Summary
The goal of the IDAWG is to facilitate the consistent analysis of HLA and KIR data, and the sharing of those data among the immunogenomic and larger genomic communities. However, the data-management approaches currently applied by immunogenomic researchers are not widely discussed or reported in the literature, and the effect of different approaches on data-analyses is not known.
With ASHI’s support, the IDAWG developed a forty-five question survey on HLA and KIR data-generation, data-management, and data-analysis practices. Survey questions detailed the loci genotyped, typing systems used, nomenclature versions reported, computer operating systems and software used to manage and transmit data, the approaches applied to resolve HLA ambiguity, and the methods used for basic population-level analyses. Respondents were invited to demonstrate their HLA ambiguity resolution approaches in simulated data sets.By May 2012, 156 respondents from 35 nations had completed the survey . These survey respondents represent a broad sampling of the Immunogenomic community; 52% were European, 30% North American, 10% Asian, 4% South American, and 4% from the Pacific.
The project will continue in conjunction with the 17th Workshop, with the aim of developing community data-sharing standards, ambiguity resolution documentation formats, single-task data-Management tools, and, novel data-analysis methods and applications. While additional project details and plans for the 17th IHIW will be forthcoming, we welcome the input and participation in these projects from the histocompatibility and immunogenetics community.
Keywords: Meta-analysis, Statistics, HLA, Genetics
Introduction
The immunogenomics data analysis working group (IDAWG)1 is an international collaboration of histocompatibility and immunogenetics investigators who share the goal of facilitating the sharing of immunogenomic data (HLA, KIR, etc.) and fostering the consistent analysis and interpretation of those data by the immunogenomics community and the larger genomics communities. The working group was formed in advance of the 16th International HLA and Immunogenetics Workshop (IHIW) and Conference with the intent to present its project related to topics of data-management and data-analysis at the 16th IHIW and Conference. This IDAWG IHIW project is structured as an ongoing effort, and while we presented our current findings at the IHIW, the project is intended to continue in the interim time up until the 17th IHIW.
The overall goal of this project is to develop data-management tools, data reporting guidelines, and data documentation standards that are tailored to work with the HLA and KIR data-management practices in use by the immunogenetics community. The 16th IHIW Immunogenomic Data-Management Methods project proceeded in two phases, with both phases still underway and continuing beyond the 16th IHIW meeting held in Liverpool on May 28-30, 2012.
At the 16th IHIW we presented the results of a survey of the immunogenetics and immunogenomics community. The purpose of the survey is to determine current practices in:
HLA and KIR data-management and transmission,
HLA ambiguity management and resolution, and
Primary data-analysis.
As of this writing the survey is available at http://www.surveymonkey.com/s/IDAWG, and takes approximately 15 minutes to complete. The survey can be submitted anonymously, and although survey participants are invited to take part in the larger IDAWG project, information specific to individual laboratories is kept confidential; individual laboratories are not identified without providing consent. When survey participants indicate their interest, the survey is followed-up with synthetic datasets for the purpose of demonstrating HLA ambiguity resolution and primary data-analysis practices.
The survey results at the time of the 16th IHIW were presented in the project meeting. As of that time, we had received 154 responses to the survey. Forty-five respondents indicated their desire to participate in the development of standards as part of this project, and twenty-five respondents indicated their willingness to demonstrate their ambiguity resolution approaches on test-datasets that we provided. These datasets consisted of ambiguous DRB1 and DQB1 genotyping results for 400 individuals, 200 African Americans, and 200 European Americans. Within each group, 100 samples were typed using SSOP methods, and 100 using SBT methods. Allele and genotype ambiguities were recorded in the GL string format proposed for KIR genotypes (Maiers et al. 2007; discussed at: http://www.ebi.ac.uk/ipd/kir/standards.html). At the time of the project meeting, three laboratories had returned the results from this part of the project. We presented comparisons of the results of each participant’s ambiguity resolution approach, as well as the outcomes of these approaches on standard population-level analyses, at the 16th IHIW project meeting.
IHIW Project Meeting
At the 16th IHIW meeting of this project, we presented the data collected in the survey, along with preliminary conclusions on the effects of current practices on common applications of immunogenomic data, and initial recommendations for data management and analysis based on those outcomes. We continued to collect surveys at the IHIW. Project participants spoke about their own experiences with immunogenomic data-management and analysis, including challenges that they have encountered and solutions (both developed and desired) for overcoming them. We invited IHIW attendees to contribute presentations and participate in the discussion of data management and analysis recommendations and the development of the STREIS statement. Summaries of the invited presentations follow the results of the survey.
While these presentations illustrate some of the common data-management and analysis challenges encountered in the immunogenetics community and the diversity of views with regards to solutions for those challenges, they do not represent formal standards recommendations by the IDAWG or this 16th IHIW project, and this report should not be construed as a source for specific data-management and analysis recommendations. We feel that the workshop is the best forum for members of the immunogenetics community to discuss these views and issues, and that this project has greatly benefitted through the discussions that followed these presentations.
Results of the Survey of current HLA and KIR data-management practices
Laboratory demographics
At the time of the 16th IHIW meeting, there had been 154 unique, informative responses to the survey. Of the respondents that identified a geographical location, thirty-four countries (Figure 1) were represented from seven world regions (Europe, Middle East, Asia, Oceania, North America, South America and the Caribbean). The majority of respondents were from clinical laboratories, but there was also good representation from registries and academic laboratories.
Figure 1.
Gobal distribution of respondants to data management practices survey.
Gene systems typed
While essentially all surveyed laboratories genotype the HLA loci, approximately half also genotype other gene systems; of those, about half genotype the KIR loci. Other immune response genes genotyped by the surveyed laboratories include MICA/B, cytokines and a variety of other immune response loci.
HLA genotyping
Over 90% of responding laboratories genotype HLA-A, HLA-B, HLA-C, DRB1 and DQB1. DRB3/4/5, DQA1 and DPB1 are genotyped in about 50% of laboratories, while DPA1 genotyping is infrequently performed. Forty-four laboratories use serological typing for some or all of the class I loci. All of these laboratories also use DNA methods. Eighty-four laboratories use sequence-based typing (SBT) for some or all loci, but only nine use SBT exclusively. Most laboratories also use sequence-specific oligonucleotide probes (SSOP) and/or sequence-specific primers (SSP) for HLA genotyping. Eleven laboratories indicated that they use microarray methods. In many laboratories, methods vary by locus. A wide variety of software is used for allele-calling, and among the survey respondents appeared to be very method or vendor dependent; the software packages used were primarily commercial products. While the majority of laboratories have updated to HLA nomenclature version 3 (Marsh et al. 2010), many laboratories are still using version 2 (Marsh et al. 2002) for some or all loci; six laboratories indicated that they have data for some loci in version 1 nomenclature (Bodmer et al. 1990).
KIR genotyping
Thirty-nine percent of the surveyed laboratories perform KIR genotyping. The majority of the KIR genotyping is for presence/absence resolution; only three laboratories reported that they perform allele-level KIR genotyping, although thirty-five laboratories can distinguish KIR2DL5A and KIR2DL5B. Nearly all laboratories performing KIR genotyping use SSOP or SSP methods, and most of these are via commercial kits. Other KIR genotyping methods include SBT and microarray. As with HLA genotyping, laboratories utilize a variety of software depending on the typing method.
Data management
Most laboratories are using the Microsoft Windows operating system, but a wide variety of platforms are in use and many labs use more than one operating system. The majority of responding laboratories only generate data; approximately one-third also receive data. Data is managed and stored in the majority of laboratories with more than a single software system, and many different data reporting and transmission formats are widely used; many labs use more than one format for this purpose.
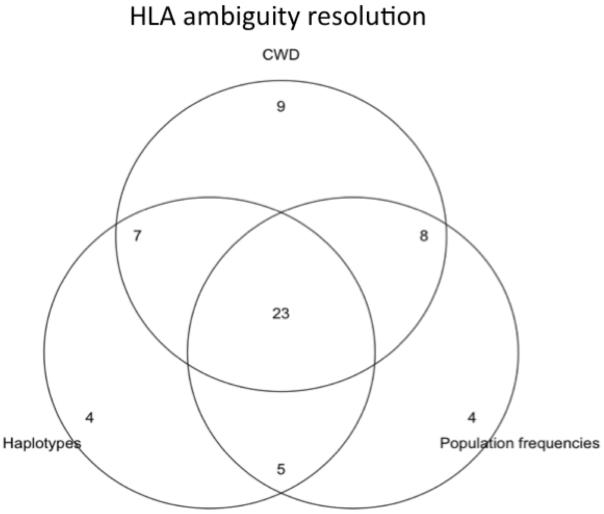
Most laboratories make some changes to genotype data prior to reporting. The most common type of genotype data post-processing is in the form of allele or genotype ambiguity resolution, which is performed in a majority of the responding laboratories. Decisions pertaining to ambiguity resolution are based primarily on a list of common and well-documented alleles (CWD)(Cano et al. 2007), as well as known haplotypic associations and previously reported allele frequencies (Figure 2). The online database of worldwide allele frequencies, allelefrequencies.net (AFND)(Middleton et al. 2003), the CWD list and the National Marrow Donor Program (NMDP) database of allele and haplotype frequencies are all widely used resources.
Figure 2.
Venn diagram illustrating the overlap of common methods used by laboratories for resolution of ambiguous HLA genotypes.
Population-level data analysis
Less than twenty percent of responding laboratories reported performing any population-level analysis other than haplotype estimation, which thirty-two laboratories perform regularly. Twenty-six of the laboratories perform analysis for conformation of genotype frequencies to expectations under Hardy-Weinberg Equilibrium (HWE) proportions, and about half of those use this analysis as a quality check. About ten percent of the laboratories report allele frequencies or perform analysis of linkage disequilibrium (LD) between alleles or loci. There is no clear choice for analytical software, with laboratories reporting to use a number of different packages, most freely available.
In summary, the results of the survey reveal that there are no standard means of managing and analyzing immunogenetic data within the immunogenetics community. Even within the same laboratory, often multiple methods on multiple platforms are used, and may vary by locus. The lack of standards for data management results in a lack of consistency between laboratories and across studies.
Impacts of HLA ambiguity resolution on analytical outcomes
Three laboratories (L1, L2, and L3) returned unambiguous genotyping results, with each applied ambiguity resolution approach yielding a different result. For example, for the African American SBT data at the DQB1 locus, L1 returned 17 alleles with a heterozygosity (h) of 0.882, L2 returned 20 alleles with h of 0.877, and L3 returned 17 alleles with h of 0.858. Eighty percent of the allele names returned by L1 and L3 were identical, whereas only 31% of allele names returned by L2 were identical to either L1 or L3. In many cases, these differences were due to variation in allele name resolution (e.g. DQB1*02:01 vs. *02:01:01 or *02:01P).
We tested the genotypic ratios in each unambiguous dataset for their adherence to expected Hardy-Weinberg equilibrium proportions. P-values for the Guo and Thompson test on the unambiguous African American SBT DQB1 data for L1, L2, and L3 were 0.00001, 0.00102, and 0.42547 respectively, suggesting that the method applied for ambiguity resolution can have a significant effect on the analytical outcomes.
Presentations by Participants
Title: An XML Export of the IMGT/HLA Database
Presenter: James Robinson
The presentation focused on the forthcoming Extensible Markup Language (XML) export of the IMGT/HLA Database (Robinson, 2000 and Robinson, 2011). The IMGT/HLA Database provides a specialist database for sequences of the human major histocompatibility complex, known as HLA and includes the official sequences for the WHO Nomenclature Committee For Factors of the HLA System. The database currently provides exports of the data in a variety of formats; this is been expanded to XML. This format defines a set of rules for encoding documents in a format that is both human and machine-readable.
A collaborative project between the HLA Informatics Group of the Anthony Nolan Research Institute and the Bioinformatics Department of the National Marrow Donor Program has developed an XML export of the data contained within the IMGT/HLA Database. The XML format combines the data included in the sequence alignments with the data available in the individual allele reports. The XML format will enable users to identify the regions within the DNA sequence, such as exons, as well as reconstruct the sequence alignments. In addition the collaborative project has developed a suite of tools for importing the data into different database schema, both open-source and proprietary, for allowing incorporation into local IT systems. The XML files will be regularly updated as part of the quarterly releases of the IMGT/HLA Database. A beta test version of the XML format and associated tools was announced and the release version will be made available from the http://hla.alleles.org/xml/ and the National Marrow Donor Program websites.
Title: Statistical Imputation of Allele-Level Multi-Locus Phased Genotypes through Structural Analysis of Ambiguous HLA
Presenter: Abeer Madbouly
Genetic matching for loci in the HLA region between a donor and a patient in hematopoietic stem cell transplantation is critical to transplant outcomes; however, methods for HLA genotyping of donors in unrelated stem cell registries yield results with allelic and phase ambiguity and do not query all clinically-relevant loci. The NMDP Bioinformatics Research Department has implemented an algorithm for resolving ambiguity through statistical imputation of HLA alleles and haplotypes in the context of matching unrelated patients and stem cell donors from the Be The Match® Registry. Allele-level HLA haplotypes can be imputed with high accuracy through the application of a set of statistical and population genetics inferences and with knowledge of haplotype frequencies and self-identified race and ethnicities. This provides a relatively inexpensive way to improve the match quality and facilitate the hematopoietic stem cell transplantation process. This method builds on haplotype frequencies estimated within registry sub-populations and exploits patterns of linkage disequilibrium across HLA haplotypes to infer high-resolution HLA assignments.
Imputation is validated on several datasets available from the registry as well as family data that, through pedigree analysis, has known phase. Validation experiments show relatively high accuracy for imputed results. We simulated ambiguity generated by several HLA genotyping methods to isolate imputation performance on several levels of resolution. Validation using simulated data also showed superior performance.
Title: Reporting of HLA typing results in unrelated stem cell donor registration
Presenter: Alexander Schmidt
In April 2010, the “G” and “P” codes for the reporting of HLA typing results were introduced (Marsh et al. 2010). While HLA alleles sharing the same nucleotide sequences for the exons encoding the peptide binding domains (in the following: the “relevant” exons) can be described using a G code, P codes include alleles that encode identical peptide binding domains.
Most HLA typing worldwide is carried out for the purpose of stem cell donor registration. In this setting, it is widespread practice to analyze only the relevant exons and to waive A) the exclusion of null alleles caused by mutations outside the relevant exons and B) the resolution of ambiguities caused by synonymous DNA substitutions inside the relevant exons. The approaches described by A and B prevent the use of P and G codes, respectively. Therefore, these codes are of limited practical value in the donor registration setting.
We proposed a different code, for example named “g”, which summarizes all alleles sharing the same nucleotide sequences for the relevant exons OR having nucleotide sequences that differ only by synonymous mutations within the relevant exons. An alternative formulation of this definition would be that a g code includes the union of the alleles summed up by a P code and the related null alleles.
Example: SBT of exons 2 and 3 of the HLA-A locus may result in the ambiguous typing result A*01:01:01G+A*02:01:04 | A*01:01:10+A*02:01:01G. Though preferable, the reporting of genotype lists is not yet standard in the data exchange between HLA labs, donor centers and registries. P codes are not applicable as both A*01:01:01G and A*02:01:01G include null alleles. G codes do not apply as A*01:01:10 and A*02:01:04 are not included in A*01:01:01G and A*02:01:01G, respectively. Therefore, it is current practice to use multi-allele codes for the reporting of this typing result. Using g codes, the result could be reported in a simple, correct and meaningful way as A*01:01g,02:01g. Apart from better reflecting the practice of donor registry high-throughput typing, g codes also have, in contrast to P codes, the advantage of not being compromised by newly identified null alleles. g codes have already been used in population studies based on registered German (Schmidt et al. 2009) and Polish (Schmidt et al. 2011) stem cell donors.
Title: Coping with ambiguity in HLA genotyping: lessons from the T1DGC
Presenter: Janelle Noble
The Type 1 Diabetes Genetics Consortium (T1DGC) was an international effort, conceived in 2000 and begun in 2001, to collect and genotype thousands of multiplex T1D families from around the world with the goal of comprehensive identification of all of the genes associated with the disease. The HLA region is the strongest contributor to T1D risk; therefore, all samples were genotyped for eight classical HLA loci, including DRB1, DQA1, DQB1, DPA1, DPB1, HLA-A, HLA-B, and HLA-C. Sequence-specific oligonucleotide probe “linear array” technology was the chosen genotyping method for the project, based on resolution and cost at the inception of the study.
Genotyping was performed at multiple sites, including Oakland, CA; Pleasanton, CA; Melbourne, Australia; Malmö, Sweden; and Cambridge, UK. Although instrumentation, reagents, and genotyping protocols were standardized among laboratories, the inherent ambiguity in the genotyping system necessitated the introduction of standards for genotype assignment. Consistency among genotyping centers was of paramount importance. The large number of potential alleles and the large number of probes tested precluded the generation of a standard call for every possible probe binding pattern, Instead, laboratories were advised to choose the lowest numbered allele from an ambiguity string, e.g., DQB1*02:01 instead of DQB1*02:02, when both were consistent with the primary data. Inter- and intra-laboratory quality control procedures were introduced to ensure consistency of calls both within and among laboratories (Mychaleckyj et al. 2010).
Three polymorphisms, unresolvable with linear array technology but relevant to T1D association, were later resolved with Roche 454 next-generation sequencing technology, and the calls in the final data set were adjusted to reflect the results. The results of this large global study underscore the idea that for large data sets, especially with multiple genotyping centers, consistency of genotype calling is even more important than accuracy. If the data are consistent, systematic errors in the genotyping process, or in the calling of genotypes, can be corrected globally in the final data.
Title: Haplostats.org as a Teaching Tool
Presenter: Victoria Turner
We discussed our use of Haplostats.org to teach clinicians, fellows, transplant coordinators and research colleagues about HLA complexity. The response to Haplostats.org from these groups has been uniformly positive.
Title: Analysis of Multi-loci within HLA regions and HLA/KIR combination in different Chinese ethnic groups
Presenters: Yufeng Yao and Li Shi
The management and analysis of HLA, KIR and HLA/KIR combination data in different ethnic populations in China has been performed as follows. The geographic origin, age, sex, nationalities and pedigree (unrelated through at least three generations) of each individual was ascertained before sampling in our group. For data management, all data including general information, HLA and KIR data were stored using Microsoft Excel. For data analysis, we used the PyPop software to calculate the HLA allele frequencies, Hardy-Weinberg equilibrium, Ewens-Watterson homozygosity test of neutrality, linkage disequilibrium and haplotype frequencies. We used the Mega4.0 software for Neighbor-joining tree, multidimensional scaling analysis or principal component analysis to investigate the relationship among different populations. In addition, we performed HLA/KIR combination analysis. However, we met challenges during our data analysis, such as haplotype constructiongfor multi-locus HLA/KIR combinations. Thus, we are interested in easy-use software for data management, analysis and integration, such as software to help us get the HLA/KIR combination automatically.
Title: HLA Typing at Recruitment—Is It Time to Rethink Our Strategy?
Presenter: Carolyn Katovich Hurley
To reduce the time required to identify a high resolution match, a one-step DNA sequencing strategy was used to obtain HLA-A, HLA-B and HLA-C assignments of 2746 unrelated volunteers at recruitment. The results demonstrate the high resolution of the approach and challenge several aspects of the current registry typing strategy. The diploid sequences of nearly half of individuals tested included alternative genotypes; however, the majority of the time, all but one of the alternative genotypes are rare. Assigning allele codes to the results increased the number of alternative genotypes by n2 and introduced genotypes that do not exist in the individual. Since NMDP-assigned allele codes are constantly changing to incorporate newly described alleles, the link between HLA assignments over time is difficult to comprehend. The use of secondary assays to increase the resolution of the recruitment typing assignment also has limitations since registries are unable to capture the details of the secondary assays. Even these higher resolution HLA assignments degrade over time as new alleles, not ruled out by the secondary assays, are not captured in the assignment made before their discovery. Use of the diploid exons 2-3 DNA sequence (or a QR code linked to the sequence) as the assignment will prevent the issues described above. To keep pace with current donor selection criteria and with the increasing number of new alleles, it is time to rethink our approach to recruitment typing.
Title: HLA Data on the Cloud
Presenter: Martin Maiers
A ’Silver Standard’ for HLA data collection and reporting has been described at ImmPort (immport.niaid.nih.gov, “Proposal for HLA Data Validation”) to address ambiguity reduction in the recording and reporting of HLA typing results. While standards are critical for HLA data interoperability, they are not meaningful until useful tools are developed and made available for community use. We are developing distributable tools that implement this standard. Here, we describe the development of web services that create, update, and retrieve HLA typing data in standardized formats without the need for allele codes and their inherent introduction of new ambiguities.
Title: Management of Next Generation Sequence Data Generated on the Roche 454 Platform
Presenter: Cherie Holcomb
Our lab has developed a system for HLA genotyping using the 454 Life Sciences GS FLX and GS Junior platforms coupled with the Conexio Assign ATF 454 software (Bentley et al., 2009, Holcomb et al., 2011). This massively parallel, clonal (“Next Generation”) sequencing system enables high resolution (HR), high throughput HLA typing. Genotyping is highly reliable due to the redundancy of sequencing that is inherent in the system and the quality of the software. We anticipate that future use of this method by the scientific community will result in an increase in1) publications in which reporting of full ambiguity strings becomes more common (because HR typing reduces their length), 2) the rate of discovery of new alleles and 3) a need to correct some previously reported sequences. In order to make data generated by this method readily accessible to the scientific community, it is desirable to set data standards. An examination of the Conexio ATF software reveals that: 1) new alleles are identifiable as sequences that have, at a specific site and in both directions, a mismatch with the IMGT/HLA Database, 2) the genotype ambiguity string (in various formats using combinations of delineation in columns,“+”, “or”, “,”) can be reported in text, XML or Microsoft Excel format at the 1, 2, 3 or all field level, 3) ambiguity strings can be expressed by NMDP Codes, 4) the most recent WHO HLA Nomenclature is supported, 5) the release date of the IMGT/HLA References used is reported, and 6) export of the combined consensus sequences used to make the genotype call for each locus/sample combination is semi-automated. However, export of bioinformatically inferred phase information used to make a genotype call needs to be performed amplicon-by-amplicon by copying and pasting the sequences in simple text; semi-automation of this process would be desirable. Data standards can inform and guide the development of genotyping software.
Future Directions
Recommendations for immunogenomic data management, analysis and reporting will be informed by the results of the survey and the proceedings of the project meeting. While the specific solutions presented and discussed in the IHIW meeting will not necessarily be incorporated into these recommendations, they illustrate challenges that are of great interest for the immunogenomics community and that deserve to be addressed. The ultimate goal of this project is to develop approaches to data-management and analysis that avoid further complicating these issues while adhering to current nomenclature standards.
We are applying the information provided by the community to determine the effects of the various practices in use on common applications for these data, including:
Registry Searches,
Disease-Association Studies, and
Population Studies.
In addition, the survey responses have informed the development of the STrengthening the REporting of Immunogenomic Studies (STREIS) statement, which lays out principles for immunogenomic reporting guidelines (Hollenbach et al. 2011).
The project will continue after the 16th IHIW meeting, with the aim of developing:
Community Data-Sharing Standards
Ambiguity Resolution Documentation Formats
Single-task Data-Management Tools, and
Novel Data-Analysis Methods/Applications.
While additional project details and plans for the 17th IHIW will be forthcoming, we welcome the input and participation in these projects from the histocompatibility and immunogenetics community.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grant U01AI067068 (JAH and SJM) awarded by the National Institute of Allergy and Infectious Diseases (NIAID). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the NIH. The IDAWG 16th IHIW Survey of Immunogenetic Data Management and Analysis Methods was developed and made available with the generous support of the American Society for Histocompatibility and Immunogenetics (ASHI), and we thank ASHI for their support of this project. We thank the members of the IDAWG for their participation and input in this 16th IHIW project, especially Wolfgang Helmberg, Martin Maiers, Steven GE Marsh, Derek Middleton and Carlheinz Mueller. We thank Claire Adams, Frantz Agis, Medhat Askar, Pinar Ata, Vincent Aubert, Mats Bengtsson, Graziella Carella, Nicholas Dipaola, Ronald E. Domen, Mireille Drouet, Paul Dunn, Thomas Eiermann, Robert Endres, Shirin Farjadian, Maria Edvige Fasano, Christian Gabriel, Michael Gautreaux, Franca Rosa Guerini, John Harvey, Susan Hsu, Carolyn Hurley, Khalid Al Hussein, Pavel Jindra, Frieda Jordan, Pam Kimball, Libor Kolesar, Pawinee Kupatawintu, Chantale Lacelle, N.M. Lardy, Katy Latham, Dario Ligeiro, Andrew Lobashevsky, Rami Mahfouz, Danzer Martin, Miryam Martinetti, Gunilla Martinez-Riqué, Priscila Saamara Mazini, Valeria Miotti, Mahendra Narain Mishra, Carlheinz Müller, Sonia Nesci, Jorge Neumann, Jacek Nowak, J. Gonzalo Ocejo-Vinyals, Derek O’Neill, Eduard Palou, Chryssa Papasteriades, Juha Peräsaari, Noemi Farah Pereira, Martha Perez, Tracey Rees, Lucie Richard, Chrissy H. Roberts, Iñigo Romón, Sandra Rosen-Bronson, Alexander Schmidt, Ali Sengul, David Senitzer, Li Shi, Alexandra Siorenta, Genc Sulcebe, Ingrid Tagen, Marcel Tilanus, Alberto Torio, Elizabeth Trachtenberg, Luiza Tamie Tsuneto, Victoria Turner, Marigoula Varla-Leftherioti, Michael Varney, Jose L. Vicario, Svetlana Vojvodić, Annika Wennerström, Campbell Witt, Malte Ziemann, Transplantation Diagnostics, HLA Laboratory, Institute of Transfusion Medicine, University Hospital of Essen, and First Department of Internal Medicine, Immunogenetics Lab, Thessaloniki for their participation in this project.
Footnotes
The Immunogenomics Data-Analysis Working Group is chaired by Jill A. Hollenbach and Steven J. Mack. Members of the Immunogenomics Data Analysis Working Group include (in alphabetical order): Henry A. Erlich, Michael Feolo, Marcelo Fernandez-Vina, Pierre-Antoine Gourraud, Wolfgang Helmberg, Uma Kanga, Pawinee Kupatawintu, Alexander K. Lancaster, Martin Maiers, Hazael Maldonado-Torres, Steven G.E. Marsh, Diogo Meyer, Derek Middleton, Carlheinz R. Müller, Oytip Nathalang, Myoung Hee Park, James Robinson, Richard M. Single, Brian Tait, Glenys Thomson, Ana Maria Valdes, and Michael D. Varney.
References
- Bentley G, Higuchi R, Hoglund B, et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens. 2009;74:393–403. doi: 10.1111/j.1399-0039.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer JG, Marsh SG, Albert ED, et al. Nomenclature for factors of the HLA system, 1991. WHO Nomenclature Committee for factors of the HLA system. Tissue Antigens. 1992;39:161–73. doi: 10.1111/j.1399-0039.1992.tb01932.x. [DOI] [PubMed] [Google Scholar]
- Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, Reed EF, Senitzer D, Setterholm M, Smith A, Fernandez-Vina M. Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68(5):392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Holcomb CL, Hoglund B, Anderson MW. A multi-site study using high-resolution HLA genotyping by next generation sequencing. Tissue Antigens. 2011;77:206–217. doi: 10.1111/j.1399-0039.2010.01606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach JA, Mack SJ, Gourraud PA, Single RM, Maiers M, Middleton D, Thomson G, Marsh SG, Varney MD, Immunogenomics Data Analysis Working Group A community standard for immunogenomic data reporting and analysis: proposal for a STrengthening the REporting of Immunogenomic Studies statement. Tissue Antigens. 2011;78(5):333–44. doi: 10.1111/j.1399-0039.2011.01777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiers M, Spellman S, Marsh SGE, Parham P, Rajalingam R, Reed E, Noreen H, Yu N, Cooley S. A community standard reporting format for KIR genotyping data. Human Immunology. 2007;68:S105. [Google Scholar]
- Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system. Tissue Antigens. 2002;60:407–64. doi: 10.1034/j.1399-0039.2002.600509.x. 2002. [DOI] [PubMed] [Google Scholar]
- Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database. Tissue Antigens. 2003;61(5):403–407. doi: 10.1034/j.1399-0039.2003.00062.x. http://www.allelefrequencies.net. [DOI] [PubMed] [Google Scholar]
- Mychaleckyj JC, Noble JA, Moonsamy PV, Carlson JA, Varney MD, Post J, Helmberg W, Pierce JJ, Bonella P, Fear AL, Lavant E, Louey A, Boyle S, Lane JA, Sali P, Kim S, Rappner R, Williams DT, Perdue LH, Reboussin DM, Tait BD, Akolkar B, Hilner JE, Steffes MW, Erlich HA, for the T1DGC HLA genotyping in the international Type 1 Diabetes Genetics Consortium. Clin Trials. 2010;7(1 Suppl):S75–87. doi: 10.1177/1740774510373494. PMCID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SGE. Nucleic Acids Research. 2011;39(Suppl 1):D1171–6. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- Schmidt AH, Baier D, Solloch UV, Stahr A, Cereb N, Wassmuth R, et al. Estimation of high-resolution HLA-A, -B, -C, -DRB1 allele and haplotype frequencies based on 8862 German stem cell donors and implications for strategic donor registry planning. Human Immunology. 2009;70:895. doi: 10.1016/j.humimm.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Schmidt AH, Solloch UV, Pingel J, Baier D, Böhme I, Dubicka K, et al. High-resolution human leukocyte antigen allele and haplotype frequencies of the Polish population based on 20,653 stem cell donors. Human Immunology. 2011;72:558. doi: 10.1016/j.humimm.2011.03.010. [DOI] [PubMed] [Google Scholar]