Abstract
In the last fifteen years published reports have described KIR gene-content frequency distributions in more than 120 populations worldwide. However, there have been limited studies examining these data in aggregate in order to detect overall patterns of variation at regional and global levels. Here, we present a summary of the collection of KIR gene-content data for 105 worldwide populations collected as part of the 15th and 16th International Histocompatibility and Immunogenetics Workshops, and preliminary results for data analysis.
The data were contributed by thirty-four laboratories during the four-year course of this project, including data for the HGDP-CEPH populations. Additionally, data from the 15th IHIW and data contributed to the allelefrequencies.net (AFND) database were combined with the current workshop dataset
Keywords: KIR, HLA, workshop, report
INTRODUCTION
The killer cell immunoglobulin-like receptors (KIR) are receptors expressed on natural killer (NK) cells that function to inhibit or activate NK cells. Several of the inhibitory KIR use the human leukocyte antigen (HLA) class I molecules as their ligand, while the ligands for most of the stimulatory KIR have not been identified definitively (Vilches and Parham 2002; Biron 1997; Vales-Gomez et al. 1998; Bashirova et al. 2006; Young and Uhrberg 2002; Hsu 2004; Smyth et al. 2005). The KIR gene complex is located on human chromosome 19q13.4 and is both polygenic and extremely polymorphic. While extensive allelic variability has been identified, particularly in the inhibitory genes (http://www.ebi.ac.uk/cgi-bin/ipd/kir; Robinson et al. 2010), variability in gene-content haplotypes is responsible for significant diversity both within and between populations.
In the last fifteen years published reports have described KIR gene-content frequency distributions in more than 120 populations worldwide and are available on a the public database www.allelefrequencies.net (Gonzalez-Galarza FF et al. 2011). However, there have been limited studies examining these data in aggregate in order to detect overall patterns of variation at regional and global levels (Single et al. 2007; Middleton et al. 2008; Hollenbach et al. 2010; Hollenbach et al. 2012). The KIR anthropology component (Population Global Distribution of KIR and Ligand) of the 15th and 16th International Histocompatibility workshops (IHIW) have been intended to collect and collate KIR and HLA frequency data in a diverse set of human populations in order to more closely examine worldwide variation in the KIR loci, and the relationship between KIR genes and their HLA ligands. Evidence that KIR and HLA are co-evolving was first demonstrated by Single et al. (Single et al. 2007); in the 15th IHIW KIR Anthropology component we presented further support for this notion, finding a significant correlation between KIR2DL2/L3 and its ligand, HLA-C group 1(Hollenbach et al. 2010). A primary aim of the 16th workshop project was to confirm and extend this finding in additional worldwide populations.
During the course of the 15th IHIW project, fifteen laboratories submitted KIR genotype and HLA ligand data in twenty-seven populations from six broad ethnic groups (Hollenbach et al. 2010). Data were analyzed for correlations between the frequencies of KIR and their known HLA ligands. In addition, allelic typing was performed for KIR2DL2 and KIR3DL1 in a subset of populations. Strong and significant correlations were observed between KIR2DL2/L3 genotype frequencies and the frequency of their ligand, HLA-C1. In contrast, only weak associations were seen for KIR3DL1, KIR3DS1 and the HLA-Bw4 ligand. In this case, only the HLA-B locus was considered; although some of alleles of HLA-A are known to have the Bw4 motif, these data were not available for this study. While some aspects of the correlations observed in that study differed from those reported in other populations, these data provide additional evidence of linked evolutionary histories for some KIR and HLA loci.
We planned to extend these studies during the 16th IHIW, in particular emphasizing investigation in populations not studied in the last workshop, as well as further investigation of allelic variation in the KIR. Of particular interest were non-European populations with limited admixture. Although we intended to extend allelic typing to include KIR2DL3 and KIR3DS1, as well as KIR2DL2 and KIR3DL1 in order to allow a more detailed examination of allelic variability and haplotypic associations across the KIR complex, these data were ultimately not available. Here, we present a summary of the proceedings of the workshop project and the project meeting, and the KIR gene-content data for the 105 worldwide populations that were ultimately collected for the 15th and 16th IHIW.
STATISTICAL METHODS
Carrier frequencies for the KIR loci were obtained by direct counting.
A two-dimensional clustered heat map for KIR carrier frequencies was constructed using the ‘heatmap’ function in the base ‘stats package for the R language for statistical computing (R Development Core Team 2008). Briefly, a hierarchical clustering was performed on a set of dissimilarities based on carrier frequencies for the KIR loci; both loci and populations were clustered in this manner, and frequency differences were illustrated via the default heat map color gradient.
Data were analyzed for correlations between the frequencies of KIR2DL2/L3, KIR3DL1 and their HLA ligands (HLA-C and HLA-B, respectively) using the ‘cor’ function in the R base package (Williams and Templeton 2003), as well as plotting and fitting of the regression line. In order to account for the non-independence of the study populations, testing of the statistical significance for the calculated correlation coefficients was accomplished via an empirical approach (Single et al. 2008). Briefly, resampling distributions for the correlation coefficients between KIR and HLA ligand frequencies were generated after randomly reassigning the HLA ligand status across all study populations. Permutation p-values (pperm) represent the proportion of the distribution of 10,000 permuted correlations that were greater than the true correlation.
PRELIMINARY RESULTS
At the time of writing for this report, data analysis is still ongoing. This project is intended to be part of a continuum beginning with the 15th IHIW and extending through the next (17th) IHIW. Here we present some preliminary results for the 16th IHIW project.
The data for the16th IHIW encompassed thirty-four worldwide populations and were contributed by twelve laboratories from eleven countries during the six-year course of this project, including data for the HGDP-CEPH populations (Hollenbach et al. 2012). Additionally, data from the 15th IHIW and data contributed to the Allele Frequency Net Database (AFND: allelefrequencies.net) were combined with the current workshop dataset (Table 1). In total, data for 7249 individuals from 105 worldwide populations were analyzed for this project. The global distribution of the final dataset for the 16th IHIW is shown in Figure 1. All populations were genotyped for KIR gene-content (presence/absence) for all loci. Forty populations were also genotyped for HLA-B and -C ligands.
Table 1.
Populations and sample sizes for the 16th IHIW KIR anthropology project by data source.
|
16th IHIW (population/n) |
16th IHIW/AFND (population/n) |
16th IHIW/HGDP (population/n) |
15th IHIW (population/n) |
|---|---|---|---|
| Austria/44 | Brazil_Curitiba/132 | Bantu_N/11 | SouthAfrica_Xhosa/50 |
| Gambia_Fula/62 | French_AFND/125 | Bantu_S/8 | Morocco/67 |
| Gambia_Jola/59 | Iran/200 | Biaka_Pygmies/29 | Canada_Persian/68 |
| Gambia_Mandinka/126 | Japan/47 | Mandenka/23 | Oman/98 |
| Gambia_Wolof/53 | Mexico_Mestizo_Mexico/300 | Mbuti_Pygmies/12 | HongKong/100 |
| China_Han/93 | Poland_LowerSilesia/327 | San/7 | Singapore/46 |
| China_Bulang/106 | Brazil_Parana/287 | Yoruba/23 | Thailand/100 |
| China_Nu/106 | Macedonia/214 | Mozabite/27 | Czech/121 |
| China_Zhuang/95 | Japan_Tokyo/197 | Bedouin/46 | Portugal/38 |
| China_Yugu/91 | Palestinian/42 | Orcadian_15th/90 | |
| Brazil_MatoGrossoDuSur/205 | Druze/46 | Turkey/154 | |
| Bangladesh/238 | Adygei/17 | Italy/50 | |
| Sweden/117 | French/26 | Uzbek/67 | |
| Mexico_Mestizo_Highland/257 | French_Basque/22 | Norwegian/363 | |
| Mexico_Mestizo_Jalisco/100 | North_Italian/14 | European_American_15th/255 | |
| Mexico_Mestizo_West/124 | Orcadian_HGDP/13 | Cuban_Cauc/70 | |
| Poland/60 | Russian_HGDP/24 | Brazil_Belo/90 | |
| Russia/100 | Sardinian/24 | Brazil_Rio/166 | |
| Amerind_Canada/109 | Tuscan/8 | Uruguay/41 | |
| Amerind_Cayapa/96 | Pathan/21 | ||
| Amerind_Tschila_Chiuaple/34 | Makrani/25 | ||
| Amerind_Tschila_Congoma/38 | Kalash/24 | ||
| Amerind_Shipibo/59 | Hazara/22 | ||
| Armenian/20 | Balochi/23 | ||
| Australian_aboriginal/18 | Barusho/20 | ||
| Brahui/22 | |||
| Sindhi/23 | |||
| Uygur/9 | |||
| Cambodian/11 | |||
| Dai/8 | |||
| Daur/9 | |||
| Han_HGDP/46 | |||
| Hezhen/10 | |||
| Japan_HGDP/31 | |||
| Lahu/10 | |||
| Miaozu/8 | |||
| Mongola/10 | |||
| Naxi/10 | |||
| Orogen/9 | |||
| She/10 | |||
| Tu/9 | |||
| Tujia/7 | |||
| Xibo/8 | |||
| Yakut/22 | |||
| Yizu/10 | |||
| Papuan/16 | |||
| NAN_Melanesian/21 | |||
| Karitiana/24 | |||
| Maya/24 | |||
| Pima/25 | |||
| Surui/15 | |||
| Colombian/12 |
Key for data sources:16th IHIW = population data collected specifically for the 16th IHIW KIR anthropology project; 15th IHIW = population data brought forward from the 15th IHIW project; 16th IHIW/HGDP = data for the HGDP-CEPH populations; AFND = data from the allelefrequencies.net database.
Figure 1.
Global distribution of 16th IHIW populations
All KIR and HLA genotype data collected for this project will be made publicly available on allelefrequencies.net and should serve as a valuable resource to the immunogenetics research community. Nearly 100 unique genotypes are present in the 105 populations under study for the 16th IHIW project. Over thirty of these genotypes are observed in only a single population and while some may be due to technical error, these numbers underscore the remarkable diversity in the KIR gene-content at the global level. The data were examined for an overview of global patterns of carrier frequency distributions for individual loci, demonstrating that populations cluster by carrier frequency distributions according to world regions primarily driven by different frequencies of the KIR B loci. A clustered heat map of KIR carrier frequencies (Figure 2) illustrated the general patterns associated with major world regions.
Figure 2.
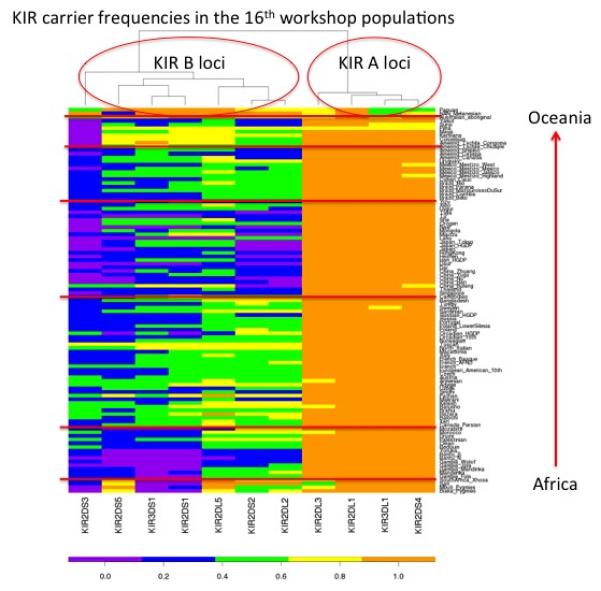
A clustered heat map of KIR carrier frequencies in the 16th IHIW populations. Populations are ordered generally by distance from Africa.
For the forty populations were also genotyped for HLA-B and HLA-C ligands, the data analyzed for correlation between KIR and HLA ligand frequencies within populations, revealing a highly significant (p<0.0001) correlation (r=0.79) between KIR2DL3 and HLA-C group frequencies is observed (Figure 3). In contrast, the correlation for KIR3DL1 and the HLA-Bw4 ligand was not significant.
Figure 3.
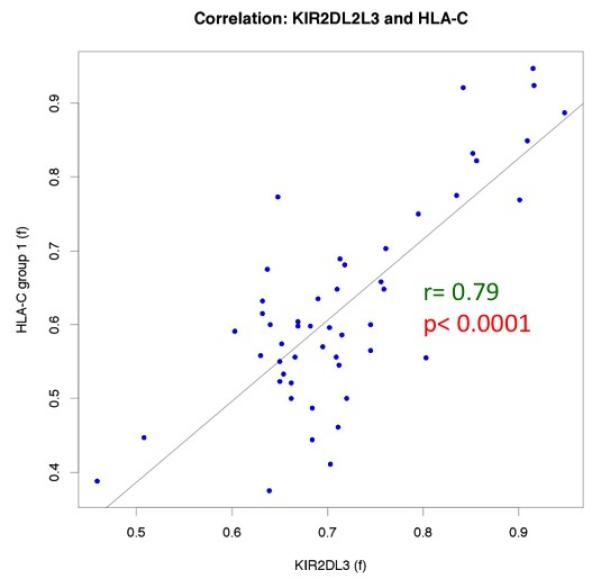
HLA-C group and KIR2DL2L3 frequencies are highly correlated within populations.
PROJECT MEETING
During the project meeting, held on Tuesday, 29 May 2012 from 08:30-12:30, several participants in the project presented further details of the populations submitted by their laboratories. Additionally, one presentation detailed the resources for KIR available on the allelefrequencies.net (AFND) website. The presenters and titles and descriptions of their presentations, are as follows:
“ KIR Gene Variation in the Cayapa-Chachis, Colorado-Tsáchila and Shipibo-Conibo Amerindian Populations from South America”
Elizabeth Trachtenberg, PhD, D(ABHI)
Three South American Amerindian populations from Ecuador and Peru were studied for KIR gene presence and absence. In total, 376 non-first degree relative samples were analyzed from the Capaya-Chachi (N=244) and the Colorado-Tsáchila (N=34 from the Chiualpe Village, and N=39 from the Congoma Village) populations. KIR2DL1, KIR2DL3, KIR3DL1, KIR2DL4, KIR3DL2 and KIR3DL3 were found at 100% frequency or close in all three populations. In agreement with our study on world-wide surveys of KIR (e.g. in the Human Genome Diversity Project - Centre d’Etude du Polymorphisme Humain (HGDP-CEPH)) showing low to absent frequency of KIR2DS3 in Amerinds, all KIR loci but KIR2DS3 was found in the three populations here; KIR2DS3 was absent in all but the Shipibo-Conibo (f=0.25) and in the Colorado-Tsáchila-Chiualpe (f=<5%). There was also more stimulatory KIR (Cen-B and Tel-B) found in the Colorado groups, which are the more isolated, with potential for more genetic drift. In haplotype analysis, higher levels of KIR B haplotypes, especially the CenB2-A and CenB2-B haplotypes, were observed in the Colorado, with concomittant lower frequencies of A/A.
“KIR Gene Variation in the CEPH Human Genome Diversity Panel” Elizabeth Trachtenberg, PhD, D(ABHI)
In this study we investigated patterns of variation in the KIR cluster in the large and well-characterized sample of worldwide human populations in the Human Genome Diversity Project - Centre d’Etude du Polymorphisme Humain (HGDP-CEPH) panel in order to better understand the patterns of diversity in the region (Hollenbach et al. 2012). Presence/absence frequencies and haplotypic associations for the KIR region is analyzed in the 52 populations comprising the panel and in accordance with major world regions (Africa, Middle East, Central Asia, East Asia, Europe, Americas and Oceania). These data represent the first overview of KIR population genetics in the well-documented HGDP-CEPH panel, and suggest different evolutionary histories and recent selection in the KIR gene cluster.
Danillo G. Augusto, PhD
“KIR diversity in admixed Brazilians”
Danillo G. Augusto, PhD
We contribute with two samples for this workshop: Curitiba (n=162) is from Southern Brazil (Curitiba, Parana State) of predominantly European ancestry and its KIR diversity was already reported (Augusto et al., 2012); the second one, Campo Grande (n=205), from Western Brazil. This population is extensively mixed by European and African ancestries, but also there is a significant Amerindian component. The KIR genotyping was performed by the PCR-SSP multiplex method (Kulkarni et al. 2010) using two pairs of primers for each gene. For KIR2DL2 and 2DL3, an additional pair of primers for each gene(Vilches et al. 2007) was included to confirm the results. For Curitiba, the HLA typing was performed by PCR-SSOP and previous reported (Braun-Prado 2000). For Campo Grande, HLA class I typing was performed applying the LABType® SSO reagent kits (One Lambda, USA). While Curitiba was analyzed in an anthropological study (Augusto et al. 2012b), Campo Grande was used as control of a case/control study(Augusto et al. 2012a).
“KIR Genotyping in Mexican Mestizo Populations”
Clara Gorodezky, PhD
KIR control the effectors function of NK and a subset of T cells. Beside allelic polymorphism, content of KIR genes varies among individuals producing increasing variability. To investigate the KIR gene diversity in Mexican Mestizos, we characterized the KIR gene content in Mexican belonging to three different geographical areas: State of Jalisco (100 individuals), Highlands of Mexico (257 individuals) and the West part of Mexico (124 individuals). The KIR typing was performed using the PCR-SSP, 2DP1 and 3DP1 were not tested with these primers. The primers were kindly donated by Dr Senitzer. The percentage of each KIR gene was determined by direct counting. The A and B haplotypes were deduced from the genotype data as describe elsewhere. The identification of genotype in each individual was done according to the one showed in the allelefrequencies database. The gene content, and haplotype frequencies were compared between each group using X2Y. The fourteen KIR genes tested were present in the three groups, with the frame work genes present in all individuals. Significant differences were found for 2DS4 (86%-Jalisco Vs 67.7% Highlands) and for 3DS1 (53% Jalisco Vs 37.1% -West). No significant differences were found in the AA, AB and BB haplotype distribution between individuals from the highlands and from the West of Mexico. But significant differences were found between these two groups as compared with the one from Jalisco. The frequency of AB haplotype was higher in Jalisco (50%) than in the other (West-33% & Highlands 36.9%) while the BB haplotype was decreased in Jalisco. The frequency of the AA haplotype does not show significant differences among the three groups: the highland (24.9%), the West of Mexico (25.8%), and Jalisco (30%). We found 30 different genotypes (including 1 new) in Jalisco; 47 (including 5 new) in the West and 59 (including 10 new) in the highlands. According to the number of individuals studied the higher diversity is found in the west of Mexico but considering the number of non described genotypes, the frequency is very similar for the highlands (3.8%) and the West (4%).
“KIR contribution to oral vaccine efficacy in the PROVIDE study” Janelle Noble, PhD
The PROVIDE project is a large, multi-center study aimed at addressing the immunological basis for the underperformance of oral vaccines in third world countries. We are performing both high-resolution HLA genotyping and KIR genotyping for samples enrolled for the study at the International Centre for Diarrhoeal Disease Research, Bangladesh. An introduction to the PROVIDE study was presented, along with KIR frequency data for the first 238 samples.
“KIR/HLA genotyping in Bulang, Nu, Zhuang, Yugu and an isolated Han in Yunnan province of China”
Yufeng Yao, PhD and Li Shi, PhD
The distribution of HLA, KIR genes and HLA/KIR combination in different populations in China would provide relevant immunogenetic data for the future study of viral infections, autoimmune diseases, and reproductive fitness. A total of 491 unrelated individuals in Bulang, Nu, Yugu, Zhuang ethnic groups and an isolated Han in Yunnan province were genotyped of 16 KIR genes and HLA class I genes using PCR-SSOP method with the Luminex MultiAnalyte Profiling System. HLA-A, HLA-B and HLA-C genes and HLA A-C-B haplotypes as well as KIR genes, genotypes and haplotypes were analyzed. In addition, KIR/HLA combinations were analyed and reported. Our results suggested that the HLA, KIR genes and their combinations are diverse in these populations and each population has its own characteristic distribution.
“KIR Typing of an Austrian Cohort”
Ingrid Fae, MSc
To address the question of the distribution of various KIR genes in the Austrian population we studied a population that has been used in a previous study targeting the genetic basis of body odour. The samples then were collected in a small village in Carinthia in Austrian Alps, with allegedly little population flux in the past and typed at high resolution for HLA class I and class II. The individuals were selected from 16 families; the ages were between 18 and 91. For participation in the KIR workshop of the 16th IHIW, 44 unrelated individuals were typed by SSP (Pel Freeze/Invitrogen) for the presence or absence of KIR genes.
“AFND - Resources for KIR”
This talk presented a summary of the populations collected for KIR frequencies on the AFND website, along with a demonstration of several functionalities of the website to search the data available including searches for allele and genotype frequencies, heat maps by gene, and a disease portal, among others.
Faviel Gonzalez, PhD
FUTURE DRIECTIONS
The data collected for this project represents the most comprehensive summary of global KIR gene-content variation to date and continued analysis as part of this project promises to lay a foundation for our understanding of human population diversity in the KIR complex. It is the intention of the organizers of this project to continue this work for the 17th IHIW; however, we recognize that the time has come to move beyond analysis of KIR gene-content and to work toward a more comprehensive understanding of KIR allelic polymorphism in global populations. It is our hope that as technology for KIR allele-level genotyping improves, this goal will be attainable for the 17th IHIW in 2016.
ACKNOWLEDGEMENTS
We wish to thank all participants in this project, those who contributed to discussions during the project meeting in Liverpool, and the organizers of the 16th IHIW. We thank One Lambda, Inc. (Canoga Park) for contributing SSOP Luminex reagents for this project. This work was supported by National Institutes of Health (NIH) grant U01AI067068 (JAH) awarded by the National Institute of Allergy and Infectious Diseases (NIAID). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the NIH.
REFERENCES
- Augusto DG, Lobo-Alves SC, Melo MF, Pereira NF, Petzl-Erler ML. Activating KIR and HLA Bw4 ligands are associated to decreased susceptibility to pemphigus foliaceus, an autoimmune blistering skin disease. PLoS ONE. 2012a;7(7):e39991. doi: 10.1371/journal.pone.0039991. doi:10.1371/journal.pone.0039991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto DG, Zehnder-Alves L, Pincerati MR, Martin MP, Carrington M, Petzl-Erler ML. Diversity of the KIR gene cluster in an urban Brazilian population. Immunogenetics. 2012b;64(2):143–152. doi: 10.1007/s00251-011-0565-1. doi:10.1007/s00251-011-0565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirova AA, Martin MP, McVicar DW, Carrington M. The Killer Immunoglobulin-like Receptor Gene Cluster: Tuning the Genome for Defense. Annu Rev Genomics Hum Genet. 2006 doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- Biron CA. Activation and function of natural killer cell responses during viral infections. Curr Opin Immunol. 1997;9(1):24–34. doi: 10.1016/s0952-7915(97)80155-0. [DOI] [PubMed] [Google Scholar]
- Braun-Prado K, Vieira Mion AL, Farah Pereira N, Culpi L, Petzl-Erler ML. HLA class I polymorphism, as characterised by PCR-SSOP, in a Brazilian exogamic population. Tissue antigens. 2000;56:417–427. doi: 10.1034/j.1399-0039.2000.560504.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Research. 2011;39(Database issue):D913–9. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach JA, Meenagh A, Sleator C, Alaez C, Bengoche M, Canossi A, Contreras G, Creary L, Evseeva I, Gorodezky C, Hardie RA, Karlsen TH, Lie B, Luo M, Martinetti M, Navarette C, de Oliveira DC, Ozzella G, Pasi A, Pavlova E, Pinto S, Porto LC, Santos P, Slavcev A, Srinak D, Tavoularis S, Tonks S, Trachtenberg E, Vejbaesya S, Middleton D. Report from the killer immunoglobulin-like receptor (KIR) anthropology component of the 15th International Histocompatibility Workshop: worldwide variation in the KIR loci and further evidence for the co-evolution of KIR and HLA. Tissue Antigens. 2010;76(1):9–17. doi: 10.1111/j.1399-0039.2010.01459.x. doi:10.1111/j.1399-0039.2010.01459.x. [DOI] [PubMed] [Google Scholar]
- Hollenbach JANI, Ladner MB, Single RM, Trachtenberg EA. Killer Cell Immunoglobulin-like Receptor (KIR) Gene-Content Variation in the HGDP-CEPH Populations. Immunogenetics. 2012;64:719–737. doi: 10.1007/s00251-012-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K. 13th IHWS NK/KIR Joint Report. HLA 2004: Immunobiology of the Human MHC. 2004.
- Kulkarni S, Martin MP, Carrington M. KIR genotyping by multiplex PCR-SSP. Methods Mol Biol. 2010;612:365–375. doi: 10.1007/978-1-60761-362-6_25. doi:10.1007/978-1-60761-362-6_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D, Meenagh A, Serrano-Vela JI, Moscoso J, Arnaiz-Villena A. Different Evolution Of Inhibitory And Activating Killer Immuoglobulin Receptors (KIR) In Worldwide Human Populations. The Open Immunology Journal. 2008;1:42–50. [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- Robinson J, Mistry K, McWilliam H, Lopez R, Marsh SGE. IPD-the Immuno Polymorphism Database. Nucleic Acids Research. 2010;38:D863–9. doi: 10.1093/nar/gkp879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd J, Kidd KK, Carrington M. Global diversity and evidence for co-evolution of KIR and HLA genes. Nature Genetics. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- Single RM, Martin MP, Meyer D, Gao X, Carrington M. Methods for assessing gene content diversity of KIR with examples from a global set of populations. Immunogenetics. 2008;60(12):711–725. doi: 10.1007/s00251-008-0331-1. doi:10.1007/s00251-008-0331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth MJ, Cretney E, Kelly JM, Westwood JA, Street SE, Yagita H, Takeda K, van Dommelen SL, Degli-Esposti MA, Hayakawa Y. Activation of NK cell cytotoxicity. Mol Immunol. 2005;42(4):501–510. doi: 10.1016/j.molimm.2004.07.034. [DOI] [PubMed] [Google Scholar]
- Vales-Gomez M, Reyburn HT, Erskine RA, Strominger J. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci U S A. 1998;95(24):14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70(5):415–422. doi: 10.1111/j.1399-0039.2007.00923.x. doi:10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- Vilches C, Parham P. KIR: Diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- Williams SM, Templeton AR. Race and genomics. N Engl J Med. 2003;348(25):2581–2582. doi: 10.1056/NEJM200306193482521. author reply 2581-2582. doi:10.1056/NEJM200306193482521. [DOI] [PubMed] [Google Scholar]
- Young NT, Uhrberg M. KIR expression shapes cytotoxic repertoires: a developmental program of survival. Trends Immunol. 2002;23(2):71–75. doi: 10.1016/s1471-4906(01)02113-5. [DOI] [PubMed] [Google Scholar]


