Abstract
In this study, we developed a structure-based approach to identify Helitrons in four lepidopterans and systematically analysed Helitrons in the silkworm genome. We found that the content of Helitrons varied greatly among genomes. The silkworm genome harboured 67 555 Helitron-related sequences that could be classified into 21 families and accounted for ∼4.23% of the genome. Thirteen of the families were new. Three families were putatively autonomous and included the replication initiator motif and helicase domain. The silkworm Helitrons were widely and randomly distributed in the genome. Most Helitron families radiated within the past 2 million years and experienced a single burst of expansion. These Helitron families captured 3724 gene fragments and contributed to at least 1.4% of the silkworm full-length cDNAs, suggesting important roles of Helitrons in the evolution of the silkworm genes. In addition, we found that some new Helitrons were generated by combinations of other Helitrons. Overall, the results presented in this study provided insights into the generation and evolution of Helitron transposons and their contribution to transcripts.
Keywords: Helitron, silkworm, gene fragment acquisition, evolution, transcript
1. Introduction
Helitrons, originally discovered in the genomes of the model organisms Arabidopsis, rice, and Caenorhabditis elegans, are classified as DNA transposable elements (TEs).1 However, their sequence structures and mechanisms of transposition are completely different from those of other DNA transposons. They are thought to transpose via a rolling circle mechanism, because some intact Helitrons encode proteins that include the replication initiator (Rep) motif and helicase domain. These two genetic elements are necessary for bacterial IS91 (insertion sequence 91) transposition through a rolling circle mechanism.2 Although Helitrons do not have terminal inverted repeats or target site duplications, they preferentially insert into the dinucleotide AT and are characterized by a TC dinucleotide at the 5′-end, a CTRR motif at the 3′-end, and often a palindromic sequence near the 3′-terminus.
In recent years, Helitrons have been identified in almost all eukaryotic genomes. They constitute 0–5% of total genomic DNA in some model organisms. For example, they comprise >2% of the genome in C. elegans,1 >0.5% in frog,3 ∼3% in Nematostella vectensis,4 <0.1% in Aspergillus nidulans,5 ∼3% in bat,6 ∼2% in maize,7,8 and 1–5% in fruit flies.9 Furthermore, Helitron content is often highly variable among closely related species. For instance, they occupy 1–5% of genomic DNA in different fruit flies and 0.03–2.09% in different rice species.9–11
Helitrons vary greatly in sequence length, even within the same Helitron family, in part because different gene fragments are captured by these elements. More than half of the Helitrons in the maize B73 genome contained gene fragments whose lengths ranged from tens of base pairs (bp) to ten or more kilobase pairs (kbp).7,8 Furthermore, genes captured by Helitrons reshuffled the transcriptome of maize.12 Hence, Helitrons, creating the diversity of coding regions, can lead to the evolution of new functional genes.12,13
Although an increasing number of Helitrons are being identified in eukaryotic genomes, little is known about Helitrons in Lepidoptera. Recently, the genome sequences of three lepidopterans, Heliconius melpomene, Danaus plexippus (both Nymphalidae), and Manduca sexta (Sphingidae) were released, in addition to the previously available silkworm (Bombyx mori; Bombycidae) genome.14–16 Taken together, they provide an excellent resource for investigating Helitrons in Lepidoptera. The silkworm and M. sexta are moths, while H. melpomene and D. plexippus belong to butterfly. They diverged ∼100 million years ago (mya).15–17
The silkworm is a model insect for Lepidoptera and has important economic value for its silk and as a bioreactor. Approximately 40% of its genome consists of known TEs, with Helitrons comprising only 0.1%.18 In this study, we developed a structure-based approach to rescan the new silkworm genome assembly to identify Helitrons. We found that the silkworm genome harbours 21 Helitron families that occupy ∼4.23% of the genomic DNA. Thirteen of these families are new and three are putative autonomous elements. Estimates of insertion date and diversity for each Helitron family suggested that most Helitron families experienced a single rapid expansion within the past 2 million years (my). Strikingly, these Helitron families captured 3724 fragments from 268 genes and contributed to at least 1.4% of silkworm full-length cDNAs. A comparative analysis of Helitrons within Lepidoptera was also performed.
2. Materials and Methods
2.1. Identification and characterization of Helitrons
Genome sequences were downloaded for the following Lepidopterans as indicated: silkworm new assembly from SilkDB (http://silkworm.swu.edu.cn/silkdb); H. melpomene from the Heliconius genome project (http://butterflygenome.org/); D. plexippus V3 from MonarchBase (http://monarchbase.umassmed.edu/home.html), and M. sexta from NCBI (http://www.ncbi.nlm.nih.gov/nuccore/AIXA00000000).
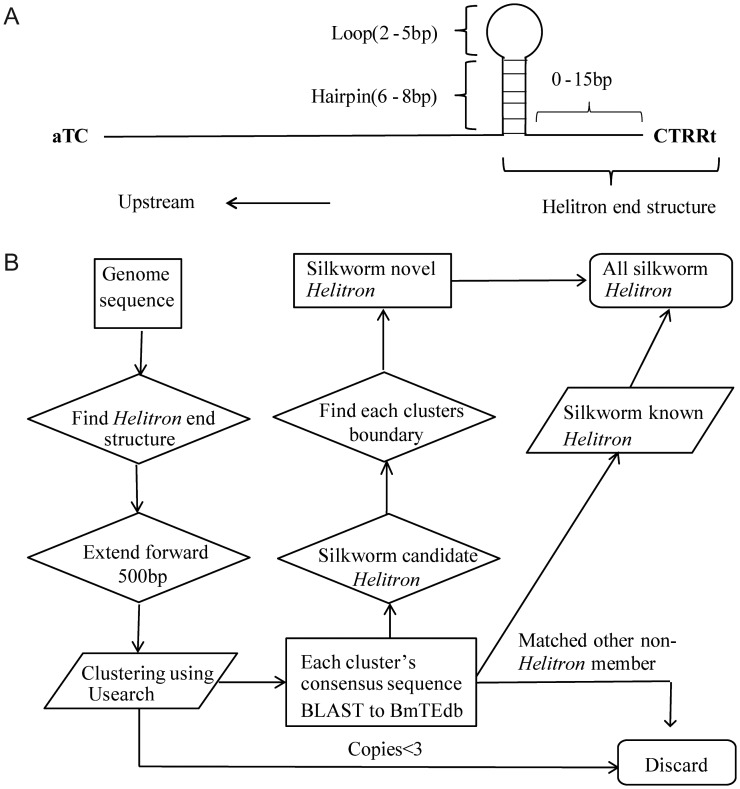
To identify Helitrons, a series of Perl scripts were written to search for Helitron sequence characteristics, similar to ‘HelSearch’.11 Briefly, the method included four steps (Fig. 1): (I) a Perl script found Helitron end structures, includes hairpins, loops, and CTRRT motifs; (II) another Perl script scanned upstream from Helitron end structure; (III) all sequences were clustered using Usearch;19 (IV) the Helitron boundaries were sought. In the step IV, we extended the sequences of each cluster in both directions using a Perl script and aligned them using MUSCLE,20 then the Helitron boundaries were manually defined. Finally, we modified Yang and Bennetzen's method of classification.8 Sequences with identities >80% in the 30 bp of both their 5′- and 3′-ends were classified as members of the same family. Full-length sequences with identity >80% were classified in the same subfamily. Our programme and readme file are available upon request.
Figure 1.
Helitrons in the silkworm. (A) Helitron structure. (B) Pipeline for Helitron identification.
To estimate copy numbers, we generated a consensus sequence for each Helitron family using DAMBE.21 We used these consensus sequences as queries for BLASTN searches of the corresponding genome database. In this step, a Helitron family was defined by E < e–6, pair-wise length >80 bp, and a minimum nucleotide identity rate of >80%. Sequences with a TC dinucleotide at the 5′-end and a CTRR motif at the 3′-end were defined as intact Helitrons.
Copy numbers of relatively long Helitron sequences (<15 kb) were estimated as follows: (i) when each end almost perfectly matched the ends of a Helitron family's consensus sequence (identity >80%, pair-wise length >80 bp for each end), and when the sequence had a TC dinucleotide and a CTRR motif at the 5′- and 3′-ends, respectively, it was defined as a Helitron copy (Supplementary Fig. S1A–C). (ii) When each end almost perfectly matched (identity >80%, pair-wise length >80 bp) non-terminal regions of a Helitron family's consensus sequence and the sequence had no or one Helitron terminal sequence (either a 5′ TC or a 3′ CTRR), it was treated as two or more Helitron fragments (Supplementary Fig. S1D–F).
To better understand the composition and structure of Helitron sequences, the AT content of each Helitron family consensus sequence was estimated using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). UNAFOLD (http://mfold.rna.albany.edu/) was used to predict the Gibbs free energy (–dG) of each Helitron.22 Finally, all Helitron families were screened against the ISfinder (http://www-is.biotoul.fr/),23 RepBase (v. 17.08),24 and NCBI non-redundant (nr) databases to identify known families. Putative autonomous Helitrons were identified by using known autonomous Helitrons downloaded from RepBase as queries and performing TBLASTN searches against all the silkworm Helitron databases.
2.2. PCR validation of predicted Helitrons
Fifteen accessions (02–320, DaZao, Ri9, 872, Ou18, Yi16, YinDuSanMian, WuLin1Hao, BH863, YingWenXing, LuoSa, RiXian2Hao, ALiKeSi, SanMianBai, and Zhong4010) representing the four main geographic strains of silkworm (Chinese, Japanese, European, and tropical) were used for insertion validation. DNA was extracted from individual pupae and moths using a standard phenol–chloroform protocol.25 A Helitron (BmHel-8) was randomly selected for insertion validation, and primers (BmHel-8-S: 5′-ATTGTCAGTGGTATCGTTGCTCC-3′, BmHel-8-A: 5′-TAAGGGAATACAATAGAGCCGTG-3′) were designed based on the flanking insertion sites.
2.3. Estimates of insertion time and expansion events
To estimate Helitron age, all full-length sequences of each Helitron family were aligned using MUSCLE,20 and the amount of nucleotide substitution (k) between each Helitron and the family consensus sequence was calculated using Kimura 2-parameter (K2P) distance.26 The age of each Helitron was estimated using the formula T = k/2r, where r = 1.56 × 10−8, the neutral rate of substitutions per year in fruit fly, which has previously been used in silkworms.27,28 Neighbor-joining trees (pair-wise deletion of gaps and K2P substitution model) for Helitron families were reconstructed using MEGA4.29 Within each Helitron family, the frequency distribution of the number of pair-wise differences between sequences was calculated with Arlequin v. 3.11.30
2.4. Distribution of Helitrons on chromosomes
All identified members of each Helitron family were mapped onto chromosomes using SilkMap (http://silkworm.swu.edu.cn/silksoft/silkmap.html), and the copy number of each family on each chromosome was counted. We divided each chromosome into 100 uniformly-sized segments and estimated each Helitron family's distribution in these fragments using a Perl script. The positions of predicted genes in scaffolds and the lengths of scaffolds were downloaded from SilkDB (http://silkworm.swu.edu.cn/silkdb);31 a Perl script was used to identify the genes near to or containing Helitrons. To determine whether Helitron insertions into genes were due to chance, a computer simulation was performed.32
2.5. Gene fragment acquisition and contribution to transcripts
Gene fragments captured by Helitrons were identified by using all identified Helitrons in a BLASTX search against the NCBI nr protein database (as of 22 January 2013). Captured gene fragments were identified if a homologue had a maximum expected value of e–10 in the silkworm, or of e–5 in a species other than silkworm. TE-related proteins were discarded.
To evaluate the contributions of silkworm Helitrons to transcripts, we used all intact Helitron sequences as queries in BLASTN analyses against the expressed sequence tag (EST) database of the silkworm (http://silkworm.swu.edu.cn/silkdb). A match was defined when the fragments had at least 99% identity and E < e−10. All matching ESTs were classified as either parental transcripts (with a similarity between the EST and the parental genes of the captured fragment greater than that between the EST and the corresponding Helitron) or Helitron transcripts.
To estimate whether the silkworm Helitrons contributed to 5′ untranslated regions (UTRs), coding regions, or 3′-UTRs of the silkworm full-length cDNAs, we downloaded the silkworm full-length cDNA database from SilkBase (http://silkbase.ab.a.u-tokyo.ac.jp/cgi-bin/index.cgi). A Perl script was written to split each full-length cDNA into 5′-UTR, coding region, and 3′-UTR. Then, we performed BLASTN analyses against these three datasets with cut-off values of at least 99% identity and E < e−10. All matching transcripts were classified as either (i) parental transcripts (with the similarity between the transcript and parental gene greater than that between the transcript and corresponding Helitron); (ii) transcripts of Helitron transposase; or (iii) chimerical transcripts composed of other genes and Helitron fragments if a full-length Helitron copy matches a cDNA sequence and there is an overlapping region (matched region) between the position of the full-length cDNA sequence and the corresponding full-length Helitron copy in the silkworm genome.
3. Results
3.1. Identification, classification, and characterization of Helitrons
We used a structure-based approach to search for Helitrons in the silkworm genome (Fig. 1). First, we searched the silkworm genome for sequences with a characteristic Helitron end structure (Fig. 1A) and found 106 766 candidate sequences. We extended the sequence of each candidate upstream, clustered all candidates, and generated a consensus sequence for each cluster. In total, we obtained 1805 consensus sequences. Each consensus sequence was used as a query in BLASTN search against BmTEdb (http://202.202.1.217/BmTEdb/), and sequences that hit to other known non-Helitron TEs were discarded. The remaining 854 consensus sequences were used in a BLASTN search against the silkworm genome. We extracted no more than 20 most-similar sequences for each consensus sequence, aligned them, and manually defined the Helitron element boundaries (Supplementary Fig. S2). Finally, sequences were classified by similarity into families and subfamilies. This pipeline (Fig. 1B) identified 21 Helitron families (Table 1) that were designated BmHel-1 through BmHel-21.
Table 1.
Summary information for the Helitron families in four lepidopteran species
| Species | Family | Subfamily | Length (bp) | Copies | AT (%) | −dG | Annotationref |
|---|---|---|---|---|---|---|---|
| B. mori | BmHel-1 | 198–781 | 514 | 56.6 | 33.8 | Bm_283BmTEdb | |
| BmHel-2 | 822–9452 | 79 | 63.3 | 141.7 | bm_691BmTEdb | ||
| BmHel-3 | BmHel-3a | 196–8700 | 1386 | 63.9 | 30.5 | Helisimi33 | |
| BmHel-3b | 206–10479 | 1995 | 69.4 | 144.5 | Helisimi33 | ||
| BmHel-4 | 290–9930 | 661 | 64.0 | 69.0 | Helianu33 | ||
| BmHel-5 | 3121–6696 | 80 | 68.7 | 535.3 | Heliminu33 | ||
| BmHel-6 | 594–738 | 25 | 69.5 | 82.8 | Novel | ||
| BmHel-7 | 126–10099 | 6007 | 61.9 | 35.3 | Novel | ||
| BmHel-8 | 105–10644 | 14656 | 71.9 | 55.4 | Novel | ||
| BmHel-9 | 608–1256 | 11 | 68.9 | 52.6 | Novel | ||
| BmHel-10 | 455–935 | 21 | 63.6 | 72.0 | Novel | ||
| BmHel-11 | 96–9788 | 12206 | 65.4 | 21.9 | Novel | ||
| BmHel-12 | 152–10623 | 3138 | 59.3 | 62.6 | Novel | ||
| BmHel-13 | 285–8964 | 3428 | 66.9 | 43.3 | Novel | ||
| BmHel-14 | 258–2723 | 3431 | 69.9 | 69.7 | Novel | ||
| BmHel-15 | 142–10468 | 8297 | 67.1 | 71.2 | Lep134 | ||
| BmHel-16 | 136–10228 | 6537 | 65.9 | 25.9 | Novel | ||
| BmHel-17 | 288–683 | 645 | 66.9 | 57.5 | Novel | ||
| BmHel-18 | 113–9945 | 3768 | 64.7 | 26.8 | Novel | ||
| BmHel-19 | 300–2462 | 149 | 58.0 | 138.4 | Novel | ||
| BmHel-20 | 296–2355 | 144 | 65.9 | 43.8 | HeligloriaAi33 | ||
| BmHel-21 | 149–6073 | 377 | 67.9 | 82.1 | HeligloriaAii33 | ||
| H. mel | HmHel-1 | 134–10671 | 6148 | 70.1 | 122.6 | Lep134 | |
| HmHel-2 | 120–10381 | 6827 | 66.3 | 103.2 | Helitron-517 | ||
| HmHel-3 | 127–11371 | 3940 | 66.9 | 52.2 | Helitron-4,717 | ||
| HmHel-4 | 134–10068 | 5665 | 59.5 | 44.1 | Helitron-1517 | ||
| HmHel-5 | 268–7417 | 4398 | 64.8 | 191.9 | Helitron-6,1117 | ||
| HmHel-6 | 134–15055 | 15359 | 47.5 | 72.8 | Novel | ||
| HmHel-7 | 276–7883 | 7103 | 60.5 | 563.1 | Helitron-1317 | ||
| HmHel-8 | 289–373 | 74 | 64.5 | 53.5 | Helitron-1617 | ||
| HmHel-9 | 217–1404 | 621 | 67.8 | 37.4 | Helitron-117 | ||
| HmHel-10 | 192–5100 | 888 | 70.7 | 103.5 | Helitron-917 | ||
| D. ple | DpHel-1 | 111–4232 | 1010 | 66.6 | 82.0 | Novel | |
| M. ext | MsHel-1 | 156–9446 | 6386 | 58.5 | 133.5 | Lep134 | |
| MsHel-2 | 134–6559 | 2975 | 57.6 | 53.3 | Novel | ||
| MsHel-3 | MsHel-3a | 134–7735 | 3270 | 66.1 | 49.5 | Novel | |
| MsHel-3b | 119–9845 | 5632 | 60.4 | 33.0. | Novel | ||
| MsHel-4 | 120–10147 | 697 | 60.3 | 91.1 | Novel | ||
| MsHel-5 | 120–3351 | 1957 | 62.1 | 49.2 | Novel | ||
| MsHel-6 | 122–9236 | 2574 | 65.5 | 31.4 | Novel | ||
| MsHel-7 | 107–3867 | 2691 | 62.3 | 119.2 | Novel |
–dG, average Gibbs energy (kcal/mol) for each Helitron family consensus sequence.
The silkworm Helitron families were annotated based on homology. Using a consensus sequence for each Helitron family as queries, we searched the BmTEdb, ISfinder, RepBase, and NCBI nr databases and found that eight families (BmHel-1, 2, 3 4, 5, 15, 20, and 21) had been previously identified. The other 13 families had no matches to any known Helitron (Table 1).
To estimate the abundance of these 21 families, we searched the silkworm genome. We identified 67 555 Helitrons in total, which constitute about 19.7 Mb (∼4.23%) of the silkworm genome. The insertion sites of these Helitrons into accession numbers (NCBI) are shown in Supplementary Table S1. Similar to previous reports on Helitrons,7,8 the silkworm Helitron size varied greatly both among and within a family; sizes ranged from 96 to 10 644 bp (Table 1). There were 202 very long (from 6000–10 644 bp) Helitron copies. The internal sequences of these Helitrons had at most 50% identity, but their ends (100 bp) had at least 80% identity. This pattern could be caused by different DNA sequences being captured either by the Helitrons or by the insertion of other TEs into the Helitrons. We identified 19 580 intact Helitrons, with a TC dinucleotide at the 5′-end and a CTRR motif at the 3′-end; they made up 10.7 Mb (∼2.30%) of the silkworm genome. Of these, 15 272 (∼78%) had at least 80% identity and 8615 (∼44%) had at least 90% identity.
The Helitron families were AT rich, with AT contents ranging from 56.61% to 71.9%. The average AT content of the silkworm genome is ∼62%. Four Helitron families (BmHel-1, 7, 12, and 19) had AT contents that did not exceed the genome average (Table 1). Almost all of the silkworm Helitron families had high predicted –dG values, indicating that most silkworm Helitrons can form stable secondary structures.
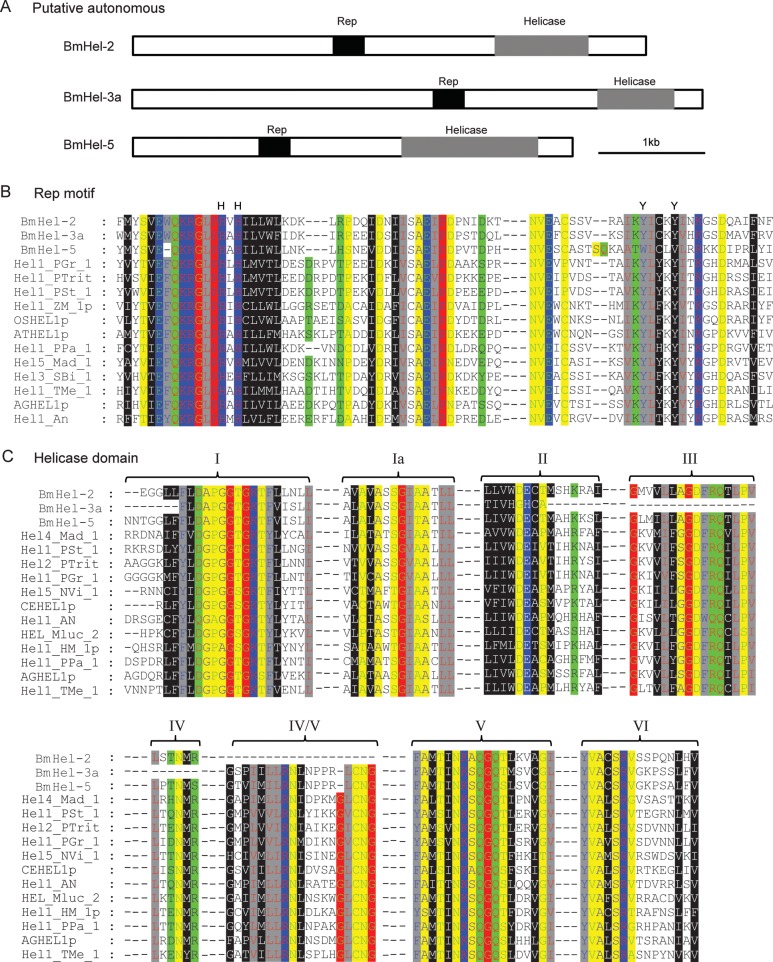
Putative autonomous silkworm Helitrons were founded based on homology. An autonomous Helitron should encode a Rep/helicase protein, because both the Rep motif and DNA helicase domain are necessary for transposition. All 141 known autonomous Helitrons were downloaded from RepBase and screened against all intact silkworm Helitrons. Three silkworm Helitron families (BmHel-2, 3, and 5) were putatively autonomous (Fig. 2). These candidate autonomous elements encoded complete open reading frames (ORFs), in addition to a Rep motif and helicase domain. For example, the SilkDB accession numbers of transposase for BmHel-2, BmHel-3, and BmHel-5 were BGIBMGA003354-TA, BGIBMGA012372-TA, and BGIBMGA008616-TA, respectively. Furthermore, two of the three putative autonomous families had EST evidence; BGIBMGA003354-TA matched the EST BY927485 (identity, 0.98; length, 760 bp) and BGIBMGA012372-TA matched ESTs BB983132 (identity, 0.94; length, 681 bp), BY932007 (identity, 0.99; length, 702 bp), CK528421 (identity, 0.95; length, 632 bp), and BY916909 (identity, 0.96; length, 657 bp). Thus, we concluded that these elements could be active in the silkworm genome.
Figure 2.
Three predicted putative autonomous elements in the silkworm based on protein domain. (A) A schematic representation of the putative autonomous Helitrons; Rep, rolling circle replication initiator motif; Helicase, region similar to SF1 superfamily of DNA helicases. (B) Alignment of REP motifs between silkworm and 12 other species. (C) Alignment of eight conserved motifs of the SF1 superfamily of DNA helicases.
3.2. Validation of predicted Helitrons
A BmHel-8 insert site was selected for PCR verification in 15 silkworm accessions representing four main geographic strains. The results indicated that BmHel-8 was present in most of the strains, but absent in YinDuSanMian, YingWenXing, LuoSa, and RiXian2Hao (Supplementary Fig. S3). This polymorphism indicated that the Helitron was not fixed in the silkworm genome and verified the efficacy of our approach.
3.3. Helitron abundance in other lepidopteran genomes
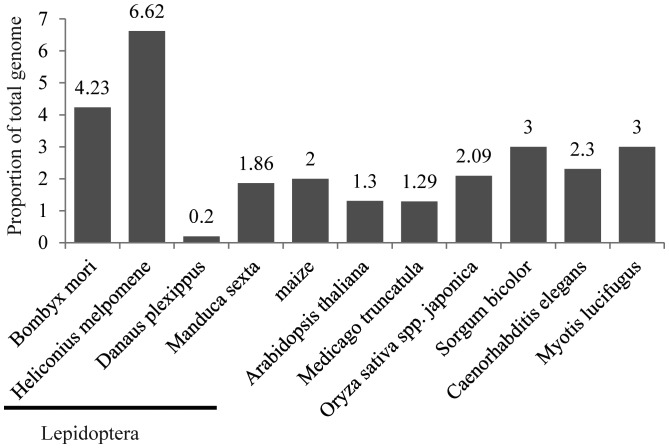
To investigate whether Helitrons were pervasive in lepidopteran, three other recently released lepidopteran genomes were searched for Helitrons. Helitron abundance varied greatly among these genomes (Table 1 and Fig. 3). For instance, H. melpomene had 10 Helitron families that comprised ∼6.62% (17.1/260 Mb) of the genome. Manduca sexta harboured seven Helitron families that made up ∼1.86% (7.23/388 Mb) of the genome. However, D. plexippus had only one Helitron family that represented only ∼0.20% (0.48/237 Mb) of the genome. The locations of each Helitron in these three genomes are listed in Supplementary Table S2 (H. melpomene), Supplementary Table S3 (D. plexippus), and Supplementary Table S4 (M. sexta).
Figure 3.
Helitron abundances in different organisms. Underlined Helitron contents were identified in this study, while others came from previous studies.1,3–11
3.4. Distributions of Helitrons on chromosomes
The silkworm Helitrons were distributed on all 28 silkworm chromosomes and were uniformly distributed among chromosomes (P> 0.05; Supplementary Fig. S4). We also found that most silkworm Helitrons had no distinct insertion bias within chromosomes (Supplementary Fig. S5). When we examined whether the Helitrons preferentially inserted into or near genes, we discovered that their frequencies within introns and >1 kb from genes were significantly higher than expected (Supplementary Fig. S6), suggesting the silkworm Helitrons preferential insertion into these regions.
3.5. Insertion times and expansion patterns
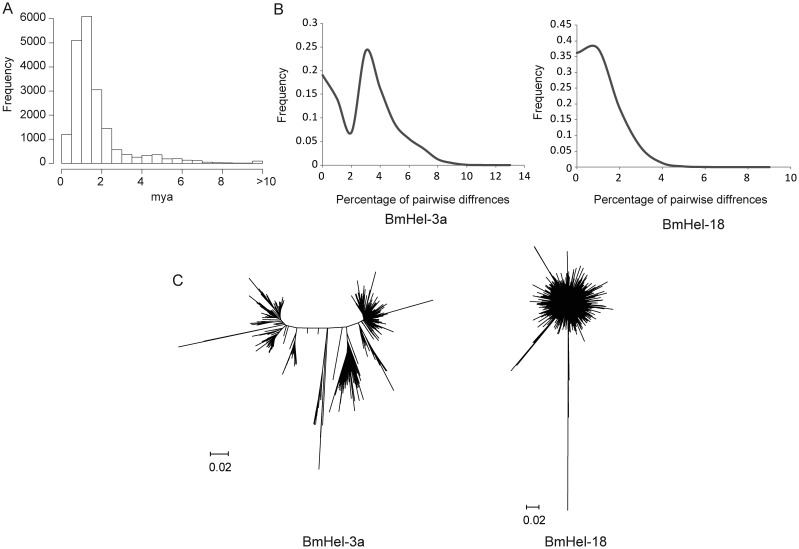
We estimated the age of each intact Helitron by first estimating k between each intact Helitron and its family consensus sequence based on K2P distances.28 The range was 0–0.69, but 15 443 copies (∼79% of the 19 580 copies) had k≤ 0.06. Insertion dates based on these k-values ranged from 0 to >10 mya (Fig. 4A), but most expansion events appeared to have happened within the most recent 2 my (corresponding to k = ∼0.06).
Figure 4.
Evolutionary history of Helitrons in the silkworm. (A) Helitron amplification dates. (B) Distributions of pair-wise nucleotide diversity among full-length elements of BmHel-3a (with a bimodal distribution, suggesting more than one round of amplification) and BmHel-18 (with a unimodal distribution, suggesting one amplification burst). (C) Phylogenetic trees of BmHel-3a (bimodal pair-wise nucleotide diversity and more than one clade) and BmHel-18 (unimodal pair-wise nucleotide diversity and a single clade).
To investigate the history of Helitron expansion in silkworms, pair-wise nucleotide diversities of intact Helitrons were calculated and histograms were drawn for each Helitron family. Most histograms were wave-like (Fig. 4B and Supplementary Fig. S7). These histograms indicated that each family may have experienced a rapid population expansion (burst) during its evolutionary history.35,36 Thirteen families (BmHel-4, 7, 8, 10, 11, 12, 13, 14, 15, 16, 17, 20, and 21) of silkworm Helitrons had unimodal distributions, two (BmHel-2 and 3) had bimodal distributions, and the other six families displayed multimodal distributions (Supplementary Fig. S7), indicating that these Helitron families had experienced one, two, or multiple expansions, respectively.
To further investigate the histories of these Helitron families, phylogenetic trees were reconstructed (Fig. 4C and Supplementary Fig. S8). Families with unimodal histograms formed star-shaped clades, indicating a rapid amplification from a single master element. Those with bi- or multimodal distributions had more than one clade, providing evidence for amplification bursts at different times. Most silkworm Helitron families experienced a single evolutionary radiation.
3.6. Gene fragment acquisition and contribution to transcripts
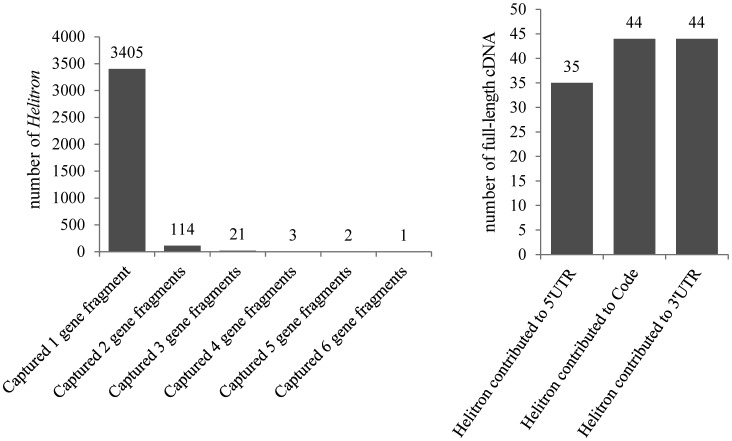
To estimate the numbers of gene fragments captured by silkworm Helitrons, we performed a BLASTX search against the NCBI nr protein database. More than 18% (3546/19 580) of the intact elements captured one or more gene fragments (Fig. 5A). The number of captured gene fragments ranged from one to six. Most intact Helitrons (∼96%) captured no more than one gene fragment. A total of 3724 gene fragments from 268 genes were captured (Supplementary Table S5). Examples of genes captured by Helitrons are shown in Supplementary Fig. S9.
Figure 5.
Silkworm Helitrons within genes. (A) Distribution of the number of gene fragments captured by silkworm Helitrons. (B) Silkworm Helitrons contributed to coding regions and to 5′- and 3′-UTRs of full-length cDNA.
To investigate whether these Helitrons had transcriptional activity, we performed a BLASTN search against the silkworm EST database and discarded parental transcripts. We found that 1317 (∼6.7%) intact Helitrons had transcriptional activity, contributing to 1210 ESTs (Supplementary Table S6). Among these ESTs, five matched the transposases of BmHel-2 (BY927485) and BmHel-3a (BB983132, BY932007, CK528421, and BY916909), while 1205 matched 1317 intact Helitrons (Supplementary Table S6). We could not distinguish between Helitron transcripts and transcripts composed of Helitron fragments plus other genes (the 1205 ESTs), because most ESTs were too short.
To estimate whether the Helitrons contributed to 5′-UTRs, ORFs, and 3′-UTRs of full-length silkworm cDNAs, we performed BLASTN searches and discarded parental and Helitron transposase transcripts. The intact Helitrons contributed to the 5′-UTRs of 35 full-length cDNAs, to the ORFs of 44, and to the 3′-UTRs of 44 (Fig. 5B; Supplementary Table S7–9). These donated fragments contributed to 123 full-length cDNAs, which represented ∼1.4% (123 of 8,654) of the silkworm full-length cDNAs. Examples are shown in Supplementary Fig. S10.
3.7. New Helitron creation through combinations of different Helitrons
By clustering of all silkworm Helitron family consensus sequences with an all-versus-all BLAST search, we found that some distinct Helitrons had merged to form new Helitrons. Three examples are shown in Fig. 6: BmHel-6 was formed from forward-oriented segments of BmHel-7 and BmHel-8; BmHel-9 comprised forward BmHel-7 and reverse BmHel-8 segments; and BmHel-10 united forward segments of BmHel-7 and dBmHel-11.
Figure 6.
Possible mechanism of new Helitron generation through Helitron sequence acquisition and new end creation.
4. Discussion
4.1. Identification and characterization of silkworm Helitrons
TEs in higher eukaryotic genomes are identified in two main ways: homology-based and structure-based methods. Homology-based methods are biased toward detecting previously identified families; its major limitation is that it cannot detect TEs that are very distinct in sequence from known TEs. In contrast, structure-based method use prior knowledge about the common structural features and can effectively identify unique TEs. However, a precondition for this method is that TEs have conserved sequence structures. Homology- and structure-based methods have been developed to identify Helitrons. The homology-based HelitronFinder has been used to identify Helitrons in the maize genome,7 but its applicability to other organisms is limited by the fact that Helitrons vary greatly among organisms. HelSearch is a structure-based programme.11
We developed a new structure-based pipeline to identify Helitrons in the silkworm genome. This method was fast and effective. We used Usearch to cluster sequences with Helitron end structures rather than BLASTALL, which greatly sped the clustering but also generated a consensus sequence for each cluster. In addition, we discarded false-positive matches to non-Helitron TEs when all candidate Helitron sequences were used to BLAST against the known silkworm TEs. A total of 951 such consensus sequences were discarded. Finally, the computational requirements of this method were very low; an ordinary PC machine could complete all of the work. Our pipeline was structure-based like HelSearch, so both methods should have similar efficacy.
Although the silkworm genome is rich in various types of TEs,18 few Helitrons have been identified. We performed a genome-wide structure-based scan for Helitrons and identified 21 Helitron families with a total of 67 555 copies. These Helitron families comprised ∼4.23% of the silkworm genome, a proportion higher than in other organisms. For instance, Helitrons occupied >2% of the genome sequence in C. elegans, >0.5% in frog, ∼3% in the N. vectensis, <0.1% in A. nidulans, ∼1.3% in Arabidopsis thaliana, ∼1.29% in Medicago truncatula, ∼3% in bat, and ∼2% in maize.1,3–8,11 Furthermore, our estimated proportion was much higher than the value of ∼0.1% previously reported for silkworm.18 This discrepancy may be because the previous study used an homology-based search. Helitrons vary greatly among organisms,9,37 so an homology-based search could greatly underestimate Helitron content.
We found that 13 of 21 Helitron families were new, while eight had been previously published in the BmTEdb or RepBase database or in papers.33,34 One silkworm Helitron-like family (Bm_1607) published in the BmTEdb was not identified in this study, because it does not have typical Helitron characteristics, such the 5′-TC dinucleotide and 3′-CTRR motif. These results, together with PCR verification, indicated that our approach was reliable and efficient at identifying Helitrons, but it could not identify structurally atypical Helitron families.
We found that the silkworm genome had 19 580 intact Helitrons, many more than the 1930 intact Helitrons in maize, the 281 in Arabidopsis and C. elegans, the 230 in Medicago, the 651 in rice, and the 608 in sorghum.11 Their abundance and high sequence identities (>90% in 8615 of 19 580 sequences, or ∼44%) in the silkworm genome implied a recent amplification history. Two Helitron families (BmHel-2 and -3) exhibited features of putative autonomous families. BmHel-5 (BmHel1p) was previously identified as putatively autonomous,38 suggesting that autonomous Helitrons may exist in the silkworm. However, BmHel-2 and -5 had very small copy numbers, just 79 and 80 copies, respectively. In contrast, some silkworm Helitron families with many copies did not have features of putative autonomous elements. A possible reason will be discussed below.
4.2. Helitron abundances in other lepidopterans
We analysed Helitron abundances in three recently released lepidopteran genomes using our approach. Helitron abundance varied strikingly among lepidopterans. For instance, there were 10 Helitron families that constituted ∼6.62% of the genome in H. melpomene. This proportion was much larger than the previously reported 5.37%,16 because we identified an additional big Helitron family (HmHel-6) that comprised ∼1.17% of the genome. However, we did not find the low-copy-number families of Helitron-like-2, 8, and 10, because they lacked the 5′-TC dinucleotide and 3′-CTRR motif. Unexpectedly, only one family, representing ∼0.20% of the D. plexippus genome, was identified. Thus, different lepidopteran genomes contained very different numbers of Helitrons, consistent with previous reports that Helitron content was highly variable even among closely-related species. For instance, Helitrons make up 1–5% of genomic DNA in different fruit fly species,9 0–3% in mammals.6,39
Different Helitron abundances could be caused by three factors: (i) different rates of Helitron expansion or deletion in different lineages; (ii) vertical transfer with frequent diversification and extinction; and (iii) horizontal acquisition of Helitrons. For example, there were 21 Helitron families in the silkworm genome, but only one in the D. plexippus genome. These results could be caused by horizontal transfer. A previous study reported that five Helitron families (BmHel-3a, 4, 5, 20, and 21) experienced horizontal transfer in the silkworm.33 Furthermore, we found that an intact homologue of BmHel-16 was also present in the Cotesia sesamiae Kitale bracovirus genome. A phylogenetic analysis indicated that the silkworm BmHel-16 was more closely related to the C. sesamiae bracovirus copy than that to sequences from other Lepidoptera (Supplementary Fig. S8), suggesting horizontal transfer. However, BmHel-16 and the C. sesamiae bracovirus sequence had only 79% identity, implying that the horizontal transfer happened long ago.
4.3. Distribution of Helitrons on chromosomes
Previous studies indicated that Helitrons preferentially insert into gene-poor regions.11 For instance, Helitrons in Arabidopsis were rich in pericentromeric regions. Similarly, Helitrons in the C. elegans genome were most abundant in the terminal regions of each chromosome, which are often in the heterochromatin state. However, the silkworm Helitrons were randomly distributed on chromosomes. If Helitrons generally insert into heterochromatin regions, their random distribution in the silkworm genome is expected, because silkworm chromosomes are holocentromeres.
We also found that the numbers of Helitrons that inserted into introns and >1 kb away from genes were higher than expected. The reasons for this observation are not clear. A previous study proved that genes captured by Helitrons reshuffled the transcriptome of maize.12 Thus, preferential accumulation in intron regions could drive gene evolution through gene capture and exonization of Helitrons.
4.4. Massive expansions and diversity patterns
Our results indicated that major expansion events of silkworm Helitrons occurred in the past 2 my (Fig. 4A). Similarly, about 87% of BaShos insertions occurred within the most recent 5 my in A. thaliana and ∼71% of Hel1-105 elements and 69% of Hel-106 elements inserted within the past 1 my in maize.7,40
To investigate history of the silkworm Helitron amplifications, we estimated pair-wise nucleotide diversity and phylogenetic trees of the full-length Helitrons (Fig. 4B and C). Most Helitron families experienced single expansions. However, the mechanism was not clear. As discussed above, we did not find putative autonomous Helitron copies in some high-copy-number families (e.g. BmHel-8, 11, and 15). In contrast, some low-copy-number families (BmHel-2 and 5) appeared to be putative autonomous elements. These results were reminiscent of miniature inverted-repeat TEs (MITEs) that were highly transposable because of transposases encoded by distantly related and self-restrained autonomous elements in rice,41 a mechanism known as cross-mobilization. Thus, some non-autonomous Helitrons might move using transposases encoded by autonomous Helitrons. Whether this is true in silkworm Helitrons remains to be investigated.
Why some silkworm Helitrons experienced bursts of expansion is not clear. Most Helitrons probably remain inactive for most of their evolutionary histories, and they may be suddenly activated by ‘genome shock’, as observed in rice MITEs. For instance, mPing is known to be activated by irradiation, cell culture, and recent domestication.42–44 The silkworm was domesticated from wild Chinese silkworms about 5000 years ago.45,46 Whether some silkworm Helitrons were activated by domestication is an interesting question.
4.5. Gene fragments acquisition and contribution to transcripts
Helitrons vary greatly in sequence length, even within a family. One explanation is that these elements capture different gene fragments.7,8,12,47 Although some molecular mechanisms for gene capture have been proposed,48–51 clear experimental evidence for a particular mechanism is lacking. In this study, we found that 3546 intact Helitrons (>18% of all intact Helitrons) had captured one or more gene fragments, for a total of 3724 captured fragments. The average number of captured gene fragments per intact Helitron was 1.08, similar to the value (1.81) for maize Helitrons.8
Furthermore, we found that ∼6.7% of intact silkworm Helitrons (1317 of 19 580) had EST evidence. Based on a homology search against silkworm full-length cDNAs, we found that these intact Helitrons contributed to about 123 full-length cDNAs (∼1.4% of the published total) by donating one or more exons. A recent study suggested that ∼9% of maize Helitrons had EST evidence and could generate abundant transcripts through alternative splicing.12 Thus, Helitrons may play important roles in the evolution of silkworm transcripts.
4.6. Generation of new Helitrons
Previous studies indicated that Helitrons could acquire new sequences by recognizing either a new 3′ termination site or a new 5′ start site.11,52 A hairpin was proposed to serve as a stop signal during Helitron transposition. When this hairpin is destroyed by unknown mechanism, a new hairpin-like sequence could be acquired, perhaps from nearby Helitrons, to generate chimeric elements.11 Interestingly, we found that two Helitrons could combine to produce a new Helitron (Fig. 6). Similarly, in maize, Helitron_mc2 was composed of ZmHelA5 and Helitron_mc.52 Thus, new Helitrons can be generated in different ways, making them the most diverse class of transposons.
5. Conclusions
In present study, we developed a structure-based approach to identify Helitrons in a genome and analysed their presence in four Lepidoptera species. Helitron abundance and the number of families varied greatly among these insect genomes. One plausible explanation is that horizontal transfer caused these differences. A systematic analysis of silkworm Helitrons revealed that they accounted for ∼4.23% of the genome, much more than the previously reported ∼0.1%.18 A total of 21 Helitron families were identified in the silkworm, and 13 were new families. Most Helitron families expanded within the past 2 my in a single radiation. Furthermore, we found that Helitrons contributed to at least 1.4% of silkworm full-length cDNAs, indicating their important roles in the evolution of the silkworm genes. In addition, existent Helitrons could generate new families by combining. Our results provided insights into the generation and evolution of Helitron transposons as well as their contribution to transcripts.
Authors' contribution
Z.Z. and M.J.H. designed the study. Y.H.S., M.S.X., H.Y.L., H.H.Z., and M.J.H. analysed the data. Z.Z. provided the platform for analysis. Z.Z. and M.J.H. drafted and revised the manuscript. All authors read and approved the final manuscript.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the Hi-Tech Research and Development (863) Program of China (2013AA102507), a grant from Natural Science Foundation Project of CQ CSTC (cstc2012jjB80007), and the Doctorial Innovation Fund of Southwest University (kb2010016).
Supplementary Material
Acknowledgements
We are grateful to two anonymous reviewers for their valuable comments and critiques, which helped us to greatly improve the paper. We thank Dr Fang-Yin Dai for help in collecting domesticated silkworm samples and the members of Z.Z.'s laboratory group for their help with data analysis and helpful discussions.
Footnotes
Edited by Dr Takashi Ito
References
- 1.Kapitonov V.V., Jurka J. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA. 2001;98:8714–19. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendiola M.V., Bernales I., de la Cruz F. Differential roles of the transposon termini in IS91 transposition. Proc. Natl. Acad. Sci. USA. 1994;91:1922–26. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapitonov V.V. Helitron-N1_XT, a family of non-autonomous Helitron from frog. Repbase Reports. 2006;6:494–6. [Google Scholar]
- 4.Putnam N.H., Srivastava M., Hellsten U., et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 5.Galagan J.E., Calvo S.E., Cuomo C., et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–15. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 6.Pritham E.J., Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc. Natl Acad. Sci. USA. 2007;104:1895–900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du C., Fefelova N., Caronna J., He L., Dooner H.K. The polychromatic Helitron landscape of the maize genome. Proc. Natl Acad. Sci. USA. 2009;106:19916–21. doi: 10.1073/pnas.0904742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L., Bennetzen J.L. Distribution, diversity, evolution, and survival of Helitrons in the maize genome. Proc. Natl Acad. Sci. USA. 2009;106:19922–7. doi: 10.1073/pnas.0908008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapitonov V.V., Jurka J. Helitrons in fruit flies. Repbase Reports. 2007;7:127–32. [Google Scholar]
- 10.Zuccolo A., Sebastian A., Talag J., et al. Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol. Biol. 2007;7:152. doi: 10.1186/1471-2148-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., Bennetzen J.L. Structure-based discovery and description of plant and animal Helitrons. Proc. Natl Acad. Sci. USA. 2009;106:12832–7. doi: 10.1073/pnas.0905563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbaglia A.M., Klusman K.M., Higgins J., Shaw J.R., Hannah L.C., Lal S.K. Gene capture by Helitron transposons reshuffles the transcriptome of maize. Genetics. 2012;190:965–75. doi: 10.1534/genetics.111.136176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A., Rafalski A. Gene duplication and exon shuffling by Helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 14.Xia Q., Zhou Z., Lu C., et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–40. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 15.Zhan S., Merlin C., Boore J.L., Reppert S.M. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–85. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium T.H.G. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–8. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimaldi D., Engel M.S. Evolution of the insects. New York: Cambridge University Press; 2005. [Google Scholar]
- 18.Osanai-Futahashi M., Suetsugu Y., Mita K., Fujiwara H. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect. Biochem. Mol. Biol. 2008;38:1046–57. doi: 10.1016/j.ibmb.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 20.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia X., Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 2001;92:371–3. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 22.Markham N.R., Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 23.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–6. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements , Cytogenet. Genome Res. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 25.Nagaraja G.M., Nagaraju J. Genome fingerprinting of the silkworm, Bombyx mori, using random arbitrary primers , Electrophoresis. 1995;16:1633–8. doi: 10.1002/elps.11501601270. [DOI] [PubMed] [Google Scholar]
- 26.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J. Mol. Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 27.Li W.-H. Molecular evolution. Sunderland, Massachusetts: Sinauer Associates; 1997. [Google Scholar]
- 28.Xia Q., Guo Y., Zhang Z., et al. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx) Science. 2009;326:433–6. doi: 10.1126/science.1176620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 30.Excoffier L., Laval G., Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 31.Duan J., Li R., Cheng D., et al. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010;38:453–6. doi: 10.1093/nar/gkp801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han M.J., Shen Y.H., Gao Y.H., Chen L.Y., Xiang Z.H., Zhang Z. Burst expansion, distribution and diversification of MITEs in the silkworm genome. BMC genomics. 2010;11:520. doi: 10.1186/1471-2164-11-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas J., Schaack S., Pritham E.J. Pervasive horizontal transfer of rolling-circle transposons among animals. Genome Biol. Evol. 2010;2:656–64. doi: 10.1093/gbe/evq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coates B.S., Hellmich R.L., Grant D.M., Abel C.A. Mobilizing the genome of Lepidoptera through novel sequence gains and end creation by non-autonomous Lep1 Helitrons. DNA Res. 2012;19:11–21. doi: 10.1093/dnares/dsr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zerjal T., Joets J., Alix K., Grandbastien M.A., Tenaillon M.I. Contrasting evolutionary patterns and target specificities among three Tourist-like MITE families in the maize genome. Plant Mol. Biol. 2009;71:99–114. doi: 10.1007/s11103-009-9511-0. [DOI] [PubMed] [Google Scholar]
- 36.Rogers A.R., Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992;9:552–69. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 37.Sweredoski M., DeRose-Wilson L., Gaut B.S. A comparative computational analysis of nonautonomous Helitron elements between maize and rice. BMC genomics. 2008;9:467. doi: 10.1186/1471-2164-9-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coates B.S., Sumerford D.V., Hellmich R.L., Lewis L.C. A Helitron-Like Transposon Superfamily from Lepidoptera Disrupts (GAAA)(n) microsatellites and is responsible for flanking sequence similarity within a microsatellite family. J. Mol. Evol. 2010;70:275–88. doi: 10.1007/s00239-010-9330-6. [DOI] [PubMed] [Google Scholar]
- 39.Smit A.F.A. 2006. Repeat Masker edition, Release 20061006, Repbase Update.
- 40.Hollister J.D., Gaut B.S. Population and evolutionary dynamics of Helitron transposable elements in Arabidopsis thaliana. Mol. Biol. Evol. 2007;24:2515–24. doi: 10.1093/molbev/msm197. [DOI] [PubMed] [Google Scholar]
- 41.Yang G., Nagel D.H., Feschotte C., Hancock C.N., Wessler S.R. Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science. 2009;325:1391–4. doi: 10.1126/science.1175688. [DOI] [PubMed] [Google Scholar]
- 42.Jiang N., Bao Z., Zhang X., et al. An active DNA transposon family in rice. Nature. 2003;421:163–7. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 43.Nakazaki T., Okumoto Y., Horibata A., et al. Mobilization of a transposon in the rice genome. Nature. 2003;421:170–2. doi: 10.1038/nature01219. [DOI] [PubMed] [Google Scholar]
- 44.Naito K., Cho E., Yang G.J., et al. Dramatic amplification of a rice transposable element during recent domestication. Proc. Natl Acad. Sci. USA. 2006;103:17620–5. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshitake N. Phylogenetic aspects on the origin of Japanese race of the silkworm, Bombyx mori L. J. Sericol. Sci. Jpn. 1968;37:83–7. [Google Scholar]
- 46.Sun W., Yu H., Shen Y., Banno Y., Xiang Z., Zhang Z. Phylogeny and evolutionary history of the silkworm. Sci. China Life Sci. 2012;55:483–96. doi: 10.1007/s11427-012-4334-7. [DOI] [PubMed] [Google Scholar]
- 47.Feschotte C., Pritham E.J. A cornucopia of Helitrons shapes the maize genome. Proc. Natl. Acad. Sci. USA. 2009;106:19747–8. doi: 10.1073/pnas.0910273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feschotte C., Wessler S.R. Treasures in the attic: rolling circle transposons discovered in eukaryotic genomes. Proc. Natl Acad. Sci. USA. 2001;98:8923–4. doi: 10.1073/pnas.171326198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennetzen J.L. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 2005;15:621–7. doi: 10.1016/j.gde.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 50.Brunner S., Pea G., Rafalski A. Origins, genetic organization and transcription of a family of non-autonomous Helitron elements in maize. Plant J. 2005;43:799–810. doi: 10.1111/j.1365-313X.2005.02497.x. [DOI] [PubMed] [Google Scholar]
- 51.Lal S., Oetjens M., Hannah L.C. Helitrons: Enigmatic abductors and mobilizers of host genome sequences. Plant Sci. 2009;176:181–6. [Google Scholar]
- 52.Dong Y., Lu X., Song W., et al. Structural characterization of Helitrons and their stepwise capturing of gene fragments in the maize genome. BMC Genomics. 2011;12:609. doi: 10.1186/1471-2164-12-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.