Abstract
Halloysite is aluminosilicate clay with a hollow tubular structure with nanoscale internal and external diameters. Assessment of halloysite biocompatibility has gained importance in view of its potential application in oral drug delivery. To investigate the effect of halloysite nanotubes on an in vitro model of the large intestine, Caco-2/HT29-MTX cells in monolayer co-culture were exposed to nanotubes for toxicity tests and proteomic analysis. Results indicate that halloysite exhibits a high degree of biocompatibility characterized by an absence of cytotoxicity, in spite of elevated pro-inflammatory cytokine release. Exposure-specific changes in expression were observed among 4081 proteins analyzed. Bioinformatic analysis of differentially expressed protein profiles suggest that halloysite stimulates processes related to cell growth and proliferation, subtle responses to cell infection, irritation and injury, enhanced antioxidant capability, and an overall adaptive response to exposure. These potentially relevant functional effects warrant further investigation in in vivo models and suggest that chronic or bolus occupational exposure to halloysite nanotubes may have unintended outcomes.
Keywords: halloysite, intestinal epithelia, enterocytes, proteomics, label-free quantitative mass spectrometry
Introduction
Halloysite is natural aluminosilicate clay with a hollow tubular structure that enables loading and sustained release of biomacromolecules and drugs. The nanotubes are about 0.5–2 μm long with a 10–20 nm inner luminal diameter and can be loaded with saturated solutions of proteins, DNA, antibiotics and other drugs drawn into the tubes by capillary forces (Joussein et al., 2005; Price et al., 2001; Shchukin et al., 2005). This enables long-term bioavailability of drugs (ciprofloxacin, gentamicin and tetracycline) and microbial antiseptics (iodine, brilliant green and amoxicillin). The surface of halloysite nanotubes is silicon oxide that is considered a biocompatible material. However, more detailed studies of halloysite clay toxicity are necessary to demonstrate its suitability for drug delivery formulations.
Halloysite biomedical applications include its use for stem cell attachment, cancer cell isolation, bone implants, teeth fillers, cosmetics and controlled drug delivery (Hughes and King, 2010; Kelly et al., 2004; Kommireddy et al., 2006; Veerabadran et al., 2007; Vergaro et al., 2012; Wei et al., 2012). The first published paper on halloysite use as a nanocontainer demonstrated 10–20 h sustained release of tetracycline, khellin and NAD (Price et al., 2001), and was followed by multiple publications on halloysite controlled drug release. Halloysite functionalized with selectin protein was used as target-specific coatings to capture leukemic and epithelial cancer cells (Hughes and King, 2010). Intracellular uptake by cells of different origins (cervical adenocarcinoma or breast cancer cells) and cytoviability tests demonstrated relative halloysite cytocompatibility and potential as a bio-friendly cargo nanocontainer for biomaterials (Vergaro et al., 2010). In that previous study, cell viability decreased to 80% of the control after exposure to 500 μg ml–1 halloysite (a very high concentration of c. 4 × 108 nanotubes per ml). Those clay nanotubes were further modified through: (i) increased lumen diameter resulting in an increased drug loading capacity up to 30–40%; and (ii) encapsulation of the clay tubes in organic polymeric shells with tube-end stoppers resulting in increased control over the release kinetics of loaded drugs in the range of 20–100 h (Abdullayev et al., 2012). Halloysite clay has been considered to be a ’green‘ environmentally safe material, and the nanotubes are available in tons from natural deposits.
Because halloysite nanotubes are being considered for oral drug delivery (Forsgren et al., 2010; Kawakami et al., 2012; Suresh et al., 2010), it is critical to assess their potential gastrointestinal effects. Besides the study mentioned above, few in vitro studies have examined the effects of halloysite exposure, and those that did demonstrated variable effects, including significant declines in viability in Hela (cervical) and MCF-7 (breast) cancer cell lines (Vergaro et al., 2012) after 24 h, but no effect on 3T3 (fibroblast) cells over 48 h (Suh et al., 2011). To provide a culture system more relevant to halloysite ingestion, the present study used a well-characterized in vitro Caco-2/HT29-MTX co-culture model simulating the large intestine. Caco-2 and HT29-MTX adenocarcinoma cell lines are derived from intestinal absorptive and mucus-secreting goblet cell (Lesuffleur et al., 1991) types, respectively. Their combination provides a physiologically relevant co-culture system characterized by tight junctions and considerable mucus secretion (Hilgendorf et al., 2000; Mahler et al., 2009a, 2009b; Walter et al., 1996) that covers the entire monolayer. We have recently used this model to study the effects of carbon nanotube exposure (Lai et al., 2013), and that paper illustrates the distinct polarization and tight-junction formation achieved in the current investigation.
Using this co-culture model, cytotoxic and proinflammatory end-point assays, and an innovative, label-free quantitative mass spectrometric (LFQMS) platform recently developed in our laboratory (Lai et al., 2011), we present significant effects of halloysite exposure on cellular protein expression, in the absence of overt toxicity, and link these changes to specific pathways and molecular functions that have physiological relevance. This represents the first investigation of halloysite effects on intestinal epithelial cells in vitro, providing novel evidence regarding the inflammatory and functional effects of halloysite clay nanotubes on the intestinal barrier.
Materials and Methods
Reagents and Nanoparticles
DL-Dithiothreitol (DTT), urea, triethylphosphine, iodoethanol and ammonium bicarbonate were purchased from Sigma-Aldrich (St. Louis, MO, USA). LC-MS grade 0.1% formic acid in acetonitrile and 0.1% formic acid in water were purchased from Burdick & Jackson (Muskegon, MI, USA). Modified sequencing grade porcine trypsin was obtained from Princeton Separations (Freehold, NJ, USA). Halloysite clay powder was obtained from Applied Minerals Inc., NY, USA and used without further purification. It contained 95% halloysite clay nanotubes and the remaining few percent were kaolin and silica (see Table 1).
Table 1.
Physicochemical properties of halloysite clay nanotubes
| Outer diameter (nm) | 50–100 |
| Inner diameter (nm) | 10–20 |
| Length (μm) | 0.2–2.0 |
| Zeta potential (mV) | –42.6 |
| C content (atomic %) | 6.74 |
| N content (atomic %) | 1.25 |
| O content (atomic %) | 50.17 |
| Al content (atomic %) | 20.33 |
| Si content (atomic %) | 19.25 |
| Ca content (atomic %) | 0.10 |
| Fe content (atomic %) | 0.69 |
| Cu content (atomic %) | 1.47 |
Characterization of Halloysite Nanotubes
Halloysite samples were mixed with ethanol and sonicated in a water bath (Branson, Danbury, CT, USA) for 10–15 min until well dispersed. For each sample, a droplet of the suspension was placed on a copper grid and dried at room temperature. Transmission electron microscopy imaging and energy dispersive spectroscopy (EDS) elemental analysis were performed using a Hitachi HD 2000 STEM. The zeta potential of the suspended halloysite in water was measured using electrophoretic light scattering (ZetaSizer Nano; Malvern Instruments, Malvern, Worcestershire, United Kingdom). A 1-mg sample of the halloysite clay nanotubes was analyzed for the presence of endotoxin by nanoComposix (San Diego, CA, USA Malvern, Worcestershire WR14 1XZ United Kingdom) using the kinetic turbidity Limulus Amebocyte Lysate (LAL) assay.
Cell Culture and Exposures
Caco-2, human colorectal adenocarcinoma cells (ATCC HTB-37, Lot 57863838) were maintained in Eagle's minimal essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS) (ATCC catalog numbers 30-2003 & 30-2020) and incubated at 37 °C – 5% CO2. HT29-MTX, human colon adenocarcinoma cells treated with methotrexate (Lesuffleur et al., 1993), were obtained from Dr Thécla Lesuffleur (INSERM, Paris, France) and were maintained in Dulbecco's Modified Eagle's Medium (DMEM) with glutamax and 10% heat inactivated FBS (Invitrogen, Carlsbad CA, USA) at 37 °C – 5% CO2. Both cell lines were sub-cultured at 80–90% confluence. Caco-2 (passage 45) and HT29-MTX cells (passage 29) were cultured to obtain several Corning T75 flasks. At 80–90% confluence, the cells were trypsinized and the flasks were pooled. Cells were counted using a hemocytometer and then mixed together at a 75: 25 Caco-2 to HT29-MTX ratio and placed in the HT29-MTX medium with 1% penicillin-streptomycin. Combined cells were mechanically mixed and 1.5 mL was added to six-well Transwell™ inserts to obtain 0.4 × 106 cells per well (8.9 × 104 cells per cm2) (Hilgendorf et al., 2000).
Cells were incubated at 37 °C – 5% CO2 for 14 days prior to exposure, replacing the medium every other day, to achieve full, post-confluent differentiation, polarization and tight-junction formation. Halloysite clay nanotubes were weighed and added to autoclavable glass tubes with screw caps; water was added to make a 5 mg ml–1 suspension. Prior to cell exposure, the halloysite suspension was sonicated in a water bath sonicator for 5 min and vortexed for 20–30 s. Immediately, a 160-μl aliquot was diluted in 3.84 ml of HT29-MTX medium (DMEM) to a final concentration of 200 μg ml–1. This stock was then diluted to exposure concentrations of 1, 10 and 100 μg ml–1. These halloysite doses were chosen to correspond to those used in a previous study (Vergaro et al., 2010).
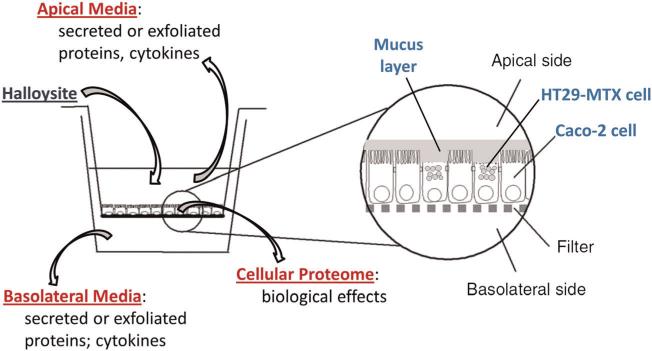
The final halloysite mixtures were gently sonicated, vortexed quickly and 1.5 ml placed in the top (apical) compartment of the six Transwell™ plate containing the co-cultured cells, as depicted in Fig. 1. The bottom (basolateral) compartment contained 2.5 ml of fresh medium.
Figure 1.
Caco-2/HT29-MTX intestinal cell co-culture system for modeling gastrointestinal halloysite exposure.
Control groups received only fresh medium in both compartments. The treated cells were incubated for 6 h at 37 °C – 5% CO2. This exposure duration was used to simulate the transient nature of gastrointestinal exposure to ingested nanotubes. The authors acknowledge that this short time frame of exposure may not have captured the potential cytotoxicity that might have been better assessed at a later time point. To assess the cell number at exposure, replicate co-cultured cells were trypsinized and counted using a hemocytometer (n = 4) after 14 days. Cell counts indicated a total of ~750 000 cells per Transwell™ insert.
Cell Viability, Membrane Integrity and Permeability Assays
In assays that produce insoluble formazan dyes (such as the MTT assay), NTs can attach to the insoluble MTT formazan product disrupting the distinguishing, colorimetric reaction (Worle-Knirsch et al., 2006). Therefore, assays that produce soluble dyes such as XTT are preferred. Accordingly, cell viability was determined using the Cell Proliferation Kit (Roche, Indianapolis, IN, USA) according to manufacturer's instructions. Membrane leakage of LDH into the apical media from potentially damaged or dead cells was assessed using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega, Fitchburg, WI, USA) according to the manufacturer's instructions. Apically applied Triton™ X-100 treatment (0.05%) was included as a positive control and barrier function was assessed by measuring unidirectional paracellular flux of Lucifer yellow from the apical to basolateral compartments. This assay was conducted via the Lucifer Yellow Permeability Assay (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's instructions, using Transwell™ cultures as described above.
Chemokine/Cytokine Analysis
To determine the level of irritation or proinflammatory response associated with halloysite exposure after 6 h, cell culture medium was collected from apical and basolateral compartments of the Transwell™ plates and evaluated for the following chemokines: monocyte chemotactic protein-1 (MCP-1), a proinflammatory cytokine produced and released in response to various stimuli, such as bacterial invasion and interleukin-8 (IL-8) another proinflammatory chemokine. In addition, granulocyte macrophage-colony-stimulating factor (GM-CSF), a cytokine that amplifies an inflammatory response and promotes tissue repair and tumor necrosis factor-alpha (TNF-α), a prominent component of intestinal proinflammatory activities were evaluated. Assays were carried out by enzyme-linked immunosorbent assay (ELISA) Quantikine Colorimetric kits (R&D Systems, Inc., Minneapolis, MN, USA) according to the manufacturer's instructions. All four of these chemokines/cytokines are known to be expressed constitutively by intestinal epithelial cells (Jung et al., 1995) as substantiated by the Gene Expression Omnibus (GEO) database (Edgar et al., 2002), which stores individual gene expression profiles from curated data sets in its repository. Group means for cell viability, membrane integrity, permeability and chemokine/cytokine analyses were compared by one-way ANOVA using post hoc pairwise multiple comparisons [Holm–Sidak method (Holm, 1979)].
Proteomics and Bioinformatic Analysis
After incubation of the exposed cells, the media from both compartments were individually removed, placed into separate labeled microfuge tubes, snap frozen in liquid nitrogen and placed on dry ice until transfer to a -80°C freezer for cytokine analysis. The Transwell™ membranes containing adherent cells were quickly rinsed 3 times in ice-cold 250 mM sucrose, snap frozen in liquid nitrogen, placed on dry ice and then stored at –80°C. For Caco-2/HT29-MTX lysate preparation for LFQMS, 500 μl of lysis buffer (8 M urea, 10 mM DTT, freshly prepared) was added to each sample. All cells were incubated at 35 °C for 1 h with agitation and transferred to a test tube. Cell lysates were centrifuged at 15 000 g for 20 min at 4 °C to remove insoluble materials and the supernatant transferred to a new tube. Fully solubilized cell protein samples were then stored at –80°C until LFQMS analysis.
Protein concentration was determined using the Bradford Protein Assay using Bio-Rad (Hercules, CA, USA) protein assay dye reagent concentrate. An aliquot containing 100 μg of each cell lysate sample was adjusted to 200 μl with 4 M urea and then reduced and alkylated by triethylphosphine and iodoethanol, as described previously (Lai et al., 2008). A 150-μl aliquot of a 20 μg ml–1 trypsin solution was added to the sample and incubated at 35 °C for 3 h, after which another 150 μl of trypsin was added, and the solution incubated at 35 °C for 3 h (n = 5 per observation). Exactly 20 μg of each tryptic digest sample (n = 5) were injected randomly as two technical replicates onto a C18 reversed phase column (TSK gel ODS-100V, 3 μm, 1.0 × 150 mm) at a flow rate of 50 μl min–1 as part of the Surveyor autosampler and MS HPLC system (Thermo-Electron, Waltham, MA, USA) coupled to a Thermo-Finnigan linear ion-trap (LTQ) mass spectrometer. The mobile phases A and B were 0.1% formic acid in water and 50% ACN with 0.1% formic acid in water, respectively. The gradient elution profile was as follows: 10% B (90% A) for 7 min and 10–67.1% B (90–32.9% A) for 163 min, 67.1–100% B (32.9–0% A) for 10 min. The spectral data were collected in the ’data dependent MS/MS‘ mode with the ESI interface using a normalized collision energy of 35%. Dynamic exclusion settings were repeat count 1, repeat duration 30 s, exclusion duration 120 s and exclusion mass width 0.6 m/z (low) and 1.6 m/z (high). A blank was injected between each sample to clean and balance the column and to eliminate carryover. The acquired data were searched against the International Protein Index (IPI) database (ipi.HUMAN.v3.83) using SEQUEST (v. 28 rev. 12) algorithms in Bioworks (v. 3.3). General parameters were set to: peptide tolerance 2.0 amu, fragment ion tolerance 1.0 amu, enzyme limits set as ’fully enzymatic – cleaves at both ends’ and missed cleavage sites set at 2. Peptide and protein identifications were validated by PeptideProphet (Keller et al., 2002) and ProteinProphet (Nesvizhskii et al., 2003) in the Trans-Proteomic Pipeline (TPP, v. 3.3.0; http://tools.proteomecenter.org/software.php). Only proteins with a probability ≥ 0.9000 and peptides with a probability ≥ 0.8000 were reported. Protein abundance was determined using IdentiQuantXL™ (Lai et al., 2011). Briefly, after chromatogram alignment and peptide retention time determination, a weighted mean m/z of each peptide was calculated and a tab delimited file was created to extract peptide intensity using MASIC (Monroe et al., 2008). Peptides were then filtered according to intensity coefficients of variation (CV) across all samples and intensity correlation, for those identifying a particular protein. Protein abundance was then calculated from all qualified corresponding peptides matched to that protein.
Comparison of the abundance of individual protein dose-group means generated by LFQMS was performed within the IdentiQuantXL™ platform using one-way ANOVA and Pairwise Multiple Comparisons (Holm–Sidak method). The critical F-ratio significance for ANOVA was set at P < 0.01 and a pairwise comparison at P < 0.05. The False Discovery Rate (FDR) (Storey, 2002) was estimated using Q-value software. To interpret the biological relevance of the differential protein expression data and to compare biological effects across different halloysite exposure levels, protein lists and their corresponding expression values (fold change) were uploaded onto the Ingenuity Pathway Analysis (IPA) software server (http://www.ingenuity.com) and analyzed using the Core Analysis module to rank the proteins into top biological functions and canonical pathways, and to discover upstream regulators that may explain the observed differential protein expression.
Results
Characterization of Halloysite Nanotubes
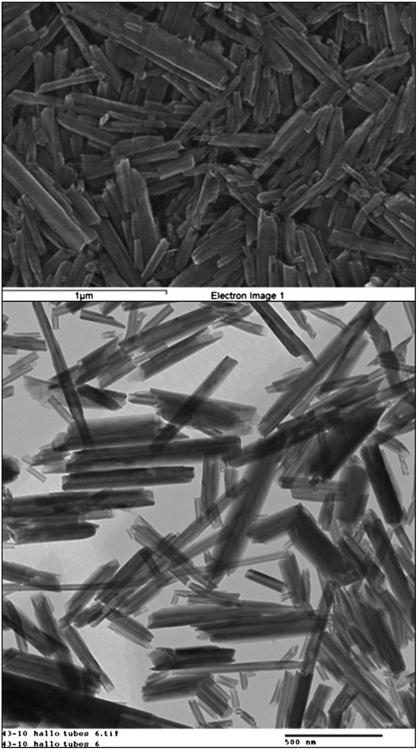
The nanotubes used in this study ranged from 0.2–2.0 μm in length and 50–100 nm in outer diameter (Table 1 and Fig. 2A, B). The lumen diameter was 10–20 nm such that its volume comprises 15–20% of the total nanotube volume. Periodicity in the individual layer packing determined with X-ray analysis is 0.72 nm, which corresponds to the dehydrated halloysite. The Brunauer–Emmett–Teller (BET) surface area of the halloysite is 50 ± 4 m2/g1.
Figure 2.
Transmission electron microscopic images of two different concentrations of the halloysite nanotubes used in this investigation, confirming their moderately high aspect ratio, dimension range and hollow interior.
Results from the elemental analysis of energy dispersive spectra also confirmed an abundance of oxygen, aluminum and silicon in the halloysite nanotubes (Table 1). The hydrodynamic sizes of the halloysite tubes dispersed in water at pH 6.5 is in the range 250–300 nm (effective diameter corresponding to the 3D radius of gyration of elongated particle gives kind of average size). Halloysite zeta potential in water (Table 1) was –42.6 mV, thus confirming that electrochemical behavior of halloysite in water has a similarity with silica nanoparticles. The outer surface of these clay nanotubes consists of SiO2 and the innermost lumen surface is Al2O3. At pH range 4-9, the nanotube's aluminal interior is positively charged whereas the tube's outside surface, relevant to silica, is negatively charged. Zeta potential analysis of the culture media was non-determinant owing to screening of the nanotubes by constituent ions and biomolecules.
Endotoxin analysis revealed a level of 0.01885 EU ml–1, permitting us to eliminate this as a factor that may have influenced the results.
Cell Viability, Membrane Integrity and Permeability Assays
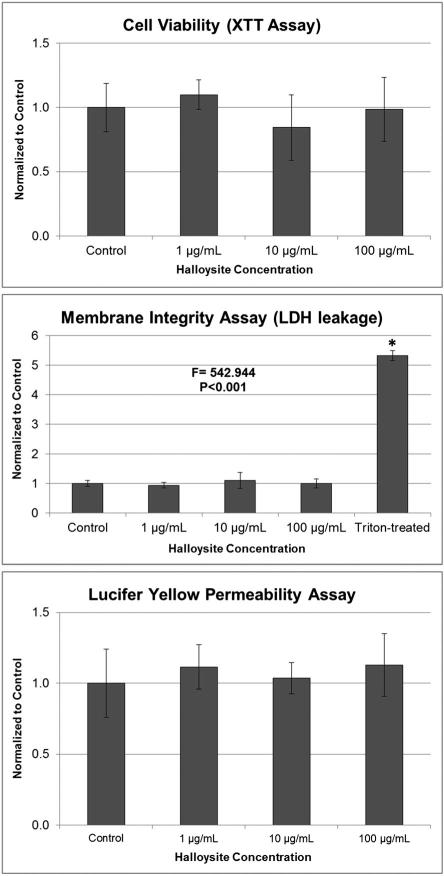
Halloysite exposure for 6 h had no effect at any concentration on cell viability, membrane integrity as measured by LDH leakage or barrier permeability (Fig. 3). Membrane integrity was disrupted only in the Triton™ X-100-treated positive control, as expected.
Figure 3.
Effect of halloysite exposure on Caco-2/HT29-MTX cells: cell viability after 6 h determined by the XTT assay; membrane integrity (LDH leakage) after 6 h determined by the CytoTox 96 assay. Triton X-100 was included as a positive control. ANOVA P < 0.001, *post-hoc Holm–Sidak P < 0.001 vs. halloysite expoures and control; and epithelial barrier integrity (apical to basolateral compartment paracellular permeability) after 6 h determined by the Lucifer Yellow assay. All values in the three graphs are means ± standard deviation (SD) for n = 5, with optical densities normalized to the control.
Chemokine/Cytokine Analysis
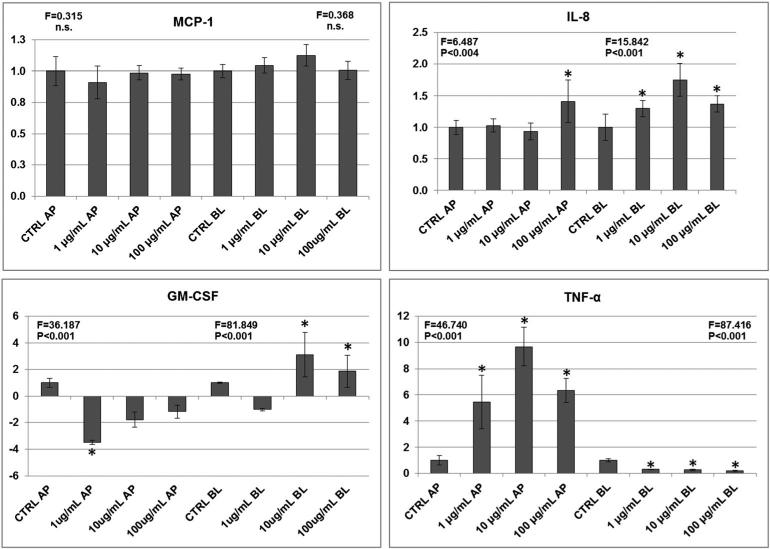
As illustrated in Fig. 4, the secretion of several chemokines and cytokines into both apical and basolateral media was altered by halloysite exposure. While the chemokine MCP-1 was unaffected, IL-8 was significantly elevated by the 100 μg ml–1 exposure in the apical (luminal) and by all three halloysite concentrations in basolateral (interstitial) media (1.5-fold at 10 μg ml–1). In contrast, with GM-CSF and TNF-α, polarized secretion was clearly evident with halloysite exposure. GM-CSF secretion was significantly reduced by 3.5-fold in the apical media by the 1 μg ml–1 exposure and significantly elevated in the basolateral media at 10 and 100 μg ml–1. TNF-α secretion in the apical media was elevated by all exposures, maximally at nearly 10-fold by the 10 μg ml–1 exposure, whereas secretion on the basolateral side was significantly reduced by all exposures.
Figure 4.
Effect of 6-h halloysite exposure on mean ± standard deviation (SD) cytokine release (n = 5) by the Caco-2/HT29-MTX cells into apical (AP) and basolateral (BL) media compartments. The y-axis in each graph represent fold-differences when ELISA results are normalized to Control group values. *P < 0.05 based on post-hoc pairwise multiple comparisons (Holm–Sidak method) after ANOVA, NS, not significant.
Proteomics
LFQMS analysis identified and quantified 4081 non-redundant Caco-2/HT29-MTX proteins. These are unique database hits representing individual proteins, splice variants, or isoforms whose relative abundance ranged across five orders of magnitude. A complete list of these proteins along with mass spectral data including identification parameters and group mean quantities/statistics is presented in Table S1. Of the 4081 proteins analyzed, significant quantitative expression changes as a result of halloysite exposure were observed in 473. Lists of proteins altered by each exposure (1, 10 and 100 μg ml–1) along with their quantitation data can be found in Tables S2–4. The average CV for control, 1, 10 and 100 μg ml–1 groups was 21.4%, 13.9%, 18.3% and 11.9%, respectively. In the 100 μg ml–1 group, 3137 proteins had a CV < 15%.
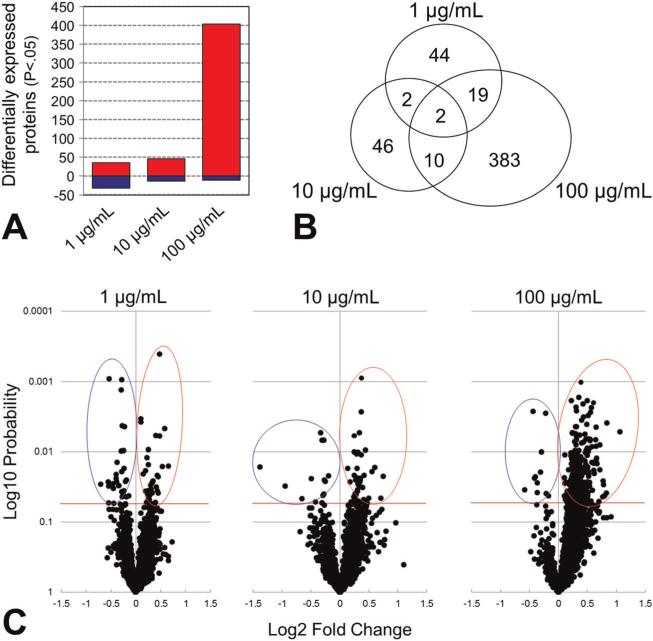
As illustrated graphically in Fig. 5A and C, 100 μg ml–1 halloysite exposure significantly altered the expression of 415 proteins, 404 of which were up-regulated. In contrast, the 1 and 10 μg ml–1 exposures altered the expression of only 67 and 60 proteins, respectively. At 1 μg ml–1, 35 proteins were up-regulated and 32 were down-regulated, whereas 46 were up-regulated and only 14 were down-regulated at 10 μg ml–1. Table 2 shows the top 10 protein fold differences (both up- and down-regulation) at each exposure (P < 0.05). These lists indicate that the greatest fold-changes in expression were +2.1 [histone-lysine N-methyltransferase (MLL)] and -2.6 WD repeat-containing protein 66 (WDR66), whereas most were considerably less, and all demonstrate that the halloysite effects on differential protein expression were moderate, in spite of their statistical significance.
Figure 5.
Caco-2/HT29-MTX protein expression data determined by label-free quantitative mass spectrometry: (A) bar graphs illustrating the number of proteins up-regulated (red) and down-regulated (blue) by each halloysite exposure. (B) Venn diagram illustrating the lack of overlap in differentially expressed proteins across the three exposures. (C) Volcano plots of all 4081 proteins identified and quantified, illustrating their magnitude, significance, and direction of differential expression observed at each halloysite exposure. The horizontal red line in each graph signifies P < 0.05.
Table 2.
Ten most halloysite-mediated up- and down-regulated Caco-2/HT29-MTX cell proteins (P < 0.05) based on fold-difference
| Fold 1 μg m l–1 | Fold 10 μg m l–1 | Fold 100 μg m l–1 |
|---|---|---|
| 1.6 Structural maintenance of chromosomes protein | 1.7 Methylosome subunit pICIn | 2.1 Histone-lysine N-methyltransferase, isoform 1 |
| 1.5 Eukaryotic translation initiation factor 4 gamma 2 | 1.6 Laminin subunit beta-1 | 1.8 Chromatin target of PRMT1 protein, isoform 1 |
| 1.4 Chromatin accessibility complex protein 1 | 1.5 Eukaryotic translation initiation factor 4 gamma 2 | 1.8 5′-AMP-activated protein kinase subunit gamma-1 |
| 1.4 D-tyrosyl-tRNA(Tyr) deacylase 1 | 1.5 Keratin 25D | 1.7 Low MW phosphotyrosine protein phosphatase |
| 1.4 High mobility group protein B3 | 1.4 Dynein light chain Tctex-type 1 | 1.7 Protein KIAA1967 (Deleted in breast cancer gene 1 protein) |
| 1.4 Protein transport protein Sec61 subunit alpha | 1.4 Mitogen-activated protein kinase 14, isoform CSBP2 | 1.7 StAR-related lipid transfer protein 10 |
| 1.4 Signal recognition particle 9 kDa protein | 1.4 Phosphate carrier protein, mitochondrial, isoform A | 1.7 Alcohol dehydrogenase class-3 |
| 1.4 Tudor domain-containing protein 1 | 1.3 60S ribosomal protein L19 | 1.6 ADP-ribosylation factor 6 |
| 1.3 Centrosomal protein 110kDa | 1.3 6-phosphogluconolactonase | 1.6 Calpain-9, isoform 1 |
| 1.3 Copine-1 | 1.3 Brain-specific angiogenesis inhibitor 1-associated protein | 1.6 HLA class II histocompatibility antigen, DRalpha chain |
| -1.6 Serine/threonine-protein phosphatase 2A | -2.6 WD repeat-containing protein 66 | -1.5 Putative PDZ domain-containing protein 1P |
| -1.5 Polyhomeotic 2 homolog | -1.9 60S ribosomal protein L9 | -1.4 Signal recognition particle 14 kDa protein |
| -1.5 PDZ domain-containing protein 1P | -1.6 FACT complex subunit SSRP1 | -1.3 Serum paraoxonase/arylesterase 2, isoform 1 |
| -1.5 Eukaryotic translation initiation factor 1 | -1.4 26S proteasome non-ATPase regulatory subunit | -1.3 Phosphoglucomutase-2 |
| -1.4 U6 snRNA-associated Sm-like protein LSm3 | 12-1.3 Mucin-17 | -1.3 ATP-dependent RNA helicase DHX40, isoform 1 |
| -1.4 Polymerase I and transcript release factor | -1.3 Erlin-2 | -1.3 Coiled-coil domain-containing protein 120 |
| -1.4 Phosphoglucomutase-2 | -1.3 ADP-ribosylation factor GTPase-activating protein 2 | -1.2 PDZ and LIM domain protein 5 |
| -1.4 BH3-interacting domain death agonist | -1.2 Vesicular integral-membrane protein VIP36 | -1.2 Cadherin-2 |
| -1.3 Syntaxin-binding protein 3 | -1.2 Serine/threonine-protein kinase ULK1 | -1.2 Arsenical pump-driving ATPase |
| -1.3 Signal recognition particle 14 kDa protein | -1.2 Glucosamine-6-phosphate isomerase | -1.2 Cytochrome b5 reductase 3 |
Gray-shaded proteins are altered by more than one exposure.
Bioinformatics
Given the heterogeneity of differential expression and lack of overlap across halloysite exposures, to get a better understanding of the biological effects of the protein changes, we conducted a bioinformatic analysis of the protein alteration profiles associated with each exposure using Ingenuity Pathway Analysis. This assessment enabled us to identify the top (i) biological functions and their predicted activation states (Table 3), (ii) canonical pathways (Table 4) and (iii) upstream regulators (Table 5) that were associated with differentially expressed proteins in a statistically significant manner and relevant to barrier epithelia.
Table 3.
Top biological functions and their activation states mapped to differentially expressed Caco-2/HT29-MTX cell proteins
| Exposure | Category | Annotation | P-value | Predicted activation state |
Activation z-score | Proteins |
|---|---|---|---|---|---|---|
| 1 μg ml–1 | NA | NA | NA | NA | NA | NA |
| 10 μg ml–1 | Cellular Growth & Proliferation | proliferation of tumor cell lines | 0.03 | Increased | 2.034 | BID,C9orf86,EIF4B,IGBP1,ITGA6,MAPK14,MYCBP,PPP1R13B,PXN,RHOC |
| 100 μg ml–1 | Infectious Disease | viral infection | 0.003 | Increased | 5.73 | APIS1, APC2, BAX, CCAR1, CDC42, CHMP2A, CHMP4A, COPA, CTBP1, CYCS, DDX5, DDX6, DEK, DHX15, DLST, DNAJB1, DNM2, EIF2AK2, EIF2B5, ERI3, ETF1, FLOT2, GSTOI, GTF2I, GYG1, HMCN2, HMGCS1, MAP4, MAPK14, MDN1, PAK1, PICALM, PIK3C2G, PRDX2, PSMA2, PSMA3, PSMA5, PSMD2, PSMD4, RPS14, RPS27A, RPS5, SCARB2, SF3B2, SRRT, SRSF1, SSB, SUB1, SUMO2, TXN, UAP1, UBQLN4, VAMP3, VTN, WASF2 |
| Infectious Disease | infection of cells | 0.003 | Increased | 5.50 | C1QBP, CCAR1, CDC42, COPA, CTBP1, CYCS, DHX15, DLST, DNAJB1, DNM2, EIF2AK2, EIF2B5, ERI3, ETF1, HMCN2, HMGCS1, MAP4, MDN1, PAK1, PICALM, PSMA2, PSMA3, PSMA5, PSMD4, SF3B2, SRRT, SSB, SUB1, SUMO2, UAP1, UBQLN4, WASF2 | |
| Embryonic Development | size of embryo | 0.004 | Increased | 3.84 | ARHGAP1, CDC42, CFL1, CYCS, DLD, DNM1L, HSPG2, KIF5B, LMNB1, POU2F1, SNX1, TRA2B, TXNRD1, VCL, WASF2 | |
| Cellular Growth and Proliferation | proliferation of cells | 0.02 | Increased | 2.77 | ABCB4, ACP1, AHSA1, AKR1B10, AKR1C1/AKR1C2, APEX1, ARHGAP1, ARL1, BAX, C1QBP, C9orf86, CACYBP, CALR, CAPN2, CDC42, CDH2, CDHR5, CEACAM6, CFL1, COMT, COPS4, CRIP2, CTBP1, CTNND1, DDB1, DDX5, DECR1, DLST, DNAJB1, DNM1L, DNM2, EIF2AK2, EIF4G2, FDFT1, FLOT2, GFRA3, GLRX, GNPNAT1, GTF2I, H2AFX, HPRT1, HSPG2, KRT10, LAMB1, LIMA1, LMNB1, MAPK14, MCM3, MLL, MST4, MTHFD1, MYH10, NT5E, NUMA1, PAK1, PCYT1A, PDZK1, PKP3, PPL, PPP1CA, PPP1R12A, PRDX2, PRDX3, PSMD2, RAB1A, RAN, RBM3, RPS3A, S100A6, SELENBP1, SERPINH1, SF1, SF3B2, SMC3, SND1, SRRT, SRSF1, STARD10, STX6, SUMO2, TPM1, TPM3, TRIM28, TRPC5, TSN, TXN, TXNRD1, UCHL3, USP10, USP8, VTN, WARS, WASF2 | |
| Cellular Assembly and Organization | quantity of filaments | 0.01 | Increased | 2.43 | CFL1, HSPG2, LIMA1, MAP4, SERPINH1, VTN | |
| Organismal Development | growth of organism | 0.01 | Increased | 2.38 | ADH5, ARHGAP1, BAX, CDC42, DLD, FDFT1, HSD17B4, KIF5B, PLEKHA1, POU2F1, SNX1, SRRT, SSB, SUMO2, TXN, TXNRD1, VCL, WASF2 | |
| Nucleic Acid Metabolism | hydrolysis of nucleotide | 0.0003 | Increased | 2.34 | ARFGAP1, CALR, CDC42, NT5E, NUDT5, PSMC2, RAN, RANGAP1, RUVBL1 | |
| Cell Cycle | cytokinesis | 0.004 | Increased | 2.21 | ARF6, CAP1, CDC42, CFL1, KIF4B, MYH10, SEPT11, TPM1 | |
| Cellular Assembly and Organization | formation of lamellipodia | 0.01 | Increased | 2.18 | ACTR3, C1QBP, CAPZB, CDC42, SNX1, VCL, WASF2 | |
| Cellular Assembly and Organization | organization of cytoplasm | 0.001 | Increased | 2.17 | ACP1, ACTR3, ARF6, ARHGAP1, ARL1, BAX, C1QBP, CALR, CANX, CAP1, CAPN2, CAPZB, CDC42, CDH2, CFL1, COMT, CTBP1, CTNND1, DNM1L, DNM3, DYNLL1, GOLGB1, HPRT1, KIF5B, LAMB1, MAP4, MYH10, NDRG1, NSFL1C, NUMA1, PAK1, PICALM, PPP1R12A, PRDX3, RAN, SEPT11, SNX1, STX6, TPM1, TPM3, USP8, VCL, VTN, WASF2, YWHAH | |
| Cellular Assembly and Organization | organization of cytoskeleton | 0.001 | Increased | 2.17 | ACP1, ACTR3, ARF6, ARHGAP1, BAX, C1QBP, CALR, CANX, CAP1, CAPN2, CAPZB, CDC42, CDH2, CFL1, COMT, CTNND1, DNM1L, DNM3, DYNLL1, HPRT1, KIF5B, LAMB1, MAP4, MYH10, NDRG1, NUMA1, PAK1, PICALM, RAN, SEPT11, SNX1, TPM1, TPM3, VCL, VTN, WASF2, YWHAH |
NA, not applicable.
Table 4.
Top canonical pathways mapped* to differentially expressed Caco-2/HT29-MTX cell proteins
| Exposure | Canonical pathway | -log (P-value) | % of pathway mapped | Proteins |
|---|---|---|---|---|
| 1 μg ml–1 | NA | NA | NA | NA |
| 10 μg ml–1 | NA | NA | NA | NA |
| 100 μg ml–1 | Glutathione redox reactions II | 3.45 | 100% | GSR▲, TXNDC12▲, GLRX▲ |
| Ascorbate recycling (cytosolic) | 2.98 | 67% | GLRX▲, GSTO1▲ | |
| Serine biosynthesis | 2.68 | 50% | PSAT1▲, PSPH▲ | |
| 2-ketoglutarate dehydrogenase complex | 2.68 | 40% | DLST▲, DLD▲ | |
| UDP-N-acetyl-D-galactosamine biosynthesis II | 3.46 | 33% | GNPNAT1▲, GALE▲, UAP1▲ | |
| Thioredoxin Pathway | 2.47 | 33% | TXN▲, TXNRD1▲ | |
| UDP-N-acetyl-D-glucosamine biosynthesis II | 2.30 | 33% | GNPNAT1▲, UAP1▲ | |
| Granzyme B signaling | 3.73 | 25% | NUMA1▲, CYCS▲, LMNB1▲, DFFA▲ | |
| Lipid antigen presentation by CD1 | 3.33 | 20% | CALR▲, ARF6▲, CANX▲, AP2A2▲ |
Only those pathways significantly (P < 0.01) mapped to at least two proteins representing ≥ 20% of pathway protein constituents are listed. NA, not applicable.
Table 5.
Top upstream regulators mapped to differentially expressed Caco-2/HT29-MTX cell proteins
| Exposure | Upstream regulator | Molecule type | Predicted activation state | Activation z-score* | P-value# | Regulated target proteins from dataset |
|---|---|---|---|---|---|---|
| 1 μg ml–1 | NA | NA | NA | NA | NA | NA |
| 10 μg ml–1 | NA | NA | NA | NA | NA | NA |
| 100 μg ml–1 | NFE2L2 | transcription regulator | Activated | 4.015 | 0.00004 | ABCB4, AKR1B10, DYNLL1, EIF4G2, GSR, GSTO1, HMBS, HPRT1, OAT, PGD, PSAT1, PSMA5, PSMB3, RAN, RUVBL1, TXN, TXNRD1, UGDH |
| IL5 | cytokine | Activated | 3.162 | 0.01 | AK3, HSPH1, LMNB1, NDRG1, PSAT1, RBM3, S100A6, SDF2L1, SRI, UCHL3 | |
| XBP1 | transcription regulator | Activated | 2.800 | 0.001 | CALR, GOLGB1, HLA-DRA, PCYT1A, S100A6, SDF2L1, SEC31A, SMC3, SRP9, TXN | |
| TGFB1 | growth factor | Activated | 2.717 | 0.02 | AKR1C1/AKR1C2, BAX, CAB39, CAP1, CDH2, CFL1, CRIP2, CTNND1, DDX5, DYNLL1, GARS, GYG1, HADH, HSPG2, KPNA3, KRT10, MAP4, MAPK14, NT5E, PDLIM5, PPID, PSPH, RAB1A, RBM3, S100A6, SERPINH1, SRI, TPM1, TPM3, TXNRD1, USH1C, VAPB, VAT1, VCL | |
| MYC | transcription regulator | Activated | 2.475 | 0.000001 | APEX1, BAX, C1QBP, CANX, CAPN2, CAPZB, CDC42, CDH2, CRIP2, CYCS, DDX5, GSR, HSPH1, LIMA1, MAP4, MCM6, MTHFD1, NDRG1, PHB2, PPID, PRDX2, PRDX3, PSAT1, RPL19, RPL22, RPL27, RPL6, RPL7, RPL7A, RUVBL1, S100A6, SDCBP, SERPINH1, SUMO2, THOP1, TXN, VAMP3, VARS | |
| ATF4 | transcription regulator | Activated | 2.219 | 0.01 | APEX1, GARS, NDRG1, PSAT1, PSPH, WARS | |
| HSF1 | transcription regulator | Activated | 2.180 | 0.0004 | CCT4, DNAJB1, HSBP1, HSPA4, HSPA7, HSPH1, TMPRSS3 | |
| miR-124-3p | mature microRNA | Inhibited | -2.969 | 0.01 | CTNND1, DNM2, ERH, FAM129B, HADH, HEBP2, LMNB1, MAPK14, VAMP3 | |
| miR-1 | mature microRNA | Inhibited | -2.630 | 0.0004 | CAP1, DDX5, DHX15, DNAJB1, GNPNAT1, OAT, PGM2, PICALM, SRSF9, TPM1, TPM3 | |
| miR-155-5p | mature microRNA | Inhibited | -2.335 | 0.00001 | DHX40, DNAJB1, NT5E, PDLIM5, PICALM, PPL, SDCBP, SYNE2, TBCA, TXNDC12, TXNRD1, VAMP3 |
z-score measures the consistency of a protein expression pattern with the activation or inhibition of a given “upstream regulator”
P-value measures the significance of the overlap between proteins regulated by an “upstream regulator” and differentially expressed proteins in the data set. NA, not applicable.
Discussion
The cell viability, membrane integrity and permeability assay results are consistent with previous observations where only long-term (> 24 h) exposure to nanoclays decreased the viability of Hela and MCF-7 (Vergaro et al., 2010), HepG2 (Lordan et al., 2011), and another study where Cloisite (R) Na(+) nanoclay cytotoxicity was absent in human monocytic U937 cells when nanoparticles were coated with FBS (Lordan and Higginbotham, 2012), an effect attributed to greater dispersibility. Similarly, Li et al. (2010) observed minor cytotoxicity of Cloisite nanoclay in Chinese hamster ovary cells below 1000 μg ml–1 after 12-h exposures, and a dose-dependent effect after 24-h incubation. In general, it seems that at low doses, nanoclay has low cytotoxicity. When toxic effects are observed, they are related to clay nanotube functionalization or compounds filling them (Sharma et al., 2010; Shi et al., 2011; Vergaro et al., 2012; Zapor and Dzierzawska, 2012).
In general, nanoparticles exert proinflammatory effects in lung and immune tissues that is characterized by, at least in part, increased cytokine/chemokine production (Chang, 2010; Shannahan et al., 2012a). However, exposure of intestinal epithelia to nanomaterials in vitro has been shown to have little effect on proinflammatory cytokine release (Bergin and Witzmann, 2012), and in a previous study using this co-culture system (Lai et al., 2013), altered cytokine release in response to functionalized carbon nanotube exposure up to 10 μg ml–1 was negligible, in spite of significant changes in cellular protein expression. Proinflammatory effects of halloysite exposure have not been reported, nor does it appear that such consequences have been investigated in intestinal cells. In contrast, halloysite has been shown to exert anti-inflammatory effects in murine primary peritoneal macrophages in vitro (Cornejo-Garrido et al., 2012) and allergic inflammation in vivo in mice (Park et al., 2008). Alternatively, bacterial infection of enterocytes is known to cause the polarized, basolateral secretion of the cytokines measured in this study (Jung et al., 1995), especially IL-8 (Laurent et al., 1997) or both IL-8 and MCP-1 (Kim et al., 2002). Typical bacilli are rod shaped and are about 2.0 μm long and 500 nm in diameter, somewhat similar to the range of halloysite dimensions used in this study. As bioinformatic analysis of differential expression shows (see below), proteins linked to cell infection were up-regulated by the 100 μg ml–1 exposure. Together, these observations suggest that the intestinal cells may respond to halloysite as if infected by bacteria. Although the halloysite nanotube interaction with the apical membrane and potential internalization is beyond the scope of the present study, much larger, high-aspect ratio nanoparticles, such as multi-walled carbon nanotubes, do not seem to enter enterocytes in culture (Clark et al., 2011; Lai et al., 2013), so it is possible the same is true for the halloysite nanotubes. This notion warrants further study.
As halloysite exposure concentration increased, up-regulation of specific proteins also increased, while protein down-regulation was ameliorated. This suggests unique cellular effects at these different exposures and infers the development of an adaptive, cytoprotective generalized stress response (Osburn and Kensler, 2008) (see below in Upstream Regulators section) with an increasing halloysite concentration. For instance, five proteins commonly associated with cell stress were significantly up-regulated only at 100 μg ml–1, heat shock factor-binding protein 1, activator of 90-kDa heat shock protein ATPase, heat shock 70-kDa proteins 4 and 7, and heat-shock protein 105 kDa.
In terms of overall impact, compared with the 100 μg ml–1 exposure that altered the expression of over 10% of the proteins analyzed, halloysite exposures that were one and two orders of magnitude lower had a comparatively little effect. The notion of differential dose-related protein regulation is further supported by the Venn diagram, Fig. 5B, which illustrates the discernible scarcity of overlap in those proteins whose expression was altered by each exposure. Only two proteins, brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAIAP2L1) and eukaryotic translation initiation factor 4 gamma 2 (EIF4G2), were common to all three exposures, and all three were moderately up-regulated. BAIAP2L1 [also known as insulin receptor tyrosine kinase substrate (IRTKS)] is a ubiquitously expressed protein involved in the formation of clusters of actin bundles (Millard et al., 2007), is known to be up-regulated in inflammatory conditions (Galligan et al., 2007) and plays a role in the reorganization of the actin cytoskeleton in response to bacterial infection (Vingadassalom et al., 2009). EIF4G2, another ubiquitous protein, is also known as death-associated protein 5 (DAP-5) and p97. This protein is abundantly expressed in proliferating cells and is recruited to ribosomes after growth factor stimulation to promote protein synthesis and cell proliferation (Lee and McCormick, 2006). In contrast, inducible expression of EIF4G2 (p97) has been shown to reduce overall protein synthesis by acting as a general repressor of translation (Imataka et al., 1997). Its moderate up-regulation at all exposures may reflect similar responses with different cellular outcomes in the enterocytes at different levels of halloysite nanoclay exposure. Both BAIAP2L1 and EIF4G2 have been shown to be up-regulated in HT29 cells undergoing endoplasmic reticular stress (Saito et al., 2009), one facet of an adaptive response. Of those proteins undergoing the greatest fold change, histone-lysine N-methyltransferase (MLL) mediates the methylation of ’Lys-4’ of histone H3 (H3K4me), a specific tag for epigenetic transcriptional activation (Ansari et al., 2009). Histone H3 lysine 4 methylation generally correlates with active transcriptional expression (Santos-Rosa et al., 2002), so the two-fold increase in MLL expression may signify higher transcriptional activity at 100 μg ml–1, which may be related to the observed up-regulation of 404 proteins at this exposure.
Consistent with the overall lack of overlap across exposure groups, only two differentially expressed proteins appeared in more than one exposure, phosphoglucomutase-2 (PGM2) and signal recognition particle 14 kDa protein (SRP14), which were down-regulated by 1 and 100 μg ml–1 halloysite exposures. Also known as phosphopentomutase, PGM2 participates in the pentose phosphate pathway and in purine metabolism by catalyzing the conversion of the nucleoside breakdown products ribose-1-phosphate and deoxyribose-1-phosphate to the corresponding 5-phosphopentoses. SRP14 is a component of the signal recognition particle (SRP) assembly that targets secretory proteins to the rough endoplasmic reticulum membrane (Lütcke, 1995). The SRP is composed of SRP9, SRP14, SRP19, SRP54, SRP68 and SRP72 (Walter and Blobel, 1980), where SRP9 and SRP14 are required for RNS binding. All six of these SRP components were identified and quantified in this study, including the signal recognition particle receptor subunit alpha. However, only SRP9 and SRP14 were differentially expressed, and surprisingly, SRP9 was up-regulated by 1 and 100 μg ml–1 halloysite exposures, compared with SRP14's down-regulation.
With respect to biological functions associated with differential expression, proteins whose expression was altered by the 1 μg ml–1 halloysite exposure failed to associate significantly with any increased or decreased activation states. Proteins altered by the 10 μg ml–1 exposure were associated with ’proliferation of tumor cell lines‘, a result consistent with the cell viability data. Proteins altered by the 100 μg ml–1 exposure were associated with ’infection of cells‘ (32 proteins) and ’viral infection of cells‘ (55 proteins) along with 9 other functional annotations variably associated with increased ’cell growth and proliferation‘ and ’cellular assembly and organization’. These latter annotations include increased activation states in nucleotide hydrolysis and cytokinesis, both emblematic of growth (‘growth of organism‘) and cell division.
Activation of lamellipodia (filopodia) formation in colon cancer cells increases their migratory and invasive properties (Makrodouli et al., 2011) and is associated with significant cytoskeletal (actin) rearrangement (Khurana et al., 2008). However, it is also an important cell migration mechanism in wound healing in intestinal epithelia (Arciero et al., 2011). Whether this response to high-dose halloysite exposure reflects actual injury, which our cytotoxicity data do not support, or related to the use of cancer cells in the in vitro model requires additional study. The fact that activation states associated with cell infection were increased and mapped to a significant number of proteins, suggests that an injury response, or at least an irritation response (based on IL-8 and TNF-α responses in Fig. 4), may have been underway at 6 h. Perhaps the halloysite size and shape, along with its protein corona (Shannahan et al., 2012b), may elicit such a broad response in the Caco-2/HT29-MTX cells under these conditions. This speculation requires additional investigation. Nevertheless, in terms of biological function, a significant response by the intestinal cell-lines in co-culture to 100 μg ml–1 halloysite exposure is clear, whereas the low doses had little effect, if any, in this regard.
Canonical pathway analysis detected no significant associations at the 2 lowest exposures. However, proteins altered by 100 μg ml–1 halloysite exposure mapped to 9 pathways, all of which were up-regulated. Three of these pathways, glutathione redox reactions II, ascorbate recycling and the thioredoxin pathway, represent an interrelated antioxidant system in which the component enzymes are capable of regenerating several antioxidant compounds, including ascorbic acid, as well as reducing oxidized proteins and reactive oxygen species (ROS) (Nordberg and Arnér, 2001). Serine biosynthesis pathway up-regulation is consistent with enhanced cell growth and proliferation suggested by the biological function analysis.
The ’lipid antigen presentation by CD1‘ pathway was mapped (P < 0.0005) by 4 up-regulated proteins. The expression and extracellular presentation of lipid antigens to immune cells (e.g. T cells) by CD1 molecules in intestinal cells may play a role in natural host defense against bacterial colonization and invasion at the mucosal barrier (Dougan et al., 2007; Sugita et al., 2000). Up-regulation of components of this pathway may relate to the ’cell infection‘ function activated by halloysite exposure and alluded to earlier.
Ingenuity Pathway Analysis was used to predict which upstream regulators may have been activated or inhibited by halloysite exposure and to help explain the bioactivity of halloysite nanotubes in the Caco-2/HT29-MTX co-culture system. In this case, ’upstream regulator‘ refers to any molecule that can affect the expression of a protein or set of related proteins in a regulatory pathway, including transcription factors, cytokines, microRNAs, receptors and kinases. Neither the 1 nor 10 μg ml–1 exposures resulted in the identification of upstream regulators that were significantly related to differentially expressed proteins. However, proteins whose expression was altered by 100 μg ml–1 halloysite nanotube exposure were mapped to 10 different upstream regulators, 7 of which were predicted to be activated and 3 inhibited.
Activated regulators included the antioxidant transcription factor NFE2L2 (nuclear factor erythroid 2-related factor 2 or Nrf2), a transcription activator that binds to antioxidant response elements in the promoter regions of target genes (Itoh et al., 1997) and is involved in the coordinated up-regulation of genes in response to oxidative stress in the intestine (McMahon et al., 2001) and a generalized cytoprotective adaptive response to toxic insult (Osburn and Kensler, 2008). Several of the activated regulators are associated with cell growth and proliferation. For instance, in contrast to antioxidant response, NFE2L2 is also associated closely with increased cell growth (Ohta et al., 2008). Two other growth-related factors, MYC and TGFB1 also were identified.
In terms of upstream regulators with inhibited activation states, 3 microRNAs were identified and protein alterations mapped to them that suggested their activity was inhibited by 100 μg ml–1 halloysite exposure. These included miR-124-3p, miR-1 and miR-155-5p. MicroRNAs (miRNAs) are key posttranscriptional regulators of gene expression involved in diverse biological pathways, acting through translational repression, mRNA decay, or both (Lee and Shin, 2012). Assessment of miRNA responses to nanoparticle exposure in mammalian or human systems has not yet been reported, nor have indirect methods such as we present here been used previously. Our protein profiles suggest that nanoparticle exposure may act through altered expression of these novel regulators and should be taken into account in future studies.
The present investigation was conducted to examine the effect of halloysite clay nanotube exposure and represents the first investigation of halloysite effects on intestinal barrier epithelia in co-culture in vitro. The major new findings of this study include: (i) significant proinflammatory effects at halloysite exposures as low as 1 μg ml–1 and (ii) significant changes in protein expression at 100 μg ml–1. Based on these findings, halloysite clay nanotubes appear unlikely to have toxic effects at moderate levels of exposure. However, significant exposure-specific changes in expression were observed among the 4081 proteins analyzed, particularly at the high concentration. Bioinformatic analysis of differentially expressed protein profiles generated by 100 μg ml–1 exposure suggest that halloysite stimulates processes related to cell growth and proliferation, subtle responses to cell infection, irritation and injury, enhanced antioxidant capability, all characteristic of an overall adaptive response to exposure. These potentially relevant functional effects warrant further investigation in both in vitro and in vivo models and suggest that chronic or high concentration bolus exposure to halloysite nanotubes might have unintended outcomes.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the technical assistance of Dr Pu-Chun Ke, Clemson University, who participated in the halloysite characterization, and Heather Ringham and Meixian Fang for their assistance with the cell culture, exposures, sample preparation and toxic endpoint assays. This work was supported by NIH grants RC2ES018025 and R01GM085218 (FAW) and the Integrated Nanosystems Development Institute (INDI) at Indiana University – Purdue University Indianapolis.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
Four additional data files in PDF format contain the extensive lists of all the proteins identified and quantified along with mass spectral data (Table S1) and proteins altered by halloysite exposure of 1, 10 and 100 μg ml–1 along with their quantitation data (Tables S2–4). This material may be found in the online version of this article.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abdullayev E, Joshi A, Wei W, Zhao Y, Lvov Y. Enlargement of halloysite clay nanotube lumen by selective etching of aluminum oxide. ACS Nano. 2012;6:7216–7226. doi: 10.1021/nn302328x. [DOI] [PubMed] [Google Scholar]
- Ansari KI, Mishra BP, Mandal SS. MLL histone methylases in gene expression, hormone signaling and cell cycle. Front. Biosci. 2009;14:3483–3495. doi: 10.2741/3466. [DOI] [PubMed] [Google Scholar]
- Arciero JC, Mi Q, Branca MF, Hackam DJ, Swigon D. Continuum model of collective cell migration in wound healing and colony expansion. Biophys. J. 2011;100:535–543. doi: 10.1016/j.bpj.2010.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergin IL, Witzmann FA. Nanoparticle toxicity by the gastrointestinal route: evidence and knowledge gaps. Int. J. Biomed. Nanosci. Nanotechnol. 2012 doi: 10.1504/IJBNN.2013.054515. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. The immune effects of naturally occurring and synthetic nanoparticles. J. Autoimmun. 2010;34:J234–J246. doi: 10.1016/j.jaut.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Clark KA, O'Driscoll C, Cooke CA, Smith BA, Wepasnick K, Fairbrother DH, Lees PSJ, Bressler JP. Evaluation of the Interactions Between Multiwalled Carbon Nanotubes and Caco-2 Cells. J. Toxicol. Environ. Health A. 2011;75:25–35. doi: 10.1080/15287394.2011.589105. [DOI] [PubMed] [Google Scholar]
- Cornejo-Garrido H, Nieto-Camacho A, Gómez-Vidales V, Ramírez-Apan MT, del Angel P, Montoya JA, Domínguez-López M, Kibanova D, Cervini-Silva J. The anti-inflammatory properties of halloysite. Appl. Clay Sci. 2012;57:10–16. [Google Scholar]
- Dougan S, Kaser A, Blumberg R. CD1 expression on antigen-presenting cells T cell activation by cd1 and lipid antigens. In: Moody DB, editor. Current Topics in Microbiology and Immunology. Vol. 314. Springer; Berlin: 2007. pp. 113–141. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren J, Jamstorp E, Bredenberg S, Engqvist H, Stromme M. A ceramic drug delivery vehicle for oral administration of highly potent opioids. J. Pharm. Sci. 2010;99:219–226. doi: 10.1002/jps.21814. [DOI] [PubMed] [Google Scholar]
- Galligan CL, Baig E, Bykerk V, Keystone EC, Fish EN. Distinctive gene expression signatures in rheumatoid arthritis synovial tissue fibroblast cells: correlates with disease activity. Genes Immun. 2007;8:480–491. doi: 10.1038/sj.gene.6364400. [DOI] [PubMed] [Google Scholar]
- Hilgendorf C, Spahn-Langguth H, Regardh CG, Lipka E, Amidon GL, Langguth P. Caco-2 versus Caco-2/HT29-MTX co-cultured cell lines: permeabilities via diffusion, inside- and outside-directed carrier-mediated transport. J. Pharm. Sci. 2000;89:63–75. doi: 10.1002/(SICI)1520-6017(200001)89:1<63::AID-JPS7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand. Stat. Theory Appl. 1979;6:65–70. [Google Scholar]
- Hughes AD, King MR. Use of naturally occurring halloysite nanotubes for enhanced capture of flowing cells. Langmuir. 2010;26:12155–12164. doi: 10.1021/la101179y. [DOI] [PubMed] [Google Scholar]
- Imataka H, Olsen HS, Sonenberg N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997;16:817–825. doi: 10.1093/emboj/16.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase ii detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Joussein E, Petit S, Churchman J, Theng B, Righi D, Delvaux B. Halloysite clay minerals - A review. Clay Miner. 2005;40:383–426. [Google Scholar]
- Jung HC, Eckmann L, Yang SK, Panja A, Fierer J, Morzycka-Wroblewska E, Kagnoff MF. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J. Clin. Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Ebara M, Izawa H, Sanchez-Ballester NM, Hill JP, Ariga K. Supramolecular approaches for drug development. Curr. Med. Chem. 2012;19:2388–2398. doi: 10.2174/092986712800269254. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kelly HM, Deasy PB, Ziaka E, Claffey N. Formulation and preliminary in vivo dog studies of a novel drug delivery system for the treatment of periodontitis. Int. J. Pharm. 2004;274:167–183. doi: 10.1016/j.ijpharm.2004.01.019. [DOI] [PubMed] [Google Scholar]
- Khurana S, Tomar A, George SP, Wang Y, Siddiqui MR, Guo H, Tigyi G, Mathew S. Autotaxin and lysophosphatidic acid stimulate intestinal cell motility by redistribution of the actin modifying protein villin to the developing lamellipodia. Exp. Cell Res. 2008;314:530–542. doi: 10.1016/j.yexcr.2007.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kim JS, Jung HC, Oh YK, Song IS, Kim CY. Differential expression and polarized secretion of CXC and CC chemokines by human intestinal epithelial cancer cell lines in response to Clostridium difficile toxin A. Microbiol. Immunol. 2002;46:333–342. doi: 10.1111/j.1348-0421.2002.tb02704.x. [DOI] [PubMed] [Google Scholar]
- Kommireddy DS, Sriram SM, Lvov YM, Mills DK. Stem cell attachment to layer-by-layer assembled TiO2 nanoparticle thin films. Biomaterials. 2006;27:4296–4303. doi: 10.1016/j.biomaterials.2006.03.042. [DOI] [PubMed] [Google Scholar]
- Lai X, Bacallao RL, Blazer-Yost BL, Hong D, Mason SB, Witzmann FA. Characterization of the renal cyst fluid proteome in autosomal dominant polycystic kidney disease (ADPKD) patients. Proteomics Clin. Appl. 2008;2:1140–1152. doi: 10.1002/prca.200780140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X, Blazer-Yost BL, Clack JW, Fears SL, Mitra S, Ntim SA, Ringham HN, Witzmann FA. Protein expression profiles of intestinal epithelial co-cultures after low-level exposure to functionalized carbon nanotubes. Int. J. Biomed. Nanosci. Nanotechnol. 2013 doi: 10.1504/IJBNN.2013.054508. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X, Wang L, Tang H, Witzmann FA. A novel alignment method and multiple filters for exclusion of unqualified peptides to enhance label-free quantification using peptide intensity in LC-MS/MS. J. Proteome Res. 2011;10:4799–4812. doi: 10.1021/pr2005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent F, Eckmann L, Savidge TC, Morgan G, Theodos C, Naciri M, Kagnoff MF. Cryptosporidium parvum infection of human intestinal epithelial cells induces the polarized secretion of C-X-C chemokines. Infect. Immun. 1997;65:5067–5073. doi: 10.1128/iai.65.12.5067-5073.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Shin C. MicroRNA–target interactions: new insights from genome-wide approaches. Ann. N. Y. Acad. Sci. 2012;1271:118–128. doi: 10.1111/j.1749-6632.2012.06745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, McCormick F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 2006;25:4008–4019. doi: 10.1038/sj.emboj.7601268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuffleur T, Barbat A, Luccioni C, Beaumatin J, Clair M, Kornowski A, Dussaulx E, Dutrillaux B, Zweibaum A. Dihydrofolate reductase gene amplification-associated shift of differentiation in methotrexate-adapted HT-29 cells. J. Cell Biol. 1991;115:1409–1418. doi: 10.1083/jcb.115.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuffleur T, Porchet N, Aubert JP, Swallow D, Gum JR, Kim YS, Real FX, Zweibaum A. Differential expression of the human mucin genes MUC1 to MUC5 in relation to growth and differentiation of different mucus-secreting HT-29 cell subpopulations. J. Cell Sci. 1993;106(Pt 3):771–783. doi: 10.1242/jcs.106.3.771. [DOI] [PubMed] [Google Scholar]
- Li PR, Wei JC, Chiu YF, Su HL, Peng FC, Lin JJ. Evaluation on cytotoxicity and genotoxicity of the exfoliated silicate nanoclay. ACS Appl. Mater. Interfaces. 2010;2:1608–1613. doi: 10.1021/am1001162. [DOI] [PubMed] [Google Scholar]
- Lordan S, Higginbotham CL. Effect of serum concentration on the cytotoxicity of clay particles. Cell Biol. Int. 2012;36:57–61. doi: 10.1042/CBI20100587. [DOI] [PubMed] [Google Scholar]
- Lordan S, Kennedy JE, Higginbotham CL. Cytotoxic effects induced by unmodified and organically modified nanoclays in the human hepatic HepG2 cell line. J. Appl. Toxicol. 2011;31:27–35. doi: 10.1002/jat.1564. [DOI] [PubMed] [Google Scholar]
- Lütcke H. Signal Recognition Particle (SRP), a ubiquitous initiator of protein translocation. Eur. J. Biochem. 1995;228:531–550. doi: 10.1111/j.1432-1033.1995.tb20293.x. [DOI] [PubMed] [Google Scholar]
- Mahler GJ, Esch MB, Glahn RP, Shuler ML. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol. Bioeng. 2009a;104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- Mahler GJ, Shuler ML, Glahn RP. Characterization of Caco-2 and HT29-MTX cocultures in an in vitro digestion/cell culture model used to predict iron bioavailability. J. Nutr. Biochem. 2009b;20:494–502. doi: 10.1016/j.jnutbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Makrodouli E, Oikonomou E, Koc M, Andera L, Sasazuki T, Shirasawa S, Pintzas A. BRAF and RAS oncogenes regulate Rho GTPase pathways to mediate migration and invasion properties in human colon cancer cells: a comparative study. Mol. Cancer. 2011;10:118. doi: 10.1186/1476-4598-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J. Cell Sci. 2007;120(Pt 9):1663–72. doi: 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- Monroe ME, Shaw JL, Daly DS, Adkins JN, Smith RD. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput. Biol. Chem. 2008;32:215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nordberg J, Arnér ESJ. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 2001;31:1287–1312. doi: 10.1016/s0891-5849(01)00724-9. [DOI] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- Osburn WO, Kensler TW. Nrf2 signaling: An adaptive response pathway for protection against environmental toxic insults. Mutat. Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Lee CW, Lee MY. Inhibitory effect of ore minerals on the allergic inflammation in mouse. J. Korean Soc. Appl. Bi. 2008;51:269–275. [Google Scholar]
- Price R, Gaber BP, Lvov Y. In-vitro release characteristics of tetracycline HCl, khellin and nicotinamide adenine dineculeotide from halloysite; a cylindrical mineral. J. Microencapsul. 2001;18:713–722. doi: 10.1080/02652040010019532. [DOI] [PubMed] [Google Scholar]
- Saito S, Furuno A, Sakurai J, Sakamoto A, Park H-R, Shin-ya K, Tsuruo T, Tomida A. Chemical genomics identifies the unfolded protein response as a target for selective cancer cell killing during glucose deprivation. Cancer Res. 2009;69:4225–4234. doi: 10.1158/0008-5472.CAN-08-2689. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. http://www.nature.com/nature/journal/v419/n6905/suppinfo/nature01080_S1.html. [DOI] [PubMed] [Google Scholar]
- Shannahan JH, Kodavanti UP, Brown JM. Manufactured and airborne nanoparticle cardiopulmonary interactions: a review of mechanisms and the possible contribution of mast cells. Inhal. Toxicol. 2012a;24:320–339. doi: 10.3109/08958378.2012.668229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannahan JH, Brown JM, Chen R, Ke PC, Lai X, Mitra S, Witzmann FA. Comparison of nanotube-protein corona composition in cell culture media. Small. 2012b doi: 10.1002/smll.201202243. DOI:10.1002/smll.201202243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Schmidt B, Frandsen H, Jacobsen NR, Larsen EH, Binderup ML. Genotoxicity of unmodified and organo-modified montmorillonite. Mutat. Res. 2010;700:18–25. doi: 10.1016/j.mrgentox.2010.04.021. [DOI] [PubMed] [Google Scholar]
- Shchukin DG, Sukhorukov GB, Price RR, Lvov YM. Halloysite nanotubes as biomimetic nanoreactors. Small. 2005;1:510–513. doi: 10.1002/smll.200400120. [DOI] [PubMed] [Google Scholar]
- Shi YF, Tian Z, Zhang Y, Shen HB, Jia NQ. Functionalized halloysite nanotube-based carrier for intracellular delivery of antisense oligonucleotides. Nanoscale Res. Lett. 2011;6:608. doi: 10.1186/1556-276X-6-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J. Roy. Stat. Soc. B. 2002;64:479–498. [Google Scholar]
- Sugita M, Peters PJ, Brenner MB. Pathways for lipid antigen presentation by CD1 molecules: nowhere for intracellular pathogens to hide. Traffic. 2000;1:295–300. doi: 10.1034/j.1600-0854.2000.010401.x. [DOI] [PubMed] [Google Scholar]
- Suh YJ, Kil DS, Chung KS, Abdullayev E, Lvov YM, Mongayt D. Natural nanocontainer for the controlled delivery of glycerol as a moisturizing agent. J. Nanosci. Nanotechnol. 2011;11:661–665. doi: 10.1166/jnn.2011.3194. [DOI] [PubMed] [Google Scholar]
- Suresh R, Borkar SN, Sawant VA, Shende VS, Dimble SK. Nanoclay drug delivery system. Int. J. Pharmaceut. Sci. Nanotech. 2010;3:901–905. [Google Scholar]
- Veerabadran NG, Price RR, Lvov YM. Clay nanotubes for encapsulation and sustained release of drugs. Nano. 2007;2:115–120. [Google Scholar]
- Vergaro V, Abdullayev E, Lvov YM, Zeitoun A, Cingolani R, Rinaldi R, Leporatti S. Cytocompatibility and uptake of halloysite clay nanotubes. Biomacromol. 2010;11:820–826. doi: 10.1021/bm9014446. [DOI] [PubMed] [Google Scholar]
- Vergaro V, Lvov YM, Leporatti S. Halloysite clay nanotubes for resveratrol delivery to cancer cells. Macromol. Biosci. 2012;12:1265–1271. doi: 10.1002/mabi.201200121. [DOI] [PubMed] [Google Scholar]
- Vingadassalom D, Kazlauskas A, Skehan B, Cheng HC, Magoun L, Robbins D, Rosen MK, Saksela K, Leong JM. Insulin receptor tyrosine kinase substrate links the E. coli O157:H7 actin assembly effectors Tir and EspF(U) during pedestal formation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6754–6759. doi: 10.1073/pnas.0809131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter E, Janich S, Roessler BJ, Hilfinger JM, Amidon GL. HT29-MTX/Caco-2 cocultures as an in vitro model for the intestinal epithelium: in vitro-in vivo correlation with permeability data from rats and humans. J. Pharm. Sci. 1996;85:1070–1076. doi: 10.1021/js960110x. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Abdullayev E, Hollister A, Mills D, Lvov YM. Clay nanotube/poly(methyl methacrylate) bone cement composites with sustained antibiotic release. Macromol. Mater. Eng. 2012;297:645–653. [Google Scholar]
- Worle-Knirsch JM, Pulskamp K, Krug HF. Oops they did it again! Carbon nanotubes hoax scientists in viability assays. Nano Lett. 2006;6:1261–1268. doi: 10.1021/nl060177c. [DOI] [PubMed] [Google Scholar]
- Zapor L, Dzierzawska K. Toxicity of nanofillers for polymer composites – in vitro studies. 30th International Congress on Occupational Health; 2012. http://icoh.confex.com/icoh/2012/webprogram/Paper7372.html. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.