Abstract
Type III secretion is a tightly controlled virulence mechanism utilized by many gram negative bacteria to colonize their eukaryotic hosts. To infect their host, human pathogenic Yersinia spp. translocate protein toxins into the host cell cytosol through a preassembled Ysc-Yop type III secretion device. Several of the Ysc-Yop components are known for their roles in controlling substrate secretion and translocation. Particularly important in this role is the YopN and TyeA heterodimer. In this study, we confirm that Y. pseudotuberculosis naturally produce a 42 kDa YopN-TyeA hybrid protein as a result of a +1 frame shift near the 3 prime of yopN mRNA, as has been previously reported for the closely related Y. pestis. To assess the biological role of this YopN-TyeA hybrid in T3SS by Y. pseudotuberculosis, we used in cis site-directed mutagenesis to engineer bacteria to either produce predominately the YopN-TyeA hybrid by introducing +1 frame shifts to yopN after codon 278 or 287, or to produce only singular YopN and TyeA polypeptides by introducing yopN sequence from Y. enterocolitica, which is known not to produce the hybrid. Significantly, the engineered 42 kDa YopN-TyeA fusions were abundantly produced, stable, and were efficiently secreted by bacteria in vitro. Moreover, these bacteria could all maintain functionally competent needle structures and controlled Yops secretion in vitro. In the presence of host cells however, bacteria producing the most genetically altered hybrids (+1 frameshift after 278 codon) had diminished control of polarized Yop translocation. This corresponded to significant attenuation in competitive survival assays in orally infected mice, although not at all to the same extent as Yersinia lacking both YopN and TyeA proteins. Based on these studies with engineered polypeptides, most likely a naturally occurring YopN-TyeA hybrid protein has the potential to influence T3S control and activity when produced during Yersinia-host cell contact.
Introduction
Invertebrate and vertebrate hosts are potentially subject to a myriad of bacterial infections. Scores of these infectious agents are Gram-negative bacterial pathogens that colonize their eukaryotic hosts through a virulence strategy that involves having a type III secretion system (T3SS) as the centrepiece [1,2]. Similar systems also function in the biosynthesis of the flagellum motility organelle and in establishing mutualistic interactions between bacteria and their eukaryotic hosts. At least in pathogenic bacteria, target cell contact triggers a pre-assembled needle-like T3SS consisting of ~25 proteins spanning the bacterial envelope to become competent for delivery of newly synthesized effector toxins direct from the bacterial interior into the host cell cytosol in a one- or two-step process that presumably involves effector transit through a translocon pore formed in the host cell membrane [3]. At least three types of protein substrates are known to be secreted by a T3SS [4]; early substrates are those that contribute to the final phase of polymerizing the external needle appendage, middle substrates are pore-forming translocator proteins that bridge the gap between the protruding needle and host cell surface, thereby facilitating the passage of late substrates into the host cell interior. These late substrates are the effector toxins that harbour diverse enzymatic activities to manipulate host-cell signalization. This can affect many aspects of cell and host physiology – for instance immune system responsiveness, to promote bacterial survival in the host and host-to-host transmission [5].
This functional demarcation of substrate classes implies that their production and subsequent secretion is needed only at discrete phases during T3S activity. To ensure this concise temporal and spatial control, multiple layers of regulatory control are needed [1,6-10]. Common to all T3SSs appears to be a substrate switching mechanism which, following assembly of the needle extension, triggers a change in substrate secretion from early needle components to the middle translocators and late effectors. This notion is based on a plethora of studies that have dissected aspects of the complex crosstalk between YscU-like, YscP-like and YscI-like protein families that are highly conserved in both flagella and non-flagella T3SSs [11-24].
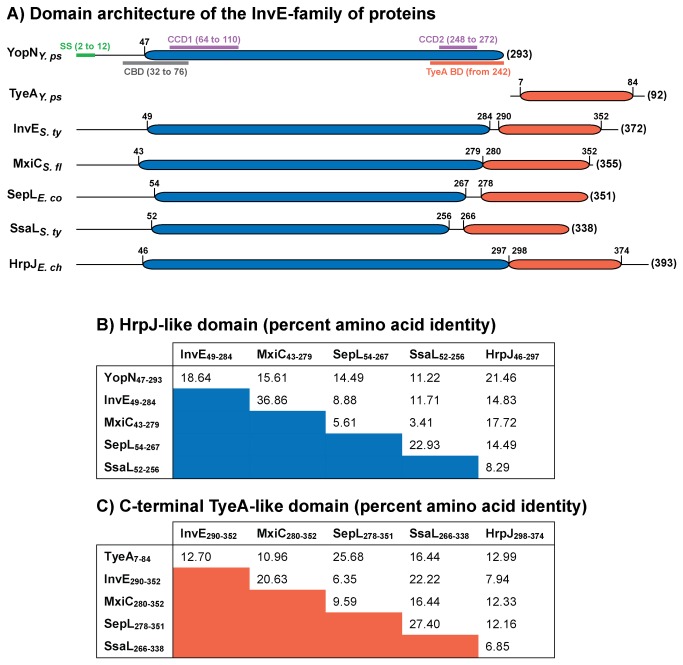
It is also anticipated that a secretion order may exist among the middle and late secretion substrates. This is based on the assertion that a translocon pore should form in the host cell plasma membrane prior to the secretion of the translocated toxins. Indeed, accumulating genetic studies are providing evidence that in some bacteria middle substrates are prioritized for secretion over late substrates. A growing heterogeneous family of proteins headlined by InvE of S. enterica Typhimurium are being reported for their roles in ensuring translocator secretion before effector secretion in their respective bacteria. InvE directly recognizes translocator-chaperone complexes that may prioritize their secretion [25,26]. Alternatively, the C-terminus of SepL may specifically bind effector substrates to stall their T3S from enteropathogenic Escherichia coli [27-29] or MxiC may bind the system ATPase creating a blockade that similarly inhibits effector secretion by Shigella flexneri [19,30,31]. No matter how it is achieved, these studies identify intrinsic mechanisms for orchestrating hierarchical secretion among the T3S translocator and effector substrates. A Conserved Domain Database (CDD) [32] search revealed a distinct HrpJ-like domain (denoted pfam07201) architecture in all of them (Figure 1A), although only a modest amount of sequence identity is shared between them [33]. For example, amino acid identity within the HrpJ-like domain is highest (36.86%) between InvE and MxiC, but then sharply drops away for the others (Figure 1B).
Figure 1. Domain architecture and sequence identity among the InvE-family of T3SS proteins.
YopN and TyeA from human pathogen Yersinia sp. are two distinct polypeptides (A). In several other T3SSs, homologues to both YopN and TyeA exist as a single polypeptide (for example, InvE, MxiC, SepL, SsaL and HrpJ). Numbers in parentheses indicate the full length (in amino acids) of each protein. Other numbers indicate the bordering amino acids that demarcate YopN homology (blue shade) that is defined Pfam as a HrpJ-like domain (pfam07201), TyeA homology (orange shade) or functionally relevant regions of YopN (various coloured solid lines). The schematic illustration of YopN and TyeA homology domains within the InvE-family was derived from comprehensive multiple sequence alignments coupled to a Conserved Domain Database (CDD) [32,33]. SS, secretion signal [80]; CBD, T3S chaperone (YscB-SycN heterodimer) binding domain [92]; CCD1 and CCD2, coiled-coil domain 1 and 2 [61]; TyeA BD, TyeA binding domain [61,92]. Percent amino acid sequence identity between the InvE family of proteins was determined by BLASTP analysis for the N-terminal HrpJ-like domain (equivalent to YopN) (B) and the C-terminal TyeA-like domain (C). Representative sequences were retrieved from the NCIB genome database archived with the following GI reference numbers shown in parentheses: Y. ps, Yersinia pseudotuberculosis YopN (48634); Y. ps TyeA (48635); S. ty, Salmonella enterica Typhimurium InvE (16766203); S. fl, Shigella flexneri MxiC (12329090); E. co, Escherichia coli SepL (215267040); S. ty SsaL (16419933); E. ch, Erwinia chrysanthemi HrpJ (28628125).
InvE-family homologues were also reported within the plasmid encoded Ysc-Yop T3SS carried by the infamous Yersinia pestis, the etiological agent of plague, and the less aggressive foodborne enteropathogens Y. enterocolitica and Y. pseudotuberculosis. Intriguingly, this homology was partitioned over two proteins; YopN with a HrpJ-like domain displayed moderate identity to the N-terminus and TyeA followed with modest identity over the C-terminus of each InvE-family member (Figure 1A) [33]. The region of YopN containing the HrpJ-like domain was most identical at the amino acid level to HrpJ (21.46%) (Figure 1B), while TyeA amino acid sequence most closely resembled the C-terminal region of SepL (25.68%) (Figure 1C). The YopN and TyeA proteins do function as a 42kDa YopN-TyeA complex to control Yop substrate secretion [34-36]. Moreover, YopN function is required for the polarized translocation of T3S effectors into the host eukaryotic cell [35,37,38]. Curiously, Y. pestis but not Y. enterocolitica were observed to produce a singular 42 kDa YopN-TyeA hybrid polypeptide; a consequence of a +1 frame shift that occurs during translation of the 3’ -prime end of yopN mRNA. The produced hybrid protein was competent for general T3S control [39].
The mechanisms of Yop secretion control in Yersinia are complex and require input from multiple contributing proteins that function at different levels and in response to different environmental cues [9,10,24,40-44]. This study had the goal to further investigate the biological significance of the YopN-TyeA hybrid given the documented roles played by YopN and TyeA in Yop secretion control and their homology to the InvE-family. To do so, we first confirmed the natural production and T3S of the singular YopN-TyeA hybrid in Y. pseudotuberculosis. Next, an in cis site directed mutagenesis approach generated Y. pseudotuberculosis that either produced predominately the YopN-TyeA hybrid by introducing +1 frame shifts to yopN after codons 278 or 287, or produced only singular YopN and TyeA polypeptides by introducing yopN sequence from Y. enterocolitica. Like parental Yersinia, mutants that produced solely the YopN-TyeA hybrid maintained T3SS assembly and function in vitro and could also successfully establish systemic colonization during competitive infections of mice. In light of this functionality, a possible mechanism for regulating the natural formation of the YopN-TyeA hybrid was explored.
Materials and Methods
Bacterial Strains, Plasmids and Growth Conditions
Strains and plasmids used in this study are listed in Table 1. Routine bacterial culturing of E. coli and Y. pseudotuberculosis was performed at 37°C and 26°C respectively, typically in Luria Bertani (LB) broth. When examining protein expression and secretion from Yersinia, strains were grown in brain heart infusion (BHI) broth, both in minus calcium (BHI supplemented with 5 mM EGTA and 20mM MgCl2 – T3S permissive medium) and in plus calcium (2.5mM CaCl2 – T3S non-permissive medium) conditions. In both cases, bacteria were grown in the presence of 0.025% (v/v) Triton X-100. This treatment detached Yops prone to associate to the bacterial surface, thereby ensuring that our T3S analysis would include all Yops secreted beyond the bacterial envelope [45]. When appropriate, antibiotics at the following concentrations were used to select for plasmid maintenance during culturing: Carbinicillin (Cb) 100µg/ml, Chloramphenicol (Cm) 25µg/ml, and Kanamycin (Km) 50µg/ml.
Table 1. Strains and plasmids used in this study.
| Strains and plasmids | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strain | ||
| E. coli | ||
| DH5 | F−, recA1, endA1, hsdR17, supE44, thi-1, gyrA96, relA1 | Vicky Shingler |
| S17-1λpir | recA, thi, pro, hsdR- M +, SmR, <RP4:2-Tc:Mu:Ku:Tn7>TpR | [86] |
| Y. pseudotuberculosis | ||
| YPIII/pIB102 | yadA::Tn5, KmR (wild type) | [47] |
| YPIII/pIB75 | pIB102, yscU in frame deletion of codons 25-329, KmR | [87] |
| YPIII/pIB75-26 | pIB102, yscU and lcrQ double mutant, KmR | [45] |
| YPIII/pIB202 | pIB102, yscF in frame deletion of codons 11-69, KmR | [88] |
| YPIII/pIB619 | pIB102, yopB and yopD full length deletion, KmR | [89] |
| YPIII/pIB82 | pIB102, near full length deletion of yopN, KmR | [90] |
| YPIII/pIB801a | pIB102, tyeA in frame deletion of codons 19-59, KmR | This study |
| YPIII/pIB8201a | pIB102, in frame double deletion of yopN and tyeA, KmR | This study |
| YPIII/pIB8214 | pIB102, yopN allele with a missense mutation at codon 286 (LysAAA→IleATA) to give a YopNYps→Yen, KmR | This study |
| YPIII/pIB8205 | pIB102, yopN allele with a +1 frameshift deletion mutation (‘T’) after codon 278 to give a YopN278(F+1)TyeA chimera, KmR | This study |
| YPIII/pIB8206 | pIB102, yopN allele with a +1 frameshift deletion mutation (‘T’) after codon 278 and the conservative mutations at codons 283 and 284 (GlnCAG→CAA and ArgAGG→CGT) that partially disrupts the presumed tyeA Shine-Dalgarno sequence to give a YopN278(F+1), SDTyeA chimera, KmR | This study |
| YPIII/pIB8210 | pIB102, yopN allele with a +1 frameshift deletion mutation (‘A’) after codon 287 to give a YopN287(F+1)TyeA chimera, KmR | This study |
| YPIII/pIB8211 | pIB102, yopN allele with a +1 frameshift deletion mutation (‘A’) after codon 287 and the conservative mutations at codons 283, 284 and 285 (SerTCA→TCC, GluGAG→GAA and GlyGGT→GGC) that partially disrupts the presumed tyeA Shine-Dalgarno sequence to give a YopN287(F+1), SDTyeA chimera, KmR | This study |
| YPIII170/pIB102 | In cis polar mutation of YPK_3687 in the parental background, CmR, KmR | This study |
| YPIII170/pIB8201a | In cis polar mutation of YPK_3687 in the yopN and tyeA background, CmR, KmR | This study |
| YPIII170/pIB8214 | In cis polar mutation of YPK_3687 in the YopNYps→Yen-producing background, CmR, KmR | This study |
| YPIII170/pIB8205 | In cis polar mutation of YPK_3687 in the YopN278(F+1)TyeA-producing background, CmR, KmR | This study |
| YPIII170/pIB8206 | In cis polar mutation of YPK_3687 in the YopN278(F+1), SDTyeA-producing background, CmR, KmR | This study |
| YPIII170/pIB8210 | In cis polar mutation of YPK_3687 in the YopN287(F+1)TyeA-producing background, CmR, KmR | This study |
| YPIII170/pIB8211 | In cis polar mutation of YPK_3687 in the YopN287(F+1), SDTyeA-producing background, CmR, KmR | This study |
| YPIII/pIB8215 | pIB102, yopN allele with a conservative mutation at codon 278 (PheTTT→PheTTC) to give a YopNF278F, KmR | This study |
| YPIII/pIB8216 | pIB102, yopN allele with a missense mutation at codon 279 (TrpTGG→PheTTC) to give a YopNW279F, KmR | This study |
| YPIII/pIB8217 | pIB102, yopN allele with a deletion of codon 278 to give a YopNΔ278F, KmR | This study |
| YPIII/pIB8218 | pIB102, yopN allele with a deletion of codon 279 to give a YopNΔ279W, KmR | This study |
| Y. enterocolitica | ||
| 8081/pYVe8081 | clinical isolate, biotype 1b (serotype 0:8) | [91] |
| Plasmid | ||
| pTZ57R/T | PCR cloning and sequencing vector, CbR | Thermo Scientific |
| pMMB208 | Expression vector, CmR | [49] |
| pAA269 | pMMB208 with full-length yopN and tyeA including native upstream SD sequences, CmR | This study |
| pAA271 | pMMB208 with chimeric yopN 278(F+1), SD tyeA including native upstream SD sequences, CmR | This study |
| pAA304 | pMMB208 with full-length yopN and tyeA-flag® including native upstream SD sequences, CmR | This study |
| pAA305 | pMMB208 with full-length yopN Yps→Yen and tyeA-flag® including native upstream SD sequences, CmR | This study |
| pAA306 | pMMB208 with chimeric yopN 278(F+1)tyeA-flag® including native upstream SD sequences, CmR | This study |
| pAA307 | pMMB208 with chimeric yopN 278(F+1), SD tyeA-flag® including native upstream SD sequences, CmR | This study |
| pAA308 | pMMB208 with chimeric yopN 287(F+1)tyeA-flag® including native upstream SD sequences, CmR | This study |
| pAA309 | pMMB208 with chimeric yopN 287(F+1), SD tyeA-flag® including native upstream SD sequences, CmR | This study |
| pUA066 | pNQ705-derived mutagenesis vector for the construction of a polar insertion in YPK_3687, CmR | This study |
| pDM4 | Suicide vector with oriR6K, sacB, CmR | [46] |
| pAA256 | SalI/XbaI PCR fragment of tyeA with a in frame deletion of codons 19-59 in pDM4, CmR | This study |
| pSF019 | SalI/XbaI PCR fragment flanking upstream of yopN and downstream of tyeA in pDM4, CmR | This study |
| pAA251 | SalI/XbaI PCR fragment of yopN with a missense mutation at codon 286 (LysAAA→IleATA) in pDM4, CmR | This study |
| pAA242 | SalI/XbaI PCR fragment of yopN with a +1 frameshift deletion mutation (‘T’) after codon 278 in pDM4, CmR | This study |
| pAA243 | SalI/XbaI PCR fragment of yopN with a +1 frameshift deletion mutation (‘T’) after codon 278 and the conservative mutations at codons 283 and 284 (GlnCAG→CAA and ArgAGG→CGT) in pDM4, CmR | This study |
| pAA247 | SalI/XbaI PCR fragment of yopN with a +1 frameshift deletion mutation (‘A’) after codon 287 in pDM4, CmR | This study |
| pAA248 | SalI/XbaI PCR fragment of yopN with a +1 frameshift deletion mutation (‘T’) after codon 278 and the conservative mutations at codons 283, 284 and 285 (SerTCA→TCC, GluGAG→GAA and GlyGGT→GGC) in pDM4, CmR | This study |
| pAA252 | SalI/XbaI PCR fragment of yopN with a conservative mutation at codon 278 (PheTTT→PheTTC) in pDM4, CmR | This study |
| pAA253 | SalI/XbaI PCR fragment of yopN with a missense mutation at codon 279 (TrpTGG→PheTTC) in pDM4, CmR | This study |
| pAA254 | SalI/XbaI PCR fragment of yopN with a deletion of codon 278 in pDM4, CmR | This study |
| pAA255 | SalI/XbaI PCR fragment of yopN with a deletion of codon 279 in pDM4, CmR | This study |
Mutant Construction
The various mutated yopN alleles were created by the overlap PCR method using the various primer pairs listed in Table S1. PCR fragments were cloned directly into pTZ57R/T using the InsTAclone PCR cloning strategy (Thermo Scientific) and each mutation confirmed by sequence analysis (Eurofins MWG Operon, Ebersburg, Germany). Confirmed DNA fragments were then lifted into the pDM4 suicide mutagenesis vector [46] following SalI-XbaI restriction. E. coli S17-1λpir harbouring the different mutagenesis constructs were used as the donor strains in independent conjugations with Y. pseudotuberculosis parent (YPIII/pIB102) [47]. Appropriate allelic exchange events were monitored by Cm sensitivity and sucrose resistance. All mutants were confirmed by a combination of PCR and sequence analysis. Significantly, each variant was introduced in cis on the Y. pseudotuberculosis virulence plasmid to ensure expression occurred in the context of native regulatory elements.
To generate a polar mutation in the YPK_3687 locus of various Y. pseudotuberculosis YPIII derived strains, we used the pUA066 mutagenesis vector. The pUA066 construct is based on pNQ705 and was generated by digestion with SalI/XbaI and then ligation of a DNA fragment that was PCR amplified with the primer pair combination of pFpNQ066 and pRpNQ066 (Table S1) using DNA template derived from a boiled lysate of Y. pseudotuberculosis IP32953. Conjugal transfer of pUA066 into Yersinia involved a mating with E. coli S17-1λpir carrying the mutagenesis vector. Disruption of YPK_3687 occurred via a single homologous recombination cross-in of pUA066. Verification of the disruption utilised PCR and a series of primer combinations including a pair intended to amplify the entire YPK_3687 open reading frame and another combination designed to amplified the 5-prime end of the YPK_3687, including the upstream flanking region, and part of the integrated pUA066 vector.
Analysis of In Vitro Yop Synthesis and Secretion
Analysis of Yop synthesis and secretion by Y. pseudotuberculosis followed the procedure as previously described [45]. Samples of culture suspensions were taken to represent the total protein fraction, whereas the cleared bacteria-free supernatant corresponds to the secreted Yops fraction. Primary rabbit polyclonal antibodies recognizing YopN, YopD, YopE and DnaK were all a gift of Hans Wolf-Watz (Umeå University, Sweden), while those recognizing TyeA were a gift of Gregory Plano (University of Miami, USA). Detection used anti-rabbit antiserum conjugated with horse radish peroxidase (GE Healthcare, Buckinghamshire, United Kingdom) and Thermo Scientific Pierce ECL 2 Western Blotting Substrate to detect individual protein bands by western blotting.
Intracytoplasmic Stability Assay
Intrabacterial protein stability was assessed by the method of Feldman and colleagues using Cm as the de novo protein synthesis inhibitor [48]. Protein fractions were analyzed by SDS-PAGE and Western blot. Steady state accumulated YopN or YopN-TyeA hybrid was detected by treatment of the PVDF membrane with rabbit polyclonal YopN antiserum, in combination with horseradish peroxidase conjugated anti-rabbit antibodies (Amersham Biosciences) and a homemade luminol-based detection kit.
Generation of Constructs for Ectopic Expression of YopN and TyeA
Lysates of Yersinia parent and mutant bacteria was used in PCR to amplify the overlapping yopN and tyeA alleles on a single DNA fragment using the primer pair combinations listed in Table S1. Fragments were digested with BamHI and EcoRI prior to ligation with similarly digested pMMB208 [49]. Confirmed clones were stored in E. coli S17-1λpir, which was also used as donor in conjugal matings to mobilise the expression constructs into the ΔyopN, tyeA double mutant (YPIII/pIB8201a).
Low Calcium Growth Measurements
The ability of Yersinia to grow at 37°C under high- and low-Ca2+ conditions was performed by measuring absorbance at 600nm (A600) of bacterial cultures grown in liquid Thoroughly Modified Higuchi’s (TMH) medium (minus Ca2+) or TMH medium supplemented with 2.5 mM CaCl2 (plus Ca2+) [50]. Growth phenotypes were compared to parental Y. pseudotuberculosis (YPIII/pIB102), which is defined as calcium dependent (CD), since it is unable to grow in the absence of Ca2+ at 37°C, and Yersinia lacking the yscU and lcrQ alleles (YPIII/pIB75-26) which is termed temperature sensitive (TS) reflecting its inability to grow at 37°C [45].
YscF Surface Localization and Chemical Crosslinking
Overnight cultures from Yersinia strains were grown with shaking at 26°C in 2 ml of BHI broth supplemented with 2.5mM CaCl2. Subsequently, 0.1 volumes of bacterial suspension were sub-cultured into 3 ml fresh media and incubated for 3 hour at 37°C. After each culture was standardized by A600, 1 ml volumes were harvested by centrifugation at 8000g for 5 min at 4°C. Each bacterial pellet was gently resuspended in 1 ml of cold 20 mM HEPES, 2.5 mM CaCl2 (pH 8). Bacterial surface proteins were cross-linked for 30 min at ambient temperature with the non-cleavable, membrane-impermeable, amine-reactive cross-linker Pierce bis(sulfosuccinimidyl)suberate (BS3) (Thermo Scientific) at a final concentration of 5 mM. Cross-linking reactions were quenched for 15 min by addition of Tris-HCl (pH 8.0) to a final concentration of 20 mM. Cell fractions were collected by centrifugation at 12200g for 5 min at 4°C. Bacterial pellets were then resuspended in 100 µl of 1x SDS-PAGE loading buffer (50mM Tris-HCl, pH 6.8, 2% SDS, 0.1% Bromophenol blue, 10% Glycerol, 5% β-Mercaptoethanol) and analyzed by 18% acrylamide SDS PAGE and immunoblotting with rabbit anti-YscF polyclonal antiserum (a gift from Hans Wolf-Watz) that underwent several rounds of immunoadsorption with purified YscF to enhance its monospecificity.
Non-Polarized Secretion During Target Cell Contact
Cultivation and infection of HeLa cell monolayers was performed using our standard methods [51,52]. After 3 hours post-infection, 500 µl from the overlaying DMEM media was carefully collected, clarified by centrifugation for 10 min at 4 °C, and the bacterial-free supernatant representing the secreted protein fraction was added to 4x SDS-PAGE sample buffer (200mM Tris-HCl, pH 6.8, 8% SDS, 0.4% Bromophenol blue, 40% Glycerol, 20% β-Mercaptoethanol). To detect total protein levels, the infected HeLa cells were harvested directly into 125 µl of 4x SDS-PAGE loading buffer. Equivalent volumes of the total and soluble fractions were subjected to SDS-PAGE and western blotting. Comparable loading was confirmed by using mouse monoclonal antibodies specific for the eukaryotic protein β–actin (Clone AC-74, Sigma-Aldrich). Yop levels were detected using rabbit polyclonal anti-YopE and anti-YopD antisera. By comparing the amount of protein secreted into the extracellular media (soluble fraction) to the total synthesized protein induced upon bacteria-host cell contact (total whole cell lysates fraction), the proportion of YopE and YopD secreted into the media and thus the degree of non-polarized secretion can be estimated. The assay does not measure effector injection capacities, so the degree of polarized translocation of the YopE cytotoxin directly into the host cell cytosol remains unknown. Placebo controls utilized mock infections with bacteria in the absence of cell monolayers and cell monolayers in the absence of bacteria.
Bacterial Viability in the Presence of Eukaryotic Cells
A modified method of Bartra and co-workers [53] as described in earlier studies [45,54,55] was used to establish bacterial viability in the presence of murine macrophage-like J774 cells. In essence, bacteria lacking a fully functional T3SS are more readily phagocytosed and are therefore more susceptible to the antimicrobial effects of J774 cells. This reduced viability was determined by performing colony forming unit (CFU) counts for relevant bacterial strains in infected eukaryotic cell lysates.
Mouse Co-Infections and Competitive Index Measurements
Disruption by polar insertion of the gene encoding for a 349 amino acid inner membrane oligo-dipeptide/nickel ABC transporter permease (annotated as YPTB0523 in Y. pseudotuberculosis IP32953) has no measurable effect on Yersinia virulence in the mouse model neither in single strain infections nor in competitive infections with the isogenic wild-type strain (UA, unpublished). Therefore, this mutation was introduced into our mutants by a single cross-over of the pUA066 mutagenesis plasmid. As well as creating a polar mutation in the equivalent gene in Y. pseudotuberculosis YPIII (annotated as YPK_3687), integration of the mutagenesis plasmid conferred to these newly generated double mutants a CmR marker for counter-selection against CmS parental bacteria. Retention of the pIB102 virulence plasmid was verified with our standard in vitro Ysc-Yop synthesis and secretion assay. Comparable growth rates (monitored by A600) and corresponding CFU counts of all bacteria were also performed.
Female eight-week-old BALB/c mice (Taconic, Denmark) were given food and water ad libitum. Then groups of five mice were deprived of food and water 16 h prior to oral infection. For infection, bacteria were grown overnight in 50 ml LB broth at 26°C, then pelleted and serially diluted in sterile tap water supplemented with 150 mM NaCl. Serial dilutions were plated to record CFU/ml and their corresponding A600 measured to establish the volume of culture needed to inoculate 50 ml of sterile drinking water with 2.5 x 109 viable mutant bacterial cells (CmR) and 2.5 x 109 viable parental bacterial cells (CmS). Mice were allowed to drink from this inoculated water for 6 hours. Measurement of CFU was again performed to calculate the amount of CmR bacteria in the inoculation water, which was expressed as an input percentage of the total inoculated dose (CmS + CmR). At day 4 post infection, spleens were harvested aseptically in sterile PBS, homogenized, and plated for bacterial CFU analysis to determine the amount of viable CmR bacteria, and this was expressed as an output percentage of the total recovered population. In turn, the competitive index was determined as the ratio of percent CmR output versus percent CmR input.
Ethics Statement
The infection studies were performed in strict accordance with the Swedish Bioethical Guidelines for care and use of laboratory animals. The protocol was approved by The Umeå Committee on the Ethics of Animal Experiments (Permit Number: A-60-10).
Results
Y. pseudotuberculosis Naturally Produce and Secrete a YopN-TyeA Hybrid
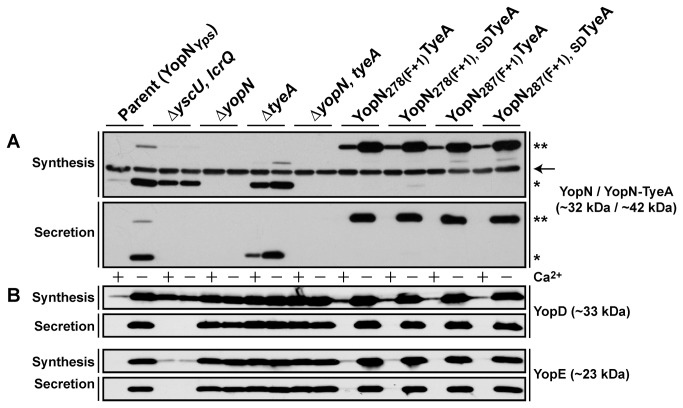
Y. pestis can produce and secrete a singular polypeptide consisting of a ~42 kDa hybrid of YopN and TyeA that was the result of a +1 frame shift during translation of the 3´-end of the yopN mRNA [39]. This hybrid was also a substrate of the Ysc-Yop T3SS. In contrast, a similar hybrid was not produced by Y. enterocolitica because any +1 frame-shift along the yopN mRNA would result in a premature stop codon immediately upstream of, and in the same reading frame as translated tyeA mRNA [39]. However, the yopN nucleotide sequences from Y. pseudotuberculosis and Y. pestis are identical (Figure 2). This would suggest that Y. pseudotuberculosis could also naturally produce a YopN-TyeA product. To examine for this, bacteria were grown in BHI broth restrictive (with Ca2+) or permissive (without Ca2+) for T3S to examine the in vitro synthesis and secretion profile of YopN. During growth in T3S permissive conditions, parental Y. pseudotuberculosis could produce and secrete a ~32 kDa protein that is YopN (Figure 3A). Interestingly, an additional Ca2+-regulated slower migrating band of ~42 kDa in both synthesis and secretion fractions was also recognized by the anti-YopN antisera; this band is consistent with the expected mass of a YopN-TyeA hybrid protein (Figure 3A). Critically, this band was not observed in synthesis and secretion fractions derived from an isogenic mutant of Y. pseudotuberculosis lacking both yopN and tyeA or from parental Y. enterocolitica (Figure 3A).
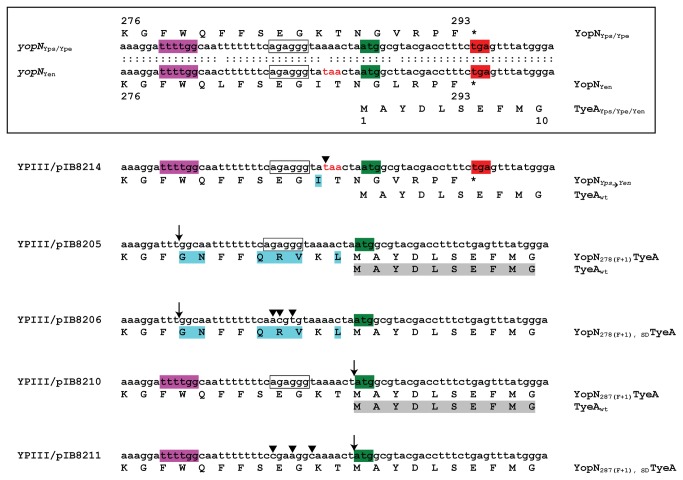
Figure 2. Region of sequence overlap between YopN and TyeA.
Comparison of the nucleotide and amino acid sequence in YopN and TyeA derived from Y. pseudtotuberculosis (Yps), Y. pestis (Ype) and Y. enterocolitica (Yen) (boxed panel). Nucleotide sequence of the sense strand is given in lower case font with identity between yopN Yps/Ype sequence and yopN Yen sequence indicated by the colon symbol (:). Numbers indicated they amino acid sequence that is given in upper case font either above (for yopN Yps/Ype) or below (for yopN Yen) the gene sequence. The yopN termination codon is indicated by red highlight and the tyeA initiation codon in green highlight and the upstream putative Shine-Dalgarno sequence is boxed. The first 10 amino acid residues of TyeA are identical in all three Yersinia species. As described by others [39], the putative pausing site (‘ttttgg’) for instigating a +1 frame-shift to create a YopN-TyeA hybrid is presented in magenta highlight. The out-of-frame stop codon (‘taa’) just upstream of the tyeA start that would prevent hybrid formation via +1 frame-shifting in Y. enterocolitica is given in red font. Shown below the boxed panel are the mutations used to modulate YopN-TyeA hybrid formation in Y. pseudotuberculosis. The first mutation was a missense mutation (▼) at codon 286 (LysAAA→IleATA) to introduce an out-of-frame ‘taa’ stop codon that abolished hybrid formation (YPIII/pIB8214; YopNYps→Yen). The second mutation was a +1 frameshift deletion mutation (removal of ‘T’) after codon 278 (↓) to give a YopN278(F+1)TyeA chimera (YPIII/pIB8205). The third mutation was a +1 frameshift deletion mutation (‘T’) after codon 278 (↓) combined with conservative mutations (▼) at codons 283 and 284 (GlnCAG→CAA and ArgAGG→CGT) that partially disrupts the presumed tyeA Shine-Dalgarno sequence to give a YopN278(F+1), SDTyeA chimera (YPIII/pIB8206). The fourth mutation was a +1 frameshift deletion mutation (removal of ‘A’) after codon 287 (↓) to give a YopN287(F+1)TyeA chimera (YPIII/pIB8210). The fifth mutation was the same +1 frameshift deletion mutation (removal of an ‘A’) after codon 2878 (↓) combined with conservative mutations (▼) at codons 283, 284 and 285 (SerTCA→TCC, GluGAG→GAA and GlyGGT→GGC) that partially disrupts the presumed tyeA Shine-Dalgarno sequence to give a YopN287(F+1), SDTyeA chimera (YPIII/pIB8211). Altered amino acid sequence in YopN prior to the tyeA initiation codon is indicated in blue highlight. Gray highlight reflects the cessation of TyeA production as a singular polypeptide courtesy of disrupting its upstream Shine-Dalgarno sequence.
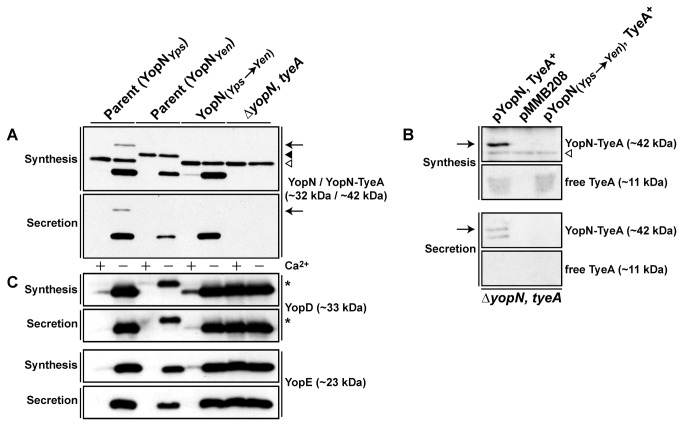
Figure 3. Analysis of naturally produced YopN-TyeA hybrid synthesis and secretion by Y. pseudotuberculosis.
Overnight cultures of Y. pseudotuberculosis were sub-cultured into BHI medium in the presence (+) or absence (-) of calcium ions at 26°C for 1 hour and at 37°C for 3 hours. Protein in the total bacterial suspension (Synthesis) and free in the cleared culture supernatant (Secretion) were collected, fractionated by 12% acrylamide SDS-PAGE, wet-blotted onto PDVF membrane and then detected using rabbit polyclonal anti-YopN (A), anti-TyeA (B) and also anti-YopD and anti-YopE (C) antibodies. The arrow (←) is pointing toward the ~42 kDa YopN-TyeA hybrid. The open (∇) arrowhead identifies non-specific protein bands uniquely recognised by the anti-YopN and anti-TyeA antisera in protein samples derived from Y. pseudotuberculosis. The closed (▼) arrowhead indicates a non-specific protein band recognised by the anti-YopN antiserum in protein samples derived from Y. enterocolitica. The asterisk (*) highlights the altered mobility of the YopD product derived from Y. enterocolitica. In A and C, lanes are represented by: Parent (YopNYps), Y. pseudotuberculosis YPIII/pIB102; Parent (YopNYen), Y. enterocolitica 8081/pYVe8081; YopNYps→Yen, Y. pseudotuberculosis YPIII/pIB8214; ΔyopN, tyeA, YPIII/pIB8201a. In B, lanes are Y. pseudotuberculosis ΔyopN, tyeA (YPIII/pIB8201a) also containing pYopN, TyeA+ (pAA304), empty vector (pMMB208) or pYopN(Yps→Yen), TyeA+ (pAA305). Approximate molecular mass values shown in parentheses were deduced from primary amino acid sequences.
In an effort to confirm natural YopN-TyeA chimeric production, initially we used anti-TyeA polyclonal antibodies to directly detect in cis production of native singular TyeA (~11 kDa) or native TyeA produced as a hybrid (~42 kDa). However, in our hands this was unsuccessful (data not shown), possibly due to low level production or a high rate of TyeA turnover. To circumvent this, we ectopically expressed the native yopN and tyeA alleles from an IPTG inducible promoter harboured on the pMMB208 expression plasmid (pAA304). Despite uncoupling regulatory control from the Ysc-Yop regulators, the gene synteny remained identical to that present on the virulence plasmid. From lysates derived from the ΔyopN, tyeA null mutant ectopically co-producing native YopN and TyeA, a ~42 kDa product in both synthesis and secreted fractions could be detected with anti-TyeA (Figure 3B). Additionally, the anti-TyeA antibodies also detected a diffuse band representing the free ~11 kDa TyeA product in the synthesis fraction only (Figure 3B).
To further confirm the contributions of both yopN and tyeA sequence in this hybrid, using site-directed mutagenesis the 3-prime yopN nucleotide sequence of Y. pseudotuberculosis was manipulated to generate the substitution K286I that resembled the yopN allele from Y. enterocolitica, which does not naturally produce the YopN-TyeA hybrid (Figure 2) [39]. The resulting mutant producing the YopNYpsYen variant failed to produce or secrete a ~42 kDa product either when produce in cis (Figure 3A) or in trans when produced under the control of an IPTG inducible promoter harboured on the pMMB208 expression plasmid (pAA305) (Figure 3B). However, the free ~32 kDa product of singular YopN (Figure 3A) and ~11 kDa product of free TyeA (Figure 3B) were synthesized as normal. Interestingly, the inability to produce the ~42 kDa YopN-TyeA product in bacteria producing YopNYpsYen did not negate the ability of these bacteria to maintain Ca2+-dependent control over the synthesis and secretion of middle and late Yop substrates, such as YopD and YopE respectively (Figure 3C). In contrast, complete removal of the yopN and/or tyeA alleles lead to the constitutive synthesis and secretion of YopD and YopE (Figure 3C and data not shown). Moreover, bacteria lacking tyeA could not maintain steady state levels of YopN (Figure 4A), suggesting that YopN stability and function depends on the presence of TyeA. We also confirmed that steady state levels of YopNYpsYen were equivalent to native YopN (Figure 4A).
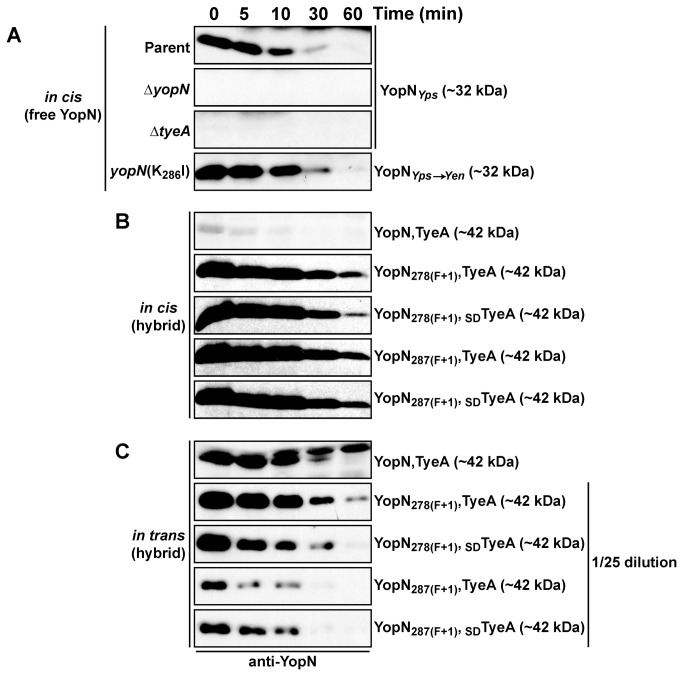
Figure 4. Intrabacterial stability of pre-formed pools of genetically engineered YopN-TyeA chimeras.
Bacteria were first cultured for 1 hour in non-inducing (plus 2.5 mM CaCl2) BHI broth at 37°C either without (A and B) or with 0.4 mM IPTG (C). The protein synthesis inhibitor chloramphenicol (50 µg/ml) was added at time point 0 minutes (min). Samples were then collected at this and subsequent time points. Protein levels associated with pelleted bacteria were detected by Western blot using polyclonal anti-YopN antiserum to detect singular YopN produced in cis (A) or YopN produced as a hybrid with TyeA derived from in cis production (B) or IPTG inducible ectopic in trans production (C). Note that the majority of samples in C were diluted by a factor of 25 to reduce the amount of material subjected to gel fractionation. In A, samples are derived from: Parent (YopNYps), YPIII/pIB102; ΔyopN, YPIII/pIB82; ΔtyeA, YPIII/pIB801a; YopNYps→Yen (YopNK286I), YPIII/pIB8214. In B, samples are derived from: Parent (YopNYps), YPIII/pIB102; YopN 278(F+1)TyeA, YPIII/pIB8205; YopN 278(F+1), SDTyeA, YPIII/pIB8206; YopN 287(F+1)TyeA, YPIII/pIB8210; YopN 287(F+1), SDTyeA, YPIII/pIB8211. In C, samples are derived from Y. pseudotuberculosis ΔyopN, tyeA (YPIII/pIB8201a) also containing pYopN, TyeA+ (pAA304), pYopN278(F+1), TyeA+ (pAA306), pYopN278(F+1), SD, TyeA+ (pAA307), pYopN287(F+1), TyeA+ (pAA308), or pYopN287(F+1), SD, TyeA+ (pAA309). Approximate molecular mass values shown in parentheses were deduced from primary amino acid sequences.
Taken together, these data are all consistent with the ability of Y. pseudotuberculosis to naturally produce a ~42 kDa YopN-TyeA singular polypeptide presumably as a result of a +1 frame shift during translation of the 3´-end of yopN mRNA. Moreover, this product undergoes Ca2+-regulated secretion via the Ysc-Yop T3SS. This corroborates occurrence of a similar sized product produced and secreted by the Ysc-Yop system in Y. pestis [39].
Stable Production of Genetically Engineered YopN-TyeA Chimeras in Y. pseudotuberculosis
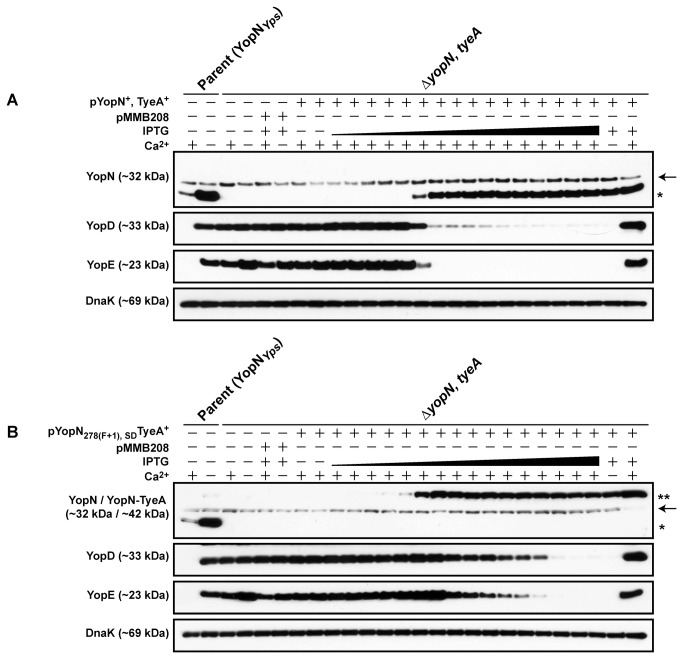
In prokaryotes (including viruses) and eukaryotes, programmed frame-shifting events are an important translational control mechanism for regulating the production of diverse functioning proteins [56-60]. The ~42 kDa hybrid protein naturally produced by Y. pestis and Y. pseudotuberculosis involved a frame-shifting event that fused the translation of yopN to overlapping tyeA, the products of which are essential mediators of T3S control. Although the levels of hybrid production and secretion are significantly lower than when produced as separate entities, we wondered if this hybrid is biologically relevant for T3S function in Yersinia. In order to investigate this, we utilized site directed mutagenesis to engineer in cis mutations in yopN that resulted in the artificial production of predominantly YopN-TyeA chimeras by Y. pseudotuberculosis. The first mutation was a +1 frame-shift directly introduced after yopN codon 278 by removal of a single ‘T’ nucleotide. This generated bacteria that produced a YopN-TyeA fusion – designated YopN 278(F+1)TyeA – that consisted of native YopN amino acid until residue 278, followed by an altered sequence between residues 279 and 287, prior to the switch to TyeA specific coding sequence (Figure 2). This means that the extreme YopN C-terminus encompassing residues 288 to 293 are replaced by unadulterated N-terminal TyeA sequence. Similarly, a second strain was generated by introducing a +1 frame shift after yopN codon 287 by removal of an ‘A’ nucleotide located immediately upstream of the tyeA start codon. The result was a bacterium able to produce a YopN-TyeA fusion termed YopN 287(F+1)TyeA, which incorporated native YopN sequence until residue 287, but was then followed by TyeA sequence. Once again, the extreme six residue YopN C-terminus was replaced by the beginning of TyeA (Figure 2). As these two mutants still left upstream of tyeA an uncharacterised but intact putative Shine Dalgarno (SD) sequence, albeit displaced by n−1 in the second mutant, they could conceivably still produce trace amounts of TyeA as a single (free) polypeptide entity. This was addressed by generating two additional mutants in which this putative SD sequence was conservatively ‘scrambled’ as much as possible without altering the yopN coding sequence. This resulted in two new mutants designated YopN 278(F+1), SDTyeA and YopN 287(F+1), SDTyeA respectively (Figure 2).
The stability of these four chimeras in the presence of endogenous proteases was examined. The larger ~42 kDa products synthesized in cis were easily detectable with anti-YopN antisera and remained as stable as the smaller ~32 kDa singular YopN polypeptide produced by parental Y. pseudotuberculosis (compare Figure 4B with Figure 4A). Additionally, all larger synthetic ~42 kDa variants accumulated in greater abundance, in contrast to the natural hybrid product that was barely detectable (Figure 4B). At this stage we have no firm grasp on why this might be the case. To determine whether the engineered YopN-TyeA(~42 kDa) variants displayed similar stability to the naturally formed hybrid produced by the parental strain, it was therefore necessary to establish a series of expression constructs that placed the various overlapping yopN and tyeA alleles PCR amplified from parent and mutant bacteria under an IPTG promoter on pMMB208. Ectopic in trans expression in the ΔyopN, tyeA double mutant now afforded sufficiently elevated production levels to detect stability of the natural hybrid (Figure 4C). Although it was necessary to load 25 times less protein material derived from the synthetic YopN-TyeA chimeric strains (i.e. diluted by a factor of 25) compared to the parental strain, their stability was essentially comparable to the native hybrid with the exception of YopN287(F+1), TyeA that was a little less stable (Figure 4C).
Since free TyeA could be functional and bias the behavior of individual synthetic YopN-TyeA hybrids, it was also necessary to explore its status in the constructed strains. Antibodies raised against TyeA recognized the in cis produced ~42 kDa band representing artificially produced chimeric YopN-TyeA hybrids, but not the ~11 kDa band of free TyeA from these mutants or from parental bacteria (data not shown). To circumvent this, the pMMB208-derived expression constructs described for the stability assays (see Fig 4C) were again used to measure TyeA synthesis and secretion. Using anti-TyeA antibodies, we could once more detect high levels of the ~42 kDa band when ectopically expressed in Yersinia lacking yopN and tyeA (Figure S1). In contrast, the ~11 kDa band of free TyeA was clearly detected only when co-expressing the native yopN and tyeA alleles in the synthesis fraction, with possibly very low level expression of free TyeA detectable from the two constructs expressing the hybrids YopN 287(F+1)TyeA and YopN 287(F+1), SDTyeA (Figure S1). Thus, if any free TyeA is produced in the four engineered chimeric strains, it is so low as to be essentially undetectable by western blot and consequently would likely not interfere with the function of YopN that is produced as part of the YopN-TyeA hybrid.
Hence, it was evident from this series of experiments that we successfully genetically manipulated Y. pseudotuberculosis to specifically produce a range of stable YopN-TyeA chimeras suitable to investigate their functional relevance to Yersinia biology.
Secreted YopN-TyeA Hybrids Maintain In Vitro Yops Secretion Control
The current working hypothesis suggests that a tetra-complex of YopN, together with the cognate T3S chaperones YscB and SycN, as well as TyeA act together as a secretion plug located at the cytoplasmic face of the inner membrane to prevent entry of Yop substrates into the secretion channel [34-36,38]. When the T3S apparatus is competent for secretion, environmental cues such as target cell contact or calcium depletion are anticipated to alter conformation of the YscF needle in a way that permits secretion of YopN. Once the secretion plug is removed, the T3SS can engage with and secrete the raft of middle and late Yop substrates. Thus, to investigate the impact of YopN-TyeA chimera production on T3SS activity, we began by investigating the degree to which the YscF needle component was secreted and polymerized at the bacterial surface – the final step in the assembly of an active Ysc-Yop T3SS. In our assay, visualization of YscF polymerization was aided by the presence of the non-membrane permeable chemical crosslinker BS3. With the exception of the yscF null mutant used as an antibody specificity control, monomeric YscF that was located in the bacterial cytoplasm and protected from the membrane impermeable crosslinker was detected in all samples (Figure S2). Parental bacteria could also secrete YscF that was readily cross-linked by BS3 to form higher order structures indicative of the T3S needle (Figure S2). In contrast, surface-located YscF was completely absent in the T3SS-defective full-length yscU, lcrQ deletion mutant, even though cytoplasmic located monomeric YscF protected from the non-membrane permeable crosslinker was visualized (Figure S2). Critically, YopN-TyeA chimera production by bacteria did not impact on their ability to produce higher order YscF structures at the bacterial surface (Figure S2). Hence, chimeric-produce bacteria assemble the Ysc-Yop T3SS that is competent for secretion of early substrates such as the YscF needle component.
Next we examined if the YopN-TyeA chimeras could be secreted by the assembled T3SS during bacterial growth in BHI broth restrictive (plus Ca2+) and permissive (minus Ca2+) for T3S. Having already confirmed by western blot the presence of both YopN and TyeA sequence in the synthetic hybrids, for convenience we used only anti-YopN antisera in subsequent western blot analyses of their synthesis and secretion profiles. Parental bacteria produced and secreted both YopN alone (~32 kDa) and a YopN-TyeA hybrid (~42 kDa) (Figure 5A). Once again, it was evident that the engineered ~42 kDa YopN-TyeA hybrids accumulated to greater levels than did the smaller ~32 kDa singular YopN polypeptide (Figure 5A). As noted earlier [34,61,62], a ΔtyeA null mutant has lost control of T3S activity, producing and secreting YopN during growth in both low and high calcium media (Figure 5A). Interestingly, the ΔtyeA null mutant also produced a smaller YopN-TyeA20-59 hybrid product, consistent with the reduced size of truncated and inactivated TyeA (Figure 5A). Secretion was T3SS-dependent because a strain devoid of the YscU – an integral inner membrane component of the Ysc-Yop T3SS – failed to secrete YopN. Interestingly, YopN-TyeA hybrid producing bacteria did not cause any deviation in the synthesis and secretion profiles of the so-called middle (e.g. YopD) and late (e.g. YopE) Yop substrates, since they were all comparable to parental bacteria (Figure 5B). On the other hand, the single ΔyopN and ΔtyeA mutants along with the double ΔyopN, tyeA mutant had all lost general control with Yop substrate synthesis and secretion being constitutive regardless of the calcium concentration (Figure 5B). Thus, it appears that engineered YopN-TyeA hybrids all have the capacity to maintain tight control over Yop secretion reminiscent of when they are produced as two separate polypeptides [34-36]. This happens despite the higher steady-state accumulation of each individual hybrid. At this stage, we can only speculate that the reason for increased protein levels involves some aspect of translation efficiency and/or product stability not measurable by assays utilized in this study.
Figure 5. Analysis of YopN-TyeA hybrid synthesis and secretion.
Overnight cultures of Y. pseudotuberculosis were sub-cultured into BHI medium in the presence (+) or absence (-) of calcium ions at 26°C for 1 hour and at 37°C for 3 hours. Protein in the total bacterial suspension (Synthesis) and free in the cleared culture supernatant (Secretion) were collected, fractionated by 12% acrylamide SDS-PAGE, wet-blotted onto PDVF membrane and then detected using rabbit polyclonal anti-YopN (A) and also anti-YopD and anti-YopE (B) antibodies. The arrow (→) point towards a non-specific protein band recognised by the anti-YopN antiserum. The single asterisk (*) highlights the single YopN polypeptide, while the double asterisk (**) indicates the larger YopN-TyeA hybrid protein. Lanes: Parent (YopNYps), YPIII/pIB102; ΔyscU, lcrQ double mutant, YPIII/pIB75-26; ΔyopN null mutant, YPIII/pIB82; ΔtyeA null mutant, YPIII/pIB801a; ΔyopN, tyeA double mutant, YPIII/pIB8201a; YopN 278(F+1)TyeA, YPIII/pIB8205; YopN 278(F+1), SDTyeA, YPIII/pIB8206; YopN 287(F+1)TyeA, YPIII/pIB8210; YopN 287(F+1), SDTyeA, YPIII/pIB8211. Approximate molecular mass values shown in parentheses were deduced from primary amino acid sequences.
Deregulated defects in Yop secretion control correspond to aberrant growth patterns in low calcium at elevated temperature. Therefore, in parallel we measured growth of our Yersinia mutants in TMH growth medium (low calcium) and supplemented with 2.5 mM CaCl2 (high Ca2+) at 37 °C. Growth of parental bacteria followed a typical calcium-dependent profile, where growth was observed only in the presence of calcium (Figure S3). Significantly, this was similar to the growth profiles of all four YopN-TyeA hybrid producing bacteria (Figure S3), corroborating their intact Yops secretion control. In contrast, the single ΔyopN and ΔtyeA mutants along with the double ΔyopN, tyeA mutant that no longer had control over Yops synthesis and secretion, were all rendered completely temperature sensitive for growth regardless of a high or low Ca2+ concentration (Figure S3). Altogether, these data suggest that YopN-TyeA hybrids maintain yop regulatory control, at least during growth under these standard laboratory conditions.
YopN-TyeA Hybrid Function in Effector Translocation
Although recently challenged by a study proposing a two-step translocation model [63], Yop effector delivery into target eukaryotic cells has long been considered a one-step polarized mechanism that avoids wasteful effector substrate secretion into the extracellular environment [43,64,65]. In fact, yopN or tyeA mutant bacteria that have lost the ability to control Yop secretion in vitro also secrete Yops in a non-polarized fashion into the extracellular milieu when in contact with eukaryotic cells. As a result, subsequent yopN and tyeA mutant effector injection capacities are reduced [34,61,62,64,66]. Hence, the degree of non-polarized Yops secretion during host cell contact by Y. pseudotuberculosis producing hybrid YopN-TyeA polypeptides was measured. We compared two different fractions from infected HeLa cell monolayers; the first was the clarified extracellular supernatant (non-polarized secreted protein fraction) and the second was whole cell lysates (total protein fraction associated with bacteria, HeLa cells and in the supernatant). Very little Yops were detected in the supernatant fraction of HeLa cell infections with parental Y. pseudotuberculosis, despite high levels of protein available in the total protein pool (Figure 6). This observation reflects the central tenet that Yops are directly delivered into cells and are seldom released free into the environment. This contrasts with the yopN and/or tyeA deletion mutants that liberate far greater amounts of Yop material free into the extracellular environment (Figure 6), which is indicative of their reduced effector injection capacities as described previously [34,61,62,64,66]. For reasons currently unknown, Y. pseudotuberculosis lacking tyeA display greater de-repression than does the single yopN mutant. For bacteria producing engineered YopN 278(F+1)TyeA and YopN 278(F+1), SDTyeA hybrid polypeptides, their capacity for Yops translocation was inferior as evidenced by the slight elevation in non-polarized Yops secretion into the extracellular environment during infection of tissue culture cell monolayers (Figure 6). In contrast, bacteria producing either YopN 287(F+1)TyeA or YopN 287(F+1), SDTyeA still maintained polarized Yops secretion suggesting that these bacteria deliver Yops into HeLa cells with efficiencies reminiscent of parental bacteria (Figure 6). Hence, all four hybrid-producing bacteria maintain far superior control over T3SS activity than do bacteria lacking yopN and/or tyeA. The reduction observed for YopN 278(F+1)TyeA and YopN 278(F+1), SDTyeA hybrid-producing bacteria is consistent with these variants producing a YopN module having the most altered C-terminal sequence (i.e. after codon 278; see Figure 2). Critically, this fault in target cell contact stimulated T3S control is not evident when examining low Ca2+-dependent induction in vitro in standard laboratory growth medium (see Figure 5).
Figure 6. Polarized translocation of YopE by YopN-TyeA hybrid producing bacteria.
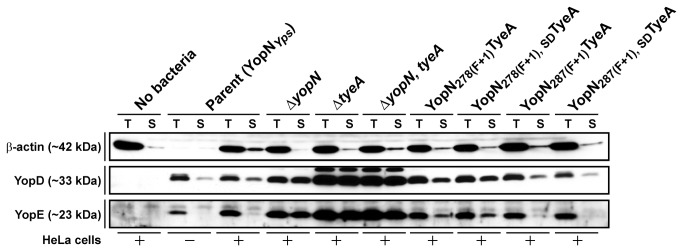
HeLa cells were infected with parental and mutated Y. pseudotuberculosis strains. The cell-free culture supernatant (S) and total cellular material (T) was then analysed for YopE and YopD by ECL-Western blot using rabbit anti-YopE and anti-YopD serum. The extent of eukaryote cell cytosolic material in each fraction was indicated by a western blot probing for host derived β–actin. Lanes: No bacteria, Mock infection with HeLa cell monolayer alone: Parent (YopNYps), YPIII/pIB102 either in the absence (−) or presence (+) of a HeLa cell monolayer; ΔyopN null mutant, YPIII/pIB82; ΔtyeA null mutant, YPIII/pIB801a; ΔyopN, tyeA double mutant, YPIII/pIB8201a; YopN 278(F+1)TyeA, YPIII/pIB8205; YopN 278(F+1), SDTyeA, YPIII/pIB8206; YopN 287(F+1)TyeA, YPIII/pIB8210; YopN 287(F+1), SDTyeA, YPIII/pIB8211. Approximate molecular mass values shown in parentheses were deduced from primary amino acid sequences.
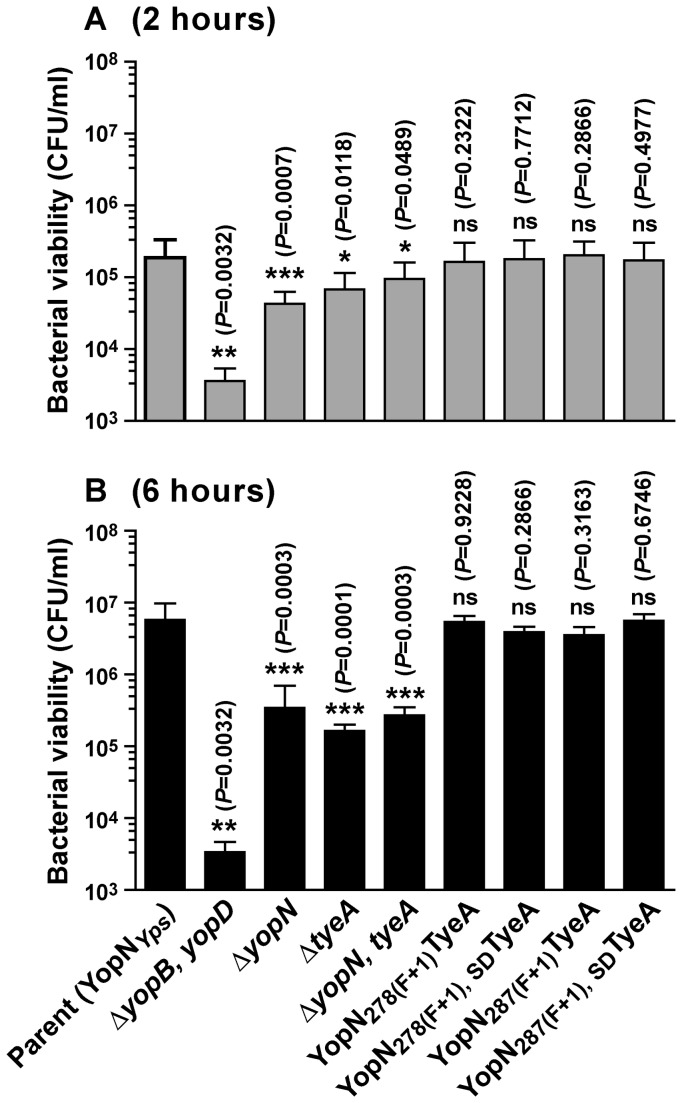
In parallel, we measured the capacity of our YopN-TyeA hybrid producing bacteria to resist phagocytosis and killing by J774A.1 macrophage-like immune cells [45,53-55], which is a hallmark of Ysc-Yop T3S activity [67]. In principal, any bacteria with a compromised T3SS will be phagocytosed by immune cells, exposing these internalized bacteria to potent and effective anti-microbial killing strategies. In contrast, an active T3SS will protect bacteria from phagocytosis so they can proliferate extracellularly. Bacterial infections were observed up to 6h post-infection. At 2h and 6h post-infection, the viability of bacteria associated with host cells was determined by measuring colony forming units (CFU). Importantly, the translocation defective and growth restricted ΔyopB, yopD null mutant cannot resist immune cell phagocytosis and is efficiently killed, which dramatically restricts the recovery of viable bacteria at 2h (Figure 7A, P =0.0032, **) and again at 6h post-infection (Figure 7B, P =0.0032, **). While not to the same extent as the ΔyopB, yopD null mutant, removal of yopN and/or tyeA is also a serious impediment to sustaining bacterial viability in the face of immune cell activity at both early (Figure 7A, P <0.05, * and ***) and late time points (Figure 7B, P <0.005, ***), corroborating severe defects in polarized secretion of effector Yops (see Figure 6) [34,61,62,64,66]. On the other hand, all four YopN-TyeA hybrid producing bacteria efficiently resisted phagocytosis and killing by J774A.1 macrophage-like immune cells at both early and late time-points to a similar degree as parental bacteria (Figure 7, P>0.05, no significant difference). This suggests that the deficiencies in polarized secretion observed for YopN 278(F+1)TyeA and YopN 278(F+1), SDTyeA producing bacteria does not impact negatively on their resistance to immune cell engulfment and killing. When considered altogether, these in vitro-based assays suggest that the YopN-TyeA hybrids can support T3SS function.
Figure 7. Formation of YopN-TyeA hybrids does not compromise in vitro T3SS activity.
Y. pseudotuberculosis strains were used to infect murine macrophage-like J774-1 cells. Bacterial cells with a compromised T3SS were more rapidly phagocytosed and killed by these immune cells. Bacterial viability as measured by CFU/ml was determined at 2 hours (A) and 6 hours (B) post-infection and is expressed as a mean of 4 independent assays ± the standard deviation. Strains: Parent (YopNYps), YPIII/pIB102; ΔyopB, yopD double mutant, YPIII/pIB619; ΔyopN null mutant, YPIII/pIB82; ΔtyeA null mutant, YPIII/pIB801a; ΔyopN, tyeA double mutant, YPIII/pIB8201a; YopN 278(F+1)TyeA, YPIII/pIB8205; YopN 278(F+1), SDTyeA, YPIII/pIB8206; YopN 287(F+1)TyeA, YPIII/pIB8210; YopN 287(F+1), SDTyeA, YPIII/pIB8211. Data sets were analyzed using the non-parametric two-tailed Mann-Whitney U-test. Analysis was performed using GraphPad Prism version 5.00 for Windows. Differences between mutants and parent (yopN wt) with a p-values < 0.05 were considered significant (*, ** and ***). ns – not statistically different.
Virulence Attenuation of Yersinia Producing YopN-TyeA Hybrids
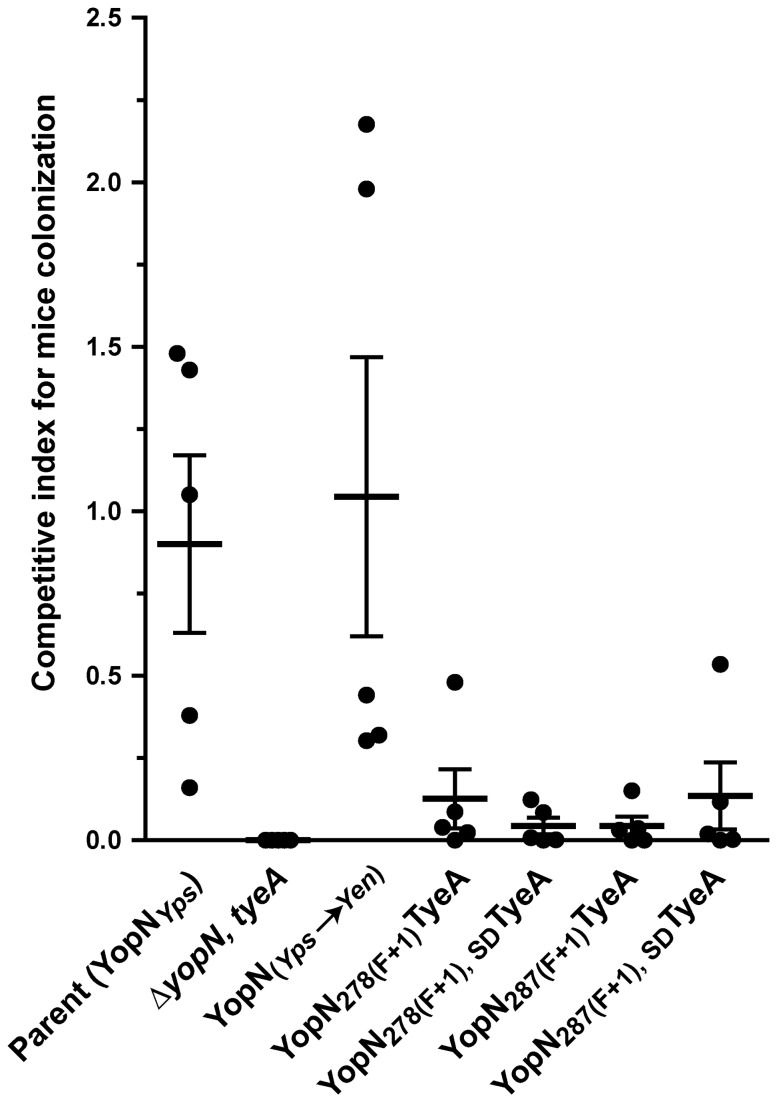
If the YopN-TyeA hybrid can fully support Ysc-Yop T3S function, then bacteria producing these should compete equally well with parental bacteria for survival during co-infection of mice. To facilitate these competition infection experiments, we utilised our prior knowledge that CmR bacteria containing a polar mutation within the gene encoding for a inner membrane oligo-dipeptide/nickel ABC transporter permease (annotated as YPTB0523 in Y. pseudotuberculosis IP32953) successfully competes with parental bacteria for equal colonization of organ tissues in orally infected mice (UA, unpublished). Therefore, we introduced this polar mutation into the orthologous YPK_3687 locus (as annotated in Y. pseudotuberculosis YPIII) of our temperature sensitive ΔyopN, tyeA mutant as well as all four regulatory competent YopN-TyeA hybrid producing bacteria and the YopNYpsYen producing bacteria (that can no longer naturally produce any hybrid). This gave rise to six new strains that now are all CmR to serve as a convenient selective marker to distinguish them from the CmS parental bacteria during the process of determining CFU counts derived from spleens dissected on day 4 from groups of five mice orally co-infected with a known input ratio of both parent (CmS) and mutant (CmR) bacteria. As a control, we also co-infected with parental bacteria (CmS) and the isogenic mutant containing only the additional polar mutation introduced into the YPK_3687 gene (CmR). As anticipated from unpublished data, a competitive index (CI) value of 0.9 confirms that this YPK_3687 polar mutation in parental bacteria (yopN wt), does not compromise the ability of these CmR bacteria to compete with CmS parent (also yopN wt) for systemic spreading and spleen colonization (Figure 8 and Table S2) (UA, unpublished). On the other hand, the CmR ΔyopN, tyeA mutant fared extremely poorly in competition with the CmS parent containing the wild type yopN allele (Figure 8 and Table S2; P=0.0079, **). At least in part, the very low CI score of 0.00008 for the ΔyopN, tyeA mutant reflects its inability to grow at body temperature. On the other hand, YopNYpsYen producing bacteria possessed a CI score of 1.04 (Figure 8 and Table S2; P=0.8413). This suggests that while singular YopN and TyeA are being produced, it matters not whether these bacteria also produce the larger hybrid form. Interestingly, the YopN 278(F+1)TyeA, YopN 278(F+1), SDTyeA, YopN 287(F+1)TyeA and YopN 287(F+1), SDTyeA hybrid producing bacteria presented CI values of 0.096 (P=0.0317, *), 0.032 (P=0.0079, **), 0.059 (P=0.0159, *) and 0.135 (P=0.0317, *) respectively, which were all significantly lower than parental control bacteria (Figure 8 and Table S2). Significantly, only two of these hybrid producing bacteria were compromised in polarized secretion (see Figure 6). Hence, these sensitive competitive survival co-infection experiments revealed that all four YopN-TyeA hybrids are not the functional equal of YopN and TyeA produced as independent polypeptides; an observation missed when using in vitro based assays that evidently lack the discriminatory sensitivity to resolve subtle biologically relevant imperfections in T3SS activity.
Figure 8. Competitive index for mice colonization.
Y. pseudotuberculosis mutants with defective yopN alleles as well as parental bacteria (yopN +) were manipulated to confer resistance to chloramphenicol by virtue of introducing a polar mutation into the YPK_3687 allele. These strains were used together with parental bacteria (CmlS) to co-infect groups of five mice via intentional contamination of their drinking water. Bacteria recovered from extracted spleens were measured by CFU/ml after four days of infection. The competitive indices (CI) were determined according to the footnotes in Table S2. Each symbol (•) reflects the CI derived from an individual mouse and the horizontal line is the mean of five mice ± the standard error. Strains: Parent (YopNYps), YPIII170/pIB102; YopNYps→Yen, Y. pseudotuberculosis YPIII170/pIB8214; ΔyopN, tyeA double mutant, YPIII170/pIB8201a; YopN 278(F+1)TyeA, YPIII170/pIB8205; YopN 278(F+1), SDTyeA, YPIII170/pIB8206; YopN 287(F+1)TyeA, YPIII170/pIB8210; YopN 287(F+1), SDTyeA, YPIII170/pIB8211. Note that all strains harbour a polar insertion in YPK_3687 (i.e. strain designation ‘YPIII170’).
We were curious to identify a reason for the slight virulence attenuation of the YopN-TyeA hybrid producing bacteria. The fact that the hybrids YopN 278(F+1)TyeA and YopN 278(F+1), SDTyeA displayed a subtle increase in non-polarized Yop secretion (see Figure 6) hinted that the fine-tuning of Yop secretion control is a reason for virulence attenuation. To investigate this, an in vitro regulatory assay was designed that had an enhanced discriminatory power over traditional T3S assays. Two IPTG-inducible expression constructs based upon pMMB208 were generated; the first contained native full-length and overlapping yopN and tyeA alleles (pAA269) and the second with the engineered yopN 278(F+1), SD tyeA allele (pAA271) whose hybrid product caused the most virulence attenuation (see Figure 8 and Table S2). Using the fact that the ΔyopN, tyeA double mutant is deregulated for Yop synthesis, even at the non-permissive high Ca2+ conditions (see Figure 5), we examined how efficient the two expression constructs were at restoring feedback inhibitory control i.e. preventing Yops synthesis at high Ca2+ conditions. We did this by progressively titrating into the growth medium increasingly higher concentrations of IPTG. It was very evident that as soon as ectopic singular YopN (~32 kDa) and TyeA (not shown) expression was detectable (using as little as 0.01 mM IPTG) cessation of YopE and to a lesser extent YopD synthesis occurred concomitantly (Figure 9A). In contrast, although ectopic YopN 278(F+1), SDTyeA hybrid (~42 kDa) protein was detectable at an even lower IPTG concentration (using as little as 0.04 mM IPTG), complete cessation of YopE synthesis, and to a lesser extent YopD synthesis, required at least a 5-fold higher IPTG concentration than was used for native YopN and TyeA expression (Figure 9B). However, this delay in Yop synthesis inhibition cannot be explained by insufficient accumulation of YopN 278(F+1), SDTyeA, which was at least the equivalent of maximal levels of singular YopN even at low IPTG doses. Hence, we can only assume that the action of the hybrid in instigating repression – presumably by resetting the YopN secretion plug in the channel – is comparatively sluggish. Thus, we believe hybrid producing mutants are routinely less fit in infected animals because they are unable to respond rapidly to coordinate changes in Ysc-Yop synthesis and secretion in accordance with environmental flux encountered when in the host animal.
Figure 9. Minimal level of YopN and TyeA required for regaining Yop synthesis control.
A deregulated yopN and tyeA double mutant of Y. pseudotuberculosis (YPIII/pIB8201a) expressing either an IPTG inducible variant of native YopN and TyeA (A; pYopN+, TyeA+) or an engineered YopN-TyeA hybrid (B; pYopN 278(F+1), SDTyeA+) was grown at 37 °C in T3S restrictive (+Ca2+) or T3S permissive (−Ca2+) conditions. Protein associated with pelleted bacteria was fractionated by 12% acrylamide SDS–PAGE and detected by Western blot using polyclonal anti-YopN, anti-YopD and anti-YopE antiserum. As a bacterial loading control we probe levels of the cytoplasmic molecular chaperone DnaK with anti-DnaK antibodies. Variable ectopic expression of native YopN and TyeA or the engineered hybrid variant was achieved via the incremental increase in the final concentration of IPTG added to the growth media (0.001 mM, 0.002 mM, 0.003 mM, 0.004 mM, 0.005 mM, 0.01 mM, 0.02, mM, 0.025 mM, 0.03 mM, 0.035 mM, 0.04 mM, 0.045 mM, 0.05 mM, 0.1 mM, 0.2 mM and 0.3 mM respectively). The `−´ symbol indicates the absence of IPTG, while `+´ indicates a final concentration of 0.4 mM. Strains: Parent (YopNYps), YPIII/pIB102; ΔyopN, tyeA double mutant, YPIII/pIB8201a; The single asterisk (`*´) indicates singular YopN, while the double asterisk (`**´) represents the YopN-TyeA hybrid variant. The arrow (→) is pointing toward a non-specific protein band recognized by the anti-YopN antiserum. Approximate molecular mass values shown in parentheses were deduced from primary amino acid sequences.
Establishing a Frame-shifting Mechanism for YopN-TyeA Hybrid Production
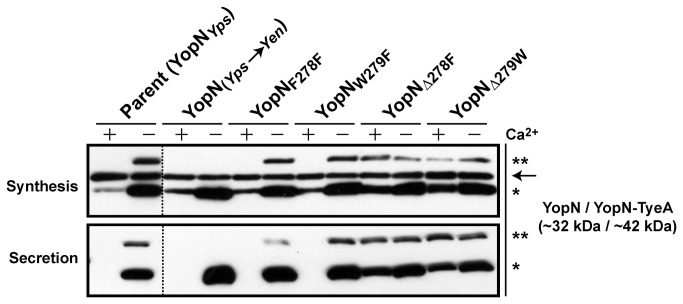
The mechanism for formation of the naturally occurring YopN-TyeA hybrid in Y. pestis was proposed to be a +1 translational frame-shifting event instigated by a putative ribosomal pausing site ‘UUU-UGG’ encompassing codons F278 and W279 within the 3´-end of yopN mRNA [39]. Given the existence of identical yopN sequence in Y. pseudotuberculosis and Y. pestis (see Figure 2), one might assume for this potential frame-shifting mechanism to be shared between the two species. However, this could not be confirmed by mass spectroscopy because our numerous attempts to determine the protein sequence of native YopN-TyeA were fruitless (data not shown), a situation also experienced by others [39]. Therefore, we proceeded to target the putative ‘UUU-UGG’ ribosomal pausing sequence by site-directed mutagenesis in Y. pseudotuberculosis. Four yopN mutations were generated; the first a silent FUUU FUUC mutation to give YopNF278F, the second a missense WUGG FUUC mutation to give YopNW279F, while the remaining two were clean deletions of codon 278 and 279 to give YopNΔ278F and YopNΔ279W, respectively. All four mutants were grown in BHI media that was either T3S–restrictive (plus Ca2+) or T3S–permissive (minus Ca2+). Critically, all four bacteria retained the ability to synthesize and secrete a YopN-TyeA chimeric protein of ~42 kDa (Figure 10). However, it was evident that bacteria producing YopNΔ278F or YopNΔ279W had lost the capacity to maintain control of Ysc-Yop T3S, since product was constitutively made and secreted regardless of Ca2+ concentration (Figure 10). Hence, the codons 278 and 279 are relevant for YopN activity, but on their own are not solely responsible for the frame-shifting event. This must mean that the frame-shifting mechanism is more complex than previously appreciated, requiring more than just a ribosomal pausing site. In fact, probably no single feature is alone responsible, with additional architectural features neighbouring the site bound to make necessary contributions.
Figure 10. Analysis of Yop synthesis and secretion in YopN mutants manipulated at codons 278 or 279.
Overnight cultures of Y. pseudotuberculosis were sub-cultured into BHI medium in the presence (+) or absence (-) of calcium ions at 26°C for 1 hour and at 37°C for 3 hours. Protein in the total bacterial suspension (Synthesis) and free in the cleared culture supernatant (Secretion) were collected, fractionated by 12% acrylamide SDS-PAGE, wet-blotted onto PDVF membrane and then detected using rabbit polyclonal anti-YopN. The arrow (→) is pointing toward a non-specific protein band recognized by the anti-YopN antiserum. The single asterisk (*) highlights the single YopN polypeptide, while the double asterisk (**) indicates the larger YopN-TyeA hybrid protein. Lanes: Parent (YopNYps), YPIII/pIB102; YopNYps→Yen, YPIII/pIB8214; YopNF278F, YPIII/pIB8215; YopNW279F, YPIII/pIB8216; YopNΔ278F, YPIII/pIB8217; YopNΔ279W, YPIII/pIB8218. Approximate molecular mass values shown in parentheses were deduced from primary amino acid sequences.
Discussion
The InvE family of T3S regulators primarily exist as singular proteins [33]. However, a subset of these present in the Ysc-Yop evolutionary clade of T3SSs exist as two separate polypeptides i.e. in the form of YopN- and TyeA-like proteins [2]. This study revealed that Y. pseudotuberculosis YopN and TyeA can be synthesised as a singular YopN-TyeA stable polypeptide, corroborating existence of a naturally occurring YopN-TyeA hybrid first observed in Y. pestis [39]. In cis mutants of Y. pseudotuberculosis engineered to synthesize solely the YopN-TyeA hybrid proved a useful tool to thoroughly probe function. At least in vitro, YopN-TyeA hybrids are efficiently secreted and can themselves control the low Ca2+-dependent T3S of other Yops. However, shortcomings of this secretion control first revealed during bacteria-target eukaryotic cell contact, impact negatively on their ability to survive during in vivo co-infection of mice. Hence, while YopN-TyeA fusions are functional in calcium regulation of Yops secretion, these alleles are not as good as wild type YopN and TyeA produced as discrete polypeptides. As yet, we have no molecular comprehension for why some type III secretion systems prefer two polypeptides and others prefer one. More apparent was that our standard in vitro assays routinely used to assess defects in Yop regulatory control can lack the sensitivity to detect subtle abnormalities. It follows though that any such abnormalities identifiable in vitro, no matter how subtle, are most likely meaningful in the context of bacterial colonization and survival in vivo during an animal model infection.
Although many frame-shifting events are errors in mRNA translation processing that result in mRNA decay and partly completed non-functional products, programmed frame-shifting can be an important translational control mechanism for regulating the production or diversity of protein entities [56-60]. For good reasons, natural YopN-TyeA hybrid production in Y. pestis was thought to involve a +1 translational frame-shift brought about by a UUU-UGG ribosomal pausing site at codon positions 278 and 279 of yopN mRNA [39]. This raised the possibility of this being a genuine ‘programmed’ frame-shift that had evolved to modulate YopN-TyeA hybrid levels for a physiological purpose. In reality however, disruption of this putative UUU-UGG ribosomal pausing site in yopN of Y. pseudotuberculosis had no detrimental impact on YopN-TyeA hybrid formation (see Figure 10). This observation could mean that this sequence is not the actual ribosomal pausing site. However, this could not be confirmed as numerous attempts by us and others [39] failed to determine directly the amino acid sequence of purified YopN-TyeA hybrid. On the other hand, it remains feasible that the frame-shifting mechanism utilizes this ribosomal pausing site, but requires influence from architectural features neighbouring the site. As SD sites internal to the open reading frame influence translational pausing [68-70], it is conceivable that composition and location of the putative SD sequence of tyeA relative to the upstream pausing site and the downstream tyeA initiation codon could affect the extent of YopN-TyeA formation. In addition, frame-shifting can be heavily influenced by the mRNA secondary structure downstream of the pausing site [71,72], and can also be controlled by codon usage and the relative abundance of a given tRNA [73,74]. Thus, a number of epigenetic features have potential to influence frame-shifting events that lead to YopN-TyeA formation.
Epigenetic regulatory elements aside, it is also established that polyamines can enhance +1 frame-shifting [75]. Polyamines are small polycationic molecules ubiquitous in almost all life-forms [76,77]. They have a natural affinity for binding to RNA, which affords them the opportunity to alter protein synthesis in ways that influence multiple cellular functions [76,77]. The prospect that polyamines could influence the frame-shifting event leading to YopN-TyeA hybrid production is tantalising given how they are already linked to controlling T3SS activity in some bacteria [78,79]. Thus, it is apparent that the frame-shifting event leading to YopN-TyeA formation is probably multifaceted; dissecting this mechanism will need to address numerous possible influences.
Engineering a +1 frameshift after codon 278 or codon 287 had the purpose of coercing the production of a YopN-TyeA hybrid. Remarkably, these bulkier hybrids are accumulated to higher levels than the single YopN product, are efficiently secreted and, to varying degrees, also support the T3S control of other Yops. In our minds, the secretion of YopN-TyeA underlines the tolerance that T3SSs display for the secretion of diverse substrates. It seems that the YopN-TyeA polypeptide still boasts a recognisable N-terminal secretion signal [80], can still be piloted for secretion by cognate heterodimeric T3S chaperone composed of YscB and SycN [36,81], and can still unfold itself in a manner that must ensure both controlled and efficient secretion via the Ysc-Yop T3SS [82], and only when the bacteria senses the appropriate environmental signals.
In reality, functional hybrids contain a C-terminal YopN sequence between residues 278 to 286 that barely resembles native YopN, and the residues beyond 287 no longer exist as they are replaced by the TyeA N-terminus (see Figure 2). In particular, a +1 frame-shift after 278 appeared to produce a more defective hybrid – as evidenced by decreased polarized secretion following cell contact and virulence attenuation – then when introducing a +1 frame-shift after codon 287. Consistent with this, the former hybrids contain several more amino acid modifications in the YopN C-terminal sequence. We speculate that these extra changes might be the reason for a more defective hybrid entity. This could be either as a direct consequence of reducing activity of the YopN module or through generating a less flexible TyeA tail module. Since specific deletion of either codon 278 or 279 produced YopN variants with inferior control of Yop synthesis and secretion (see Figure 10), this suggests that the native YopN C-terminal segment has a key role in mediating T3S control. This will be addressed in our future work.
Independent methods have also indicated that at least a portion of YopN can be translocated into eukaryotic cells, in a process that is negatively influenced by the presence of functional TyeA [38,66]. With no known enzymatic activity or intracellular molecular target, the relevance of translocated YopN remains obscure. This study did not attempt to investigate if any of the YopN-TyeA hybrids are translocated into HeLa cell monolayers. If a legitimate role for YopN translocation is identified, the YopN-TyeA hybrids could be used to investigate by what mechanism the presence of a C-terminal TyeA appendage quenches this YopN activity.
As already hinted above, it is also possible that TyeA function within these engineered hybrids is affected. For example, hybrid secretion places this TyeA component outside of the cell, effectively depleting its cytoplasmic pools. Past conjecture has surrounded the secretory status of native TyeA [34,61,62]. However, the current paradigm for Yop secretion control places TyeA in the cytoplasm, functioning as an intracellular anchor facilitating the plugging of the secretory channel by YopN [41]. It is also probable that TyeA fused to the C-terminus of YopN becomes less supple, losing the required flexibility to perform its YopN anchoring role. Structural flexibility of TyeA may be needed for other protein-protein interactions. Aside from YopN, TyeA has been shown to interact with YopD [34,61] and more recently YscG and a hypothetical protein annotated as YPCD1.16C in Y. pestis (pYV0009 in Y. pseudotuberculosis IP32953) [83]. However, the significance of these reported interactions is unclear; it is not known whether any of them assist TyeA in anchoring the YopN plug to control Yops secretion. Interestingly, the well-studied YopD protein has a role both inside and outside of the bacteria [55,84], theoretically making it available to either non-secreted or secreted TyeA. At any rate, altering the context of TyeA function beyond YopN may explain the reduced fitness in mice (see Figure 8 and Table S2) and the sluggish secretion control (see Figures 6 and 9) of bacteria producing the 278(F+1) series of hybrids. Our YopN-TyeA hybrids, or variants thereof, where cytoplasmic TyeA depletion is forced, could open up unexplored avenues to study the mechanism of TyeA-dependent anchoring of YopN in addition to revealing the biological consequences of these other TyeA-dependent protein-protein interactions.
Another point is that free TyeA produced as an independent polypeptide may affect YopN-TyeA hybrid function. For example, this native TyeA could engage with the YopN component of the hybrid, potentially contributing to the small phenotypic differences we have observed in this study. For this reason we generated two extra hybrids (i.e. the SD-minus YopN 278(F+1), SDTyeA and YopN 287(F+1), SDTyeA mutants) having a ‘scrambled’ sequence aimed at disrupting a probable SD site of tyeA to limit its production. However, western blotting confirmed that all four hybrids (i.e. regardless of the SD sequence being intact or disrupted) produced very little to no detectable free TyeA. First of all, these data cannot substantiate whether the nucleotides ‘AGAGGG’ (see Figure 2) do actually represent a bona fide SD of tyeA. In addition, we found no correlation between native free TyeA production and the modest phenotypic defects displayed by the YopN-TyeA chimeras. Therefore, it seems unlikely that a free native TyeA bias (i.e. in the SD+ mutants of YopN278(F+1), TyeA and YopN287(F+1), TyeA) can account for the defects in functionality of the hybrids.
Analysis of the InvE family of proteins is adding credence to the concept of a secretion hierarchy among the middle translocator substrates and the late effector secretion substrates [19,25-31]. This is attractive because it fits nicely with the original tenet that the translocon pore should form in the host cell plasma membrane before substrates destined for translocation through this pore are actually secreted. As described in a recent review [41], there is some evidence that Yersinia preferentially secretes YopB and YopD translocator substrates in Ca2+ rich media that otherwise prevents Yop effector secretion. In these situations (i.e. prior to cell contact or in the presence of elevated calcium), it is the YopN/SycN/YscB/TyeA complex that inhibits effector secretion in order to prioritise translocator secretion [38,66]. This cannot be true of all situations however, since unstimulated Y. pseudotuberculosis contains on their surface both Yop translocators and Yop effectors in equal measure that proceed to form translocation-competent Yop complexes [63]. Moreover, our western blotting experiments probing for levels of both the YopD translocator and the YopE effector in defined yopN-tyeA mutants did not reveal any preferential secretion of YopD in Ca2+ replete conditions in vitro (e.g. see Figure 5). Hence, if hierarchical secretion does exist in Y. pseudotuberculosis, YopN and TyeA apparently do not orchestrate it. Interestingly, it appeared that basal levels of YopD are always subtly higher than YopE both in vitro (see Figure 9) and during cell contact (see Figure 6). This is reminiscent of recent studies purporting to a hierarchical Yops expression profile instigated through the translation-inhibitory effects of YopD/LcrH complexes differentially bound to the various 5´-untranslated regions of yop mRNAs [84,85]. Arguably therefore, any preference for prioritising translocator secretion may have its genesis in regulating cytoplasmic pools of pre-made Yops.
In summary, this study was unable to show any direct support for YopN and TyeA in orchestrating hierarchical secretion among the Yop translocator and effector substrates. However, it was clear that no matter how subtle the regulatory defects were in controlling Yop synthesis and secretion in vitro, they had a direct impact on the in vivo fitness of Yersinia bacteria. Our outcomes also lead us to posit that work aimed at further unravelling the specific mechanisms of feedback inhibition and post-transcriptional control of yops expression through YopD function could have fertile consequences for understanding how Yersinia might consider prioritising different Yop substrates for secretion.
Supporting Information
Analysis of free TyeA synthesis and secretion in synthetic YopN-TyeA chimeric mutants. Overnight cultures of Y. pseudotuberculosis were sub-cultured into BHI medium in the absence of calcium ions at 26°C for 1 hour and at 37°C for 3 hours. At the time of temperature up-shift, 0.4 mM IPTG was added to all cultures. Protein in the total bacterial suspension (Synthesis) and free in the cleared culture supernatant (Secretion) were collected, fractionated by 15% acrylamide SDS-PAGE, wet-blotted onto PDVF membrane and then detected using rabbit polyclonal anti-TyeA antibodies. The arrow (→) point towards a non-specific protein band recognized by the anti-TyeA antiserum. The single asterisk (*) highlights the larger YopN-TyeA hybrid protein. Lanes are Y. pseudotuberculosis ΔyopN, tyeA (YPIII/pIB8201a) also containing pYopN, TyeA+ (pAA304), empty vector (pMMB208), pYopN278(F+1), TyeA+ (pAA306), pYopN278(F+1), SD, TyeA+ (pAA307), pYopN287(F+1), TyeA+ (pAA308), or pYopN287(F+1), SD, TyeA+ (pAA309). Approximate molecular mass values shown in parentheses were deduced from primary amino acid sequences.
(TIF)
YopN-TyeA hybrid-producing bacteria spawn external YscF multimers. Yersinia strains were grown in non permissive T3S media (plus Ca2+). Where indicated (+), the membrane-impermeable chemical cross-linker BS3 was added to the bacteria. After being quenched with Tris-HCl, bacteria pellets were solubilized in sample buffer and then protein fractionated by 12% acrylamide SDS-PAGE. After wet-transfer to PVDF, YscF was detected with immune-absorbed monospecific anti-YscF antiserum. Non-cross-linked monomeric YscF was observed in all lanes except the ΔyscF null mutant control. Cell-surface YscF multimers were observed in all lanes except for the ΔyscF null mutant control as well as the YscF+, but T3SS-defective, ΔyscU, lcrQ null mutant control. The predicted molecular mass of monomeric YscF is given in parenthesis, while approximate sizes of protein molecular weight standards are given to the right. Strains: Parent (YopNYps), YPIII/pIB102; ΔyopN null mutant, YPIII/pIB82; ΔtyeA null mutant, YPIII/pIB801a; ΔyopN, tyeA double mutant, YPIII/pIB8201a; YopN 278(F+1)TyeA, YPIII/pIB8205; YopN 278(F+1), SDTyeA, YPIII/pIB8206; YopN 287(F+1)TyeA, YPIII/pIB8210; YopN 287(F+1), SDTyeA, YPIII/pIB8211; ΔyscF null mutant, YPIII/pIB202; ΔyscU, lcrQ double mutant, YPIII/pIB75-26.
(TIF)
Low calcium response growth phenotypes of Y. pseudotuberculosis producing YopN-TyeA hybrids. Bacteria were grown at 37°C in TMH medium supplemented with 2.5 mM CaCl2 (plus Ca2+; A) or non-supplemented (minus Ca2+; B). Two different growth phenotypes were detected: TS – bacteria are sensitive to elevated temperature regardless of the presence or absence of calcium (ΔyopN and/or ΔtyeA null mutants) and, CD – calcium dependent growth (all remaining strains). Strains: Parent (YopNYps), YPIII/pIB102; ΔyopN null mutant, YPIII/pIB82; ΔtyeA null mutant, YPIII/pIB801a; ΔyopN, tyeA double mutant, YPIII/pIB8201a; YopN 278(F+1)TyeA, YPIII/pIB8205; YopN 278(F+1), SDTyeA, YPIII/pIB8206; YopN 287(F+1)TyeA, YPIII/pIB8210; YopN 287(F+1), SDTyeA, YPIII/pIB8211.
(TIF)
Oligonucleotides used in this study.
(PDF)
Competitive index for mice colonization.
(PDF)
Acknowledgments
This work was performed within the virtual framework of the Umeå Center for Microbial Research Linnaeus Program and Molecular Infection Medicine Sweden. We express gratitude to Hans Wolf-Watz (Umeå University, Umeå, Sweden) for the gifts of antiserum specific to DnaK and various Ysc/Yop antigens and Gregory Plano (University of Miami, FL, USA) for the gift of anti-TyeA antiserum.
Funding Statement
This work was supported by grants from the Swedish Research Council (ÅF, MSF) (http://www.vr.se/), Foundation for Medical Research at Umeå University (MSF) and J C Kempe Memorial Fund (AAA, TRC, UA). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of manuscript.
References
- 1. Büttner D (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev 76: 262-310. doi: 10.1128/MMBR.05017-11. PubMed: 22688814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Troisfontaines P, Cornelis GR (2005) Type III secretion: more systems than you think. Physiology (Bethesda) 20: 326-339. doi: 10.1152/physiol.00011.2005. PubMed: 16174872. [DOI] [PubMed] [Google Scholar]
- 3. Edgren T, Forsberg A, Rosqvist R, Wolf-Watz H (2012) Type III secretion in Yersinia: injectisome or not? PLOS Pathog 8: e1002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Francis MS (2010) Type III secretion chaperones: a molecular toolkit for all occasions. In: Durante P, Colucci L. Handbook of molecular chaperones: roles, structures and mechanisms. Hauppauge, NY: Nova Science Publishers, Inc. pp. 79-147. [Google Scholar]
- 5. Galán JE (2007) SnapShot: effector proteins of type III secretion systems. Cell 130: 192 PubMed: 17632065. [DOI] [PubMed] [Google Scholar]
- 6. Brutinel ED, Yahr TL (2008) Control of gene expression by type III secretory activity. Curr Opin Microbiol 11: 128-133. doi: 10.1016/j.mib.2008.02.010. PubMed: 18396449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osborne SE, Coombes BK (2011) Expression and secretion hierarchy in the nonflagellar type III secretion system. Future Microbiol 6: 193-202. doi: 10.2217/fmb.10.172. PubMed: 21366419. [DOI] [PubMed] [Google Scholar]
- 8. Deane JE, Abrusci P, Johnson S, Lea SM (2010) Timing is everything: the regulation of type III secretion. Cell Mol Life Sci 67: 1065-1075. doi: 10.1007/s00018-009-0230-0. PubMed: 20043184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flores-Kim J, Darwin AJ (2012) Links between type III secretion and extracytoplasmic stress responses in Yersinia . Front Cell Infect Microbiol 2: 125 PubMed: 23087910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Francis MS (2011) Secretion systems and metabolism in the pathogenic Yersiniae. In: Kidd SP. Stress response in pathogenic bacteria. Wallingford, Oxfordshire, UK: CABI Publishing; pp. 185-220. [Google Scholar]
- 11. Magdalena J, Hachani A, Chamekh M, Jouihri N, Gounon P et al. (2002) Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J Bacteriol 184: 3433-3441. doi: 10.1128/JB.184.13.3433-3441.2002. PubMed: 12057936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tamano K, Katayama E, Toyotome T, Sasakawa C (2002) Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J Bacteriol 184: 1244-1252. doi: 10.1128/JB.184.5.1244-1252.2002. PubMed: 11844752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg Å et al. (2003) YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol 185: 2259-2266. doi: 10.1128/JB.185.7.2259-2266.2003. PubMed: 12644497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Agrain C, Callebaut I, Journet L, Sorg I, Paroz C et al. (2005) Characterization of a Type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica . Mol Microbiol 56: 54-67. doi: 10.1111/j.1365-2958.2005.04534.x. PubMed: 15773978. [DOI] [PubMed] [Google Scholar]
- 15. Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM et al. (2006) Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441: 637-640. doi: 10.1038/nature04822. PubMed: 16738660. [DOI] [PubMed] [Google Scholar]
- 16. Lorenz C, Schulz S, Wolsch T, Rossier O, Bonas U et al. (2008) HpaC controls substrate specificity of the Xanthomonas type III secretion system. PLOS Pathog 4: e1000094 PubMed: 18584024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wood SE, Jin J, Lloyd SA (2008) YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J Bacteriol 190: 4252-4262. doi: 10.1128/JB.00328-08. PubMed: 18424518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Björnfot AC, Lavander M, Forsberg A, Wolf-Watz H (2009) Autoproteolysis of YscU of Yersinia pseudotuberculosis is important for regulation of expression and secretion of Yop proteins. J Bacteriol 191: 4259-4267. doi: 10.1128/JB.01730-08. PubMed: 19395493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cherradi Y, Schiavolin L, Moussa S, Meghraoui A, Meksem A et al. (2013) Interplay between predicted inner-rod and gatekeeper in controlling substrate specificity of the type III secretion system. Mol Microbiol 87: 1183-1199. doi: 10.1111/mmi.12158. PubMed: 23336839. [DOI] [PubMed] [Google Scholar]
- 20. Sal-Man N, Deng W, Finlay BB (2012) EscI: a crucial component of the type III secretion system forms the inner rod structure in enteropathogenic Escherichia coli . Biochem J 442: 119-125. doi: 10.1042/BJ20111620. PubMed: 22087554. [DOI] [PubMed] [Google Scholar]
- 21. Monjarás Feria J, García-Gómez E, Espinosa N, Minamino T, Namba K et al. (2012) Role of EscP (Orf16) in injectisome biogenesis and regulation of type III protein secretion in enteropathogenic Escherichia coli . J Bacteriol 194: 6029-6045. doi: 10.1128/JB.01215-12. PubMed: 22923600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thomassin JL, He X, Thomas NA (2011) Role of EscU auto-cleavage in promoting type III effector translocation into host cells by enteropathogenic Escherichia coli . BMC Microbiol 11: 205. doi: 10.1186/1471-2180-11-205. PubMed: 21933418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV et al. (2008) Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453: 124-127. doi: 10.1038/nature06832. PubMed: 18451864. [DOI] [PubMed] [Google Scholar]
- 24. Frost S, Ho O, Login FH, Weise CF, Wolf-Watz H et al. (2012) Autoproteolysis and intramolecular dissociation of Yersinia YscU precedes secretion of its C-terminal polypeptide YscU. PLOS ONE 7(11):e49349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kubori T, Galán JE (2002) Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J Bacteriol 184: 4699-4708. doi: 10.1128/JB.184.17.4699-4708.2002. PubMed: 12169593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim JS, Jang JI, Eom JS, Oh CH, Kim HG et al. (2013) Molecular characterization of the InvE regulator in the secretion of type III secretion translocases in Salmonella enterica serovar Typhimurium. Microbiology 159: 446-461. doi: 10.1099/mic.0.061689-0. PubMed: 23288540. [DOI] [PubMed] [Google Scholar]
- 27. Deng W, Li Y, Hardwidge PR, Frey EA, Pfuetzner RA et al. (2005) Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun 73: 2135-2146. doi: 10.1128/IAI.73.4.2135-2146.2005. PubMed: 15784556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O’Connell CB, Creasey EA, Knutton S, Elliott S, Crowther LJ et al. (2004) SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol Microbiol 52: 1613-1625. doi: 10.1111/j.1365-2958.2004.04101.x. PubMed: 15186412. [DOI] [PubMed] [Google Scholar]
- 29. Wang D, Roe AJ, McAteer S, Shipston MJ, Gally DL (2008) Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol Microbiol 69: 1499-1512. doi: 10.1111/j.1365-2958.2008.06377.x. PubMed: 18673458. [DOI] [PubMed] [Google Scholar]
- 30. Botteaux A, Sory MP, Biskri L, Parsot C, Allaoui A (2009) MxiC is secreted by and controls the substrate specificity of the Shigella flexneri type III secretion apparatus. Mol Microbiol 71: 449-460. doi: 10.1111/j.1365-2958.2008.06537.x. PubMed: 19017268. [DOI] [PubMed] [Google Scholar]
- 31. Martinez-Argudo I, Blocker AJ (2010) The Shigella T3SS needle transmits a signal for MxiC release, which controls secretion of effectors. Mol Microbiol 78: 1365-1378. PubMed: 21143311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41: D348-D352. doi: 10.1093/nar/gks1243. PubMed: 23197659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pallen MJ, Beatson SA, Bailey CM (2005) Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol 5: 9. doi: 10.1186/1471-2180-5-9. PubMed: 15757514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng LW, Schneewind O (2000) Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells. J Bacteriol 182: 3183-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferracci F, Schubot FD, Waugh DS, Plano GV (2005) Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol Microbiol 57: 970-987. doi: 10.1111/j.1365-2958.2005.04738.x. PubMed: 16091038. [DOI] [PubMed] [Google Scholar]
- 36. Joseph SS, Plano GV (2013) The SycN/YscB chaperone-binding domain of YopN is required for the calcium-dependent regulation of Yop secretion by Yersinia pestis . Front Cell Infect Microbiol 3: 1 PubMed: 23355975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forsberg Å, Viitanen AM, Skurnik M, Wolf-Watz H (1991) The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis . Mol Microbiol 5: 977-986. doi: 10.1111/j.1365-2958.1991.tb00773.x. PubMed: 1857212. [DOI] [PubMed] [Google Scholar]
- 38. Cheng LW, Kay O, Schneewind O (2001) Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica . J Bacteriol 183: 5293-5301. doi: 10.1128/JB.183.18.5293-5301.2001. PubMed: 11514512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferracci F, Day JB, Ezelle HJ, Plano GV (2004) Expression of a functional secreted YopN-TyeA hybrid protein in Yersinia pestis is the result of a +1 translational frameshift event. J Bacteriol 186: 5160-5166. doi: 10.1128/JB.186.15.5160-5166.2004. PubMed: 15262954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee VT, Mazmanian SK, Schneewind O (2001) A program of Yersinia enterocolitica type III secretion reactions is activated by specific signals. J Bacteriol 183: 4970-4978. doi: 10.1128/JB.183.17.4970-4978.2001. PubMed: 11489848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dewoody RS, Merritt PM, Marketon MM (2013) Regulation of the Yersinia type III secretion system: traffic control. Front Cell Infect Microbiol 3: 4 PubMed: 23390616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M et al. (1996) Modulation of virulence factor expression by pathogen target cell contact. Science 273: 1231-1233. doi: 10.1126/science.273.5279.1231. PubMed: 8703058. [DOI] [PubMed] [Google Scholar]
- 43. Rosqvist R, Magnusson KE, Wolf-Watz H (1994) Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J 13: 964-972. PubMed: 8112310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schiano CA, Lathem WW (2012) Post-transcriptional regulation of gene expression in Yersinia species. Front Cell Infect Microbiol 2: 129 PubMed: 23162797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Amer AA, Åhlund MK, Bröms JE, Forsberg A, Francis MS (2011) Impact of the N-terminal secretor domain on YopD translocator function in Yersinia pseudotuberculosis type III secretion. J Bacteriol 193: 6683-6700. doi: 10.1128/JB.00210-11. PubMed: 21965570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Milton DL, O’Toole R, Horstedt P, Wolf-Watz H (1996) Flagellin A is essential for the virulence of Vibrio anguillarum . J Bacteriol 178: 1310-1319. PubMed: 8631707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bölin I, Wolf-Watz H (1984) Molecular cloning of the temperature-inducible outer membrane protein 1 of Yersinia pseudotuberculosis . Infect Immun 43: 72-78. PubMed: 6317574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feldman MF, Müller S, Wüest E, Cornelis GR (2002) SycE allows secretion of YopE-DHFR hybrids by the Yersinia enterocolitica type III Ysc system. Mol Microbiol 46: 1183-1197. doi: 10.1046/j.1365-2958.2002.03241.x. PubMed: 12421321. [DOI] [PubMed] [Google Scholar]
- 49. Morales VM, Bäckman A, Bagdasarian M (1991) A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97: 39-47. doi: 10.1016/0378-1119(91)90007-X. PubMed: 1847347. [DOI] [PubMed] [Google Scholar]
- 50. Straley SC, Bowmer WS (1986) Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect Immun 51: 445-454. PubMed: 3002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rosqvist R, Forsberg Å, Wolf-Watz H (1991) Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun 59: 4562-4569. PubMed: 1937815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Francis MS, Wolf-Watz H (1998) YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: evidence of a structural domain necessary for translocation. Mol Microbiol 29: 799-813. doi: 10.1046/j.1365-2958.1998.00973.x. PubMed: 9723919. [DOI] [PubMed] [Google Scholar]
- 53. Bartra S, Cherepanov P, Forsberg A, Schesser K (2001) The Yersinia YopE and YopH type III effector proteins enhance bacterial proliferation following contact with eukaryotic cells. BMC Microbiol 1: 22. doi: 10.1186/1471-2180-1-22. PubMed: 11696238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Costa TR, Amer AA, Fällman M, Fahlgren A, Francis MS (2012) Coiled-coils in the YopD translocator family: A predicted structure unique to the YopD N-terminus contributes to full virulence of Yersinia pseudotuberculosis . Infect Genet Evol 12: 1729-1742. doi: 10.1016/j.meegid.2012.07.016. PubMed: 22910185. [DOI] [PubMed] [Google Scholar]
- 55. Costa TR, Amer AA, Farag SI, Wolf-Watz H, Fällman M et al. (2013) Type III secretion translocon assemblies that attenuate Yersinia virulence. Cell Microbiol 15: 1088-1110. doi: 10.1111/cmi.12100. PubMed: 23279117. [DOI] [PubMed] [Google Scholar]
- 56. Dinman JD (2012) Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA 3: 661-673. doi: 10.1002/wrna.1126. PubMed: 22715123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buchan JR, Stansfield I (2007) Halting a cellular production line: responses to ribosomal pausing during translation. Biol Cell 99: 475-487. doi: 10.1042/BC20070037. PubMed: 17696878. [DOI] [PubMed] [Google Scholar]
- 58. Baranov PV, Gesteland RF, Atkins JF (2002) Recoding: translational bifurcations in gene expression. Gene 286: 187-201. doi: 10.1016/S0378-1119(02)00423-7. PubMed: 11943474. [DOI] [PubMed] [Google Scholar]
- 59. Namy O, Rousset JP, Napthine S, Brierley I (2004) Reprogrammed genetic decoding in cellular gene expression. Mol Cell 13: 157-168. doi: 10.1016/S1097-2765(04)00031-0. PubMed: 14759362. [DOI] [PubMed] [Google Scholar]
- 60. Farabaugh PJ (1996) Programmed translational frameshifting. Annu Rev Genet 30: 507-528. doi: 10.1146/annurev.genet.30.1.507. PubMed: 8982463. [DOI] [PubMed] [Google Scholar]
- 61. Iriarte M, Sory MP, Boland A, Boyd AP, Mills SD et al. (1998) TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. EMBO J 17: 1907-1918. doi: 10.1093/emboj/17.7.1907. PubMed: 9524114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sundberg L, Forsberg A (2003) TyeA of Yersinia pseudotuberculosis is involved in regulation of Yop expression and is required for polarized translocation of Yop effectors. Cell Microbiol 5: 187-202. doi: 10.1046/j.1462-5822.2003.00267.x. PubMed: 12614462. [DOI] [PubMed] [Google Scholar]
- 63. Akopyan K, Edgren T, Wang-Edgren H, Rosqvist R, Fahlgren A et al. (2011) Translocation of surface-localized effectors in type III secretion. Proc Natl Acad Sci U S A 108: 1639-1644. doi: 10.1073/pnas.1013888108. PubMed: 21220342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Persson C, Nordfelth R, Holmström A, Håkansson S, Rosqvist R et al. (1995) Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol 18: 135-150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. PubMed: 8596454. [DOI] [PubMed] [Google Scholar]
- 65. Lee VT, Anderson DM, Schneewind O (1998) Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol Microbiol 28: 593-601. doi: 10.1046/j.1365-2958.1998.00822.x. PubMed: 9632261. [DOI] [PubMed] [Google Scholar]
- 66. Day JB, Ferracci F, Plano GV (2003) Translocation of YopE and YopN into eukaryotic cells by Yersinia pestis yopN, tyeA, sycN, yscB and lcrG deletion mutants measured using a phosphorylatable peptide tag and phosphospecific antibodies. Mol Microbiol 47: 807-823. doi: 10.1046/j.1365-2958.2003.03343.x. PubMed: 12535078. [DOI] [PubMed] [Google Scholar]
- 67. Viboud GI, Bliska JB (2005) Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol 59: 69-89. doi: 10.1146/annurev.micro.59.030804.121320. PubMed: 15847602. [DOI] [PubMed] [Google Scholar]
- 68. Li GW, Oh E, Weissman JS (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484: 538-541. doi: 10.1038/nature10965. PubMed: 22456704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen H, Bjerknes M, Kumar R, Jay E (1994) Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res 22: 4953-4957. doi: 10.1093/nar/22.23.4953. PubMed: 7528374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF (1988) Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli . EMBO J 7: 1503-1507. PubMed: 2457498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hansen TM, Reihani SN, Oddershede LB, Sørensen MA (2007) Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc Natl Acad Sci U S A 104: 5830-5835. doi: 10.1073/pnas.0608668104. PubMed: 17389398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kontos H, Napthine S, Brierley I (2001) Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol Cell Biol 21: 8657-8670. doi: 10.1128/MCB.21.24.8657-8670.2001. PubMed: 11713298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McNulty DE, Claffee BA, Huddleston MJ, Porter ML, Cavnar KM et al. (2003) Mistranslational errors associated with the rare arginine codon CGG in Escherichia coli . Protein Expr Purif 27: 365-374. doi: 10.1016/S1046-5928(02)00610-1. PubMed: 12597898. [DOI] [PubMed] [Google Scholar]
- 74. Schwartz R, Curran JF (1997) Analyses of frameshifting at UUU-pyrimidine sites. Nucleic Acids Res 25: 2005-2011. doi: 10.1093/nar/25.10.2005. PubMed: 9115369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Higashi K, Kashiwagi K, Taniguchi S, Terui Y, Yamamoto K et al. (2006) Enhancement of +1 frameshift by polyamines during translation of polypeptide release factor 2 in Escherichia coli . J Biol Chem 281: 9527-9537. PubMed: 16476727. [DOI] [PubMed] [Google Scholar]
- 76. Igarashi K, Kashiwagi K (2010) Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42: 39-51. doi: 10.1016/j.biocel.2009.07.009. PubMed: 19643201. [DOI] [PubMed] [Google Scholar]
- 77. Shah P, Swiatlo E (2008) A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol 68: 4-16. doi: 10.1111/j.1365-2958.2008.06126.x. PubMed: 18405343. [DOI] [PubMed] [Google Scholar]
- 78. Jelsbak L, Thomsen LE, Wallrodt I, Jensen PR, Olsen JE (2012) Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLOS ONE 7: e36149. doi: 10.1371/journal.pone.0036149. PubMed: 22558361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou L, Wang J, Zhang LH (2007) Modulation of bacterial Type III secretion system by a spermidine transporter dependent signaling pathway. PLOS ONE 2: e1291. doi: 10.1371/journal.pone.0001291. PubMed: 18074016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goss JW, Sorg JA, Ramamurthi KS, Ton-That H, Schneewind O (2004) The secretion signal of YopN, a regulatory protein of the Yersinia enterocolitica type III secretion pathway. J Bacteriol 186: 6320-6324. doi: 10.1128/JB.186.18.6320-6324.2004. PubMed: 15342604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Day JB, Plano GV (1998) A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis . Mol Microbiol 30: 777-788. doi: 10.1046/j.1365-2958.1998.01110.x. PubMed: 10094626. [DOI] [PubMed] [Google Scholar]
- 82. Akeda Y, Galán JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437: 911-915. doi: 10.1038/nature03992. PubMed: 16208377. [DOI] [PubMed] [Google Scholar]
- 83. Yang H, Tan Y, Zhang T, Tang L, Wang J et al. (2013) Identification of novel protein-protein interactions of Yersinia pestis type III secretion system by yeast two hybrid system. PLOS ONE 8: e54121. doi: 10.1371/journal.pone.0054121. PubMed: 23349800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen Y, Anderson DM (2011) Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol Microbiol 80: 966-980. doi: 10.1111/j.1365-2958.2011.07623.x. PubMed: 21481017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Anderson DM, Ramamurthi KS, Tam C, Schneewind O (2002) YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J Bacteriol 184: 1287-1295. doi: 10.1128/JB.184.5.1287-1295.2002. PubMed: 11844757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Simon R, Priefer U, Pühler A (1983) A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1: 784-791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 87. Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H et al. (2002) Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol 184: 4500-4509. doi: 10.1128/JB.184.16.4500-4509.2002. PubMed: 12142420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg A et al. (2003) YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol 185: 2259-2266. doi: 10.1128/JB.185.7.2259-2266.2003. PubMed: 12644497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Edqvist PJ, Bröms JE, Betts HJ, Forsberg Å, Pallen MJ et al. (2006) Tetratricopeptide repeats in the type III secretion chaperone, LcrH: Their role in substrate binding and secretion. Mol Microbiol 59: 31-44. doi: 10.1111/j.1365-2958.2005.04923.x. PubMed: 16359316. [DOI] [PubMed] [Google Scholar]
- 90. Rosqvist R, Forsberg Å, Rimpiläinen M, Bergman T, Wolf-Watz H (1990) The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol 4: 657-667. doi: 10.1111/j.1365-2958.1990.tb00635.x. PubMed: 2191183. [DOI] [PubMed] [Google Scholar]
- 91. Portnoy DA, Moseley SL, Falkow S (1981) Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun 31: 775-782. PubMed: 7216474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Schubot FD, Jackson MW, Penrose KJ, Cherry S, Tropea JE et al. (2005) Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis . J Mol Biol 346: 1147-1161. doi: 10.1016/j.jmb.2004.12.036. PubMed: 15701523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of free TyeA synthesis and secretion in synthetic YopN-TyeA chimeric mutants. Overnight cultures of Y. pseudotuberculosis were sub-cultured into BHI medium in the absence of calcium ions at 26°C for 1 hour and at 37°C for 3 hours. At the time of temperature up-shift, 0.4 mM IPTG was added to all cultures. Protein in the total bacterial suspension (Synthesis) and free in the cleared culture supernatant (Secretion) were collected, fractionated by 15% acrylamide SDS-PAGE, wet-blotted onto PDVF membrane and then detected using rabbit polyclonal anti-TyeA antibodies. The arrow (→) point towards a non-specific protein band recognized by the anti-TyeA antiserum. The single asterisk (*) highlights the larger YopN-TyeA hybrid protein. Lanes are Y. pseudotuberculosis ΔyopN, tyeA (YPIII/pIB8201a) also containing pYopN, TyeA+ (pAA304), empty vector (pMMB208), pYopN278(F+1), TyeA+ (pAA306), pYopN278(F+1), SD, TyeA+ (pAA307), pYopN287(F+1), TyeA+ (pAA308), or pYopN287(F+1), SD, TyeA+ (pAA309). Approximate molecular mass values shown in parentheses were deduced from primary amino acid sequences.
(TIF)
YopN-TyeA hybrid-producing bacteria spawn external YscF multimers. Yersinia strains were grown in non permissive T3S media (plus Ca2+). Where indicated (+), the membrane-impermeable chemical cross-linker BS3 was added to the bacteria. After being quenched with Tris-HCl, bacteria pellets were solubilized in sample buffer and then protein fractionated by 12% acrylamide SDS-PAGE. After wet-transfer to PVDF, YscF was detected with immune-absorbed monospecific anti-YscF antiserum. Non-cross-linked monomeric YscF was observed in all lanes except the ΔyscF null mutant control. Cell-surface YscF multimers were observed in all lanes except for the ΔyscF null mutant control as well as the YscF+, but T3SS-defective, ΔyscU, lcrQ null mutant control. The predicted molecular mass of monomeric YscF is given in parenthesis, while approximate sizes of protein molecular weight standards are given to the right. Strains: Parent (YopNYps), YPIII/pIB102; ΔyopN null mutant, YPIII/pIB82; ΔtyeA null mutant, YPIII/pIB801a; ΔyopN, tyeA double mutant, YPIII/pIB8201a; YopN 278(F+1)TyeA, YPIII/pIB8205; YopN 278(F+1), SDTyeA, YPIII/pIB8206; YopN 287(F+1)TyeA, YPIII/pIB8210; YopN 287(F+1), SDTyeA, YPIII/pIB8211; ΔyscF null mutant, YPIII/pIB202; ΔyscU, lcrQ double mutant, YPIII/pIB75-26.
(TIF)
Low calcium response growth phenotypes of Y. pseudotuberculosis producing YopN-TyeA hybrids. Bacteria were grown at 37°C in TMH medium supplemented with 2.5 mM CaCl2 (plus Ca2+; A) or non-supplemented (minus Ca2+; B). Two different growth phenotypes were detected: TS – bacteria are sensitive to elevated temperature regardless of the presence or absence of calcium (ΔyopN and/or ΔtyeA null mutants) and, CD – calcium dependent growth (all remaining strains). Strains: Parent (YopNYps), YPIII/pIB102; ΔyopN null mutant, YPIII/pIB82; ΔtyeA null mutant, YPIII/pIB801a; ΔyopN, tyeA double mutant, YPIII/pIB8201a; YopN 278(F+1)TyeA, YPIII/pIB8205; YopN 278(F+1), SDTyeA, YPIII/pIB8206; YopN 287(F+1)TyeA, YPIII/pIB8210; YopN 287(F+1), SDTyeA, YPIII/pIB8211.
(TIF)
Oligonucleotides used in this study.
(PDF)
Competitive index for mice colonization.
(PDF)