Abstract
Mouse Gasdermin A3 (Gsdma3) is the causative gene for dominant skin mutations exhibiting alopecia. Mouse has two other Gsdma3-related genes, Gsdma and Gsdma2, whereas human and rat have only one related gene. To date, no skin mutation has been reported for human GSDMA and rat Gsdma as well as mouse Gsdma and Gsdma2. Therefore, it is possible that only Gsdma3 has gain-of-function type mutations to cause dominant skin phenotype. To elucidate functional divergence among the Gsdma-related genes in mice, and to infer the function of the human and rat orthologs, we examined in vivo function of mouse Gsdma by generating Gsdma knockout mice and transgenic mice that overexpress wild-type Gsdma or Gsdma harboring a point mutation (Alanine339Threonine). The Gsdma knockout mice shows no visible phenotype, indicating that Gsdma is not essential for differentiation of epidermal cells and maintenance of the hair cycle, and that Gsdma is expressed specifically both in the inner root sheath of hair follicles and in suprabasal cell layers, whereas Gsdma3 is expressed only in suprabasal layers. By contrast, both types of the transgenic mice exhibited epidermal hyperplasia resembling the Gsdma3 mutations, although the phenotype depended on the genetic background. These results indicate that the mouse Gsdma and Gsdma3 genes share common function to regulate epithelial maintenance and/or homeostasis, and suggest that the function of human GSDMA and rat Gsdma, which are orthologs of mouse Gsdma, is conserved as well.
Keywords: Gasdermin A, duplication, knockout, transgenic, alopecia
Gene duplication is a primary source of genetic diversity in evolution. Functional divergence of duplicated genes arises from differentiation in amino acid sequence and/or gene expression pattern between duplicated genes (Ohno 1970). Such differentiation is driven by the accumulation of mutations in the coding sequence and/or cis-regulatory elements during evolution (Lynch and Conery 2000; Zhang 2003). Elucidation of the diverged functions of the duplicated genes is important to understand how organisms acquire phenotypic diversity during evolution.
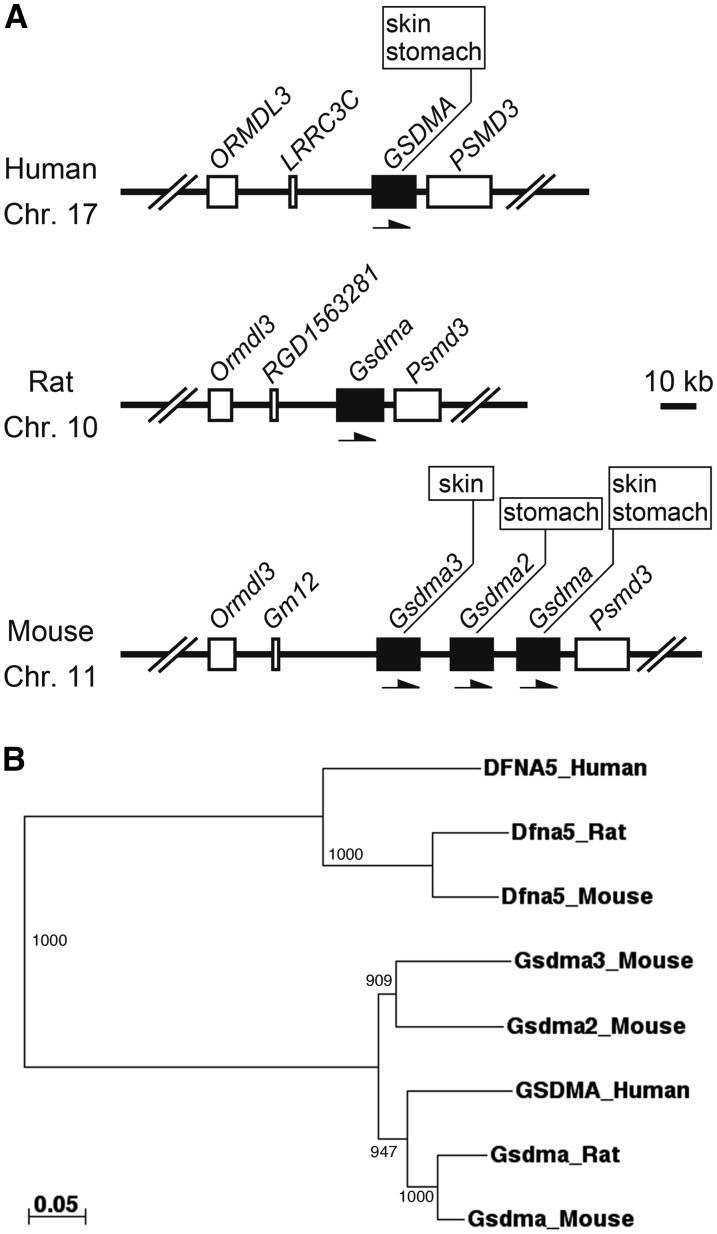
The Gasdermin (Gsdm/GSDM) gene family is composed of four paralogous genes, Gasdermin A (Gsdma/GSDMA), Gasdermin B (GSDMB), Gasdermin C (Gsdmc/GSDMC), and Gasdermin D (Gsdmd/GSDMD), in the mouse, rat, and human genomes. These genes were likely generated by two-round whole-genome duplications during vertebrate evolution (Tamura et al. 2007). The number of genes in each Gsdm/GSDM family differs among species. In mice, further tandem duplication occurred in the Gsdma, resulting in the formation of gene cluster: three Gsdma-related genes (Gsdma, Gsdma2, and Gsdma3) (Tamura et al. 2007) (Figure 1A). Phylogenetic analysis of the Gsdma cluster showed that human GSDMA has 87%, 74%, and 73% amino acid sequence similarity with mouse Gsdma, Gsdma2, and Gsdma3, respectively, indicating that human GSDMA is the counterpart of mouse Gsdma (Figure 1B) (Runkel et al. 2004). The Gsdm/GSDM family genes are differentially expressed in the epithelium from skin to gastrointestinal tract in a highly tissue-specific manner (Tamura et al. 2007). Although human GSDMA is mainly expressed in skin and stomach (Saeki et al. 2000), expression domains of mouse Gsdma cluster genes are divided into three compartments. Gsdma is expressed in the squamous epithelium from skin to the cardia of stomach, and Gsdma2 and Gsdma3 are specifically expressed in the epithelium of glandular stomach and skin, respectively (Runkel et al. 2004; Lunny et al. 2005; Tamura et al. 2007; Tanaka et al. 2007a) (Figure 1A).
Figure 1.
Gsdma orthologous genes in human, rat and mouse. (A) The genome structure of the Gsdma orthologous gene in human, rat and mouse was constructed based on Genome Reference Consortium Human Build 37, RGSC_v3.4, and Genome Reference Consortium Mouse Build 38, respectively. Transcriptional orientation of each gene is represented by an arrow. Expression domains of each Gsdma orthologous gene are shown in boxes. (B) Unrooted phylogenetic tree of Gsdma orthologous genes in human, rat, and mouse. The tree is constructed using a neighbor-joining method based on the multiple alignment generated by the ClustalW program. The numbers indicate the bootstrap values based on 1000 runs. DFNA5 orthologs are used as outgroup sequences. The scale bar indicates the number of amino acid substitutions per site.
The Gsdm/GSDM family genes encode 400−500 amino acid residues. A leucine-rich motif is well conserved in the C-terminus region across this family of genes, but there is no other known motif or domain (Tamura et al. 2007). The biologic function of the Gsdm/GSDM family genes initially was inferred from phenotypes of spontaneous and chemically-induced mouse mutants. To date, Gsdma3 has been identified as the causative gene of eight mouse alopecia mutations, all of which exhibit dominant phenotype of hyperkeratosis and hair loss (Runkel et al. 2004; Lunny et al. 2005; Tanaka et al. 2007a; Sun et al. 2009; Li et al. 2010; Lei et al. 2011; Zhou et al. 2012). On the other hand, neither a mouse nor human mutation has been reported for other members of the Gsdm/GSDM family genes.
Altered expression patterns of the GSDM family genes in human cancer cell lines hinted at their cellular function in vitro. For instance, overexpression of human GSDM family genes, except for GSDMB, induces cell-growth inhibition in cancer cell lines (Saeki et al. 2007; Saeki et al. 2009). Gene expression of GSDMA and GSDMD was frequently suppressed, and GSDMB was overexpressed in cancer cell lines and/or cancer tissue specimens (Saeki et al. 2009; Komiyama et al. 2010). These studies, together with expression patterns of the Gsdm/GSDM family genes, suggest that these genes are involved in regulation of the epithelial cell proliferation and differentiation, but their functions in vivo are still poorly understood.
In this study, we intended to clarify the in vivo functions of the Gsdm/GSDM family genes, focusing on the mouse Gsdma cluster. We generated Gsdma knockout (KO) mice and transgenic (TG) mice with the wild-type or mutant-type Gsdma transgene. These results revealed that mouse Gsdma has a function similar to that of Gsdma3, suggesting that this function also is conserved in the human and rat orthologs.
Materials and Methods
Mice
The C57BL/6J (B6) strain originally was purchased from the Jackson Laboratory (Bar Harbor, ME) and is maintained at Genetic Strains Research Center, National Institute of Genetics (NIG, Mishima, Japan). ICR strain and (C57BL/6N × DBA/2N)F1 mice were purchased from CLEA Japan (Tokyo, Japan). Mice were housed in an SPF facility (12-hr light and dark cycles). The animal experiments in this study were approved by the Animal Care and Use Committee of NIG.
Generation of Gsdma KO mice
We used the ploxFNFDT vector containing the floxed PGK-Neo-poly-A cassette, PGK-DTA-poly-A cassette, and two loxP sites that would enable global deletion of the Gsdma cluster genes in a future study. To construct the targeting vector, a 5.5-kb long arm containing exon 4–8, a 2.8-kb short arm composed of three DNA fragments containing exon 1–3 cloned from BAC RP23-395E DNA, and a LacZ-poly-A fragment were cloned from pCMV SPORT-βgal (Invitrogen Japan, Tokyo, Japan) by polymerase chain reaction (PCR). These DNA fragments were subcloned into the ploxFNFDT vector. To trace the expression of the Gsdma gene, the LacZ-poly-A fragment was inserted at the start codon in exon 2. This insertion disrupts the expression of the endogenous Gsdma gene but allows the expression of the LacZ gene controlled by the intact promoter activity of Gsdma. The targeting vector was linearized with I-Sce I and electroporated into TT2 embryonic stem (ES) cells, which are derived from a (B6 × CBA/J)F1 mouse (Yagi et al. 1993). G418-resistant ES cell clones were selected for homologous recombination by PCR and Southern blot analysis. Positive clones were aggregated with eight-cell embryos from ICR mice and transplanted into surrogate females. Male chimeras were mated with B6 females. Germline transmission of the targeting allele was confirmed by Southern blot analysis and PCR using the primer pairs: P1, 5′-AAATGGAGGGTGCAAACAAG-3′; and P2, 5′-GGGTCGTCAAGACCTGGTAA-3′. A digoxigenin-labeled probe for Southern blot analysis was synthesized by PCR using the primer pairs: F, 5′-GGATTGTGGTGGTAACGGTAG-3′; and R, 5′-CAGGACATCTTTGGGGAGTGC-3′. Signal was detected using alkaline phosphatase−conjugated antidigoxigenin antibody and CDP Star according to the manufacturer’s protocol (Roche Diagnostics Japan, Tokyo, Japan). Heterozygous mice were maintained by repeating backcrosses onto B6. We analyzed KO mice at N5 generations.
Generation of keratin 5 (K5)-Gsdma TG mice
We obtained the pKM2L-phK5 vector containing the 6.3-kb human K5 promoter from RIKEN BioResource Center (BRC, Tsukuba, Japan). cDNAs were synthesized from total RNA derived from B6 skin using PrimeScript II (TAKARA, Otsu, Japan). We amplified full-length Gsdma from skin cDNA by PCR using KOD-plus DNA polymerase (TOYOBO CO., LTD, Osaka, Japan). PCR products were directly cloned into pCR-TOPOII vector (Invitrogen Japan). After confirming the wild-type Gsdma by sequencing, the open reading frame of Gsdma with the Kozak sequence was amplified by PCR using the following primer pairs with a restriction enzyme site: F, 5’-AAAAGATCTTAAGGCGCGCCACCATGTTTGAGAATGTCACCCG-3′; and R, 5′-AAAGTCGACTTAGGAATTCTTGCTTAGCA-3′. PCR products were digested at the BglII and SalI sites. Digested PCR products were inserted into the pIRES2-EGFP vector. After confirming the open reading frame again by sequencing, the Gsdma-IRES2-EGFP fragment was replaced with the luciferase reporter gene in the pKM2L-phK5 vector at the AflII and NotI sites. For the Gsdma3Rim3-type Gsdma mutation, site-directed mutagenesis was performed by PCR using KOD-plus taq polymerase. Inverted tail-to-tail primer pairs (F, 5′-CCGGACACGCTCCCCCACCTTT-3′; R, 5′-GGAGAGGCCAGCTTAGAGGGCAC-3′) containing a point mutation were phosphorylated with T4 kinase and ATP (Toyobo Co., Ltd, Osaka, Japan). PCR products were amplified using this primer pair and pCR-TOPOII vector containing Gsdma as a template and were directly self-ligated. The Gsdma3Rim3-type Gsdma fragment confirmed by sequencing was inserted into the pKM2L-phK5 vector in the same manner. The vector was linearized at the I-SceI sites and microinjected into the pronuclei of fertilized eggs derived from (C57BL/6N × DBA/2N)F1 mice. TG mice were selected by PCR analysis using the primer pair for the human K5 promoter region (F, 5′-AGACTCAGCATAGGGCTGGA-3′; R, 5′-GGGAGAGGGTGGTATCCATT’) and the Egfp gene (F, 5′-ACGTAAACGGCCACAAGTTC-3′; R, 5′-AAGTCGTGCTGCTTCATGTG-3′). Several lines of TG mice selected by expression of the Egfp gene were maintained by repeating backcrosses onto B6.
Histology
For detection of β-galactosidase (β-gal) activity, frozen skin sections (10 μm) were fixed with 0.2% glutaraldehyde for 2 min. After washing 3 times in phosphate-buffered saline, the sections were stained in X-gal solution at 37° for 6 hr. The sections were fixed with 4% paraformaldehyde and washed in phosphate-buffered saline. Nuclear fast red was used as the counterstain. Hematoxylin and eosin staining, and immunohistological analysis, were performed as previously reported (Tanaka et al. 2007b). The following primary antibodies were used in this study: Keratin 14 (K14, 1:50, Covance, Richmond, CA), K71 (1:3200, kindly provided by Y. Shimomura) (Aoki et al. 2001), Gsdma (1:50, Santa Cruz Biotechnology, Santa Cruz, CA), EGFP (1:50, Invitrogen Japan). These antibodies were detected by using appropriate secondary antibodies conjugated with Alexa Fluor 488 (Invitrogen Japan) or Alexa Fluor 594 (Invitrogen Japan). Nuclear staining was performed using To-pro3 (Invitrogen Japan).
Results
Generation of Gsdma KO mice
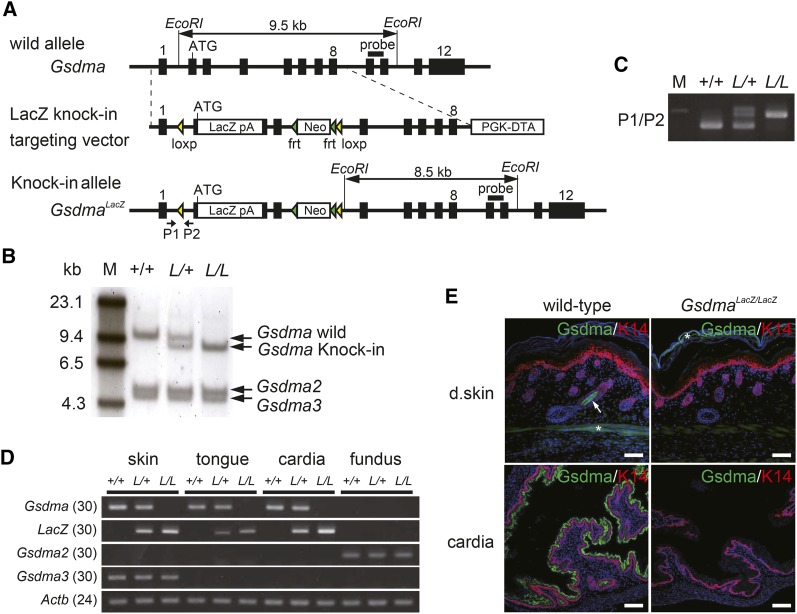
We generated Gsdma KO mice by inserting the LacZ reporter gene at the start codon of Gsdma (Figure 2A). We obtained three clones of targeted GsdmaLacZ/+ ES cells by homologous recombination and then generated a Gsdma KO strain from one of these ES cell clones. Targeted alleles in the Gsdma KO mice were confirmed by Southern blot analysis and PCR genotyping (Figure 2, B and C). We confirmed the complete loss of the Gsdma transcript in homozygous GsdmaLacZ/LacZ mice by semiquantitative reverse-transcription PCR. In addition, LacZ mRNA expression was observed in heterozygous GsdmaLacZ/+ and GsdmaLacZ/LacZ mice (Figure 2D). Gsdma was expressed in the skin, tongue, and cardia of wild-type (Gsdma+/+) mice, but it was not expressed in these tissues in GsdmaLacZ/LacZ mice (Figure 2D). Expression of the other Gsdma cluster genes, Gsdma2 and Gsdma3, was not affected in the KO homozygotes (Figure 2D). Immunostaining using anti-Gsdma antibody revealed that Gsdma protein is localized in the inner root sheath (IRS) of hair follicles and suprabasal cell layers of the cardia epithelium in wild-type mice at embryonic day (E) 18.5, but it was not detected in GsdmaLacZ/LacZ mice (Figure 2E). These results demonstrated that Gsdma is completely and specifically targeted in the Gsdma KO allele.
Figure 2.
Generation and validation of the Gsdma KO mouse. (A) Schematic diagrams of wild-allele, LacZ knock-in targeting vector, and knock-in allele of the Gsdma gene. Exons are indicated as black boxes. loxP and frt sites are indicated as yellow and green triangles, respectively. The LacZ-pA fragment was inserted into the start codon in exon 2. EcoRI fragments and the probe for Southern blot analysis are indicated by horizontal bars with double arrows and horizontal bars. Location of primers used for genotyping are indicated by allows and labeled P1 and P2. (B) Southern blot analysis of EcoRI digested genomic DNA from wild-type (+/+), heterozygous (L/+), and homozygous (L/L) mice for the LacZ knock-in allele. The probe located in exon 9−10 detects 9.5- and 8.5-kb fragments of the wild and knock-in allele, respectively. The DNA fragments containing Gsdma2 and Gsdma3 are intact. M indicates a marker. (C) PCR analysis of genomic DNA from mice with each genotype. Genotype was determined by PCR using primers P1 and P2. This yielded fragments of 410 and 530 bp for the wild and knock-in allele, respectively. (D) Semiquantitative reverse-transcription PCR analysis of Gsdma, Gsdma2, Gsdma3, and LacZ genes using cDNAs prepared from skin, tongue, cardia, and gastric fundus of each genotype. Actin-beta (Actb) was amplified as a control. The number of PCR cycles is shown in parentheses. (E) Immunohistological detection of Gsdma protein in skin and stomach of wild-type and GsdmaLacZ/LacZ mice at E18.5. K14 was used as a marker for the basal cell layer. An arrow and asterisks indicate specific and nonspecific signal, respectively. Scale bar indicates 50 μm.
Spatial expression of Gsdma and Gsdma3
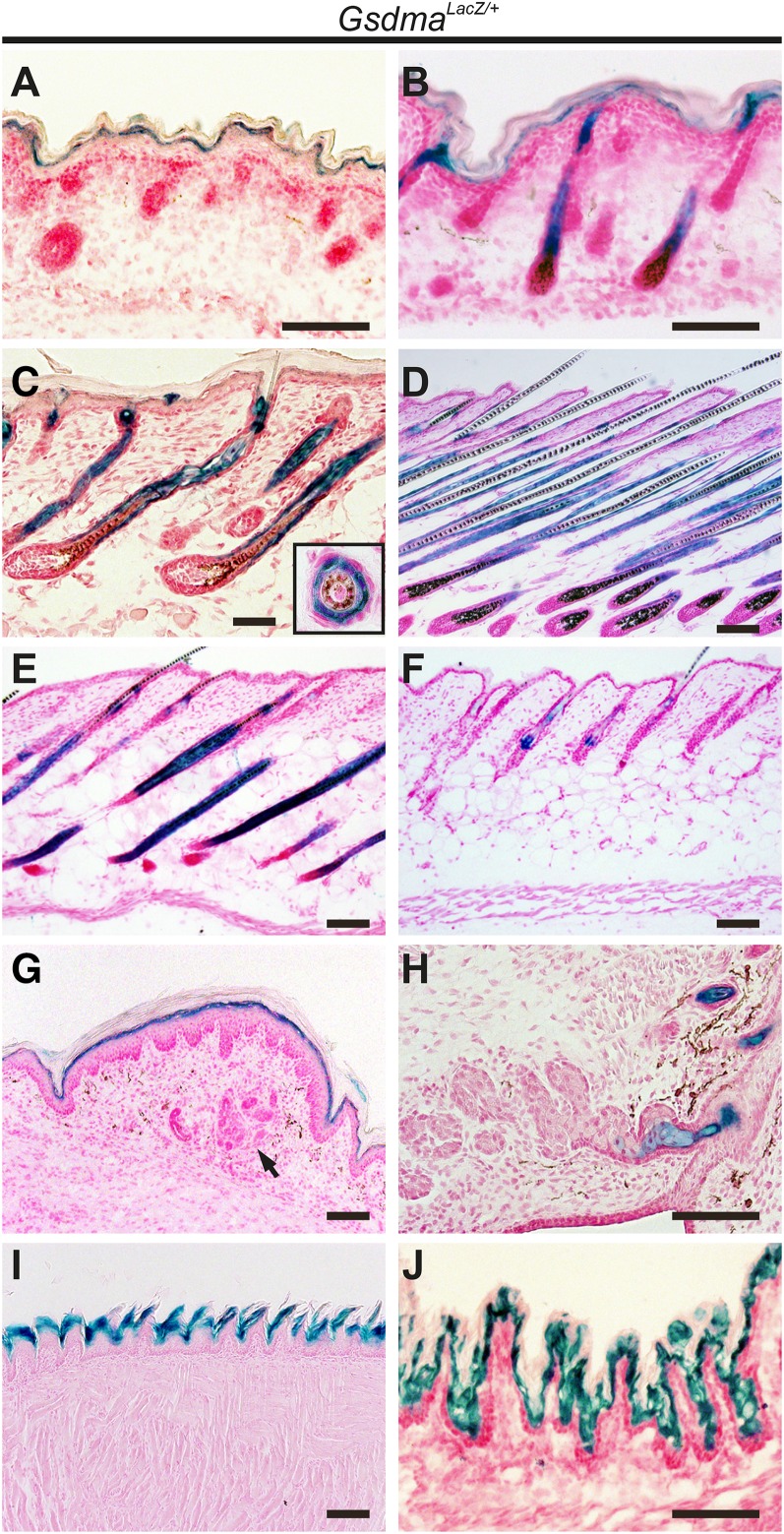
In neonatal mouse skin, both the Gsdma and Gsdma3 genes are expressed (Runkel et al. 2004; Lunny et al. 2005; Tanaka et al. 2007a). However, the spatial distribution of the mRNA and protein of both Gsdma and Gsdma3 remains elusive due to the high similarity in the nucleotide and amino acid sequences of the two genes (Runkel et al. 2004; Lunny et al. 2005; Tanaka et al. 2007a). To clarify the spatial distribution of endogenous Gsdma expression, we analyzed LacZ reporter expression in GsdmaLacZ/+ mice. X-gal staining by β-gal activity was observed in differentiated epithelium derived from ectoderm such as skin, foot pad (except for sweat gland), meibomian gland, tongue, and the cardia region of the stomach (Figure 3, A−J). In the embryonic epidermis, strong expression was detected in the suprabasal cell layer (Figure 3A). In the neonatal epidermis, expression was especially strong in the IRS of hair follicles compared with the suprabasal cell layer (Figure 3, B and C). During the first hair cycle, β-gal activity was consistently detected in IRS. These results are consistent with our previous data obtained by in situ hybridization using probes for Gsdma and Gsdma3 mRNA (Tanaka et al. 2007a).
Figure 3.
Expression of LacZ controlled by Gsdma promoter during epidermal development. X-gal−stained sections were made from GsdmaLacZ/+ skin collected during the developmental stage at E18.5 (A), anagen at P1 (B), and P5 (C); catagen at P13 (D) and P19 (E); and telogen at P19 (F). The same sections were made for footpad at P11 (G), meibomian gland at P5 (H), tongue epithelium at P9 (I) and cardia of stomach at P1 (J) in GsdmaLacZ/+ mice. An inset in (C) shows a cross section of a hair follicle at the same stage of development. An arrow (G) indicates sweat glands.
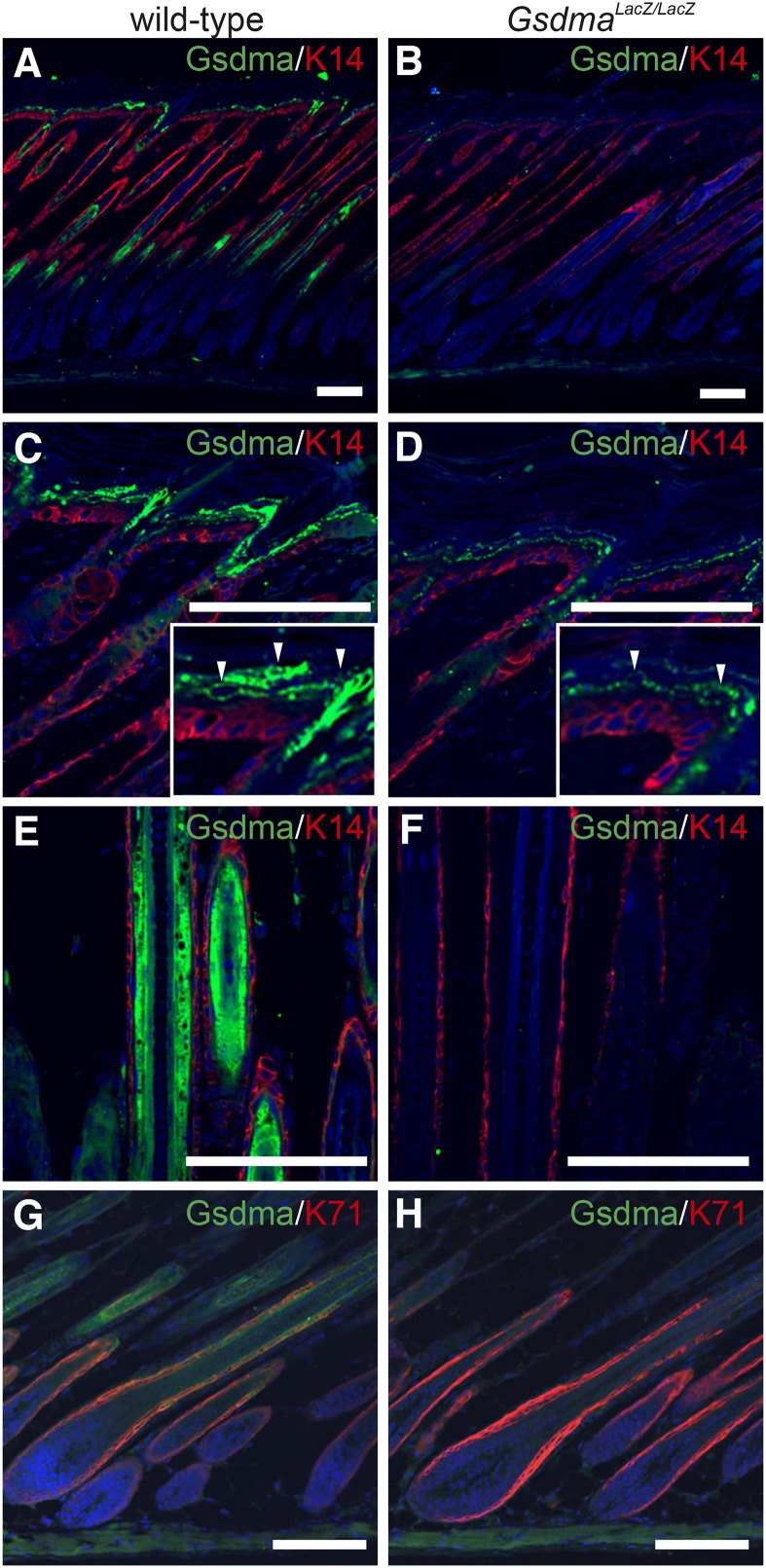
Next, to analyze the spatial distribution of Gsdma3 protein in the epidermis, we carried out immunohistochemistry using anti-Gsdma antibody, which cross-reacts with Gsdma3 protein (Supporting Information, Figure S1). The signals were detected in both the suprabasal cells and IRS of wild-type epidermis (Figure 4, A, C, E, and G) at postnatal day (P) 8. Unexpectedly, in the KO epidermis, the signals were weakly located only in the suprabasal cell layer (Figure 4, B and D) but not in the IRS of hair follicles (Figure 4, B, F, and H). The expression level of Gsdma3 in the suprabasal cell layer was the same in the wild-type and the KO epidermis at P5 by quantitative PCR analysis (Figure S2). All these data clearly indicated that Gsdma is expressed both in the suprabasal cells and IRS, whereas Gsdma3 is predominantly expressed in the suprabasal cells.
Figure 4.
Distribution of Gsdma and Gsdma3 proteins. Immunohistological detection of Gsdma and Gsdma3 proteins in skin of wild-type (A, C, E, and G) and GsdmaLacZ/LacZ (B, D, F, and H) mice at P8. Magnified images are provided to show Gsdma protein (arrowheads) in the suprabasal cell layer. K14 and K71 were used as a marker for the basal cell layer and IRS, respectively. Scale bars are 25 μm.
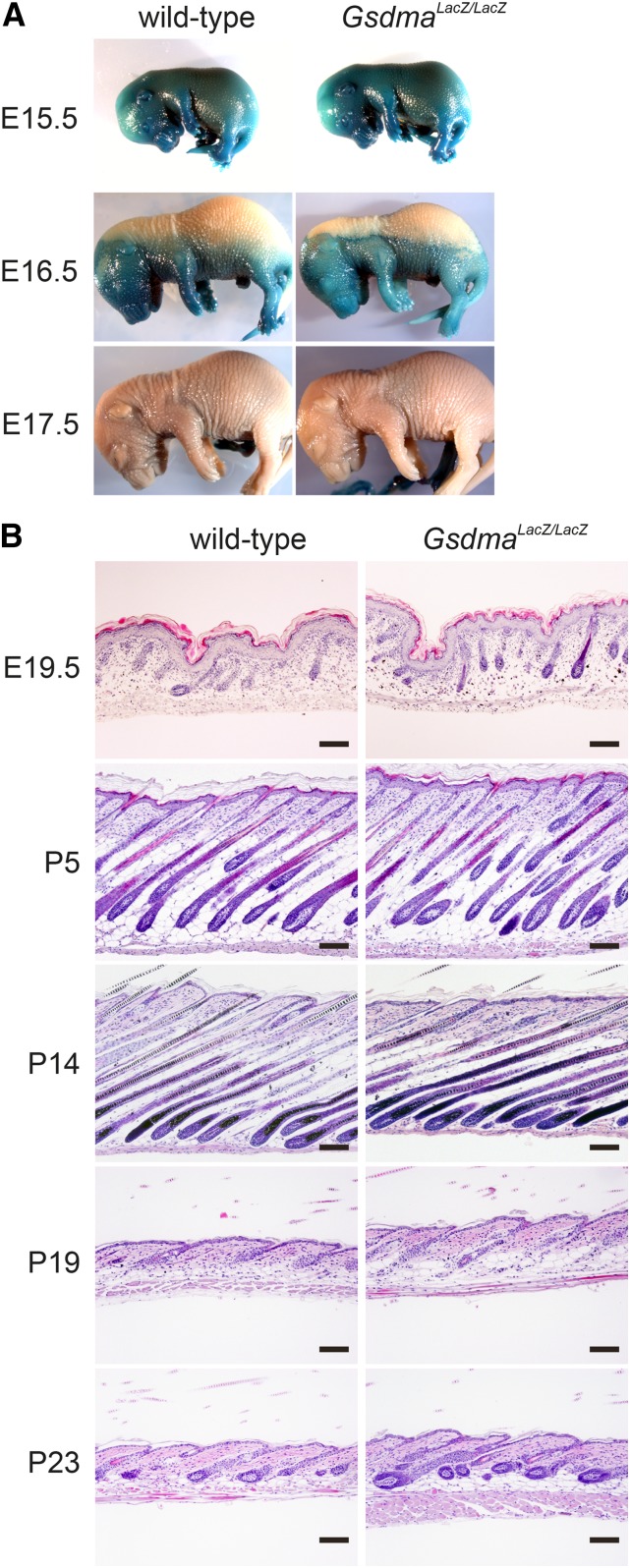
Normal development and homeostasis in Gsdma KO skin
Gsdma KO homozygotes (GsdmaLacZ/LacZ) were born at the expected Mendelian ratio following intercrossing of GsdmaLacZ/+ mice (data not shown). No obvious developmental abnormality was observed in the GsdmaLacZ/LacZ mice. Skin permeability assay showed no difference in the terminal differentiation of epidermal cells at the embryonic stage between wild-type and KO homozygotes, although Gsdma expression starts at the embryonic stage (Figure 5A). We next performed a comprehensive histologic analysis of skin during the first hair cycle. Again, no apparent morphologic difference was observed between wild-type and GsdmaLacZ/LacZ mice (Figure 5B). Finally, we extended the observation time period to 1 month after birth. Immunohistochemistry with anti-Keratin 14 antibody reacting to basal cells, anti-keratin 10 antibody reacting to suprabasal cells, and anti-Filaggrin antibody reacting to cornified cells, detected no difference in the signals of these antibodies for epidermal cells from wild-type and GsdmaLacZ/LacZ mice (Figure S3). Moreover, 1-yr-old GsdmaLacZ/LacZ mice did not exhibit any morphologic abnormalities such as alopecia or skin tumor development (data not shown). These results indicate that Gsdma is not essential for differentiation of epidermal cells and maintenance of the hair cycle.
Figure 5.
Phenotype of Gsdma KO mouse. (A) Result of skin permeability assay in wild-type and GsdmaLacZ/LacZ mice at the ages indicated. (B) HE stained sections of skin during the first hair cycle in wild-type and GsdmaLacZ/LacZ mice. Scale bars are 100 μm.
K5-Gsdma-A339T TG mice exhibit alopecia similar to the Gsdma3 mutant
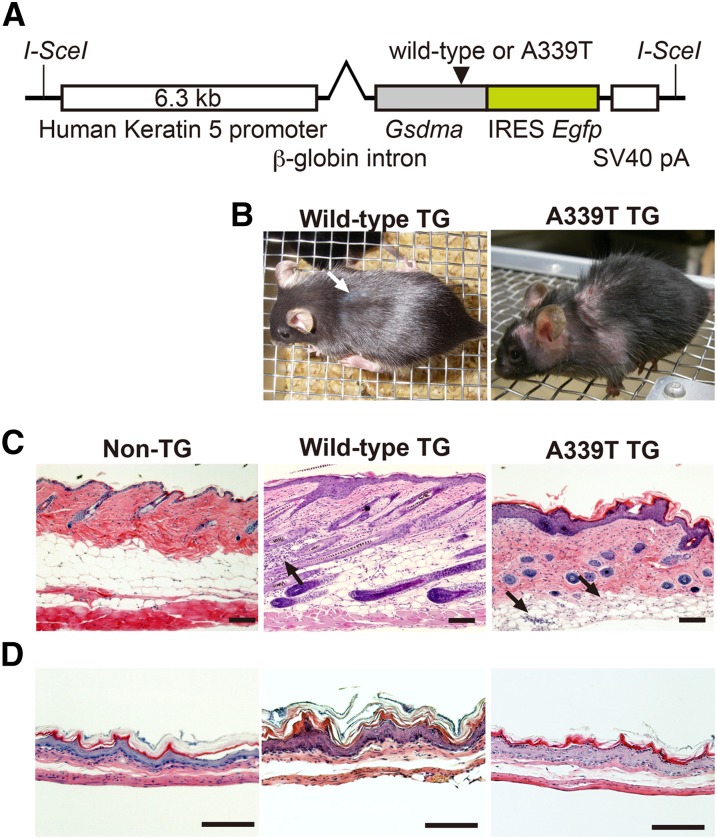
We generated TG mice that express a mutant form of Gsdma. This transgene has the Gsdma3Rim3-type point mutation, A339T, and its expression in skin is driven by human K5 promoter (Figure 6A). In parallel, we also generated another type of TG mouse that expresses wild-type Gsdma with the aim of examining the effect of Gsdma overexpression in skin.
Figure 6.
Phenotype of K5-Gsdma TG mouse. (A) Schematic diagrams of Gsdma TG vector construct with human K5 promoter. Gsdma in the construct is either the wild-type or has the Rim3-type mutation, A339T. (B) Macroscopic phenotypes of mice with wild-type or A339T mutant Gsdma transgenes at 3 months of age. A white arrow indicates patchy rough coat. Hematoxylin and eosin−stained sections of skin (C) and cardia (D) from control mouse (Non-TG), wild-type and A339T TG mice at 3 months of age. Inflammatory cells are present in the dermal fat layer (Arrows in C). Scale bars are 100 μm.
We obtained eight founder mice that expressed the mutant (A339T) Gsdma transgene and two founder mice that expressed the wild-type Gsdma transgene. Irrespective of the type of transgene construct, all these founder mice exhibited no apparent abnormalities in their skin. To examine the genetic background effect on the phenotype, we backcrossed these founder mice onto B6 mice. After three generations of backcross, one line with the wild-type Gsdma transgene started to exhibit a partial rough coat phenotype, and one line with the mutant form (A339T) of the Gsdma transgene started to exhibit alopecia resembling the Gsdma3 mutants (Figure 6B). Similar skin phenotypes to these two lines were observed in subsequent backcross generations. Notably, these phenotypes are observed only in less than 10% of the progeny with the hemizygous transgene. We further carried out histological analysis of abnormal epidermis from the descendant TG mice with the hemizygous transgenes. Epidermal hyperplasia was observed in both lines of mice (Figure 6C). These phenotypes were observed in restricted regions in which patchy EGFP expression was observed. This patchwork epidermal hyperplasia was more severe in the mouse line with the mutant (A339T) transgene than in the mouse line with the wild-type transgene (Figure 6C). Inflammatory cells were infiltrated in the dermis of both TG lines (Figure 6C). The complete loss of hair follicles was observed in 1-yr-old mice with the mutant transgene (Figure S4). These skin abnormalities resemble those of the Gsdma3 mutant mice (Runkel et al. 2004; Lunny et al. 2005; Tanaka et al. 2007a; Sun et al. 2009; Li et al. 2010; Lei et al. 2011; Zhou et al. 2012). Epithelial hyperplasia also was observed in the stomach of both TG lines (Figure 6D) and may be due to the human K5 promoter in the transgene vector construct, which is active in the basal cell layer of stratified squamous epithelium from skin to forestomach.
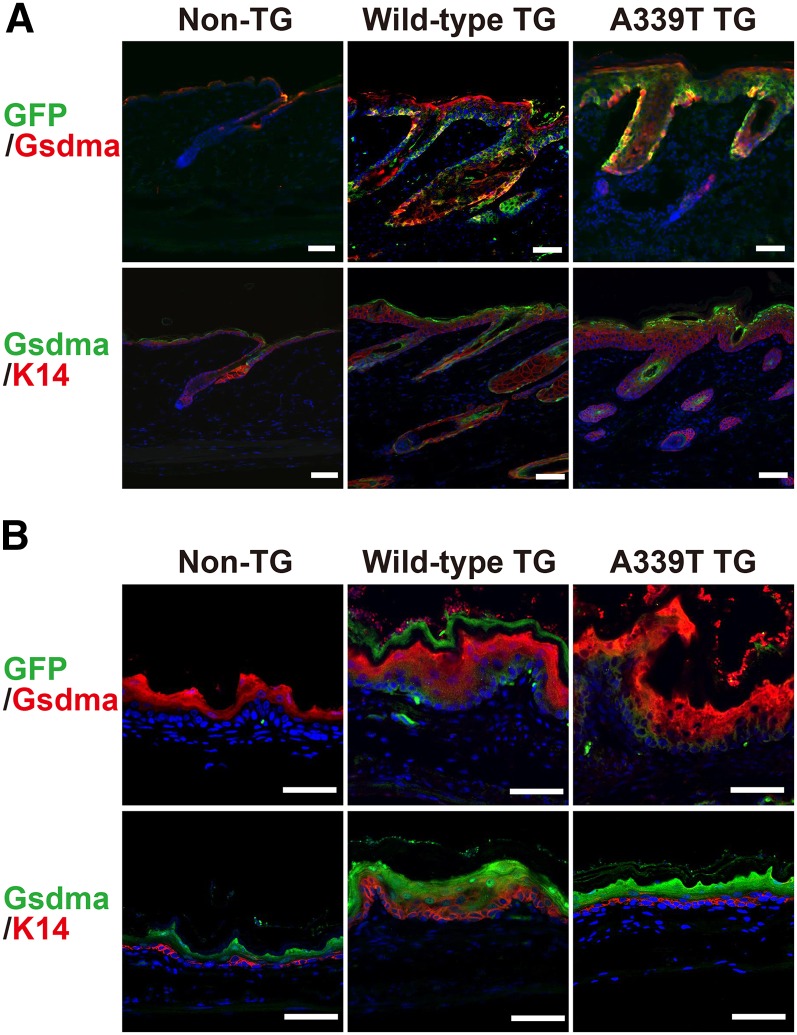
Next, we examined expression of the transgene in the affected cells by immunohistochemistry with EGFP and anti-Gsdma antibody. Overlapping patchy signals of EGFP and Gsdma protein were observed in the basal cell layer of the skin and cardia of both TG lines (Figure 7, A and B). Immunohistochemistry with K14 antibody revealed that the epidermal hyperplasia is specific to the basal cell layer in the skin and cardia of both TG lines (Figure 7, A and B). These results clearly demonstrated that the mutant form (A339T) of the Gsdma transgene and overexpression of the wild-type Gsdma transgene directly caused the epithelial hyperplasia.
Figure 7.
Mice with K5-Gsdma transgene exhibit epidermal hyperplasia. Immunohistological staining of skin (A) and cardia (B) from control mouse (Non-TG), and mice with wild-type and A339T Gsdma transgene, at 3 months of age. The sections were stained with anti-GFP and anti-Gsdma antibodies or anti-Gsdma and anti-K14 antibodies. Nuclei were stained with ToPro3. Scale bars are 50 μm.
Discussion
The Gsdma KO mice are viable and show no visible phenotype, indicating that Gsdma is not an essential gene and is not involved in differentiation of epidermal cells and maintenance of the hair cycle. By contrast, the phenotype of the TG mice indicates that Gsdma has the same function to regulate epithelial cell proliferation and/or epithelial maintenance as the mouse paralogous gene, Gsdma3. It is notable that the alopecia started to appear only in the progeny obtained by backcrossing of the original TG mice with the mixed genetic background onto the B6 strain. This may be the reason why the Gsdma mutations have not been detected in large-scale and chemically induced mutagenesis projects, since these mutagenesis projects have mostly used mice with heterozygous F1 backgrounds.
The present study, in which we used Gsdma KO mice, allowed us to observe the precise expression pattern of Gsdma3 without influence of Gsdma. Our findings clearly verify that Gsdma3 is predominantly expressed in the suprabasal cells of the interfollicular epidermis, but not in the follicular epithelium. Consistent with this result, Gsdma3 mutant mice initially exhibit hyperplasia in the interfollicular epidermis, but not in the follicular epithelium (Tanaka et al. 2007a).
The loss-of-function type mouse mutants of Gsdm family genes, such as the Gsdmd KO mice in our previous study and the Gsdma KO mice in this study, show neither morphologic anomaly such as epithelial hyperplasia nor tumor development (Fujii et al. 2008). The failure to identify a visible phenotype in the KO mice may be due to functional redundancy with the other genes. Alternatively, one explanation for the absence of phenotype in the KO mice might be that the members of the Gsdm family control susceptibility to environmental factors, such as allergens and infectious agents, or physical stress. It is known that newly duplicated genes in the mouse genome significantly enrich the functional category involved in immune defense, olfaction and drug metabolism, which are of medical importance (Cheung et al. 2003). In this regard, implication of susceptibility to environmental factors has been reported for function of the Gsdm family genes. Gsdma expression is down-regulated in Grainyhead-like 3 (Grhl3) KO mice, and the binding motif of the Grhl3 protein is conserved upstream of the transcription initiation site of the Gsdma/GSDMA gene in mouse and human, suggesting that Gsdma/GSDMA is the target gene of Grhl3 (Yu et al. 2006). Grhl3 is a master regulator of epidermal terminal differentiation, and it controls wound response in mice and drosophila (Mace et al. 2005; Ting et al. 2005; Yu et al. 2006). Furthermore, it was recently reported that polymorphisms with GSDMA and GSDMB loci are associated with asthma, atopy and intermediate phenotypes such as elevated immunoglobulin E (Lluis et al. 2011; Yu et al. 2011). Thus, we infer that members of the Gsdm family genes play some role in stress responses to environmental factors. It would be of interest to test whether the KO mice carrying the Gsdma and Gsdmd genes show some new phenotypes under stress conditions in future studies.
The molecular basis of epidermal hyperplasia observed in the skin of mice with the Gsdma transgenes is unclear, but it may be related to the inflammation observed in the skin of those mice. Transforming-growth factor β (Tgfβ) is a key cytokine involved in apoptosis and inflammation (Letterio and Roberts 1998; Siegel and Massague 2003). Overexpression of Tgfβ1 in the skin also causes inflammatory skin abnormalities (Wang et al. 1999; Liu et al. 2001; Li et al. 2004; Lu et al. 2004; Fitch et al. 2009). Moreover, TGFβ up-regulates GSDMA expression through lmo1, and the resultant GSDMA overexpression induces apoptosis in human cancer cell lines (Saeki et al. 2007). This finding is consistent with the fact that increased apoptotic cells are observed in the hair follicles of not only Tgfβ1 TG mice but also Gsdma3 mutant mice (Liu et al. 2001; Lei et al. 2011). These findings suggest that Gsdma/GSDMA has a role in regulating inflammation downstream of TGFβ/Tgfβ in vivo.
The dominant Gsdma3 alleles in mice are categorized as nonsense- and missense-mutations (Runkel et al. 2004; Lunny et al. 2005; Tanaka et al. 2007a; Sun et al. 2009; Li et al. 2010; Lei et al. 2011; Zhou et al. 2012). A mutant allele, Gsdma3Dfl, exhibits alopecia resembling Gsdma3Rim3 (Lunny et al. 2005). This mutation has a B2 element insertion in exon 7, and generates mRNA with a stop codon, which results in a C-terminally truncated Gsdma protein (Lunny et al. 2005). Deafness autosomal-dominant 5 (DFNA5) belongs to the DFNA5-Gsdm family. The N-terminus region of DFNA5 has the same amino acid sequence as the Gsdm family proteins and is referred to as the DFNA5-Gasdermin domain (Delmaghani et al. 2006; Tamura et al. 2007). Germline mutations in DFNA5 have been discovered in human families and cause autosomal-dominant hearing loss (Van Laer et al. 1998; Yu et al. 2003; Cheng et al. 2007). In many cases, the DFNA5 mutations lead to exon 8 skipping, resulting in frameshift and premature truncation of the protein (Van Laer et al. 1998; Yu et al. 2003; Cheng et al. 2007). It was reported that Dfna5 KO mice, which have a deletion of exon 8 mimicking the human mutation, produce no Dfna5 protein through nonsense-mediated RNA decay (Van Laer et al. 2005). Dfna5 KO mice display no hearing loss and do not mimic human hearing loss caused by mutations in DFNA5 (Van Laer et al. 2005). However, the expression of mutant truncated DFNA5 protein leads to cell-cycle arrest in fission yeast (Gregan et al. 2003), and the expression of an aberrant N-terminus truncated form of DFNA5 causes apoptosis in cultured human cell lines (Op de Beeck et al. 2011). Therefore, it is inferred that DFNA5-associated hearing loss is caused by a gain-of-function mutation, but not by haplo-insufficiency (Op de Beeck et al. 2011). Taken together, although the physiologic function of full-length proteins of the Gsdma/GSDMA and Dfna5 genes remains elusive, the expression of the truncated N-terminus region of the DFNA5-Gsdm domain affects cell proliferation, leading to dominant phenotypes in skin and inner ear.
Supplementary Material
Acknowledgments
We thank K. Hiratsuka and all animal facility members for maintaining the mouse strains, and H. Nakazawa, A. Okagaki, T. Fujii, and H. Komiyama for excellent technical assistance and valuable discussions. We also thank Y. Saga for providing pLoxFNFDT and pIRES2-EGFP vectors, and the BRC Gene Engineering Division for providing the pKM2L-phK5 vector. We thank Y. Shimomura of Niigata University for providing the anti-K71 antibodies. This work was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports and Science of Japan.
Footnotes
Communicating editor: J. M. Comeron
Literature Cited
- Aoki N., Sawada S., Rogers M. A., Schweizer J., Shimomura Y., et al. , 2001. A novel type II cytokeratin, mK6irs, is expressed in the Huxley and Henle layers of the mouse inner root sheath. J. Invest. Dermatol. 116: 359–365 [DOI] [PubMed] [Google Scholar]
- Cheng J., Han D. Y., Dai P., Sun H. J., Tao R., et al. , 2007. A novel DFNA5 mutation, IVS8+4 A>G, in the splice donor site of intron 8 causes late-onset non-syndromic hearing loss in a Chinese family. Clin. Genet. 72: 471–477 [DOI] [PubMed] [Google Scholar]
- Cheung J., Wilson M. D., Zhang J., Khaja R., MacDonald J. R., et al. , 2003. Recent segmental and gene duplications in the mouse genome. Genome Biol. 4: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmaghani S., del Castillo F. J., Michel V., Leibovici M., Aghaie A., et al. , 2006. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat. Genet. 38: 770–778 [DOI] [PubMed] [Google Scholar]
- Fitch E. L., Rizzo H. L., Kurtz S. E., Wegmann K. W., Gao W., et al. , 2009. Inflammatory skin disease in K5.hTGF-beta1 transgenic mice is not dependent on the IL-23/Th17 inflammatory pathway. J. Invest. Dermatol. 129: 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Tamura M., Tanaka S., Kato Y., Yamamoto H., et al. , 2008. Gasdermin D (Gsdmd) is dispensable for mouse intestinal epithelium development. Genesis 46: 418–423 [DOI] [PubMed] [Google Scholar]
- Gregan J., Van Laer L., Lieto L. D., Van Camp G., Kearsey S. E., 2003. A yeast model for the study of human DFNA5, a gene mutated in nonsyndromic hearing impairment. Biochim. Biophys. Acta 1638: 179–186 [DOI] [PubMed] [Google Scholar]
- Komiyama H., Aoki A., Tanaka S., Maekawa H., Kato Y., et al. , 2010. Alu-derived cis-element regulates tumorigenesis-dependent gastric expression of GASDERMIN B (GSDMB). Genes Genet. Syst. 85: 75–83 [DOI] [PubMed] [Google Scholar]
- Lei M., Gao X., Yang L., Yang T., Lian X., 2011. Gsdma3 gene is needed for the induction of apoptosis-driven catagen during mouse hair follicle cycle. Histochem. Cell Biol. 136: 335–343 [DOI] [PubMed] [Google Scholar]
- Letterio J. J., Roberts A. B., 1998. Regulation of immune responses by TGF-beta. Annu. Rev. Immunol. 16: 137–161 [DOI] [PubMed] [Google Scholar]
- Li A. G., Wang D., Feng X. H., Wang X. J., 2004. Latent TGFbeta1 overexpression in keratinocytes results in a severe psoriasis-like skin disorder. EMBO J. 23: 1770–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhou Y., Yang T., Wang N., Lian X., et al. , 2010. Gsdma3 is required for hair follicle differentiation in mice. Biochem. Biophys. Res. Commun. 403: 18–23 [DOI] [PubMed] [Google Scholar]
- Liu X., Alexander V., Vijayachandra K., Bhogte E., Diamond I., et al. , 2001. Conditional epidermal expression of TGFbeta 1 blocks neonatal lethality but causes a reversible hyperplasia and alopecia. Proc. Natl. Acad. Sci. USA 98: 9139–9144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluis A., Schedel M., Liu J., Illi S., Depner M., et al. , 2011. Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J. Allergy Clin. Immunol. 127: 1587–1594 e1586. [DOI] [PubMed] [Google Scholar]
- Lu S. L., Reh D., Li A. G., Woods J., Corless C. L., et al. , 2004. Overexpression of transforming growth factor beta1 in head and neck epithelia results in inflammation, angiogenesis, and epithelial hyperproliferation. Cancer Res. 64: 4405–4410 [DOI] [PubMed] [Google Scholar]
- Lunny D. P., Weed E., Nolan P. M., Marquardt A., Augustin M., et al. , 2005. Mutations in gasdermin 3 cause aberrant differentiation of the hair follicle and sebaceous gland. J. Invest. Dermatol. 124: 615–621 [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Mace K. A., Pearson J. C., McGinnis W., 2005. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308: 381–385 [DOI] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication, George Allen and Unwin, London [Google Scholar]
- Op de Beeck K., Van Camp G., Thys S., Cools N., Callebaut I., et al. , 2011. The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein. Eur. J. Hum. Genet. 19: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel F., Marquardt A., Stoeger C., Kochmann E., Simon D., et al. , 2004. The dominant alopecia phenotypes Bareskin, Rex-denuded, and Reduced Coat 2 are caused by mutations in gasdermin 3. Genomics 84: 824–835 [DOI] [PubMed] [Google Scholar]
- Saeki N., Kuwahara Y., Sasaki H., Satoh H., Shiroishi T., 2000. Gasdermin (Gsdm) localizing to mouse Chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mamm. Genome 11: 718–724 [DOI] [PubMed] [Google Scholar]
- Saeki N., Kim D. H., Usui T., Aoyagi K., Tatsuta T., et al. , 2007. GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF-beta-dependent apoptotic signalling. Oncogene 26: 6488–6498 [DOI] [PubMed] [Google Scholar]
- Saeki N., Usui T., Aoyagi K., Kim D. H., Sato M., et al. , 2009. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer 48: 261–271 [DOI] [PubMed] [Google Scholar]
- Siegel P. M., Massague J., 2003. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer 3: 807–821 [DOI] [PubMed] [Google Scholar]
- Sun, L., X. Li, X. Du, K. Benson, N. Smart et al., 2009 Record for “Fuzzy,” updated March 30, 2009, MGI Direct Data Submission.
- Tamura M., Tanaka S., Fujii T., Aoki A., Komiyama H., et al. , 2007. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics 89: 618–629 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Tamura M., Aoki A., Fujii T., Komiyama H., et al. , 2007a A new Gsdma3 mutation affecting anagen phase of first hair cycle. Biochem. Biophys. Res. Commun. 359: 902–907 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Miura I., Yoshiki A., Kato Y., Yokoyama H., et al. , 2007b Mutations in the helix termination motif of mouse type I IRS keratin genes impair the assembly of keratin intermediate filament. Genomics 90: 703–711 [DOI] [PubMed] [Google Scholar]
- Ting S. B., Caddy J., Hislop N., Wilanowski T., Auden A., et al. , 2005. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308: 411–413 [DOI] [PubMed] [Google Scholar]
- Van Laer L., Huizing E. H., Verstreken M., van Zuijlen D., Wauters J. G., et al. , 1998. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat. Genet. 20: 194–197 [DOI] [PubMed] [Google Scholar]
- Van Laer L., Pfister M., Thys S., Vrijens K., Mueller M., et al. , 2005. Mice lacking Dfna5 show a diverging number of cochlear fourth row outer hair cells. Neurobiol. Dis. 19: 386–399 [DOI] [PubMed] [Google Scholar]
- Wang X. J., Liefer K. M., Tsai S., O’Malley B. W., Roop D. R., 1999. Development of gene-switch transgenic mice that inducibly express transforming growth factor beta1 in the epidermis. Proc. Natl. Acad. Sci. USA 96: 8483–8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T., Tokunaga T., Furuta Y., Nada S., Yoshida M., et al. , 1993. A novel ES cell line, TT2, with high germline-differentiating potency. Anal. Biochem. 214: 70–76 [DOI] [PubMed] [Google Scholar]
- Yu C., Meng X., Zhang S., Zhao G., Hu L., et al. , 2003. A 3-nucleotide deletion in the polypyrimidine tract of intron 7 of the DFNA5 gene causes nonsyndromic hearing impairment in a Chinese family. Genomics 82: 575–579 [DOI] [PubMed] [Google Scholar]
- Yu J., Kang M. J., Kim B. J., Kwon J. W., Song Y. H., et al. , 2011. Polymorphisms in GSDMA and GSDMB are associated with asthma susceptibility, atopy and BHR. Pediatr. Pulmonol. 46: 701–708 [DOI] [PubMed] [Google Scholar]
- Yu Z., Lin K. K., Bhandari A., Spencer J. A., Xu X., et al. , 2006. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol. 299: 122–136 [DOI] [PubMed] [Google Scholar]
- Zhang J., 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18: 292–298 [Google Scholar]
- Zhou Y., Jiang X., Gu P., Chen W., Zeng X., et al. , 2012. Gsdma3 mutation causes bulge stem cell depletion and alopecia mediated by skin inflammation. Am. J. Pathol. 180: 763–774 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.