Abstract
Small non-coding RNAs, microRNAs (miRNA), inhibit the translation or accelerate the degradation of message RNA (mRNA) by targeting the 3′-untranslated region (3′-UTR) in regulating growth and survival through gene suppression. Deregulated miRNA expression contributes to disease progression in several cancers types, including pancreatic cancers (PaCa). PaCa tissues and cells exhibit decreased miRNA, elevated cyclooxygenase (COX)-2 and increased prostaglandin E2 (PGE2) resulting in increased cancer growth and metastases. Human PaCa cell lines were used to demonstrate that restoration of miRNA-143 (miR-143) regulates COX-2 and inhibits cell proliferation. miR-143 were detected at fold levels of 0.41 ± 0.06 in AsPC-1, 0.20 ± 0.05 in Capan-2 and 0.10 ± 0.02 in MIA PaCa-2. miR-143 was not detected in BxPC-3, HPAF-II and Panc-1 which correlated with elevated mitogen-activated kinase (MAPK) and MAPK kinase (MEK) activation. Treatment with 10 μM of MEK inhibitor U0126 or PD98059 increased miR-143, respectively, by 187 ± 18 and 152 ± 26 fold in BxPC-3 and 182 ± 7 and 136 ± 9 fold in HPAF-II. miR-143 transfection diminished COX-2 mRNA stability at 60 min by 2.6 ± 0.3 fold in BxPC-3 and 2.5 ± 0.2 fold in HPAF-II. COX-2 expression and cellular proliferation in BxPC-3 and HPAF-II inversely correlated with increasing miR-143. PGE2 levels decreased by 39.3 ± 5.0 % in BxPC-3 and 48.0 ± 3.0 % in HPAF-II transfected with miR-143. Restoration of miR-143 in PaCa cells suppressed of COX-2, PGE2, cellular proliferation and MEK/MAPK activation, implicating this pathway in regulating miR-143 expression.
Keywords: miR-143, cyclooxygenase-2, MAPK, prostaglandin E2, pancreatic cancer
1. Introduction
Small non-coding RNAs, microRNAs (miRNAs), target the 3′-untranslated region (3′-UTR) of message RNA (mRNA) to repress translation or accelerate the degradation of the mRNA in regulating gene expression [1,2]. Functional studies have demonstrated the involvement of miRNAs in diverse biological process, including cellular development, differentiation and proliferation [2,3]. Several cancers types exhibit deregulated miRNA expression resulting in disease progression, including pancreatic cancers (PaCa) [4]. miRNAs serve as oncogenes or tumor suppressors by modulating the level of critical proteins [5]. As a tumor suppressor, miRNAs has been reported to regulate aberrant cyclooxygenase (COX) expression in several cancers [6,7].
COX exists as two isoforms, a constitutive COX-1 and an inducible COX-2, and is critical in prostaglandin E2 (PGE2) synthesis [8]. COX-2 and PGE2 mediates acute inflammation and in excess, promotes growth and survival of cancer [9,10]. Induction of COX-2 results from a variety of stimuli (cytokines, peptides, growth factors) and controlled at different levels by many signaling pathways [11]. Interestingly, PGE2 induces COX-2 expression through the prostanoid receptor EP2, with the subsequent activation of cyclic AMP response element binding protein (CREB) pathway that further increases PGE2 production [12]. Restoration of miRNA expression has been shown to abrogate COX-2 expression in cancer cells [13]. Specifically, miR-143 has been identified to target COX-2 resulting in decreased cancer cell growth and metastases [14,15].
We previously reported increased COX-2 and PGE2 contributes to growth and survival of both human PaCa tissue and cell lines [16,17]. Furthermore, PaCa exhibiting active K-Ras mutations [18,19] can suppress miR-143 repression of Ras responsive element binding protein-1 (RREB1) [20,21]. This study focuses on whether restoration of miR-143 expression can regulate COX-2 expression and inhibit PaCa cell proliferation. BxPC-3 and HPAF-II transfected with miR-143 resulted in diminished COX-2 mRNA and COX-2 protein expression. Loss of COX-2 corresponded with a decrease in PGE2 synthesis and cellular proliferation in both cell lines. This report demonstrates that the down-regulation of COX-2 expression by miR-143 results in part from the destabilization of COX-2 mRNA and inhibition of RREB1 activation through MEK/MAPK suppression.
2. Material and methods
2.1 Reagents
Actinomycin D, arachidonic acid, fetal bovine serum (FBS), dimethyl sulfoxide (DMSO), chloroform and methanol were purchased from Sigma (St. Louis, MO). COX-2 antibody and COX-2 specific inhibitor N-[2-(cyclohexyloxy)-4-nitrophenyl]-methanesulfonamide (NS-398) was purchased from Cayman Chemical (Ann Arbor, MI). CREB, p-CREB, MEK, p-MEK, MAPK, p-MAPK, p-p38MAPK and RREB1 antibodies were purchased from Cell Signaling (Danvers, MA). GAPDH antibody was acquired from Santa Cruz Biotechnology (Santa Cruz, CA). MEK inhibitors U0126 (1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio] butadiene) and PD98059 (2′-amino-3′-methoxyflavone) were acquired from Calbiochem (EMD Millipore, Billerica, MA). Horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG and enhanced chemiluminescence (ECL) reagents were obtained from (ThermoFisher Scientific, Pittsburg, PA).
2.2. Cell culture
Human PaCa cell lines AsPC-1 (CRL-1682), BxPC-3 (CRL-1687), Capan-2 (HTB-80), HPAF-II (CRL-1997), MIA PaCa-2 (CRL-1420) and Panc-1 (CRL-1469) were acquired from American Type Culture Collection (Rockville, MD). AsPC-1, BxPC-3 and HPAF-II were propagated in RPMI 1640 medium supplemented with 10% FBS and 1 % PSG antibiotic mix (100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine) (Life Technologies) at 5 % CO2 and 37 °C. Capan-2, MIA PaCa-2 and Panc-1 were propagated in DMEM supplemented with 10% FBS and 1 % PSG antibiotic mix at 10 % CO2 and 37 °C. For experiments, cells were propagated in 100-mm tissue culture dishes to confluence (5–7 days) and arrested in serum-free medium.
2.3. RNA extraction
RNA was extracted with 1 ml of Trizol reagent (Life Technologies) and 0.2 ml of chloroform at 12,000 × g for 15 min at 4 °C, precipitated with 0.5 ml of 2-propanol at 12,000 × g for 10 min at 4 °C, washed with 75% ethanol at 7,500 × g for 5 min at 4 °C, dissolved in 30 μL of RNA Storage Solution containing 1 mM sodium citrate, pH 6.4 (Life Technologies) and stored at −20 °C for subsequent analysis. RNA concentration was quantified at dual wavelengths of 260 and 280 nm on a Bio-Rad spectrophotometer (Hercules, CA).
2.4. In silico miR-143 sequence verification
Targets of miR-143 were identified using TargetScan (targetscan.org) and miRanda (microRNA.org) on-line database search. COX-2 was identified as a potential target with predicted binding of hsa-miR-143 sequence 3′-GUAGAG-5′ to COX-2 3′-UTR sequence 3′-CAUCUC-5′.
2.5. miR-143 detection
Small RNAs tagged with poly A and converted to cDNA (QuantiMir, System Biosciences, Mountain View, CA) were quantified using SYBR green kit (Bio-Rad). Primers used were miR-143 (Accession MIMAT0000435) forward sequence: 5′-TGA GAT GAA GCA CTG TAG CTC-3′ and control U6 snRNA forward sequence: 5′-CGC AAG GAT GAC ACG CAA ATT C-3′ supplied by QuantiMir. Thermal cycling conditions were 95 °C for 3 min of activation, 40 cycles of 95 °C for 15 sec denaturing and 55 °C for 60 sec of annealing/extending and 55 °C – 95 °C at 0.5 °C increments for 5 sec of melt curve analysis. Ct values were normalized to U6 control and reaction mixtures were resolved on a 1.5 % agarose gel at 90V for 60 min.
2.6. miR-143 transfection
BxPC-3 and HPAF-II were seeded at 400,000 cells/well in 6-well plates, allowed to stabilize overnight in RPMI complete media, and replenished with RPMI supplemented with 5 % FBS. Cells were treated with 0, 25, 50 and 100 nM of miR-143 (C-300611-05-0005, ThermoFisher Scientific) delivered in 10 μL of Lipofectamine 2000 (Life Technologies) per well, incubated for 24 hrs, replenished with complete RPMI and harvested for RNA or protein after 72 hrs.
2.7. COX-2 mRNA by real time PCR
RNA extracts were reverse transcribed and cDNAs amplified using TaqMan Gold RT-PCR kit (Applied Biosystems, Foster City, CA). COX-2 (accession NM_000963) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control were quantified by real-time PCR analysis using a Bio-Rad IQ5. COX-2 primer used was sense 5′-GCT TTA TGC TGA AGC CCT ATG A-3′ and antisense 5′-TCC AAC TCT GCA GAC ATT TCC-3′ with corresponding universal probe 2 (Cat. 04684982001, Roche, Indianapolis, IN). Human GAPDH primer and probe set was acquired from Applied Biosystems. Thermal cycling conditions were 48 °C for 15 min of activation, 95 °C for 10 min of amplification and 40 cycles of 95 °C at 15 sec denaturing and 60 °C at 60 seconds annealing/extending.
2.8. Protein expression/Western blotting
Protein were harvested using RIPA lysis buffer (ThermoFisher Scientific), diluted with 2X LDS buffer containing SDS (Life Technologies) and denatured at 95 °C for 10 min. Protein lysates were subjected to a variable 4 - 20% Precise Tris-Glycine gel (ThermoFisher Scientific) for 45 min at 200 V and transferred onto a nitrocellulose membrane for 75 min at 100 V. Membranes were washed with Tris buffered saline (TBS, Sigma), blocked with 5% dried non-fat milk (Bio-Rad) in 1% tween-TBS and probed with antibody raised against p-CREB (1:1000), p-MEK (1:1000), p-MAPK (1:1000), RREB1 (1:1000), p-p38MAPK (1:1000) or COX-2 (1:1000) with GAPDH (1:2500) as a visual loading control. Bands were visualized by secondary antibody IgG-linked horseradish peroxidase conjugate (1:2500) and ECL and quantified using ChemiDoc XRS imaging software (Bio-Rad).
2.9. PGE2 assay
PGE2 from serum-starved, confluent cells cultured in 24-well plates challenged with 5 μM of arachidonic acid stabilized with fatty acid-free BSA (Sigma) were quantified using Prostaglandin E2 Express EIA kit and 6-keto Prostaglandin F1α EIA kit (Cayman Chemical). PGE2 absorbance were measured at 405 – 420 nm and normalized to protein absorbance readings at 595 nm using the Bradford assay (Bio-Rad).
2.10. Cell proliferation assay
BxPC-3 and HPAF-II cells were seeded at 7,500 cells/well in 96-well plates and allowed to stabilize in RPMI complete media. After 24 hrs, the cells were treated with 0, 5, 10 or 20 μM of U0126 in RPMI media supplemented with 0.5 % FBS or transfected with 0, 25, 50 or 100 nM of miR-143. The cells were maintained for 48 hrs, incubated with 10 % 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma) for 4 hrs, aspirated and dissolved in DMSO for formazan product. Absorbance was measured at 560 nm with reference wavelength at 700 nm.
2.11. Statistical analyses
Data are expressed as means ± SEM from triplicate experiments. Means comparisons were performed using two-way ANOVA using Sigma Plot (SPSS, Chicago, IL) followed with a Bonferroni post-test for multiple pair-wise comparisons. Statistical significance was set at p< 0.05.
3. Results
3.1. Human PaCa cell lines express low levels of miR-143
Human PaCa cell lines AsPC-1, BxPC-3, Capan-2, HPAF-II, MIA PaCa-2 and Panc-1 were screened for miR-143 expression shown in Fig. 1A. AsPC-1, Capan-2 and MIA PaCa-2 expressed miR-143 fold levels of 0.41 ± 0.06, 0.20 ± 0.05 and 0.10 ± 0.02, respectively, whereas BxPC-3, HPAF-II and Panc-1 did not express miR-143. Since activation of Ras signaling reportedly regulates miR-143 expression [20,21], MAPK pathway activation were confirmed in Fig. 1B. BxPC-3, HPAF-II and Panc-1 exhibited elevated p-MEK, p-MAPK and RREB1 whereas AsPC-1, Capan-2 and MIA PaCa-2 expressing miR-143 exhibited diminished p-MEK, p-MAPK and RREB1. As a marker for protein kinase A (PKA) pathway activation, CREB activation did not correlate with miR-143 expression, suggesting a minimal involvement of PKA pathway in miR-143 expression.
Figure 1.
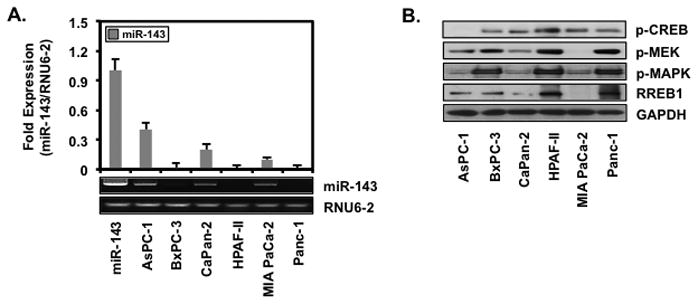
Human PaCa cell lines express low levels of miR-143
AsPC-1, BxPC-3, Capan-2, HPAF-II, MIA PaCa-2 and Panc-1 were (A) screened for miR-143 by RT-PCR and (B) Western blotted for p-CREB, p-MEK, p-MAPK, RREB1 and GAPDH.
3.2. MEK inhibitors U0126 and PD98059 increase miR-143
BxPC-3 and HPAF-II lacking miR-143 were treated with or without 10 μM of MEK inhibitors U0126 or PD98059 to substantiate the involvement of MAPK pathway in miR-143 expression. U0126 and PD98059 increased miR-143, respectively, by 187 ± 18 and 152 ± 26 fold in BxPC-3 (Fig. 2A) and 182 ± 7 and 136 ± 9 fold in HPAF-II (Fig. 2B). Since MAPK pathway activation regulates COX-2 followed with a subsequent increase in PGE2 [22], these cells were treated with 10 μM of U0126 and 100 nM of PGE2 to determine if PGE2 could reverse the induction of miR-143 expression by MAPK inhibition. PGE2 negated miR-143 expression in BxPC-3 and HPAF-II treated with U0126, indicating the involvement of COX-2 and PGE2 in miR-143 expression.
Figure 2.
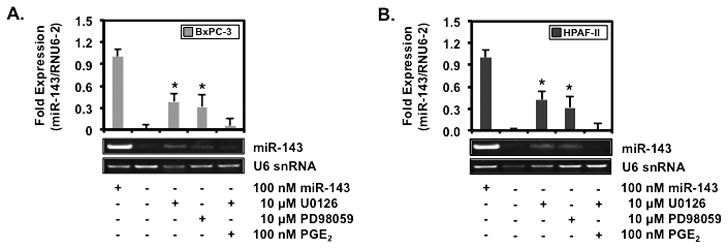
MEK inhibitors U0126 and PD98059 increase miR-143
(A) BxPC-3 and (B) HPAF-II treated with or without 10 μM of MEK inhibitor U0126, 10 μM of MEK inhibitor PD98059 or 0.1 μM of PGE2 for 6 hrs were screened for miR-143 and RNU6-2 by RT-PCR and visualized on a 1.5 % agarose gel. Ct values standardized to RNU6-2 are represented as mean fold expression ± SEM with statistical significance at p<0.05 as indicated by the asterisk (*).
3.3. miR-143 decreases COX-2 mRNA stability and inhibits p-MEK/p-MAPK/RREB1 and COX-2
To demonstrate COX-2 regulation by miR-143, BxPC-3 and HPAF-II transfected with miR-143 or control RNA were treated with 1 μM of actinomycin D to abolish de novo RNA synthesis and monitored for COX-2 mRNA expression shown in Fig. 3A and 3B, respectively. miR-143 destabilized COX-2 mRNA significantly at 60 min by 2.6 ± 0.3 fold in BxPC-3 and 2.5 ± 0.2 fold in HPAF-II compared to their control RNA counterparts. BxPC-3 and HPAF-II cells transfected with increasing concentrations of miR-143 were screened for COX-2 in verifying that accelerated destruction of mRNA decreases protein expression (Fig. 3C). COX-2 decreased with increasing concentration of miR-143 in both BxPC-3 and HPAF-II cells. Furthermore, decreased COX-2 paralleled decreased p38MAPK, MEK and MAPK activation and RREB1 expression, supporting miR-143 regulation of the MAPK pathway and COX-2 in PaCa cells.
Figure 3.
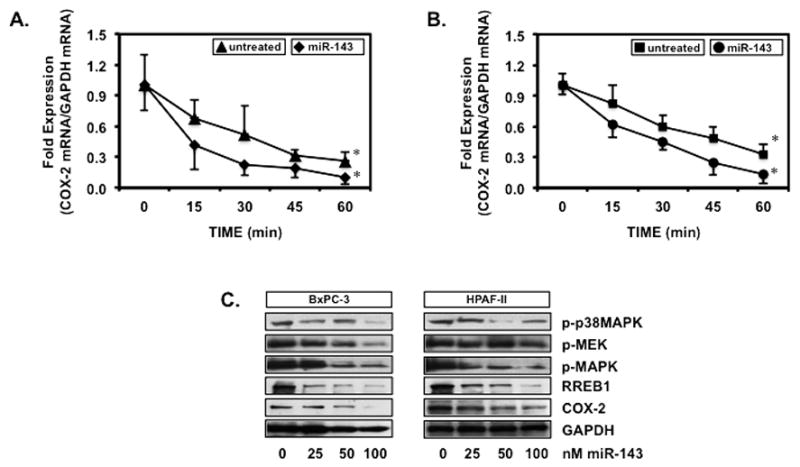
miR-143 decreases COX-2 mRNA stability and inhibits p-MEK/p-MAPK/RREB1 and COX-2
(A) BxPC-3 and (B) HPAF-II transfected with miR-143 or control RNA and dosed with 1 μM of actinomycin D were screened for COX-2 and GAPDH mRNA expression. Ct values standardized to GAPDH are represented as mean fold expression ± SEM with statistical significance at p<0.05 as indicated by the asterisk (*). (C) BxPC-3 and HPAF-II transfected with 100 nM of miR-143 were Western blotted for p-p38MAPK, p-MEK, p-MAPK, RREB1, COX-2 and GAPDH. Protein bands visualized by ECL were quantified using digital imaging software.
3.4. MEK inhibitors and miR-143 decreases PGE2 levels and inhibit cell proliferation
To corroborate that miR-143 destabilization of COX-2 mRNA resulting in decreased COX-2 reduces PGE2, BxPC-3 and HPAF-II transfected with 100 nM of random control RNA or miR-143 were challenged with 5 μM of arachidonic acid and assayed for PGE2 (Fig. 4A). miR-143 inhibited PGE2 synthesis by 39.3 ± 5.0 % in BxPC-3 and 48.0 ± 3.0 % in HPAF-II. In addition, BxPC-3 treated with 10 μM of PD98059 or 10 μM of U0126 resulted in a 57.1 ± 3.5 % and 46.1 ± 6.9 % inhibition of PGE2 synthesis, respectively. Similarly, 10 μM of PD98059 or 10 μM of U0126 inhibited PGE2 synthesis by 51.3 ± 5.8 % and 42.8 ± 5.3 % in HPAF-II cells. Because the MAPK pathway mediates cell proliferation, BxPC-3 and HPAF-II were treated with U0126 or miR-143 and assayed for proliferation shown in Fig. 4B and 4C, respectively. Increasing concentrations of U0126 suppressed BxPC-3 proliferation by 59.4 ± 6.1 % at 20 μM whereas HPAF-II proliferation was inhibited by 45.2 ± 2.8 % at 10 μM and 76.4 ± 2.3 % at 20 μM. Similarly, miR-143 inhibited BxPC-3 proliferation by 43.3 ± 6.5 % at 20 μM and HPAF-II proliferation by 36.1 ± 1.7 % at 10 μM and 47.2 ± 2.8 % at 20 μM.
Figure 4.
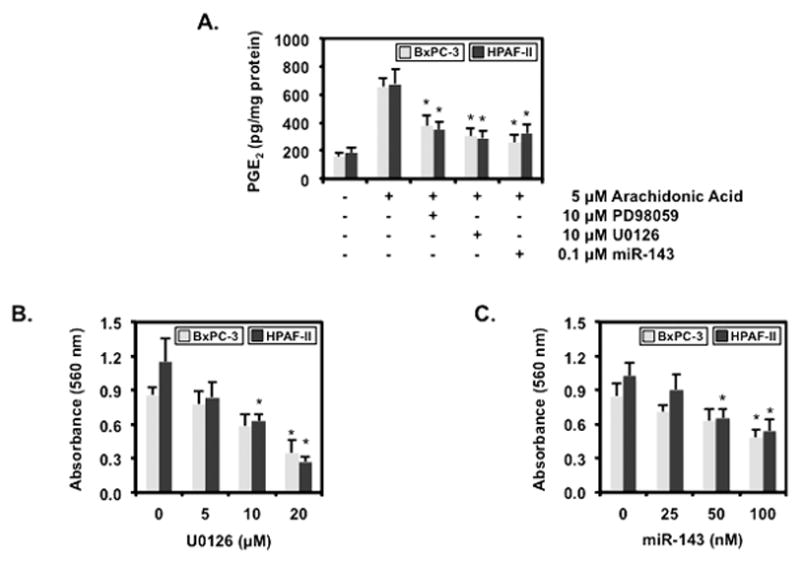
MEK inhibitors and miR-143 decreases PGE2 levels and inhibit cell proliferation
(A) BxPC-3 and HPAF-II transfected with 100 nM of random RNA or 100 nM of miR-143 and pre-treated with 10 μM of PD98059 or 10 μM of U0126 for 1 hr followed by treatment with 5 μM of arachidonic acid for 30 min were analyzed for PGE2 by ELISA. Data are expressed as mean ± SEM and a statistical significance of p<0.05 is indicated by the asterisk (*). BxPC-3 and HPAF-II (B) treated with 0, 5, 10 or 20 μM of U0126 or (C) transfected with 0, 25, 50 or 100 nM of miR-143 were assayed for proliferation by MTT. Data are expressed as mean ± SEM and a statistical significance of p<0.05 is indicated by the asterisk (*).
4. Discussion
Overall, these findings demonstrate that restoration of diminished miR-143 expression in PaCa cells results in the suppression of COX-2 expression and cellular proliferation inhibition that is, in part, mediated through the MAPK pathway. Decreased miR-143 expression in human PaCa cell lines was consistent with a previous report [23] and also observed in bladder and colon cancer [14,24]. Additionally, the absence of miR-143 correlated with a potentiated activation of MAPK signaling in the PaCa cells. Blocking MAPK pathway with UO126 or PD98059 inhibitors increased miR-143 expression whereas restoration of miR-143 levels decreased activation of p38MAPK, MEK, MAPK and RREB1 expression, demonstrating the involvement of MAPK in regulating miR-143 expression.
In several cell types, activation of MAPK cascade up-regulates COX-2 at the transcriptional and post-transcriptional level [25,26]. At the transcriptional level, Ets-1 and Elk-1 positively regulate COX-2 in RINm5F pancreatic β-cells [27]. At the post-transcriptional level, activation of the p38MAPK signaling cascade stabilizes COX-2 mRNA at the 3′-UTR [28]. COX-2 3′-UTR contains multiple copies of AU-rich element (ARE) and miRNA response element (MRE) motifs which, when bound by specific ARE-binding factors or miRNAs, influence COX-2 stability and translational efficiency [29]. Binding of miRNAs to the 3′-UTR can mediate the degradation or inhibit the translation of mRNA as reported with miR-101 inhibition of COX-2 translation and miR-16 inhibition of COX-2 expression by COX-2 3′-UTR mediated mRNA decay in colon cancer cells [5,6,30]. The restoration miR-143 in PaCa cells decreased COX-2 mRNA stability, COX-2 protein expression and subsequent decrease PGE2 levels. Alternatively, PaCa cells treated with PGE2 inhibited the induction of miR-143 expression, validating the involvement of COX-2 and PGE2 in regulating miR-143 expression.
Furthermore, bioinformatic SILAC sequence analysis of miR-143 identified 93 putative protein targets coupled with transcriptional profiling confirmed that many of these targets are regulated through translational inhibition without affecting the mRNA levels [31,32]. Among the miR-143 targets verified were COX-2 in bladder [14], K-Ras in colon [19] and ERK5 in prostate and colon [33]. The small GTPase protein K-Ras is associated with high-frequency somatic mutations that render it constitutively active, resulting in persistent signaling of downstream effectors [34,35]. K-Ras mutations mediate migratory and invasive properties in colon cancer through regulating Rho GTPase activity. GDP exchange factors GEF1 and GEF40 activate Rho catalyzing the exchange of GDP to GTP and miR-143 regulates GEF1 and GEF40 expression, thereby mediating Rho activation. In addition, K-Ras can activate RREB1 [36] that directly inhibits the transcription of the miR-143 and miR-145 cluster, resulting in the enhance Ras signaling. A feed-forward mechanism has been proposed for the repression of miR-143/miR-145 expression and activation of K-Ras and RREB1 [21,37,38]. These targets of miR-143 coincide in promoting cell growth and proliferation through the MAPK pathway. Indeed, restoration of miR-143 levels in PaCa cells decreased the activation of MEK, MAPK and RREB1 and the subsequent arrest of cancer cell proliferation. We demonstrated that miR-143 regulates PGE2 production and PGE2-mediated cellular proliferation by modulating COX-2 mRNA stability and COX-2 expression.
Since PGE2 can suppress miR-143 expression and effects, we speculate that PGE2 regulates miR-143 expression presumably through EP receptor binding and subsequent MAPK pathway activation. Our findings suggest a possible role for miR-143-based therapy targeting aberrant COX-2/PGE2 in pancreatic cancers.
Highlights.
Pancreatic cancer cells express low miR-143 levels and elevated p-MEK, p-MAPK and RREB1
MEK inhibitors U0126 and PD98059 increase miR-143 expression
miR-143 decreases COX-2 mRNA stability and expression and PGE2
miR-143 decreases p-p38MAPK, p-MEK, p-MAPK and RREB1 expression
Acknowledgments
This work was supported in part by NIH grant P01CA163200 and the Hirshberg Laboratories for Pancreatic Cancer Research.
Abbreviations
- COX-2
cyclooxygenase-2
- CREB
cyclic AMP responsive element binding protein
- miR-143
microRNA-143
- MAPK
mitogen-activated protein kinase
- MEK
MAPK kinase
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PaCa
pancreatic cancer
- PGE2
prostaglandin E2
- PKA
protein kinase A
- RREB1
Ras responsive element binding protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. MicroRNA functions. Annual Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Singh PK, Brand RE, Mehla K. MicroRNAs in pancreatic cancer metabolism. Nat Rev Gastroenterol Hepatol. 2012;9(6):334–344. doi: 10.1038/nrgastro.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.He XP, Shao Y, Li XL, Xu W, Chen GS, Sun HH, Xu HC, Xu X, Tang D, Zheng XF, Xue YP, Huang GC, Sun WH. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J. 2012;279(22):4201–4212. doi: 10.1111/febs.12013. [DOI] [PubMed] [Google Scholar]
- 7.Strillacci A, Griffoni C, Sansone P, Paterini P, Piazzi G, Lazzarini G, Spisni E, Pantaleo MA, Biasco G, Tomasi V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315(8):1439–1447. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Rouzer CA, Marnett LJ. Cyclooxygenases: structural and functional insights. J Lipid Res. 2009;50(Supp):S29–S34. doi: 10.1194/jlr.R800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakanishi M, Rosenberg DW. Multifaceted roles of PGE2 in inflammation and cancer. Semin Immunopathol. 2013;35(2):123–137. doi: 10.1007/s00281-012-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, Kaidi A. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30(3):377–386. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 11.Wang MT, Honn KV, Nie D. Cyclooxygenases, prostanoids, and tumor progression. Cancer Metastasis Rev. 2007;26(3–4):525–534. doi: 10.1007/s10555-007-9096-5. [DOI] [PubMed] [Google Scholar]
- 12.Pino MS, Nawrocki ST, Cognetti F, Abruzzese JL, Xiong HQ, McConkey DJ. Prostaglandin E2 drives cyclooxygenase-2 expression via cyclic AMP response element activation in human pancreatic cancer cells. Cancer Biol Ther. 2005;4(11):1263–1269. doi: 10.4161/cbt.4.11.2138. [DOI] [PubMed] [Google Scholar]
- 13.Moore AE, Young LE, Dixon DA. A common single-nucleotide polymorphism in cyclooxygenase-2 disrupts microRNA-mediated regulation. Oncogene. 2012;31(12):1592–1598. doi: 10.1038/onc.2011.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C, Huang L. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009;181(3):1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- 15.Noguchi S, Mori T, Hoshino Y, Maruo K, Yamada N, Kitade Y, Naoe T, Akao Y. MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Lett. 2011;307(2):211–220. doi: 10.1016/j.canlet.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi H, Li A, Dawson DW, Hines OJ, Reber HA, Eibl G. Cyclooxygenase-2 confers growth advantage to syngeneic pancreatic cancer cells. Pancreas. 2011;40(3):453–459. doi: 10.1097/MPA.0b013e31820b9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pham H, Chen M, Li A, King J, Angst E, Dawson DW, Park J, Reber HA, Hines OJ, Eibl G. Loss of 15-hydroxyprostaglandin dehydrogenase increases prostaglandin E2 in pancreatic tumors. Pancreas. 2010;39(3):332–339. doi: 10.1097/MPA.0b013e3181baecbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, Zhu L, Wang J, Yang R, Zhang Y, Ren Z, Zen K, Zhang J, Zhang CY. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28(10):1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 20.Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24(24):2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32(20):2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tung WH, Hsieh HL, Lee IT, Yang CM. Enterovirus 71 modulates a COX-2/PGE2/cAMP-dependent viral replication in human neuroblastoma cells: role of the c-Src/EGFR/p42/p44 MAPK/CREB signaling pathway. J Cell Biochem. 2011;112(2):559–570. doi: 10.1002/jcb.22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Ou Y, Wu K, Chen Y, Sun W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biol. 2012;33(6):1863–1870. doi: 10.1007/s13277-012-0446-8. [DOI] [PubMed] [Google Scholar]
- 24.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26(5):311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 25.Navarrete CM, Perez M, de Vinuesa AG, Collado JA, Fiebich BL, Calzado MA, Munoz E. Endogenous N-acyl-dopamines induce COX-2 expression in brain endothelial cells by stabilizing mRNA through a p38 dependent pathway. Biochem Pharmacol. 2010;79(12):1805–1814. doi: 10.1016/j.bcp.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Chang MS, Chen BC, Weng CM, Lee WS, Lin CH. Involvement of Ras/Raf-1/p44/42 MAPK in YC-1-induced cyclooxygenase-2 expression in human pulmonary epithelial cells. Pharmacol Res. 2009;60(4):247–253. doi: 10.1016/j.phrs.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Zhang J, Yang X, Han X. Several transcription factors regulate COX-2 gene expression in pancreatic beta-cells. Mol Biol Rep. 2007;34(3):199–206. doi: 10.1007/s11033-007-9085-3. [DOI] [PubMed] [Google Scholar]
- 28.Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20(12):4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cok SJ, Acton SJ, Morrison AR. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J Biol Chem. 2003;278(38):36157–36162. doi: 10.1074/jbc.M302547200. [DOI] [PubMed] [Google Scholar]
- 30.Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol Cancer Res. 2012;10(1):167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD, Prionidis K, Dalamagas T, Hatzigeorgiou AG. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Chaerkady R, Kandasamy K, Huang TC, Selvan LD, Dwivedi SB, Kent OA, Mendell JT, Pandey A. Identifying targets of miR-143 using a SILAC-based proteomic approach. Mol Biosyst. 2010;6(10):1873–1882. doi: 10.1039/c004401f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, Avances C, Villalba M, Culine S, Fajas L. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One. 2009;4(10):e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castellano E, Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2(3):216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 2007;67(5):2098–2106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 36.Costello LC, Levy BA, Desouki MM, Zou J, Bagasra O, Johnson LA, Hanna N, Franklin RB. Decreased zinc and downregulation of ZIP3 zinc uptake transporter in the development of pancreatic adenocarcinoma. Cancer Biol Ther. 2011;12(4):297–303. doi: 10.4161/cbt.12.4.16356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao JS, Zhang Y, Tang X, Tucker LD, Tarwater PM, Quesenberry PJ, Rigoutsos I, Ramratnam B. The Evi1, microRNA-143, K-Ras axis in colon cancer. FEBS Lett. 2011;585(4):693–699. doi: 10.1016/j.febslet.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K, Nakashima R, Kitade Y, Naoe T. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17(6):398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]


