Abstract
The unicellular alga Chlamydomonas can assemble two 10 μm flagella in one hour from proteins synthesized in the cell body. Targeting and transporting these proteins to the flagella are simplified by preassembly of macromolecular complexes in the cell body. Radial spokes are flagellar complexes that are partially assembled in the cell body before entering the flagella. On the axoneme, radial spokes are “T” shaped structures with a head of 5 proteins and a stalk of 18 proteins that sediment together at 20S. In the cell body, radial spokes are partially assembled; about half of the radial spoke proteins (RSPs) form a 12S complex. In mutants lacking a single radial spoke protein, smaller spoke subassemblies were identified. When extracts from two such mutants were mixed in vitro the 12S complex was assembled from several smaller complexes demonstrating that portions of the stepwise assembly of radial spoke assembly can be carried out in vitro to elucidate the order of spoke assembly in the cell body.
Keywords: Cilia, Flagella, Chlamydomonas, RSP3
INTRODUCTION
Flagella are complex organelles composed of hundreds of different proteins [Avidor-Reiss et al., 2004; Li et al., 2004; Pazour et al., 2005] including structural elements that form the cytoskeletal framework of the flagellum, motors that power the flagellar beat, and proteins that regulate motor activity. These motors and regulatory proteins are oligomers that are attached to the axonemal microtubules with a periodicity based on multiples of the underlying tubulin dimers. During assembly of the flagellum these components must move into the flagellar compartment and attach precisely to their specific positions on the flagellar microtubules.
In the biflagellate alga Chlamydomonas reinhardtii proteins that make up the flagella are synthesized and stored in the cell body [Rosenbaum et al., 1969] so that given the proper cues flagella can be rapidly assembled. Within 60 minutes of experimental deflagellation this cell can regenerate two 10 μm flagella [Rosenbaum et al., 1969]. Even in the absence of protein synthesis two half-length, functional flagella can be assembled [Rosenbaum et al., 1969], thereby defining the minimum size of the cytoplasmic pool as the equivalent of one full-length flagellum. The pool must contain thousands of copies of the 16 polypeptides found in outer dynein arms [Sakato and King, 2004] and of the 23 radial spokes proteins (RSPs) [Piperno et al., 1981; Yang et al., 2006]. As the flagella assemble, these polypeptides must be targeted to the flagellar base, transported to the tip where assembly occurs [Johnson and Rosenbaum, 1992; Marshall and Rosenbaum, 2001; Rosenbaum and Child, 1967; Rosenbaum et al., 1969], and attached to the nascent axonemal microtubules.
One strategy that could simplify targeting, transport, and assembly of flagellar components would be to assemble individual polypeptides into larger complexes in the cell body prior to their delivery into the flagellum. In this way one targeting sequence could direct an entire complex to the flagellum where it could attach to axonemal microtubules as a completed unit. Such preassembly would reduce both the number of individual components to be transported and the amount of assembly taking place in the flagellum. Indeed, intact dynein complexes have been found in the cell body of Paramecium [Fok et al., 1994] and C. reinhardtii [Ahmed et al., 2008; Duquesnoy et al., 2009; Fowkes and Mitchell, 1998; Omran et al., 2008], assembled in preparation to moving into the flagella.
A slightly different story has emerged for assembly of radial spokes. In the flagellum, radial spokes are composed of 23 polypeptides, RSP1-23 [Piperno et al., 1981; Yang et al., 2006] and can be isolated intact from the axoneme as 20S complexes [Yang et al., 2001]. RSP1, 4, 6, 9, and 10 form the spoke head [Piperno et al., 1981], which interacts with projections from the central pair microtubules. The remaining RSPs form a spoke stalk that tethers the head to the A tubule of the outer doublet microtubules, mediated in part by RSP3 [Luck, 1977; Diener et al., 1993]. In the cell body, at least 6 RSPs form a subassembly of the radial spoke that sediments at 12S [Qin et al., 2004]. This complex enters the flagellum and assembles onto the axoneme with other RSPs to form a 20S complex. During the continual turnover of the axoneme, 20S spoke particles are returned to the cell body and so are present in the cytoplasm along with the 12S complex [Qin et al., 2004]. Thus, the radial spoke is partially assembled in the cell body and assembly is completed on the flagellar microtubules.
In the present study the assembly state of radial spokes in the cytoplasmic pool was examined in more detail in wild-type cells as well as in several radial spoke mutants. In wild-type cells the 12S complex was found to be composed of RSP1-7 and RSP9-12. Smaller subassemblies of these RSPs were identified in the cell body of mutants deficient in either RSP2, 3 or 4. By mixing extracts from these mutants, the 12S complex could be reconstituted in vitro and spoke head proteins could be attached to headless stalks. In vitro complementation of spoke assembly in these extracts from cell bodies of mutants has helped elucidate the order of assembly of spokes and the configuration of RSPs within the spoke, in somewhat the same way as T-even phage assembly was elucidated [Wood, 1980].
MATERIALS AND METHODS
Cell Strains and Culture
Wild-type Chlamydomonas reinhardtii (CC125), the cell wall-less mutant (cw92, CC503), and paralyzed flagella mutants pf1 (CC1024), pf14 (CC1032) and pf24 (CC1384) were obtained from the Chlamydomonas Culture Center (Duke University). Most “wild-type” preparations were made using a cell wall deficient strain, cw92, to facilitate cell lysis; these preparations gave the same results as wild-type, walled cells treated with autolysin and so were considered to be wild-type. Generally, cells were grown in acetate containing media, R/2 [Kindle et al., 1989] or TAP [Gorman and Levine, 1965] on a 14 hour light:10 hour dark cycle, which gave results indistinguishable from those obtained from cells grown in minimal media, MI [Sager and Granick, 1953].
Preparation of Cytoplasmic Extracts and In Vitro Reconstitution of Complexes
Cells were harvested by several rounds of centrifugation. First, cultures were concentrated at 300 × g in 250 ml conical bottles, then cells were resuspended in medium in a 50 ml tube and sedimented again at 400 × g. After decanting, the cells were transferred in a minimal amount of 10 mM Tris pH 7.4 to 4-6 2 ml microfuge tubes and the cells were sedimented for 1 minute at 3,300 × g. The cell pellets were placed on ice and resuspended in an equal volume of 10 mM Tris. Concentrated lysis buffer (10X) and protease inhibitors (1000X) were added to bring the final concentration to: 10 mM Tris pH 7.4, 5 mM MgSO4, 25 mM KCl, 150 mM NaCl, and 0.5 mM EGTA, 1 mM dithiothreitol, 2 μg/ml aprotinin, 20 μg/ml benzamidine, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 50 μg/ml trypsin inhibitor. The viscous mixture of cells (1-5 ml) was transferred to a 5 ml Omnimixer (Sorvall) vessel and cells were disrupted on ice by two 15 second pulses of the Omnimixer on setting 4. PMSF was added to the homogenate to 1 mM. The homogenate was clarified to remove flagella and cell debris either in a microfuge or at 10,000 rpm in a SW34 rotor (Sorvall) for 10 minutes and then at 100,000 × g for 20-60 minutes in a Ti50 rotor (Beckman, Brea, CA). Ideally, the clarified extract was 20 to 40 mg/ml protein. To test for in vitro assembly of radial spoke complexes, cytoplasmic extracts of spoke mutants were mixed and incubated on ice for 45-75 minutes before loading onto sucrose gradients. When using 35S-labeled cells (see below), the extract was made by vortexing the cells (ca. 5 × 108) for 30 seconds in a microfuge tube containing one half volume of glass beads. Cells with cell walls were treated with autolysin for 10 to 60 minutes prior to homogenization. Centrifugation was performed at 4°C and the extracts were kept on ice.
Sucrose Gradients
Sedimentation centrifugation was performed using sucrose gradients containing 10 to 30% sucrose in lysis buffer. Cell extract (0.2 to 0.5 ml) was loaded on 12 ml gradients and typically was centrifuged for 17 hours at 38,000 rpm in a SW41 rotor (Beckman). Standards used to calculate S-values were BSA (4.4S), aldolase (7.35S), catalase (11.3S), and thyroglobulin (19.4S). Using these standards added to cytoplasmic extracts, the S-values of three proteins in the extract were calculated and used as internal markers in subsequent gradients.
Chromatography
Cytoplasmic extracts were analyzed on either a 1 × 95 cm column packed with Sephacryl S400 or S500 (Pharmacia, GE Healthcare Life Sciences, Uppsala, Sweden) or by FPLC on a prepacked 1.6 × 60 cm S300 column (Pharmacia). Calibration standards were inactivated Serratia marcescens or 5 kb linear DNA (void volume), fibrinogen (1.98 × 10−7 cm2/s), thyroglobulin (2.6 × 10−7 cm2/s), catalase (4.1 × 10−7 cm2/s), BSA (6.3 × 10−7 cm2/s) and dATP (bed volume). The column was equilibrated with lysis buffer. Up to 2% of the column volume was loaded and 1 ml fractions were collected. Molecular mass was calculated from the S-value and diffusion coefficient [Cole et al., 1998].
Ion exchange chromatography of radioactive proteins was performed with DEAE Sepharose (Pharmacia) in a 1 ml pipette tip. Following protein binding, the resin was washed with lysis buffer containing 300 mM NaCl and radial spoke proteins were eluted with 350 mM NaCl.
Electron Microscopy
For electron microscopy of the 12S complex, flagella were extracted with 0.5% NP40 in 10 mM HEPES pH 7.4, 5 mM MgSO4, 0.5 mM EGTA, 25 mM KCl, and 1 mM dithiothreitol (HMDEK) and the axonemes were sedimented. The supernatant was fractionated on sucrose gradients as described above except that the gradients were made in HMDEK. Fractions containing the 12S or 20S complexes were pooled and were loaded onto a 1 ml Mono Q anion exchange column (Pharmacia) in HMDEK containing 300 mM NaCl. The complexes were eluted with a linear gradient of 450 – 600 mM NaCl. Fractions were analyzed on silver stained gels, the desired fractions were dialyzed against HMDEK and were negative-stained between two layers of carbon for TEM as previously described [Qin et al., 2004]. Negatives were printed, digitized, and the images were cropped and adjusted for brightness and contrast in Photoshop (Adobe System, San Jose, CA).
Metabolic Labeling of Cells
Cells were starved for sulfur by growth in medium containing 1% of the normal concentration of sulfur. Prior to labeling the cells were washed and incubated for an hour in medium containing no sulfur. Cells, 10 ml at 5 × 107 cells/ml, were deflagellated and 50 μCi/ml 35SO4 was added. Cells were allowed to regenerate for 2 hours, incubated in autolysin for 30 min, and a cell lysate was prepared as described above.
Antibodies and Immunoblots
Polyclonal antibodies against RSP1, 3, and 6 have been previously described [Williams et al., 1986; Williams et al., 1989]. The antibodies against RSP2 and 5, though previously unpublished, were generated in parallel with the others. Antibodies were affinity purified against nitrocellulose strips of blots of axonemal proteins and/or depleted by adsorption with spokeless axonemes. These techniques were only partially successful in removing extraneous bands from the blots. Genuine bands of RSPs were confirmed by their presence in wild-type axonemes and absence from pf14 axonemes, which lack radial spokes [Luck et al., 1977; Witman et al., 1976]. Anti-RSP3 antibody was affinity purified against RSP3 isolated using Ni-NTA resin (Qiagen) either from bacteria expressing RSP3::His6 or from the flagella of pf14 cells expressing RSP3::His6. Affinity purified RSP polyclonal antibodies were diluted 1:200 and developed with either colorometric or chemiluminescent methods. The anti-RSP16 antibody was prepared against bacterially-expressed protein [Yang et al., 2005].
Electrophoresis
SDS-PAGE was performed on 8% acrylamide minigels [Laemmli, 1970]. Isoelectric focusing gels contained a 1:4 mixture of 3-10 and 5-7 ampholines and were focused for 8,000 Vh (typically 400 V for 20 h or 500 V for 16 h) followed by 1 h at 800 V [O’Farrell, 1975]. Second dimension 10% acrylamide gels (13 × 11 cm) were run according to Laemmli [1970].
RESULTS
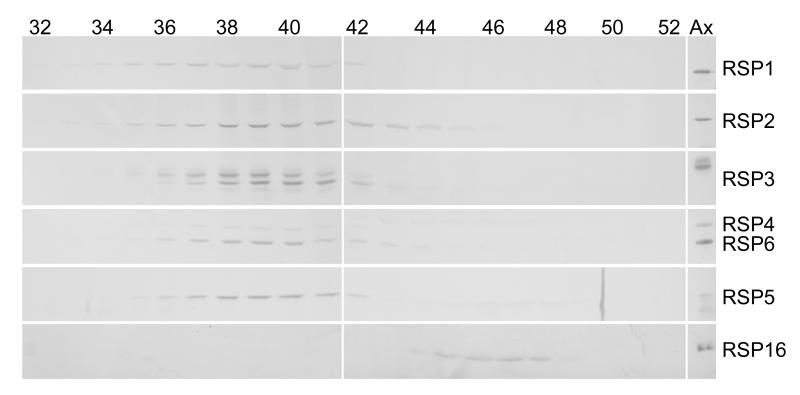
Sedimentation analysis of extracts of wild-type cell bodies previously revealed that radial spoke stalk proteins RSP2, 3 and 5 and radial spoke head proteins RSP1, 4, and 6 cosedimented in two complexes: a 20S complex, which is the fully assembled radial spoke; and a smaller subassembly of 12S [Qin et al., 2004]. To learn more about the 12S complex, cell body extracts were analyzed by gel filtration, which showed that RSP1-6 all eluted in a single peak (Fig. 1) with a diffusion coefficient of approximately 1.4 × 10−7 cm2/sec. This complex was identified as the 12S complex and not the 20S complex because it did not contain RSP16 (Fig. 1), which is characteristic of the 20S but not the 12S spoke complex [Yang et al., 2005]. RSP16 eluted from the column separate from the other RSPs, presumably as a dimer [Yang et al., 2005]; the 20S complex, which is often a very minor component in the cytoplasm, was not detected in the eluate. From the S-value and diffusion coefficient of the 12S complex, the molecular mass of the complex was calculated to be approximately 710 kD.
Figure 1.
RSP1-6 colelute from a molecular sieve column. Fractions of cell body extract eluted from a Sephadex S500 column were analyzed on immunoblots probed for RSPs as indicated on the right. The rightmost lane contains axonemes (Ax) as a standard. Column fractions are listed above the blots. Note the peak of RSP1-6 around fraction 39 (diffusion coefficient approximately 1.4 × 10−7 cm2/sec). RSP16 eluted separately, later than the other RSPs. RSP3 is known to have multiple phosphorylation forms [Piperno et al, 1981; Williams et al., 1989] and so appears as multiple bands in this and subsequent blots.
Composition and Structure of the 12S Radial Spoke Complex
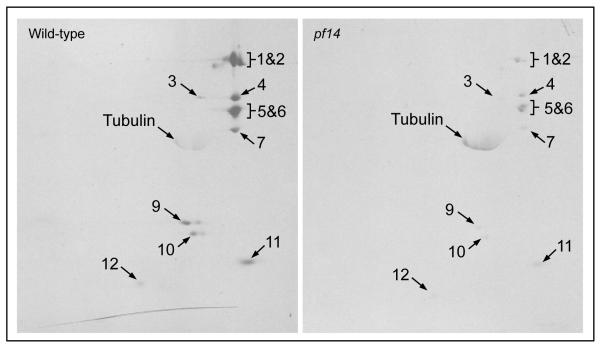
The hydrodynamic results indicate that RSP1, 2, 3, 4, 5, and 6 are contained in a complex of 710 kD in the cell body, but do not address what other proteins are present in this complex. For a more complete view of the composition of the 12S complex, wild-type cells and cells of a mutant strain (pf14) that has very little of the 12S complex (see below), were metabolically labeled with 35SO4 during flagellar regeneration. Following sucrose gradient centrifugation and DEAE chromatography of cell body extracts, proteins in the 12S complex were separated on two-dimensional gels and identified by autoradiography. As seen in Fig. 2, RSP1-6, previously identified on immunoblots as components of the 12S complex, were more prominent in extracts from wild-type than pf14 cells. In addition, five other RSPs were also identified as components of the wild-type 12S complex by their positions on the autoradiograms: RSP7, 9, 10, 11, and 12. Small proteins, such as RSP22 (LC8, 10 kD), would not have been retained on this gel.
Figure 2.
The 12S radial spoke complex is composed of at least 11 RSPs. Wild-type and pf14 cells were metabolically labeled with 35S during flagellar regeneration and the 12S radial spoke complex was isolated from cell body extracts. Autoradiography of 2-dimensional gels shows 11 RSPs were present in the complex. These RSPs are reduced in extracts from pf14 cells, which are almost devoid of the 12S complex. RSP3 labeling is low even in wild-type extracts, presumably because it contains no cysteine residues although it contains 11 methionine residues.
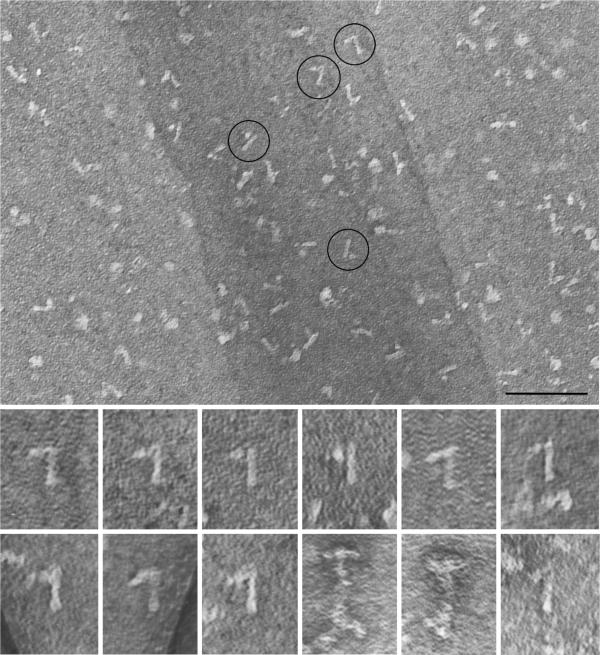
The 12S radial spoke complex was isolated from the flagellar matrix (see Materials and Methods) and was negative-stained for electron microscopy. A predominant structure in these preparations was a rod with a projection at one end forming a “7” or “L” (Fig. 3). This putative 12S complex was 28 nm (+/− 2 nm SD) long, and the width of the head was 20 nm (+/− 2 nm SD). For comparison the 20S complex was similarly purified and examined (Fig. 3, bottom right and Fig. 2 [Qin et al., 2004]). Spokes prepared by these methods were about 50 nm long with a head of 25 nm, somewhat larger than those measured in situ (40 × 20 nm [Nicastro et al., 2005]), perhaps due in part to unraveling of proteins during preparation [Qin et al., 2004]. Occasionally structures resembling those characteristic of the 12S fraction were seen in the 20S fraction (Fig. 3 lower right panel). These may represent partial disintegration of the 20S complex.
Figure 3.
The 12S spoke complex is shaped like a “7”. The 12S fractions from a gradient of soluble flagellar proteins were pooled and the RSPs were further purified by ionic exchange chromatography. Negative stained preparations showed a stalk with a projection at one end resembling a “7” or “L”. The four encircled structures are shown at twice the magnification below along with 5 other examples of the 12S complex. For comparison the last three panels show images from the 20S spoke fraction after similar purification; two images are of the intact, “T”-shaped spoke and the final image shows a complex similar to that seen in the 12S fraction. The scale bar is 100 nm.
Radial spoke complexes in radial spoke mutants
Loss of a single RSP can result in the loss of a subset of RSPs, or the entire radial spoke, from flagella [Huang, 1986; Curry and Rosenbaum, 1993; Dutcher, 1995]. The effect of several such mutations on assembly of RSP complexes in the cell body was examined. The paralyzed flagella mutant pf14 lacks radial spokes in its flagella [Piperno and Luck, 1976; Witman et al., 1976] due to a mutation in the gene encoding RSP3 [Luck et al., 1977]. No full-length RSP3 is made in these cells due to a nonsense mutation in codon 21 [Williams et al., 1989], but a small amount of truncated RSP3 is translated from a cryptic start site at codon 42, [Williams et al., 1989]. Cytoplasmic extracts of pf14 were analyzed to determine the effect of the loss of RSP3 on the integrity of the RSP complexes in the cell body.
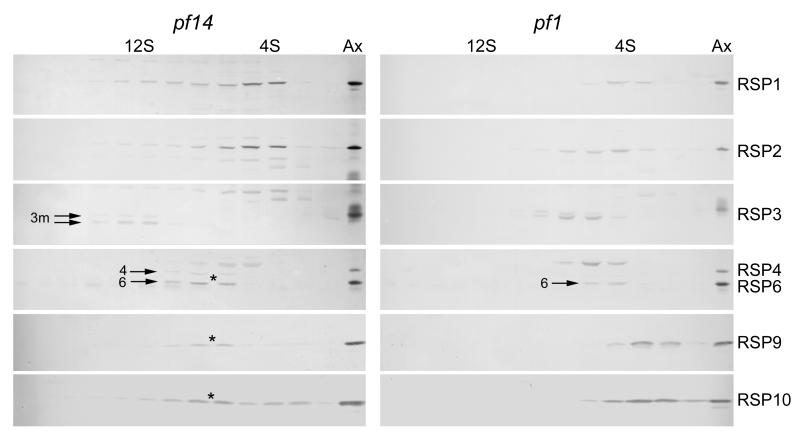
As seen in Fig. 4, in the absence of full-length RSP3 the majority of RSPs did not sediment near 12S as they did in wild-type cell extracts, indicating that RSP3 is critical for assembly of the cytoplasmic complex. The S-values and diffusion coefficients of the most prominent peaks of RSP1-6 in pf14 are listed in Table I. Also in Table I, the molecular mass of each component, estimated from the hydrodynamic data, is compared to its molecular mass calculated from its amino acid sequence. Judging from the values estimated from the hydrodynamic data of proteins isolated from pf14, RSP1 appeared to be in a monomeric state (Fig. 4 and Table I). The remaining proteins of the radial spoke head, RSP4, 6, 9, and 10 cosedimented (Fig. 4), forming a complex of 170 kD suggesting that the spoke head is intact except for the lack of RSP1. Of the stalk proteins, RSP5 appeared to be in a monomeric state (Table I), whereas RSP2 appeared to exist as a 130 kD oligomer (Table I). The other components of this RSP2 sub-complex were not identified. The small amount of truncated RSP3 (52.1 kD) that is synthesized in pf14 [Williams et al., 1989] formed a 12S complex of approximately wild-type size, which may account for some of the spreading of RSP1 and 2 into the 12S region of the gradient. Correct assembly of the truncated form of RSP3 into a 12S complex (Fig. 4) is consistent with its ability to restore motility to the flagella when overexpressed in pf14 cells [Diener et al., 1993]. RSP3, therefore, is central to the assembly of the 12S complex; without it the other RSPs that normally make up the 12S complex fall apart into smaller subassemblies or individual RSPs.
Figure 4.
The 12S radial spoke complex does not assemble in the absence of RSP3 or 4. These immunoblots of gradients of cell body extracts from pf14 (reduced amount of a truncated RSP3) and pf1 (lacking RSP4) cells show the sedimentation of the RSPs from these mutant cells. The small amount of the 12S complex in pf14 (third panel on the left) contains truncated RSP3 (3m, arrows) in both phosphorylated (upper) and not phosphorylated (lower) states. RSP4 and 6 are designated in the fourth row. The asterisks mark a peak that contains spoke head proteins RSP4, 6, 9 and 10. Pairs of adjacent gradient fractions were pooled and loaded in each lane of these gels for a more compact presentation. The axonemal markers (Ax) are from wild-type flagella and can be used to differentiate the specific RSP bands from several specious bands present in the blots. In the RSP4/6 panels, for example, the upper band (without an arrow) is not an RSP.
Table I.
Hydrodynamic properties and calculated molecular masses of cytoplasmic RSPs in pf14 and pf1 cells.
| Protein | S-value (× 10−13 sec) |
Diffusion Coefficient (× 10−7 cm2/sec) |
Molecular Mass a (kD) |
Monomer b (kD) |
|||
|---|---|---|---|---|---|---|---|
|
| |||||||
| pf14 | pf1 | pf14 | pf1 | pf14 | pf1 | ||
| RPS1 | 4.0 (0.2) c | 4.0 (0.2) | 4.0 | 4.3 | 86 | 80 | 78 |
| RSP2 | 3.8 (0.0) | 4.2 (0.1) | 2.5 | 2.1 | 130 | 170 | 76 |
| RSP3 | - | 6.4 (0.1) | - | 2.0 | - | 270 | 57 |
| RSP4 | 7.1 (0.2) | - | 3.7 | - | 170 | - | 50 |
| RSP5 | 3.2 (0.1) | 3.3 (0.1) | 6.2 | 6.0 | 44 | 47 | 48 |
| RSP6 | 7.1 (0.2) | 4.7 (0.2) | 3.5 | 3.6 | 170 | 110 | 49 |
Molecular mass calculated from the S-value and diffusion coefficient
Molecular mass of the monomer predicted from the gene sequence
Mean (standard deviation); n = 3 in most cases, 2 in a few cases
In another paralyzed mutant, pf1, the radial spoke stalk is present in the flagella, but the head is missing [Piperno et al., 1977] due to a mutation in the gene encoding RSP4 [Luck et al., 1977]. The results of gradient (Fig. 4) and gel filtration analyses of RSPs in the cell body of pf1 are presented in Table I. As in pf14, the 12S spoke complex was not present in pf1. From the hydrodynamic data, RSP1 and 5 appeared to be present as monomers and RSP2 as a small oligomer in the cell body of pf1 just as in pf14; however, RSP3 sedimented at 6.4S in a complex of 270 kD, much larger than its known molecular mass (57 kD) and consistent with its formation of homodimer [Wirschell et al., 2008] as well as binding to other RSPs. Another striking difference between pf1 and pf14 was the sedimentation of the spoke head component RSP6, which sedimented at 4.7S (versus 7.1S in pf14) in a complex of about 110 kD. The peaks of the two other spoke head proteins, RSP9 and 10, sedimented even slower than RSP6. Thus, the absence of RSP4 in pf1 drastically affects assembly of the 12S spoke complex.
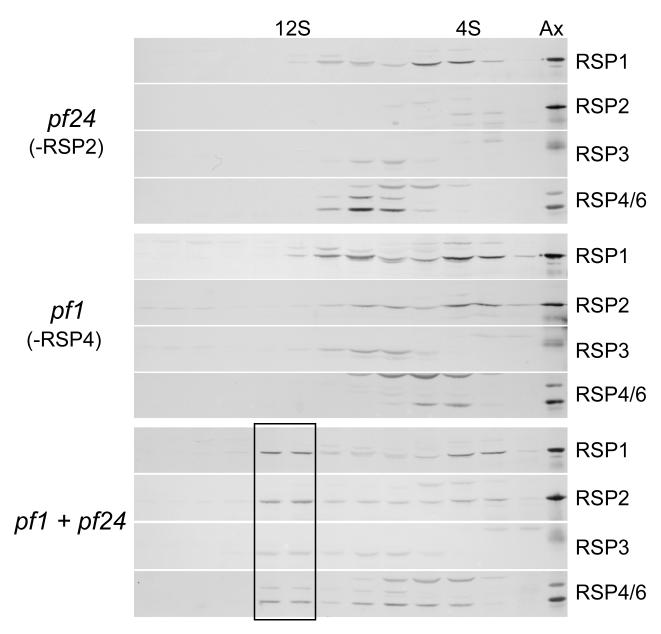
Cytoplasmic extracts of pf24, a strain that harbors a mutation in the gene encoding RSP2 [Huang, 1986; Yang et al., 2004], were also analyzed by sedimentation centrifugation. RSP1 (Fig. 5) and RSP5 (not depicted) appeared to be in a monomeric state as they did in cytoplasmic extracts from pf1 and pf14. RSP4 and 6 sedimented together in a subassembly of the head, as they did in cytoplasmic extracts from pf14. Although in gradients fractionating extracts from pf24 RSP3 partially overlapped the peak of RSP4 and 6, the peak of RSP3 was not coincident with the peak of RSP4 and 6 suggesting that these two radial spoke head proteins were detached from the RSP3 complex. Thus, in the absence of RSP2 the spoke head detaches from the remaining RSPs of the 12S complex, resulting in a smaller core subassembly that contains RSP3 together with unidentified stalk proteins.
Figure 5.
The 12S complex can assemble in vitro in a mixture of extracts from pf24 and pf1. These immunoblots show cytoplasmic extracts analyzed as in Fig. 4. The extracts from the individual mutants do not contain the 12S complex, but after mixing all of the RSPs have a secondary peak at 12S (box) indicating that this complex was assembled in vitro.
In Vitro Reconstitution of the 12S and 20S Radial Spoke Complexes
The 12S radial spoke complex partially disassembles in the absence of RSP2, 3, or 4 leaving smaller assemblages of spoke proteins in the cell body. To determine whether the 12S complex could self-assemble from such subassemblies, cell body extracts of the mutants were mixed. When cytoplasmic extracts of pf24 and pf1 were mixed, the 12S complex was assembled in vitro from the subassemblies present in the extracts (lower panels, Fig. 5). Thus, though neither of these mutants individually contained all the components of the 12S complex, when mixed, the complete complement of RSPs was present and a 12S complex similar to that found in extracts from wild-type cells was formed.
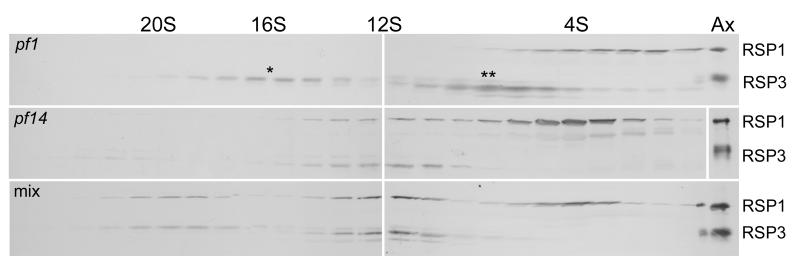
Extracts of pf1 and pf14 also were mixed to look for in vitro assembly of radial spoke complexes. As seen in the upper panel of Fig. 6, this pf1 extract was different from those shown in Figs. 4 and 5, in that it contained a prominent peak of spoke stalk protein RSP3 at 16S. This peak lacked the radial spoke head protein RSP1. Based on the presence of headless stalks in the flagella of pf1 [Piperno et al., 1977] this 16S complex is believed to be a radial spoke stalk without a head. Addition of the pf14 extract to the pf1 extract resulted in the disappearance of the 16S peak and the formation of a new peak at 20S (Fig. 6, bottom panel). Thus, the spoke head complex, contributed by the pf14 extract, was apparently attached to the 16S spoke stalk in the pf1 extract, to form the 20S radial spoke (see Discussion and Figs. 7 and 8). The 16S complex was present in two such mixing experiments, both resulting in the formation of the 20S complex. The 6.4S peak of RSP3 present in pf1 (Fig. 6, upper panel) also disappeared in the mixed extracts suggesting that it was also incorporated into a larger complex; it probably united with RSP4 and other head proteins present in pf14 to form the 12S spoke complex.
Figure 6.
Radial spoke heads can assemble onto radial spoke stalks in vitro. Cell body extracts from pf1 and pf14 cells were analyzed on sucrose gradients either separately or after mixing. Immunoblots were probed for RSP1 and 3. Note that the 16S (*) and 6.4S (**) peaks of RSP3 present in the pf1 extract disappear in the mixed extracts concomitant with the appearance of the 20S peak.
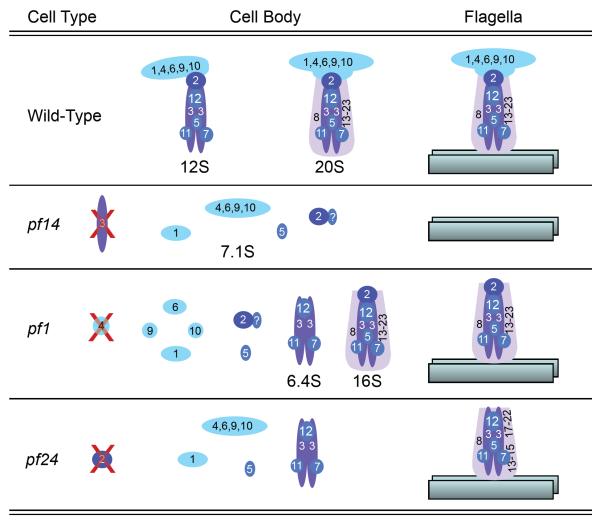
Figure 7.
This diagram illustrates the various forms of radial spoke complexes from the cytoplasm of wild-type and three radial spoke mutants. The composition of the 6.4S complex in pf1 and pf24 is not known except that it contains RSP3. The 20S and 16S complexes were not always detectable in cell extracts of wild-type and pf1 cells. The flagellar forms of spoke complexes are shown on the right, bound to an outer doublet microtubule. The stalks in the flagella of pf24 have reduced amounts of RSP16 and 23 along with RSP2 and the head proteins [Yang et al., 2005].
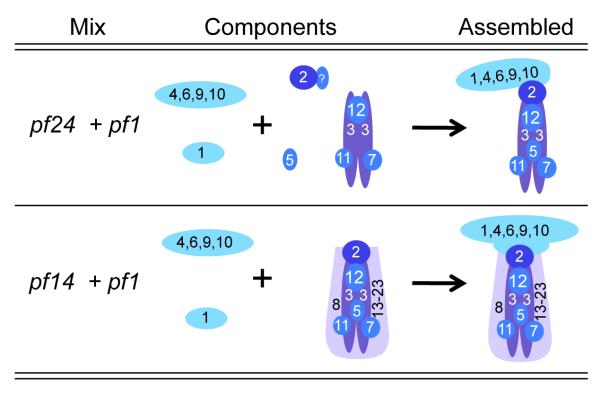
Figure 8.
This diagram illustrates the minimum assembly required to generate the 12S and 20S complexes in vitro from cytoplasmic extracts of radial spoke mutants as shown in Figs. 5 and 6.
DISCUSSION
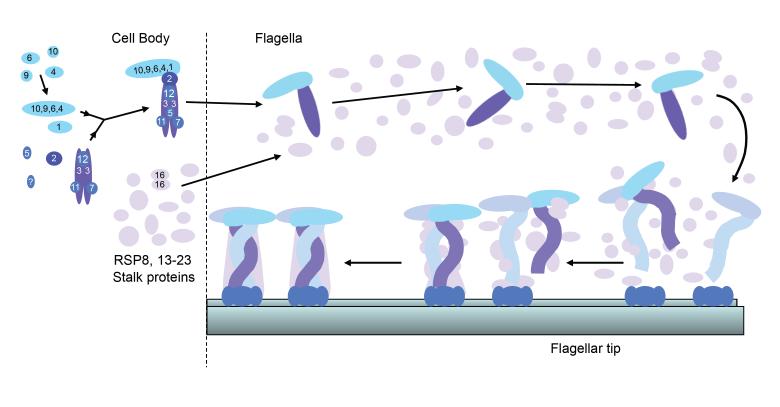
Assembly of multipeptide complexes found in an organelle such as the flagellum could occur by several different pathways. In the case of radial spokes, each of the 23 RSPs could enter the flagellum individually and sequentially assemble onto the axoneme; or, at the other extreme, intact radial spokes could assemble completely in the cell body before entering the flagellum and attaching to the outer doublet microtubules. Radial spokes appear to assemble by an intermediate pathway in which they are partially assembled in the cell body prior to entering the flagellum [Qin et al., 2004], and following transport to the flagellar tip [Johnson and Rosenbaum, 1992], these complexes unite with other RSPs to form the completed spoke on the axonemal microtubules (Fig. 9).
Figure 9.
The 12S radial spoke complex is assembled in the cell body and transported by IFT to the flagellar tip where it combines with the other RSPs to form the 20S mature spoke. Little is known about the assembly state of RSP8, 13-23 or when they attach to the 12S complex. The 20S complex is shown as a dimer of the 12S complex to illustrate how the asymmetric head of the 12S complex could give rise to the symmetric head of the mature spoke. The stalks of the two 12S complexes are shown intertwined to represent the helical quality sometimes seen in the stalk [Qin et al., 2004]. The 12S complexes may bind to a docking complex already present on the axonemal microtubules, possibly the CSC [Dymek and Smith, 2007].
12S Radial Spoke Complex
Eleven RSPs are components of a 12S subassembly of radial spokes in cell bodies of wild-type cells. This complex is an intermediate form of the radial spoke, found in the cytoplasm and matrix of the flagella [Qin et al., 2004] in transit to the tip where it combines with other spoke proteins to form the completed 20S complex [Yang et al., 2005]. The 12S complex is approximately 710 kD and comprises at least one copy each of RSP1-7 and 9-12 (summarized in Fig. 7). The total molecular mass of these RSPs is approximately 530 kD suggesting that some of the RSPs are present in more than one copy and/or other RSPs are present in the complex. Indeed, chemical crosslinking has shown that RSP3 forms a homodimer in the 12S complex [Wirschell et al., 2008]. Several other RSPs in the 12S complex are known to have homodimerization domains including RSP2, 7, and 11 [Yang et al., 2006], and so may be present in multiple copies. Furthermore, RSP20 is calmodulin [Yang et al., 2001], which binds RSP2 and may be present in multiple copies in the 12S complex [Yang et al., 2006]. Inclusion of the four dimers (RSP2, 3, 7, and 11) and four copies of calmodulin into the complex results in a calculated MW of 800 kD.
The cytoplasmic assembly state of the RSPs that were not found in the 12S complex, remains largely unknown. One exception is RSP16, an HSP40 homologue, which is present as a homodimer in the soluble fraction of flagella and subsequently is incorporated into the spoke complex [Yang et al., 2005].
Biochemically the 12S spoke complex contains all of the spoke head proteins; structurally, however, the head of the 12S complex appears to project in only one direction from the stalk. To form the “T”-shaped head characteristic of the mature spoke may require either repositioning of the head proteins relative to the stalk or the dimerization of two 12S complexes with their heads projecting in opposite directions (Fig. 9). In the 20S spoke complex, the 11 RSPs of the 12S complex generally appear to be relatively more abundant than the remaining RSPs (e.g., see Fig. 1 [Yang et al., 2006]), which is compatible with the presence of a 12S dimer in the 20S complex.
When visualized by cryo-electron microscopy, radial spokes in the flagellum are approximately 40 nm long with a head that is approximately 20 nm wide [Nicastro et al., 2005]. The 12S complex is shorter (28 nm) than the mature spoke in spite of the fact that both the spoke head and RSP3, which can bind to spokeless pf14 axonemes [Diener et al., 1993], are present. A putative spoke docking complex, which is present on the 20S complex and can bind both RSP3 and microtubules of the axoneme [Dymek and Smith, 2007], is likely responsible for at least some of the additional length specific to the 20S complex.
Subassemblies of Radial Spokes
Figure 7 summarizes the spoke proteins and sub-complexes present in cytoplasmic extracts of wild-type cells and radial spoke mutants. RSP1 and 5 were present as monomers in the cell body of mutants lacking RSP2, 3, or 4. Evidently these two RSPs are very sensitive to perturbations of radial spoke structure, losing their binding partners easily. In all three mutants there was a second, minor fraction of RSP1 that sedimented faster than the monomer. An appealing hypothesis would be that RSP1 is weakly bound to the rest of the spoke head complex so only a little RSP1 was retained during analysis of the extract; however, the minor peak of RSP1 did not cosediment precisely with the peak of RSP4 and 6 in cytoplasmic extracts of pf24 (Fig. 5). Furthermore, in pf1, in which RSP6 shifts from 7.1S to 4.7S, the minor peak of RSP1 was still present (Fig. 5), indicating that RSP1 is not interacting with either RSP6 or RSP4 (which is absent from pf1) in this peak. Neither is the minor peak of RSP1 coincident with the peak of RSP3 in pf1and pf24 (Fig. 5), so the nature of this second peak of RSP1 is not known.
In mutants lacking either radial spoke stalk protein RSP3 or the radial spoke head protein RSP4, RSP2 was found in a complex of 130-170 kD, substantially larger than a monomer (76 kD). RSP2 may dimerize [Yang et al., 2006], thereby accounting for the mass of this complex. Alternatively, some of the mass of the RSP2 complex could be due to calmodulin, RSP20 (18 kD) [Yang et al., 2001], which can bind RSP2 [Yang et al., 2004], but which would not have been detected by the methods used here.
Cytoplasmic extracts from pf14 cells revealed a radial spoke head complex of 170 kD (Table I, Fig. 7) composed of RSP4, 6, 9 and 10 (combined MW = 153 kD), but not RSP1. Interestingly, spoke head RSPs synthesized in bacteria or insect cells form a complex containing RSP1, 4, 6 and 9 but not RSP10, in vitro [Kohno et al., 2011]. The reason for the difference in these assemblages is not known, but the complex seen in pf14 probably also exists in wild-type cells. In a gradient fractionating cell extract from wild-type cells the 12S peak of RSP6 spreads, more than the other RSPs, toward the top of the gradient (Fig. 1 [Qin et al., 2004]). Spreading of the peak of this head protein may be due to the presence of the 7.1S head complex in the gradient. In cytoplasmic extracts from pf1, lacking RSP4, this head subassembly seen in pf14 was not present and RSP6 was found in a smaller form that sedimented at 4.7S. RSP6 did not appear to be bound to RSP9 or RSP10 in pf1; though there was some overlap, especially of RSP10, with RSP6, their peaks did not cosediment.
Whereas pf14 contains a sub-complex of the spoke head, pf1 contains what may be the core of the stalk and 12S complex. This 270 kD complex contains RSP3 but lacks all radial spoke head proteins, RSP2 and RSP5. Although a secondary peak of RSP2 cosedimented with the peak of RSP3 (Fig. 4), RSP2 does not appear to be a part of this complex because RSP3 sediments at the same rate in extracts that lack RSP2 (pf24). The 270 kD stalk complex is large enough to contain a homodimer of RSP3 [Wirschell et al., 2008] and the other known components of the 12S complex including RSP7, 11 and 12, but the presence of these proteins in the complex has not been assessed.
Rarely, the cytoplasm of pf1 cells contained a much larger complex that sedimented at 16S (upper panel, Fig. 6). This complex sediments at a rate similar to that of a spoke complex isolated from the flagella of pf17 [Yang et al., 2001], which like pf1, contain headless spokes. This complex was not seen in wild-type cells and does not appear to be characteristic of the normal pathway of spoke assembly; rather it is apparently a headless stalk derived from the incomplete assembly of spokes in the flagella of pf1. Like the 20S spoke found in the cell body of wild-type cells [Qin et al., 2004], the 16S complex may be present in the cytoplasm of pf1 cells due to the turnover of headless stalks in the flagella and their transport back to the cell body. The reason for the variation in abundance of the 16S complex in the cell body of pf1 (compare Figs. 4, 5 and Fig. 6) was not determined, but it may reflect the amount of turnover occurring in the flagella at a given time [Qin et al., 2004]. An analogous, but somewhat smaller, stalk complex would be expected in extracts of pf24 as well, but such a complex was never identified.
In Vitro Assembly of Radial Spokes
In vitro assembly of the 12S and 20S radial spoke complexes in cell body extracts from radial spoke mutants is summarized in Fig. 8. The 12S complex was reconstituted in a mixture of extracts from pf24 and pf1, with the missing proteins, RSP2 and 4, contributed by the complementary extracts. All other components of the 12S complex examined were found in the reconstituted 12S complex (Fig. 5). In vitro assembly of the 12S complex required that two monomers (RSP1 and 5), RSP2, and the radial spoke head subassembly containing RSP4, 6, 9, and 10, assembled onto the core stalk complex containing RSP3 (Fig. 8).
The 20S radial spoke was also reconstituted in vitro by mixing cytoplasmic extracts from pf1 and pf14, mutants that lack RSP4 and RSP3, respectively. Successful formation of the 20S complex required the addition of the radial spoke head complex (RSP4, 6, 9 and 10) contributed by the pf14 extract and RSP1 to the 16S headless stalk complex contributed by the pf1 extract (Fig. 8). This assembly appeared to occur efficiently because the 16S radial spoke complex present in the pf1 extract disappeared in the pf1/pf14 mixture when the 20S complex was formed. Although addition of the radial spoke head proteins to the 16S stalk occurred in vitro, this is not part of the normal pathway of assembly in which the spoke head is attached to a subset of stalk proteins to form a 12S complex (Fig. 9); it may be similar, however, to what happens in the flagella of pf1 during dikaryon rescue. When pf1 gametes are mated to wild-type gametes, cell pairs fuse to form dikaryons with four flagella; two of these are wild-type, the other two are paralyzed, missing spoke heads. The paralyzed flagella soon become motile presumably as spoke heads from the wild-type cytoplasm are added to the headless, pf1 stalks [Luck et al., 1977]. In a similar way, spoke heads added to headless stalks in the vitro reconstitution experiment. Mating pf1 with pf14 also produces dikaryons with four motile flagella (Diener, unpublished observation) validating the in vitro complementation experiment.
The dikaryon rescue experiment also presents an interesting question of assembly: if the spoke head proteins are normally present in the cell body as part of a 12S complex, how were spoke heads added to the headless stalks in the flagella of pf1? As described above, a spoke head complex of RSP4, 6, 9 and 10 is present in pf14 and probably wild-type cell bodies. This complex evidently can enter the flagellum apart from the rest of the 12S complex and assemble onto the stalks present in pf1 flagella. It is also interesting to note that the 12S complex never assembles in the cytoplasm of pf1, yet the radial spoke stalks assemble in the flagella of pf1. Therefore, sequences that target the spoke to the flagella, must be present in both the spoke stalk and spoke head.
In vitro assembly of the 20S complex by the coalescence of subunits smaller than the 16S complex in the mixture of cytoplasmic extracts of pf1 and pf14 seems unlikely, because such fabrication was not seen in combinations of extracts when the 16S complex was not present. Neither does the 20S complex form in the cell bodies of mutants that fail to assemble flagella (bld1, bld2, and ift88) [Qin et al., 2004] in which all the radial spoke components are presumably present in normal amounts. What prevents the 12S complex and other RSPs from forming the mature spoke complex in the cytoplasm is not known, but there may be components at the tip of the flagellum [Yang et al., 2005] or on the flagellar microtubules themselves that are necessary to integrate the various spoke constituents into the complete 20S spoke. In keeping with this hypothesis, a calmodulin-spoke complex (CSC), composed of calmodulin and three other proteins, has been identified, which may serve as a spoke-docking complex [Dymek and Smith, 2007]. This complex is bound to the 20S spoke complex extracted from axonemes with KI; furthermore, the CSC can be extracted with KI from the axonemes of the radial spokeless mutant pf14 [Dymek and Smith, 2007]. These results indicate that the CSC is an integral part of the radial spoke, yet it assembles onto the axonemal microtubules independent of the RSPs. In the cytoplasm, however, the CSC forms a complex that is not bound to RSPs [Dymek and Smith, 2008]. The attachment of the RSPs to the CSC, therefore only appears to occur in the flagella. One testable model is that assembly of the intact radial spoke can only occur when 12S spoke complexes bind to the CSC that is already on the axonemal microtubules.
Study of the subassembly complexes of radial spokes and their assembly in vitro has begun to yield information about the sequence of assembly and positioning of RSPs within the mature spoke. Assembly of multipolypeptide complexes in the cytoplasm as seen with radial spokes and outer dynein arms may simplify targeting of many flagellar proteins and facilitate assembly during flagellar growth. This modular preassembly of protein complexes may be a general mechanism used in organelle biogenesis.
ACKNOWLEDGMENTS
The authors thank Ben Williams and Mark Velleca who generated the antibodies used in this study and Hue Tran for preparing innumerable cultures of cells. We are also grateful to the many members of the Rosenbaum lab, whose thoughts and ideas have contributed to this project. This work was supported by NIH Grant #GM14642 to JLR, R37GM51173 to WSS and GM90162 to PY.
REFERENCES
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183(2):313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117(4):527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141(4):993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry AM, Rosenbaum JL. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil Cytoskeleton. 1993;24:224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- Diener DR, Ang LH, Rosenbaum JL. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J Cell Biol. 1993;123(1):183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquesnoy P, Escudier E, Vincensini L, Freshour J, Bridoux AM, Coste A, Deschildre A, de Blic J, Legendre M, Montantin G, Tenreiro H, Vojtek AM, Loussert C, Clement A, Escalier D, Bastin P, Mitchell DR, Amselem S. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am J Hum Genet. 2009;85(6):890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 1995;11(10):398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179(3):515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Smith EF. Defining a role for radial spoke/CSC interaction during axoneme assembly and ciliary motility. Molec Biol Cell. 2008;19(suppl) abstract #269. [Google Scholar]
- Fok AK, Wang H, Katayama A, Aihara MS, Allen RD. 22S Axonemal dynein is preassembled and functional prior to being transported to and attached on the axonemes. Cell Motil Cytoskeleton. 1994;29(3):215–224. doi: 10.1002/cm.970290304. [DOI] [PubMed] [Google Scholar]
- Fowkes ME, Mitchell DR. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol Biol Cell. 1998;9(9):2337–2347. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54(6):1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang BP-H. Chlamydomonas reinhardtii: A model system for the genetic analysis of flagellar structure and motility. Intl Rev Cytol. 1986;99:181–215. [Google Scholar]
- Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol. 1992;119(6):1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL, Schnell RA, Fernandez E, Lefebvre PA. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109(6):2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno T, Wakabayashi K, Diener D, Rosenbaum J, Kamiya R. Subunit interactions with the Chlamydomonas flagellar spokehead. Cytoskeleton: 2010;68:237–246. doi: 10.1002/cm.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li J, Gerdes J, Haycraft C, Fan Y, Teslovich T, May-Simera H, Li H, Blacque O, Li L, Leitch C, Lewis R, Green J, Parfrey P, Leroux M, Davidson W, Beales P, Guay-Woodford L, Yoder B, Stormo G, Katsanis N, Dutcher S. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117(4):541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Luck D, Piperno G, Ramanis Z, Huang B. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc Natl Acad Sci USA. 1977;74(8):3456–3460. doi: 10.1073/pnas.74.8.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol. 2001;155(3):405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, McIntosh JR, Baumeister W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102(44):15889–15894. doi: 10.1073/pnas.0508274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Omran H, Kobayashi D, Olbrich H, Tsukahara T, Loges NT, Hagiwara H, Zhang Q, Leblond G, O’Toole E, Hara C, Mizuno H, Kawano H, Fliegauf M, Yagi T, Koshida S, Miyawaki A, Zentgraf H, Seithe H, Reinhardt R, Watanabe Y, Kamiya R, Mitchell DR, Takeda H. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456(7222):611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G, Agrin N, Leszyk J, Witman G. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170(1):103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Luck DJL. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1977;74(4):1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Ramanis Z, Luck DJL. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88(1):73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Luck DJ. Phosphorylation of axonemal proteins in Chlamydomonas reinhardtii. J Biol Chem. 1976;251(7):2161–2167. [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol. 2004;164(2):255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Child FM. Flagellar regeneration in protozoan flagellates. J Cell Biol. 1967;34(1):345–364. doi: 10.1083/jcb.34.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas: the use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41(2):600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies with Chlamydomonas reinhardi. Ann N Y Acad Sci. 1953;56(5):831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Sakato M, King SM. Design and regulation of the AAA+ microtubule motor dynein. J Struct Biol. 2004;146(1-2):58–71. doi: 10.1016/j.jsb.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Williams BD, Mitchell DR, Rosenbaum JL. Molecular cloning and expression of flagellar radial spoke and dynein genes of Chlamydomonas. J Cell Biol. 1986;103(1):1–11. doi: 10.1083/jcb.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Velleca MA, Curry AM, Rosenbaum JL. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J Cell Biol. 1989;109(1):235–245. doi: 10.1083/jcb.109.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Zhao F, Yang C, Yang P, Diener D, Gaillard A, Rosenbaum JL, Sale WS. Building a radial spoke: flagellar radial spoke protein 3 (RSP3) is a dimer. Cell Motil Cytoskeleton. 2008;65(3):238–248. doi: 10.1002/cm.20257. [DOI] [PubMed] [Google Scholar]
- Witman GB, Fay R, Plummer J. Chlamydomonas mutants: Evidence for the roles of specific axonemal components in flagellar movement. In: Goldman R, Pollard T, Rosenbaum J, editors. Cell Motility. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1976. pp. 969–986. [Google Scholar]
- Wood W. Bacteriophage T4 morphogenesis as a model for assembly of subcellular structure. Q Rev Biol. 1980;55:353–367. doi: 10.1086/411980. [DOI] [PubMed] [Google Scholar]
- Yang C, Compton MM, Yang P. Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Molec Biol Cell. 2005;16(2):637–648. doi: 10.1091/mbc.E04-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J Cell Biol. 2001;153(6):1315–1326. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, Rosenbaum JL, Witman GB. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119(Pt 6):1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yang C, Sale WS. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryotic Cell. 2004;3(1):72–81. doi: 10.1128/EC.3.1.72-81.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yang C, Wirschell M, Davis S. Novel LC8 mutations have disparate effects on the assembly and stability of flagellar complexes. J Biol Chem. 2009;284(45):31412–31421. doi: 10.1074/jbc.M109.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]