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Natl Acad. Sci. USA. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22:577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauws E, et al. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Brownlee GG. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- Sheets MD, et al. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, et al. Transcriptome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing. Genome Res. 2011;21:1478–1486. doi: 10.1101/gr.114744.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev A, et al. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat. Struct. Mol. Biol. 2012;19:845–852. doi: 10.1038/nsmb.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, et al. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, et al. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, et al. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) Eugene, OR: M. Westerfield; 1993. [Google Scholar]
- Wilkening S, et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, et al. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl Acad. Sci. USA. 2011;108:12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.